Abstract
Thuringiensin is a thermostable secondary metabolite in Bacillus thuringiensis and has insecticidal activity against a wide range of insects. Until now, the regulatory mechanisms and genetic determinants involved in thuringiensin production have remained unclear. Here, we successfully used heterologous expression-guided screening in an Escherichia coli–Bacillus thuringiensis shuttle bacterial artificial chromosome library, to clone the intact thuringiensin synthesis (thu) cluster. Then the thu cluster was located on a 110-kb endogenous plasmid bearing insecticide crystal protein gene cry1Ba in strain CT-43. Furthermore, the plasmid, named pBMB0558, was indirectly cloned and sequenced. The gene functions on pBMB0558 were annotated by BLAST based on the GenBankTM and KEGG databases. The genes on pBMB0558 could be classified into three functional modules: a thuringiensin synthesis cluster, a type IV secretion system-like module, and mobile genetic elements. By HPLC coupling mass spectrometer, atmospheric pressure ionization with ion trap, and TOF technologies, biosynthetic intermediates of thuringiensin were detected. The thuE gene is proved to be responsible for the phosphorylation of thuringiensin at the last step by vivo and vitro activity assays. The thuringiensin biosynthesis pathway was deduced and clarified. We propose that thuringiensin is an adenine nucleoside oligosaccharide rather than an adenine nucleotide analog, as is traditionally believed, based on the predicted functions of the key enzymes, glycosyltransferase (ThuF) and exopolysaccharide polymerization protein (Thu1).
Keywords: Bacterial Genetics, Bacterial Toxins, Gene Regulation, Genetics, Metabolic Regulation, Bacillus thuringiensis, Thuringiensin, Biosynthesis Pathway, Genetic Determinants, Synthesis Cluster
Introduction
Bacillus thuringiensis is a Gram-positive insect pathogen that is extensively used in biological pest control (1). B. thuringiensis-based biopesticides are an environment-friendly product. B. thuringiensis could produce multi-insecticidal metabolites like insecticide crystal proteins (2–4), cytotoxin (3, 5), vegetative insecticidal protein (4, 6), secret insecticidal protein (7), thuringiensin (8), zwittermicin A (9), Mtx-like toxin (10), Bin-like toxin (11), etc. The diversity of toxin is paralleled by a diversity in pesticidal activity. Some B. thuringiensis strains found have novel biological activities other than insect toxicity, like parasporin, which targets human cancer cells, and some other strains are toxic to human-pathogenic protozoa (12). Specific toxins, responsible for most of those activities, have not yet been identified or characterized (13). Most of the researchers in the B. thuringiensis field pay attention to the crystal protein toxins. So far, 460 kinds of crystal protein toxins, classified into 60 classes, and 27 kinds of cytotoxin, classified into two classes, have been reported (see the Bacillus thuringiensis Toxin Nomenclature site on the World Wide Web).
Thuringiensin (β-exotoxin) is a secondary metabolite of B. thuringiensis and has insecticidal activity against a wide range of insects (14–16). From the structural formula, thuringiensin is composed of four precursors: adenosine, glucose, a phosphate group, and gluconic diacid (17). This kind of structure is seldom reported in antibiotic compounds. Historically, the consensus has been that it is an adenine nucleotide analog (17), like ATP, and this similarity makes it an inhibitor of DNA-dependent RNA polymerases (18). It is a nonspecific antibiotic insecticide and acaricide.
Until now, the regulatory mechanisms and genetic determinants involved in thuringiensin production have been unclear. Using a B. thuringiensis strain mutant library, Espinasse et al. (19) reported that an ABC transporter, which might be related to the secretion of thuringiensin, was essential for thuringiensin production.
The aim of this study was to elucidate the genetic determinant of thuringiensin in the strain CT-43. We adapted HPLC to selectively detect the characteristic peak of thuringiensin in an Escherichia coli–B. thuringiensis shuttle bacterial artificial chromosome (BAC)3 library of the thuringiensin high production strain, CT-43. The intact thu cluster was then isolated. The thuringiensin biosynthesis pathway was deduced and clarified by LCMS-IT-TOF detection and the identification of key gene thuE.
EXPERIMENTAL PROCEDURES
Materials
B. thuringiensis strain CT-43 (without a flagellum) was isolated from Chinese soil by our group and showed high production of thuringiensin. The details of bacterial strains and plasmids used in this study are listed in Table 1. All strains were grown at 28 °C in Luria-Bertani (LB) medium. Antibiotics were added at the following concentrations: ampicillin (100 μg/ml), chloromycetin (25 μg/ml), erythromycin (15 μg/ml), and kanamycin (50 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains and plasmids | Features | Source/Reference |
|---|---|---|
| DH10B | E. coli(Δmrr_hsdRMS_mcrBC) mcrA recA1 | New England Biolabs |
| BMB171 | A plasmid-cured derivative of B. thuringiensis YBT-1463, which lacks the endogenous plasmid and the thu cluster | Ref. 43 |
| CT-43 | B. thuringiensis subsp. chinensis strain, no flagellum | Ref. 20 |
| HD-2 | B. thuringiensis subsp. thuringiensis strain | Ref. 8 |
| EMB1228 | E. coli recombinant strain with BAC plasmid harboring cry1Ba gene | This work |
| EMB1234 | E. coli recombinant strain with BAC plasmid harboring cry1Ba gene | This work |
| BMB0542 | B. thuringiensis recombinant strain producing thuringiensin | This work |
| BMB0543 | B. thuringiensis recombinant strain producing thuringiensin | This work |
| BMB0545 | Mutant of strain CT-43 in which the thuE gene was knocked out | This work |
| BMB0546 | Recombinant of BMB0545 in which the knocked out thuE gene was retrocomplemented | This work |
| BMB0547 | Recombinant of BMB0545 in which there was a pHT304 plasmid | This work |
| pMD19-T | PCR product cloning vector | TaKaRa |
| pHT304 | E. coli–B. thuringiensis shuttle BAC vector | Ref. 26 |
| pHT304-ts | pHT304 derivative with a thermosensitive replicon of B. thuringiensis | Ref. 24 |
| pEMB0572 | Plasmid-interrupted thuE gene by kanamycin gene inserted in pHT304-ts | This work |
| pEMB0557 | An E. coli–B. thuringiensis shuttle BAC vector | Ref. 20 |
| pBMB0558 | Plasmid harboring cry1Ba gene of strain CT-43 | This work |
Construction of the Shuttle BAC Library of B. thuringiensis Strain CT-43
An E. coli to B. thuringiensis shuttle BAC vector, pEMB0557 (20), was used to construct the shuttle BAC library. This vector incorporated the plasmid replication origin (ori60) from the 100-kb plasmid of B. thuringiensis subsp. kurstaki strain YBT-1520, erythromycin resistance (B. thuringiensis), and chloromycetin resistance (E. coli) genes. The overnight culture of strain CT-43 was inoculated (1% v/v) into 100 ml of sterile LB and incubated at 28 °C for about 3–4 h until the cell density reached an A600 nm of 0.2–0.3. Cells were harvested by centrifugation, and agarose plugs were prepared as described (20). The genomic DNA was then extracted and separated. The separated and recovered high molecular weight genomic DNA was ligated into the cloning-ready BAC vector, pEMB0557, digested with HindIII. The ligation mixture was then transformed into B. thuringiensis host strain BMB171 by electroporation with a GenePulser electroporator (Bio-Rad).
The Screening and Identification of the BAC Library Clones
The extracted plasmid DNA of the BAC clones was digested with NotI and HindIII and separated by PFGE with a Bio-Rad CHEF III instrument that could estimate the inserted fragment sizes.
Location of the thu Cluster
To locate the thu cluster, a Southern blot assay was carried out. Plasmids were extracted from strain CT-43 and HD-2 and then transferred to an Immobilon-Ny+ nylon membrane (Millipore). Southern blotting was performed according to a standard protocol (21). Probe cry1Ba on plasmid pBMB0558 was amplified from strain CT-43. The DIG High Primer DNA Labeling and Detection Starter Kit I (Roche Applied Science) was used for Southern blotting.
The Indirect Cloning of Endogenous Plasmid, pBMB0558
According to the physical map, two BAC clones that covered the whole plasmid were selected to construct a subcloning library. The BAC clones were partially digested with EcoRI and HindIII separately and then ligated with plasmid pHT304 and transformed into E. coli DH5α. Clones were sequenced using Megabace 1000 automated sequencers (GMI). The Phred/Phrap/Consed (22) software package and DNASTAR 7.10 software package were adopted for quality assessment and sequence assembly. During the process of sequence assembly, the pBMB0558 nucleotide sequence with an average coverage of 5-fold was obtained. To confirm the position of these contigs in pBMB0558, a BAC-end sequencing technique was performed. The leaks between contigs were filled by PCR.
HPLC Analysis for Thuringiensin Detection
HPLC analysis was carried out on a system consisting of a UV-visible detector (CapLC 2487, Waters), Rheodyne manual sample injector valve 7725i, and a Waters 515 HPLC pump. 20 μl of the sample was injected into a C18 end-capped column. A 5% methanol gradient in 50 mm potassium phosphate buffer (pH 3.0) was applied for 15 min. The flow rate was 1.0 ml/min, and UV absorption was monitored at 260 nm at 25 °C. Thuringiensin was eluted at 5.5 min with a Hypersil C18 column (10 μm, 4.6 × 150 mm; Elite) and at 8.0 min with an Agilent TC-C18 column (5 μm, 4.6 × 250 mm; Agilent). The detection limit of this method for thuringiensin was 2 μg/ml.
Extraction and Preparation of Thuringiensin
To extract thuringiensin, B. thuringiensis strains were grown in LB at 200 rpm and 28 °C for 24 h. After centrifugation, the culture supernatant was collected. Acetone was added to the supernatant to 90% final concentration, and the solution was then centrifuged for 6 min at 12,000 rpm. The pellet was dissolved in 0.2 ml of ultrapure H2O, acetonitrile was added to a final concentration of 40%, and the sample was centrifuged again for 6 min at 12,000 rpm. The pellet was discarded, and the acetonitrile concentration in the supernatant was increased to 90%. The precipitate was collected by centrifugation for 6 min at 12,000 rpm at 4 °C, and the pellet was finally dissolved in 0.1 ml of HPLC elution buffer (50 mm KH2PO4, 5% methanol, pH 3.0).
Intracellular Intermediates Extracted from B. thuringiensis Strain CT-43 and thuE− Mutant BMB0545
B. thuringiensis strain CT-43 and the interrupted thuE− mutant BMB0545 were grown in LB medium. To analyze intracellular intermediates, the cells from culture filtrates were washed twice in 20 ml of sterile deionized water. The cells were collected and lysed by mechanical disruption with liquid nitrogen. The cell lysates were resuspended in 5 ml of sterile deionized water and centrifuged at 4,000 rpm for 10 min. The supernatants (5 ml) were collected for LCMS-IT-TOF analysis or stored at −20 °C.
Identification of the Intracellular Intermediates by LCMS Analysis
LCMS was performed by using an Agilent 1100 series LC/MSD trap, and the analytical column was ZORBAXSB-C18 (5 μm, 2.1 × 150 mm; Agilent). The MS operating conditions were optimized as follows: electrospray ionization (ESI) source set at the negative mode; m/z 100–1000; drying gas, 8.0 liters/min; drying gas temperature, 350 °C; and spray gas detector voltage, 30 ψ.
Identification of the Intracellular Intermediates by LCMS-IT-TOF Analysis
LCMS-IT-TOF had an ITTOFMS system (Kyoto) coupled with a high performance liquid chromatography (HPLC) system. The LC system (Shimadzu) was equipped with a solvent delivery pump (LC-20AD), an autosampler (SIL-20AC), a DGU-20A3 degasser, a photodiode array detector (SPD-M20A), a communication base module (CBM-20A), and a column oven (CTO-20AC). The separation was performed on a VP-ODS column (5 μm, 2.0 × 150 mm) using a gradient elution consisting of mobile phase B (acetonitrile/water/formic acid (80:20:0.1)). The gradient was as follows: 0–5 min, a linear gradient from 2% B to 5% B; 5–10 min, a linear gradient to 20% B; 10–15 min, a linear gradient to 20% B; 15–20 min, 2% B. The injection volume was 50 ml, the flow rate was 0.2 ml/min, and PDA detection was performed from 260 nm. The sample chamber in the autosampler was maintained at 4 °C, whereas the column was set at 40 °C. The whole analysis lasted 20 min. Mass spectral data for the metabolites were obtained using a Shimadzu ITTOF mass spectrometer. It was equipped with an ESI source operated in the negative ionization mode. Liquid nitrogen was used as nebulizing gas at a flow rate of 1.5 liters/min. The interface and detector voltages were set at 4.5 and 1.6 kV, respectively. The CDL and heat block temperatures were both 200 °C. The MS/MS spectra were produced by collision-induced dissociation of the selected precursor ions with argon as the collision gas. The ion accumulation time and relative collision energy were set at 50 ms and 50%, respectively. Data acquisition and processing were carried out using the LCMS solution version 3.41 software supplied with the instrument (23).
Interrupting the thuE Gene in the thu Cluster
The B. thuringiensis mutant strain BMB0545 was constructed via allelic exchange. The mutant allele was constructed via PCR using LA-taq polymerase. The primer sets were as follows: thuE upstream 1, 5′-GGATCCACCCTGATCATCTTGAAATGGTG-3′; thuE upstream 2, 5′-GGTACCCTTTCGATTCTGATAATCGCTGC-3′; thuE downstream 1, 5′-AAGCTTTTCCCGAAACTAGGGTTATGTTC-3′; and thuE downstream 2, 5′-CTCGAGCCAAGCATAAATCGTGATAAGGC-3′. The PCR product was cloned into pMD19-T vector (TaKaRa), and then a kanamycin coding gene was inserted between two arms of the allelic gene. Subsequently, the allelic gene interrupted by the kanamycin coding gene was cloned into a thermosensitive shuttle vector pHT304-ts (24), which had a thermosensitive replicon and was designated pEMB0572. The constructed plasmid contained 630 bp upstream and 800 bp downstream of the target gene with the DNA sequences GGATCC and CTCGAG (the recognition sequences for the restriction endonuclease BamHI and XhoI) introduced between these two flanking regions. For the ΔthuE mutation, the first 144 nucleotides were retained as well as the last 543 (this number includes the predicted stop codon) in the coding sequence. This resulted in a deletion of 3% (18 of 687 nucleotides) of thuE. The plasmid pEMB0572 was then introduced into B. thuringiensis strain CT-43 by electroporation and cultured at 43 °C and 200 rpm for 5 days. The strain with allelic double exchange was selected by PCR and named BMB0545.
Extracellular Activity Assays for Purified ThuE
The ThuE-coding gene was amplified from the genomic DNA of B. thuringiensis CT-43 by the PCR technique. The primer sets were as follows: thuE-1, 5′-GCGGATCCATGGAAAAGATATATATTGA-3′; thuE-2, 5′-CGCTCGAGTCATAGTACTTCTTCCTTAAA-3′. The amplified fragments were purified and digested with BamHI and XhoI and then subcloned into expression vector pGEX-6P-1 (Amersham Biosciences) digested with BamHI and XhoI, resulting in pEMB1101. The coding sequence was identified by sequencing. The identified plasmid pEMB1101 was transformed into E. coli strain BL21 (DE3) (Amersham Biosciences), and the positive transformants were selected on a Luria-Bertani plate containing 100 μg/ml ampicillin. subsequently, ThuE was overexpressed and purified using a GSTrap FF column (Amersham Biosciences). Purified ThuE was incubated with precursor C in reaction buffer (10 mmol/liter Tris-HCl, pH 8.5, 10 mmol/liter MgCl2, 1 mmol/liter dithiothreitol, 2.5 mm ATP). The reaction product was identified by following the HPLC and LCMS-IT-TOF assays.
Thuringiensin Bioassay
Thuringiensin was purified from the supernatant of strain CT-43 and recombinant strain BMB0542 (Table 1) separately. Bioassays were carried out by the diet incorporation method with the larvae of Helicoverpa armigera, Plutella xylostella, Musca domestica, and Meloidogyne incognita. Thuringiensin was mixed with the diet at different concentrations. Three replicates were conducted with each dilution. After 6 days of incubation at 25 °C, mortalities for each treatment were recorded. Data were analyzed by the SPSS 13.0 software. All of the bioassays were conducted three times for each treatment.
RESULTS
Genome-wide Mining for the Gene (Cluster) Responsible for Thuringiensin Synthesis in B. thuringiensis Strain CT-43
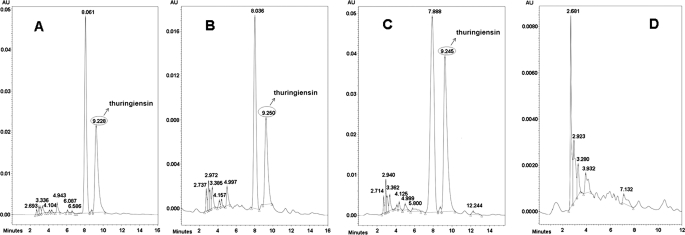
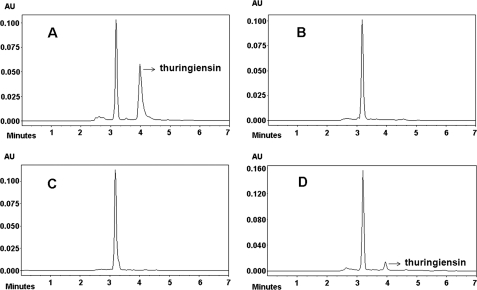
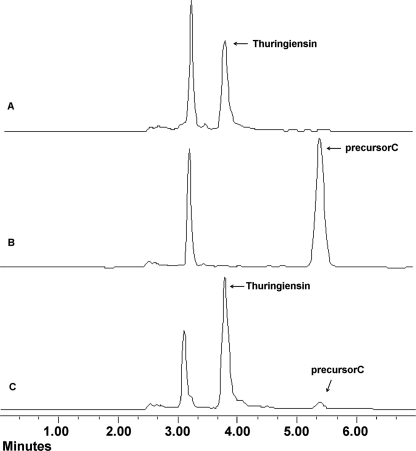
The B. thuringiensis strain CT-43 was isolated from Chinese soil by our group. The thuringiensin produced by B. thuringiensis strain CT-43 was purified. The bioassay showed that the thuringiensin was toxic to Caenorhabditis elegans, H. armigera, and P. xylostella (data not shown). To isolate the gene (cluster) responsible for thuringiensin synthesis in B. thuringiensis strain CT-43, a shuttle BAC library of this strain was established. B. thuringiensis strain BMB171, which does not produce thuringiensin (Fig. 1D), was used as the host for the library construction. An E. coli–B. thuringiensis shuttle BAC vector, pEMB0557, with a large loading capacity was constructed for this purpose (21). The average insertion size of the library was ∼80 kb, and assuming that the size of the strain CT-43 genome is 5 Mb, the library would have an 80-fold coverage of the genome. 5,000 clones were obtained in total and were divided into 250 groups. Twenty clones from each group were co-cultured and analyzed by HPLC. By this screening strategy, a characteristic peak of thuringiensin was detected in the purified supernatant (Fig. 1B) of one clone, BMB0542 (25-kb insert), and compared with that of strain CT-43 (Fig. 1A). The identity of the product was confirmed by co-injection of a sample containing equal volumes of strain CT-43 and BMB0542 (25-kb insert) supernatant mixture. The HPLC analysis showed that the suspected peak from BMB0542 (25-kb insert) overlapped the characteristic peak of thuringiensin from strain CT-43 (Fig. 1C). The single peak was collected and further identified by MS detection (supplemental Fig. S1), which showed that this compound shared the same molecular mass (701 daltons) with thuringiensin. Therefore, we deduced that the intact cluster responsible for thuringiensin synthesis was included in the DNA insert of BMB0542 (25-kb insert). Subsequently, the 25-kb inserted fragment in BMB0542 (25-kb insert) was sequenced and annotated based on data from the GenBankTM database.
FIGURE 1.
HPLC analysis of heterogeneous expression of thuringiensin in recombinant BMB0542 (25-kb insert). A, trace of the supernatant from strain CT-43 (positive control). B, trace of the supernatant from recombinant BMB0542 (25-kb insert). C, trace of the supernatant mixture from recombinant BMB0542 (25-kb insert) and strain CT-43. D, trace of supernatant from host strain BMB171 (negative control). The characteristic peak of thuringiensin is indicated by an arrow. The red lines were produced by the MS system to fix on the range of a peak and facilitate the peak area calculation by the (MS soft, Varian workstation). AU, absorbance unit.
The Minimized Assay of the Cloned 25-kb Insertion Fragment and Bioinformatics Analysis
Bioinformatics revealed that a 12-kb acyl carrier protein-dependent cluster and an insecticidal crystal protein cry1Ba gene were present on the 25-kb insert. The 12-kb acyl carrier protein-dependent cluster was suspected to be related to thuringiensin synthesis. To isolate a fragment comprising the 12-kb cluster, the 25-kb insert of BMB0542 (25-kb insert) was digested with BamHI because a BamHI site was observed to be present at either end of the 12-kb cluster. The 12-kb acyl carrier protein-dependent cluster was thus isolated, cloned into BAC vector, pEMB0557, and electroporated into B. thuringiensis host BMB171, for heterologous expression, resulting in a recombinant termed BMB0543 (12-kb insert). HPLC analysis confirmed that the 12-kb acyl carrier protein-dependent cluster could confer thuringiensin synthesis on host strain BMB171. This cluster was named the thu cluster.
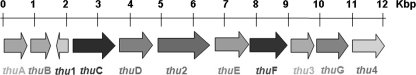
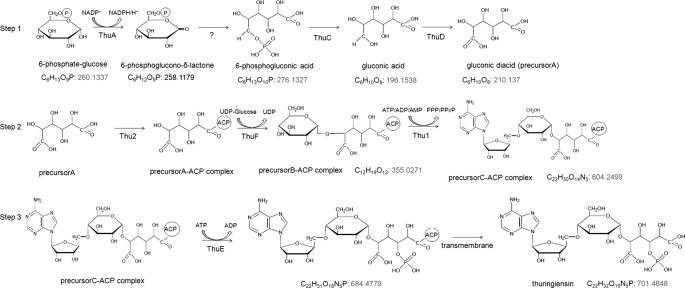
The thu cluster comprised 11 ORFs (Fig. 2). Their gene functions were predicted based on matches in the GenBankTM data base. Some genes that showed low homology to the known genes in the databases were analyzed for their functional domains and realigned; thus, their deduced functions were based only on their functional domains. The detailed results are shown in Tables 2 and 3. Based on gene function, we deduced that thuA, thuC, and thuD might encode proteins responsible for the synthesis of the key precursor, gluconic diacid (precursor A), from glucose 6-phosphate, whereas thuF and thu1 might encode proteins responsible for the assembly of thuringiensin. The predicted Thu2 protein, which comprises an adenylation (A) domain, a condensation (C) domain, and an acyl carrier protein (ACP) domain, might serve as the location for the assembly of thuringiensin. The thuE gene might encode the enzyme in charge of the final modification (phosphorylation), and thu3 might encode a protein that takes part in releasing mature thuringiensin.
FIGURE 2.
The thu cluster. thuA, glucose-6-phosphate 1-dehydrogenase gene; thuB, racemase gene; thuC, PEP-protein phosphotransferase gene; thuD, UDP-glucose-6-dehydrogenase gene; thuE, shikimate kinase gene; thuF, glycosyltransferase gene; thuG, N-acyl-d-glucosamine 2-epimerase gene; thu1, putative exopolysaccharide polymerization protein gene; thu2, non-ribosomal peptide synthetase gene; thu3, bacterial gene of unknown function; thu4, SAM-dependent methyltransferase.
TABLE 2.
Predicted proteins involved in thuringiensin production
The ORF sequences were analyzed by the protein-protein BLAST of NCBI database. The protein functions were predicted based on the identities of amino acids.
| Protein | Amino acids | Predicted protein function |
|---|---|---|
| ThuA | 120 | Glucose-6-phosphate 1-dehydrogenase |
| ThuB | 105 | Racemase |
| ThuC | 575 | PEP-protein phosphotransferase |
| ThuD | 205 | UDP-glucose-6-dehydrogenase |
| ThuE | 228 | Shikimate kinase |
| ThuF | 375 | Glycosyltransferase |
| ThuG | 226 | N-Acyl-d-glucosamine 2-epimerase |
| Thu1 | 101 | Exopolysaccharide polymerization protein |
| Thu2 | 561 | Non-ribosomal peptide synthetase |
| Thu3 | 311 | ABC transporter membrane-spanning permease |
| Thu4 | 143 | SAM-dependent methyltransferase |
TABLE 3.
NRPS/PKS-like components encoded by thu2
The ORF sequences were analyzed by the protein-protein BLAST of NCBI database. The domain functions were predicted based on the identities of amino acid.
| Proposed domain | Amino acids | Domain function |
|---|---|---|
| A | 101 | Recognition and activation |
| ACP | 60 | Acyl carrier protein |
| C | 400 | Condensation |
The thu Cluster Is Located on an Endogenous Plasmid of Strain CT-43
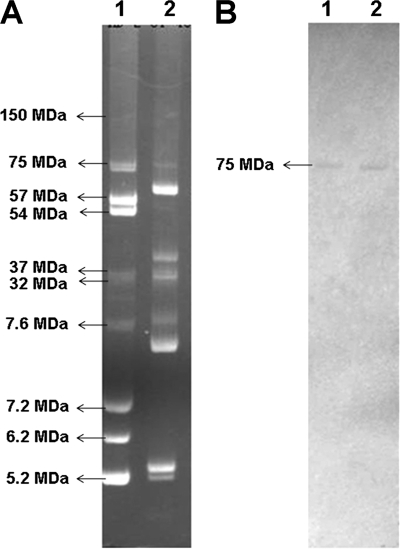
A genome walking strategy was adopted to clone the flanking sequence of the thu cluster. The known cry1Ba gene sequence at the end of the 25-kb insert was used as a probe to isolate all the target clones harboring this gene from the whole genome library. Thus, six clones harboring the cry1Ba gene, with insertion sizes ranging from 40 to 120 kb, were selected. Interestingly, sequence analysis showed that the cluster was not located on the bacterial chromosome but on an endogenous plasmid named pBMB0558. The cry1Ba gene was used as a Southern blotting probe to confirm this result, and a positive signal was detected for one of the six endogenous plasmids. The standard B. thuringiensis strain HD-2, the molecular size of whose endogenous plasmids has been established, was used as the molecular weight marker (25). The positive signal was also detected for a 75-MDa endogenous plasmid of standard B. thuringiensis strain HD-2 (Fig. 3).
FIGURE 3.
Location of the thu cluster. A, the electrophoretic profile of endogenous plasmids from B. thuringiensis strain HD-2 and CT-43. Lane 1, HD-2 (the molecular weights of endogenous plasmids are indicated); lane 2, CT-43. B, Southern blot of B. thuringiensis strain HD-2 and CT-43 using cry1Ba as probe. Lane 1, HD-2; lane 2, CT-43. The plasmid of 75 MDa in strain HD-2 and CT-43 showed a positive signal with the cry1Ba probe.
The sequencing results revealed that pBMB0558 was a circular plasmid with a molecular weight of 109,464 base pairs and 102 putative ORFs (GenBankTM accession number HM037272). The functions of the gene were predicted by BLAST based on the GenBankTM and KEGG databases. The genes on pBMB0558 could be classified into three functional modules: 1) a 12-kb thuringiensin synthesis cluster (between 30 and 42 kb), 2) a 30-kb type IV secretion system-like (0–30 kb), and 3) mobile genetic elements of about 67 kb, which included a prophage and the transposase (42–109 kb) (supplemental Fig. S2).
The Deduced Thuringiensin Biosynthesis Pathway and Assembly Process
The thuringiensin biosynthesis pathway was deduced according to the predicted genes' functions (Fig. 4). From the structural formula, it is assumed that thuringiensin is composed of four precursors: adenosine, glucose, a phosphate group, and gluconic diacid. The pathway could be divided into three steps. In step 1, the key precursor gluconic diacid (precursor A) is synthesized by the products of thuA, thuC, and thuD. The initial substrate glucose 6-phosphate is oxidized to 6-phosphoglucono-δ-lactone by a glucose-6-phosphate 1-dehydrogenase (ThuA) and subsequently hydrolyzed to 6-phosphogluconic acid. The dephosphorylation of 6-phosphogluconic acid is catalyzed by a phospholipid exchange protein-protein phosphotransferase (ThuC). The resulting gluconic acid is subsequently oxidized to gluconic diacid by UDP-glucose dehydrogenase (ThuD), which can directly oxidize a hydroxyl group to a carboxyl(ic) group. In step 2, gluconic diacid, glucose, and adenosine are assembled by thuF and thu1. First, gluconic diacid binds to the ACP region of the non-ribosomal peptide synthetase (Thu2); a hydroxyl group on the C-1 of gluconic diacid forms a thioester bond with a sulfhydryl group of the ACP protein. Second, a UDP-glucose moiety is added onto the gluconic diacid-ACP complex by a glycosyltransferase (ThuF), and a 1,5-glycosidic bond is formed between glucose and gluconic diacid. The resulting product (precursor B) is polymerized with the ribose of adenosine from ATP/ADP/AMP by an exopolysaccharide polymerization protein (Thu1). A 4,5-glycosidic bond is then formed between ribose and glucose, producing precursor C. Finally, precursor C is released from ACP. In step 3, precursor C is phosphorylated by ThuE, and the mature thuringiensin is released. The mature thuringiensin molecule can then be secreted by the cell.
FIGURE 4.
Schematic of the thuringiensin biosynthesis pathways. Step 1, synthesis of key precursor gluconic diacid (precursor A) by thuA, thuC, and thuD; step 2, assembly of gluconic diacid, glucose, and adenosine by thuF and thu1; step 3, phosphorylation of precursor C by ThuE. Numbers have been added to designate the molecular weight of each intermediate.
Identification of the Biosynthesis Pathway by LCMS-IT-TOF
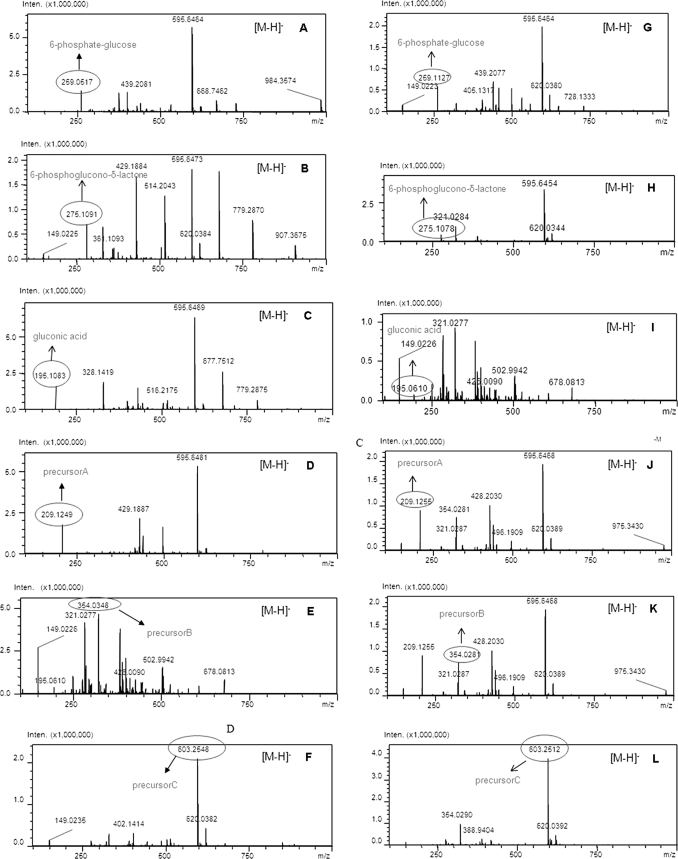
The LCMS-IT-TOF was conducted to clarify the deduced pathway of thuringiensin. The extracellular and intracellular contents of a culture of strain CT-43 were detected by LCMS-IT-TOF. In the intracellular fraction, all of the proposed intermediates could be detected (molecular weights in parentheses), i.e. to glucose 6-phosphate (m/z 259.0517), 6-phosphogluconic acid (m/z 275.1091), gluconic acid (m/z 195.1083), gluconic diacid (precursor A) (m/z 209.1249), precursor B without a hydroxy group (m/z 354.0271), and precursor C without a hydroxy group (m/z 603.2548) (Fig. 5, A–F). The correct molecular weight should be increased by 1 based on the m/z value under the [M − H]− pattern according the principle of LCMS-IT-TOF. Only the mature thuringiensin could be detected in the extracellular supernatant of strain CT-43, by both HPLC (supplemental Fig. S3) and LCMS analysis (supplemental Fig. S4). All of the proposed intermediates could not be found in the non-expressing strain BMB171, whereas the origin of structural assignments and predictions, such as glucose fragment, glucose 6-phosphate, and adenosine could be found based on the LCMS-IT-TOF data (supplemental Figs. S5 and S6).
FIGURE 5.
Intracellular metabolites produced in strain CT-43 and thuE− mutant BMB0545 detected by LCMS-IT-TOF. A–F, the LCMS-IT-TOF profile of strain CT-43. G–L, the LCMS-IT-TOF profile of thuE− mutant BMB0545. Each intracellular metabolite is indicated (m/z values are shown). Inten., ion signal strength detected by the detector.
The Biological Function of the Key Gene thuE
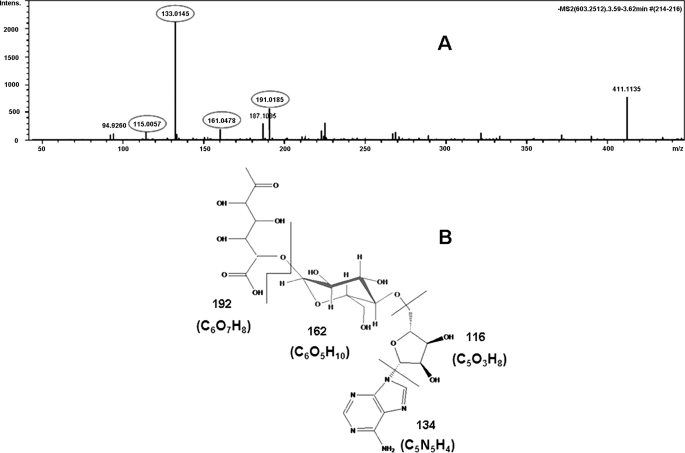
The function of the key gene thuE was confirmed by a knock-out experiment. HPLC and LCMS-IT-TOF results for the thuE− mutant BMB0545, carrying an interrupted thuE gene, demonstrated that this mutant could not produce thuringiensin (supplemental Fig. S3 and S4) in the extracellular supernatant, although it could produce all of the predicted intermediates in the cells (Fig. 5, G–L). Disruption in the thuE gene led to the complete loss of phosphorylation activity and resulted in the build-up of precursor C (m/z 603.2548) in the intracellular fraction (Fig. 5, F and L). MS/MS was further conducted to confirm the structural formula of the enriched precursor C (m/z 603.2548) in this mutant. The following components (with expected molecular weights) were detected: adenine without a hydrogen group (m/z 133.0145), ribose without a C5′-hydroxy group (m/z 115.0057), glucose without a C1′-hydroxy group and C4′-hydrogen group (m/z 161.0478), and gluconic diacid without a C1′-hydroxy group and C5′-hydrogen group (m/z 191.0185) (Fig. 6). The complementation experiment of thuE was also performed. The primer sets thuE1 and thuE2, were used to amplify the full-length thuE gene contained in a 1.5 kb DNA fragment. The double digestion product (BamHI and HindIII) of the PCR fragment was subcloned into the shuttle vector pHT304 (26). The obtained recombinant plasmid was electroporated into the thuE− mutant BMB0545, which lead to recombinant BMB0546 (thuE+). HPLC analysis of the extracellular supernatant of this recombinant revealed thuringiensin in the extracellular supernatant (Fig. 7D; compare with negative control (Fig. 7, B and C) and positive control (Fig. 7A)). Thus, the insertion of the wild-type thuE gene was able to complement the thuE− mutant BMB0545. Taken together, these results indicate that the thuE gene is responsible for the phosphorylation of thuringiensin at the last step.
FIGURE 6.
Identifying the structure of precursor C. A, MS/MS result of fragments of precursor C (m/z 603.2548, [M − H]−). Each component (with its corresponding molecular weight) is highlighted by a circle. B, the possible fractions of precursor C formed in MS/MS are separated by a line. Black numbers are the molecular weights of each component.
FIGURE 7.
HPLC analysis of the supernatant from strain CT-43, thuE− mutant BMB0545, complementary strain BMB0546 (thuE+), and BMB0547 (vector only). A, trace of the supernatant from strain CT-43. The position of the arrowhead is the characteristic peak of thuringiensin. B, trace of the supernatant from thuE− mutant BMB0545. C, trace of the supernatant from BMB0547 (vector only). D, trace of the supernatant from complementary strain BMB0546 (thuE+). The characteristic peak of thuringiensin is observed when the lack of expression of the ThuE enzyme is compensated for.
Extracellular Activity of Purified ThuE
The thuE gene was heterologously expressed in E. coli strain BL21 (DE3) by expression vector pGEX-6P-1. Then the ThuE was purified using the GSTrap FF column. The predicted precursor C was obtained by digesting purified thuringiensin with alkaline phosphatase and then identified by HPLC (Fig. 8B). The HPLC revealed a new peak with a retention of about 5.3 min. The LCMS-IT-TOF was performed to determine the presence of precursor C, whose molecular weight is 620 under the [M − H]− pattern (supplemental Fig. S7). The purified ThuE was incubated with precursor C in reaction buffer and analyzed by HPLC. The HPLC result showed the conversion of precursor C to thuringiensin after a 3-h incubation (Fig. 8C).
FIGURE 8.
HPLC analysis for extracellular activity of purified ThuE. A, trace of the supernatant from strain CT-43; B, trace of thuringiensin digested by alkaline phosphatase; C, trace of the reaction product after a 3-h incubation of purified ThuE and precursor C.
Insecticidal Activities of Thuringiensin
The bioactivity of thuringiensin extracted from strain CT-43 and BMB0542 was evaluated against the larvae of H. armigera, P. xylostella, M. domestica, and M. incognita separately. The results (Table 4) showed that thuringiensin was toxic to them. The LC50 values of thuringiensin from strain CT-43 against the larvae of H. armigera, P. xylostella, M. domestica, and M. incognita were 19.3, 0.9, 47.7, and 25.5 μg/ml, whereas the LC50 values of thuringiensin from BMB0542 were 23.2, 1.2, 49.2, and 23.7 μg/ml. The data showed that the expressed product from strain BMB0542 exhibited thuringiensin-like specific activity.
TABLE 4.
Activities of thuringiensin from strain CT-43 and BMB0542 against H. armigera, P. xylostella, M. domestica, and M. incognita
The second stage juveniles of H. armigera, P. xylostella, M. domestica, and M. incognita were tested, and LC50 values were determined on day 6.
| Origin of thuringiensin | Insect name | Regression equation | LC50a | S.E. |
|---|---|---|---|---|
| μg/ml | ||||
| Strain CT-43 | H. armigera | y = −0.85314 + 0.66321x | 19.3 | 0.15063 |
| Strain CT-43 | P. xylostella | y = 0.06337 + 1.09866x | 0.9 | 0.31076 |
| Strain CT-43 | M. domestica | y = −5.11486 + 3.04741x | 47.7 | 1.46069 |
| Strain CT-43 | M. incognita | y = −3.74985 + 2.66451x | 25.5 | 1.67528 |
| Strain BMB0542 | H. armigera | y = −1.17306 + 0.85965x | 23.2 | 0.15729 |
| Strain BMB0542 | P. xylostella | y = −0.09482 + 1.30222x | 1.2 | 0.23255 |
| Strain BMB0542 | M. domestica | y = −2.19691 + 1.29870x | 49.2 | 0.96294 |
| Strain BMB0542 | M. incognita | y = −0.62790 + 0.45646x | 23.7 | 0.75605 |
a Water was used as the negative control.
DISCUSSION
In this study, genome screening revealed the genetic determinants of a nonspecific antibiotic insecticide thuringiensin in B. thuringiensis strain CT-43.
An alternative viewpoint concerning the structure of thuringiensin was proposed in this study. Thuringiensin has historically been believed to be a thermostable adenine nucleotide analog, like ATP. If this were true, it would be an inhibitor of DNA-dependent RNA polymerases (18). The biosafety of thuringiensin to mammals is still debated due this potential toxicity (27, 28). Our results show that thuringiensin is synthesized by the polymerization of three kinds of monosaccharides: gluconic diacid, glucose, and ribose. An antibiotic substance with this structure has been seldom reported before. This new finding concerning the structure of thuringiensin will lead to a definitive establishment of its toxicity mechanism. The three-dimensional structure of thuringiensin is cryptic, and we supposed that thuringiensin possesses a unique structure. Thuringiensin, as a polymer of monosaccharides, possesses asymmetric carbon atoms. Two putative enzymes encoded by thuB and thuG in the thu cluster, a racemase and an epimerase, might be involved in the stereochemical inversion of thuringiensin. A unique ACP was also revealed in the thu cluster. There are two classic antibiotic synthesis systems: the non-ribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) systems. The key functional elements responsible for assembly in NRPS are an A domain, a C domain, and a peptidyl carrier protein. In the PKS system, an acyltransferase domain, ketoacyl synthase domain, and ACP are required (29, 30). Bioinformatic analysis showed that the Thu2 protein encoded by the thu cluster contains three domains: an A domain (101 amino acids), a C domain (400 amino acids), and an ACP domain (60 amino acids) (Table 3), showing similarities to both the NRPS and PKS systems. This kind of hybrid protein for antibiotic synthesis has not been previously reported. This combination of special functional elements might represent a novel mode of antibiotic synthesis.
During the process of pathway prediction, we considered several different ideas. Finally, the pathway was deduced based on the LCMS-IT-TOF data, function of genes, and structure of thuringiensin. The origin of structural assignments and predictions could be initially clarified based on the LCMS-IT-TOF data (supplemental Fig. S6). The origin of structural assignments should be the basal metabolism, such as Glc-6-PO4, UDP-glucose, and adenosine, in order to economize the energy. The similar enzymatic reactions were proved in other organisms. For example, from 6-phosphate-glucose to 6-phosphoglucono-lactone there needs to be a glucose-6-phosphate 1-dehydrogenase action (31–33). In the process of ATP + d-gluconic acid [dharrow] ADP + 6-phospho-d-gluconate, phosphorylation and dephosphorylation were produced (34, 35). In UDP-glucose + H2O + 2 NAD+ [dharrow] UDP-glucuronate + 2 NADH + 2 H+, an oxidation reaction is produced with a UDP-glucose-6-dehydrogenase (36, 37). Glycosyl transferase family 2 could transfer sugar from UDP-glucose, UDP-N-acetyl-galactosamine, GDP-mannose, or CDP-abequose, to a range of substrates, including cellulose, dolichol phosphate, and teichoic acids (38–40). Shikimate kinase catalyzes the phosphorylation of the 3′ C terminus during the shikimate acid pathway (41, 42). Non-ribosomal peptide synthetase mainly catalyzes the antibiotic synthesis, and the key functional elements responsible for assembly in NRPS are the AC domain and peptidyl carrier protein, whereas the key elements in PKS are the acyltransferase domain and ACP (29, 44). The current genome-wide screening work confirms the structure predicted and confirmed by chemical synthesis in the work of Kalvoda et al. (see Ref. 17 and references cited therein).
An immune mechanism related to thuringiensin was proposed in this work. A typical type IV secretion system (T4SSs), which is seldom reported in Gram-positive bacterium, was revealed adjacent to the thu cluster. Type IV secretion systems in many Gram-negative pathogens are involved in the delivery of protein and/or DNA substrates (45). The vir-encoded T4SSs typically include 12 proteins: VirB1–VirB11 and VirD4 (46). The VirD4 protein, which is responsible for the recognition and binding of the substrate, is a key gene in a type IV secretion system. Our preliminary data showed that the interruption of VirD4 in strain CT-43 resulted in absent production of thuringiensin.4 Moreover, Thu3, which is homologous to an ABC transporter membrane-spanning permease, is possibly involved in the secretion of thuringiensin. In this work, the key gene, thuE, was proved to be responsible for thuringiensin phosphorylation. This would happen during the last step of the membrane translocation process; the mature thuringiensin would then be excluded by a combination of Thu3 and T4SSs. This mechanism protects the cell from possible damage by thuringiensin.
Insecticidal factor (thuringiensin) is encoded on an endogenous plasmid related to the evolution of B. thuringiensis. Some investigators consider that B. thuringiensis has acquired insecticidal activity in the course of co-evolution with insects through a host-parasite relationship. Circumstantial evidence seems to lend support this opinion. Most of the genes encoding toxins in B. thuringiensis are plasmid-borne and are generally structurally associated with mobile elements (4, 47). This work showed that the 110-kb plasmid pBMB0558 harbored a thu cluster and a 67-kb DNA fragment related to mobile genetic elements. The ACP involved in the biosynthesis of thuringiensin is a hybrid of that seen in NRRP and PKS antibiotic synthesis systems. Therefore, it could be predicted that these endogenous plasmids were acquired by horizontal gene transfer, represent a unique genetic resource, and are part of an accessory and/or adaptive gene pool. They might play an important role in the biology and evolution of their host cells.
The endogenous plasmids of bacteria, as a unique format, harbor attractive genetic elements, which could confer special phenotypes to a host. Nevertheless, direct cloning of large molecular weight native plasmids is still a worldwide problem, particularly for endogenous plasmids. We established a novel strategy for cloning large endogenous plasmids. The identified gene on the target plasmid could act as a probe to isolate all of the target clones from a genomic library, after which the full sequence of plasmid is relatively simple to obtain. We adapted this strategy to isolate novel genes responsible for a bioactive substance carried on a plasmid (48).
Supplementary Material
Acknowledgments
We thank the National Reference Laboratory (Huazhong Agricultural University) for the test of veterinary drug residues and the Wuhan Institute of Physics and Mathematics (Chinese Academy of Sciences) for the LCMS-IT-TOF tests.
This work was supported by National Basic Research Program (973 Program) of China Grant 2009CB118902, National Natural Science Foundation of China Grants 30400003 and 30970037, National High Technology Research and Development Program (863 Program) of China Grants 2006AA02Z174 and 2006AA10A212, Genetically Modified Organisms Breeding Major Projects of China Grant 2009ZX08009-032B, and Ministry of Forestry Project (948 Program) of China Grant 2006-4-41.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) HM037272.
X.-Y. Liu, L.-F. Ruan, Z.-F. Hu, D.-H. Peng, S. -Y. Cao, Z.-N. Yu, Y. Liu, J.-S. Zheng, and M. Sun, unpublished data.
- BAC
- bacterial artificial chromosome
- ACP
- acyl carrier protein
- LCMS
- HPLC coupling mass spectrometry
- LCMS-IT-TOF
- HPLC coupling mass spectrometry, atmospheric pressure ionization with ion trap, and time-of-flight technologies
- ESI
- electrospray ionization
- NRPS
- non-ribosomal peptide synthetase
- PKS
- polyketide synthase
- AT
- acyltransferase
- contig
- group of overlapping clones
- T4SSs
- type IV secretion system.
REFERENCES
- 1.Sudakin D. L. (2003) Toxicol. Rev. 22, 83–90 [DOI] [PubMed] [Google Scholar]
- 2.Aronson A. I., Beckman W., Dunn P. (1986) Microbiol. Rev. 50, 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Höfte H., Whiteley H. R. (1989) Microbiol. Rev. 53, 242–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnepf E., Crickmore N., Van Rie J., Lereclus D., Baum J., Feitelson J., Zeigler D. R., Dean D. H. (1998) Microbiol. Mol. Biol. Rev. 62, 775–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waalwijk C., Dullemans A. M., van Workum M. E., Visser B. (1985) Nucleic Acids. Res. 13, 8207–8217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Estruch J. J., Warren G. W., Mullins M. A., Nye G. J., Craig J. A., Koziel M. G. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5389–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donovan W. P., Engleman J. T., Donovan J. C., Baum J. A., Bunkers G. J., Chi D. J., Clinton W. P., English L., Heck G. R., Ilagan O. M., Krasomil-Osterfeld K. C., Pitkin J. W., Roberts J. K., Walters M. R. (2006) Appl. Microbiol. Biotechnol. 72, 713–719 [DOI] [PubMed] [Google Scholar]
- 8.Levinson B. L., Kasyan K. J., Chiu S. S., Currier T. C., González J. M., Jr. (1990) J. Bacteriol. 172, 3172–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stabb E. V., Jacobson L. M., Handelsman J. (1994) Appl. Environ. Microbiol. 60, 4404–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crickmore N., Zeigler D. R., Feitelson J., Schnepf E., Van Rie J., Lereclus D., Baum J., Dean D. H. (1998) Microbiol. Mol. Biol. Rev. 62, 807–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wirth M. C., Walton W. E., Federici B. A. (2010) Environ. Microbiol. 12, 1154–1160 [DOI] [PubMed] [Google Scholar]
- 12.Ohba M., Mizuki E., Uemori A. (2009) Anticancer. Res. 29, 427–433 [PubMed] [Google Scholar]
- 13.Song L., Gao M., Dai S., Wu Y., Yi D., Li R. (2008) J. Invertebr. Pathol. 98, 169–176 [DOI] [PubMed] [Google Scholar]
- 14.Toledo J., Liedo P., Williams T., Ibarra J. (1999) J. Econ. Entomol. 92, 1052–1056 [DOI] [PubMed] [Google Scholar]
- 15.Tamez-Guerra P., Iracheta M. M., Pereyra-Alférez B., Galán-Wong L. J., Gomez-Flores R., Tamez-Guerra R. S., Rodríquez-Padilla C. (2004) J. Invertebr. Pathol. 86, 7–18 [DOI] [PubMed] [Google Scholar]
- 16.Tsuchiya S., Kasaishi Y., Harada H., Ichimatsu T., Saitoh H., Mizuki E., Ohba M. (2002) J. Invertebr. Pathol. 81, 122–126 [DOI] [PubMed] [Google Scholar]
- 17.Kalvoda L., Prystag M., Sorm F. (1973) Tetrahedron. Lett. 41, 4671–4674 [Google Scholar]
- 18.Sebesta K., Horská K. (1970) Biochim. Biophys. Acta. 209, 357–376 [DOI] [PubMed] [Google Scholar]
- 19.Espinasse S., Gohar M., Lereclus D., Sanchis V. (2002) J. Bacteriol. 184, 5848–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X., Peng D., Luo Y., Ruan L., Yu Z., Sun M. (2009) Appl. Microbiol. Biotechnol. 82, 765–772 [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., pp. 474–491, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 22.Ewing B., Green P. (1998) Genome Res. 8, 186–194 [PubMed] [Google Scholar]
- 23.Liu Z., Huang L., Dai M., Chen D., Wang Y., Tao Y., Yuan Z. (2008) Rapid Commun. Mass. Spectrom. 22, 1009–1016 [DOI] [PubMed] [Google Scholar]
- 24.Ji F., Zhu Y., Ju S., Zhang R., Yu Z., Sun M. (2009) FEMS Microbiol. Lett. 300, 11–17 [DOI] [PubMed] [Google Scholar]
- 25.González J. M., Jr., Brown B. J., Carlton B. C. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 6951–6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arantes O., Lereclus D. (1991) Gene 108, 115–119 [DOI] [PubMed] [Google Scholar]
- 27.Tsai S. F., Yang C., Wang S. C., Wang J. S., Hwang J. S., Ho S. P. (2004) Toxicol. Appl. Pharmacol. 194, 34–40 [DOI] [PubMed] [Google Scholar]
- 28.Tsai S. F., Yang C., Liu B. L., Hwang J. S., Ho S. P. (2006) Toxicol. Appl. Pharmacol. 216, 347–353 [DOI] [PubMed] [Google Scholar]
- 29.Finking R., Marahiel M. A. (2004) Annu. Rev. Microbiol. 58, 453–488 [DOI] [PubMed] [Google Scholar]
- 30.Chan Y. A., Podevels A. M., Kevany B. M., Thomas M. G. (2009) Nat. Prod. Rep. 26, 90–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engel H. J., Domschke W., Alberti M., Domagk G. F. (1969) Biochim. Biophys. Acta 191, 509–516 [DOI] [PubMed] [Google Scholar]
- 32.Glaser L., Brown D. H. (1955) J. Biol. Chem. 216, 67–79 [PubMed] [Google Scholar]
- 33.Miclet E., Stoven V., Michels P. A., Opperdoes F. R., Lallemand J. Y., Duffieux F. (2001) J. Biol. Chem. 276, 34840–34846 [DOI] [PubMed] [Google Scholar]
- 34.Cohen S. S. (1951) J. Biol. Chem. 189, 617–628 [PubMed] [Google Scholar]
- 35.Sable H. Z., Guarino A. J. (1952) J. Biol. Chem. 196, 395–402 [PubMed] [Google Scholar]
- 36.Axelrod J., Kalckar H. M., Maxwell E. S., Strominger J. L. (1957) J. Biol. Chem. 224, 79–90 [PubMed] [Google Scholar]
- 37.Kalckar H. M., Maxwell E. S., Strominger J. L. (1956) Arch. Biochem. Biophys. 65, 2–10 [DOI] [PubMed] [Google Scholar]
- 38.Coutinho P. M., Deleury E., Davies G. J., Henrissat B. (2003) J. Mol. Biol. 328, 307–317 [DOI] [PubMed] [Google Scholar]
- 39.Oriol R., Martinez-Duncker I., Chantret I., Mollicone R., Codogno P. (2002) Mol. Biol. Evol. 19, 1451–1463 [DOI] [PubMed] [Google Scholar]
- 40.Breton C., Imberty A. (1999) Curr. Opin. Struct. Biol. 9, 563–571 [DOI] [PubMed] [Google Scholar]
- 41.Løbner-Olesen A., Marinus M. G. (1992) J. Bacteriol. 174, 525–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng W. C., Chang Y. N., Wang W. C. (2005) J. Bacteriol. 187, 8156–8163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng D., Luo Y., Guo S., Zeng H., Ju S., Yu Z., Sun M. (2009) J. Appl. Microbiol. 106, 1849–1858 [DOI] [PubMed] [Google Scholar]
- 44.Fischbach M. A., Walsh C. T. (2006) Chem. Rev. 106, 3468–3496 [DOI] [PubMed] [Google Scholar]
- 45.Grynberg M., Li Z., Szczurek E., Godzik A. (2007) Trends Microbiol. 15, 191–195 [DOI] [PubMed] [Google Scholar]
- 46.Draper O., Middleton R., Doucleff M., Zambryski P. C. (2006) J. Biol. Chem. 281, 37628–37635 [DOI] [PubMed] [Google Scholar]
- 47.Mahillon J., Chandler M. (1998) Microbiol. Mol. Biol. Rev. 62, 725–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo S., Liu M., Peng D., Ji S., Wang P., Yu Z., Sun M. (2008) Appl. Environ. Microbiol. 74, 6997–7001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.