Abstract
The Calvin-Benson-Bassham cycle is responsible for carbon dioxide fixation in all plants, algae, and cyanobacteria. The enzyme that catalyzes the carbon dioxide-fixing reaction is ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). Rubisco from a hyperthermophilic archaeon Thermococcus kodakarensis (Tk-Rubisco) belongs to the type III group, and shows high activity at high temperatures. We have previously found that replacement of the entire α-helix 6 of Tk-Rubisco with the corresponding region of the spinach enzyme (SP6 mutant) results in an improvement of catalytic performance at mesophilic temperatures, both in vivo and in vitro, whereas the former and latter half-replacements of the α-helix 6 (SP4 and SP5 mutants) do not yield such improvement. We report here the crystal structures of the wild-type Tk-Rubisco and the mutants SP4 and SP6, and discuss the relationships between their structures and enzymatic activities. A comparison among these structures shows the movement and the increase of temperature factors of α-helix 6 induced by four essential factors. We thus supposed that an increase in the flexibility of the α-helix 6 and loop 6 regions was important to increase the catalytic activity of Tk-Rubisco at ambient temperatures. Based on this structural information, we constructed a new mutant, SP5-V330T, which was designed to have significantly greater flexibility in the above region, and it proved to exhibit the highest activity among all mutants examined to date. The thermostability of the SP5-V330T mutant was lower than that of wild-type Tk-Rubisco, providing further support on the relationship between flexibility and activity at ambient temperatures.
Keywords: Archaebacteria; Carboxylation; Enzyme Kinetics; Mutant; Photosynthesis; Protein Structure; X-ray Crystallography; CBB Cycle; Rubisco; Ribulose-1,5-bisphosphate Carboxylase/oxygenase
Introduction
The Calvin-Benson-Bassham (CBB)4 cycle contributes to the fixation of carbon for all plants, algae, and cyanobacteria. The most important enzyme in the CBB cycle is ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco: EC 4.1.1.39) (1, 2). Rubisco is responsible for the single carbon dioxide-fixing step of the cycle, in which ribulose-1,5-bisphosphate reacts with CO2 and H2O to form two molecules of 3-phosphoglycerate. However, it also catalyzes a competitive oxygenase reaction, by which ribulose-1,5-bisphosphate reacts with O2 to form one molecule of 3-phosphoglycerate and one molecule of 2-phosphoglycolate (3, 4). Moreover, a typical Rubisco is able to fix only three carbon dioxide molecules/s, an extremely low turnover rate for an enzyme (3). Rubisco is thus considered to be the rate-limiting enzyme of the CBB pathway, and has been an important target for protein engineering (3).
From sequence alignment, Rubiscos can be classified into four groups, types I, II, III, and IV (5). The classical type I and II Rubiscos function in the CBB cycle. Type I Rubisco is composed of eight large subunits (L subunits) and eight small subunits (S subunits) with tetragonal symmetry (L8S8) (6). Type II Rubisco is usually composed of only two L subunits (L2) (7, 8). In both cases, the L2 dimer is the catalytic unit of Rubisco, generating two catalytic centers (1, 9, 10). Type I Rubisco seems to be by far predominant, as it is distributed in all eukaryotes and cyanobacteria as well as in a wide range of other autotrophic bacteria. Type II Rubisco is confined to autotrophic bacteria. The eukaryotic type I enzyme cannot be functionally expressed in Escherichia coli (11, 12), whereas the cyanobacterial type I Rubisco and type II Rubisco are expressed in E. coli (13–15). Extensive structural and functional studies have been performed with the aim to clarify the structural elements that control the specificities and activity levels of Rubiscos (16), but no clear theory has been established. In terms of type IV Rubiscos, these enzymes do not display carboxylase activity due to a lack of several residues essential for Rubisco catalysis (17, 18), and are thus also called the Rubisco-like proteins. The Rubisco-like protein from Bacillus subtilis displays 2,3-diketo-5-methylthiopentyl-1-phosphate enolase activity, and functions in the methionine salvage pathway (18).
Type III Rubisco has been found only in archaea (19–21). Although the CBB cycle has not been found in archaea, the type III enzymes exhibit carboxylase activity, as in the cases of the type I and II enzymes (19). For some time now, we have been carrying out an extensive examination on the enzyme from Thermococcus kodakarensis (Tk-Rubisco). Crystallographic analyses of this enzyme have revealed that the enzyme forms an (L2)5 pentagonal ring and that the monomer structure is basically equivalent to that of the type I and II enzymes (22). Recently, it has been shown that Tk-Rubisco is involved in a new pathway involved in AMP metabolism in this organism (23). We have also elucidated that this enzyme is (i) comprised only from L subunits (24), (ii) readily expressed as a functional form in E. coli (19), (iii) not inactivated or affected by the presence of oxygen (19, 25), and (iv) so (thermo)stable that it can be expected to tolerate a high degree of mutations (26). Thus, this enzyme can be considered suitable as an alternative target of Rubisco engineering, which has been hampered by difficulties in manipulating the eukaryotic type I Rubiscos (11, 12).
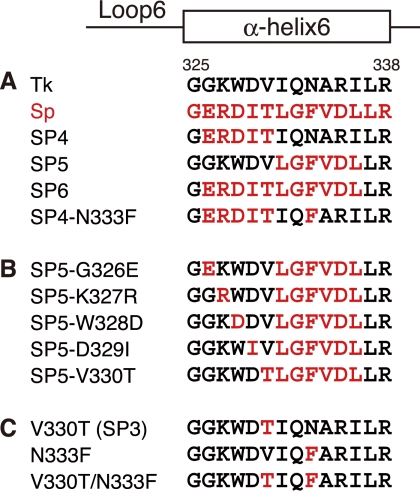
It has been found that Tk-Rubisco supported the photoautotrophic and photoheterotrophic growth of a Rubisco-deficient mutant strain of the mesophilic purple nonsulfur bacterium Rhodopseudomonas palustris number 7 (strain Δ3, in which the three native Rubisco genes have been disrupted) (25). This indicated that Tk-Rubisco can function in R. palustris cells in vivo, encouraging us to examine the enzymatic activities of Tk-Rubisco at ambient temperatures and further pursue the possibilities of improving the catalytic performance of this enzyme via protein engineering. We have carried out the mutation experiments on loop 6 of Tk-Rubisco, which harbors the active site Lys-322 interacting with the incoming gaseous substrate (27, 28), and the adjacent α-helix 6 (29). Replacement of the entire α-helix 6 of Tk-Rubisco with the corresponding region of the enzyme from spinach (Sp-Rubisco), whose activity is higher than that of Tk-Rubisco at ambient temperatures (30), resulted in a 30% increase in turnover number for the carboxylase activity at 25 °C in vitro (SP6 mutant; Fig. 1A) (29). R. palustris cells harboring the SP6 mutant also displayed a 31% increase in the specific growth rate under photoheterotrophic conditions compared with cells harboring the wild-type Tk-Rubisco. Intriguingly, the former and latter half-replacements of α-helix 6 (SP4 and SP5 mutants; Fig. 1A) did not show such improvements in Rubisco activity (29).
FIGURE 1.
Mutant design of Tk-Rubisco. Residues of Rubiscos from T. kodakarensis (Tk; GenBankTM accession number BAD86479) and spinach (Sp; accession number P00875) are colored in black and red, respectively. The secondary structures and residue numbers for Tk-Rubisco are shown over the aligned sequences. A, wild-type enzymes and mutants in our previous study (29). B, SP5 mutant derivatives. C, mutants used to investigate Val-330 and Asn-333.
To further improve the function of Tk-Rubisco, we have determined the crystal structures of the wild-type Tk-Rubisco as well as the SP4 and SP6 mutants in complex with its reaction-intermediate analog, 2-carboxy-d-arabinitol 1,5-bisphosphate (2-CABP), and examined the relationships between the structures and enzymatic activities. Based on the structural information, we designed a new mutant expected to display high performance at ambient temperatures, and examined its enzymatic activities both in vivo and in vitro.
EXPERIMENTAL PROCEDURES
Strains, Media, and Growth Conditions
The Tk-Rubisco gene (rbcTk) was isolated from T. kodakarensis KOD1, a hyperthermophilic archaeon isolated from Kodakara Island, Kagoshima, Japan (31, 32). R. palustris Δ3 is a derivative of R. palustris number 7 with its three Rubisco genes disrupted: rbcRp I-1, rbcRp I-2, and rbcRp II (25). E. coli DH5α was used for gene manipulation and plasmid construction. Gene expression was performed with E. coli BL21(DE3)CodonPlus RIL (Stratagene, La Jolla, CA) for kinetics assays, and with Rosetta2(DE3)pLysS (Novagen, Darmstadt, Germany) for crystallizations. E. coli cells were grown aerobically at 37 °C in Luria-Bertani (LB) medium with ampicillin (100 μg/ml for DH5α and BL21(DE3)CodonPlus RIL) or carbenicillin (50 μg/ml for Rosetta2(DE3)pLysS). R. palustris cells were first cultivated aerobically in the dark at 30 °C in LB medium containing 0.3% NaCl and zeocin (400 μg/ml) (heterotrophic growth) for 2 days. The harvested cells were washed and resuspended in a basal salt medium containing 5 mm NaHCO3 and 50 mm ethanol, but without yeast extract (BSA medium) (33). Cells were inoculated at an initial A660 of 0.05 into BSA medium, and photoheterotrophically incubated with a light intensity of 3,200 lux at 25 °C. Under photoheterotrophic conditions, cells were grown in a sealed glass vial filled with medium so that the head space was minimal. Specific growth rates were determined in the exponential growth phase.
DNA Manipulation
DNA manipulation was carried out using standard methods (34). Plasmid DNA was isolated with a Quantum Prep Plasmid Miniprep Kit (Bio-Rad). E. coli was transformed by the CaCl2 method. R. palustris Δ3 was transformed by electroporation. Restriction enzymes were purchased from Toyobo (Osaka, Japan) or Takara Bio (Otsu, Japan). DNA ligation was performed with a DNA ligation kit (Toyobo). KOD Plus (Toyobo) was used as a polymerase for PCR, and a GFX PCR DNA and Gel Band Purification Kit (GE Healthcare) was used to recover DNA fragments from agarose gels after electrophoresis. DNA sequencing was performed with a BigDye Terminator Cycle Sequencing Kit (version 3.1) and a model 3130xl capillary DNA sequencer (Applied Biosystems, Foster City, CA).
Site-directed Mutagenesis
Site-directed mutagenesis was carried out using a QuikChange XL Site-directed Mutagenesis Kit (Stratagene). The primers and template plasmids used are summarized in Table 1. The V330T mutant was prepared previously and reported as an SP3 mutant (29).
TABLE 1.
Primers used for site-directed mutagenesis
| Mutant | Template plasmid | Sequencea |
|---|---|---|
| SP5-G326E | pET21a(+)/SP5 | 5′-GGCAAGCTTGAGGGCGAGAAGTGGGACGTCCTC-3′ |
| 5′-GAGGACGTCCCACTTCTCGCCCTCAAGCTTGCC-3′ | ||
| SP5-K327R | pET21a(+)/SP5 | 5′-GCAAGCTTGAGGGCGGCCGCTGGGACGTCCTCGG-3′ |
| 5′-CCGAGGACGTCCCAGCGGCCGCCCTCAAGCTTGC-3′ | ||
| SP5-W328D | pET21a(+)/SP5 | 5′-GCTTGAGGGCGGCAAGGACGACGTCCTCGGCTTC-3′ |
| 5′-GAAGCCGAGGACGTCGTCCTTGCCGCCCTCAAGC-3′ | ||
| SP5-D329I | pET21a(+)/SP5 | 5′-GAGGGCGGCAAGTGGATCGTCCTCGGCTTCG-3′ |
| 5′-CGAAGCCGAGGACGATCCACTTGCCGCCCTC-3′ | ||
| SP5-V330T | pET21a(+)/SP5 | 5′-GAGGGCGGCAAGTGGGACACCCTCGGCTTCGTTGAC-3′ |
| 5′-GTCAACGAAGCCGAGGGTGTCCCACTTGCCGCCCTC-3′ | ||
| N333F | pET21a(+)/rbcTk | 5′-GTGGGACGTCATTCAGTTCGCCAGGATTCTCAGG-3′ |
| 5′-CCTGAGAATCCTGGCGAACTGAATGACGTCCCAC-3′ | ||
| V330T/N333F | pET21a(+)/V330T | 5′-GTGGGACACCATTCAGTTCGCCAGGATTCTCAGG-3′ |
| 5′-CCTGAGAATCCTGGCGAACTGAATGGTGTCCCAC-3′ |
a The underlined nucleotides correspond to the codons that have been changed.
Protein Purification
Tk-Rubisco is composed of 444 amino acid residues and its molecular mass is about 50 kDa. The expressed enzymes in E. coli were purified by heat treatment, anion exchange chromatography, and gel filtration chromatography as described previously (19), with slight modifications to prepare samples for crystallization as follows. The heat treatment condition was changed to 20 min at 85 °C. A Superdex 200 10/300 GL column (GE Healthcare) was used for the gel filtration chromatography step with a buffer containing 100 mm Bicine-NaOH (pH 8.3), 10 mm MgCl2, and 300 mm NaCl.
Crystallization
Prior to crystallization, the purified Tk-Rubisco was activated (carbamylated) by equilibrating with the carbamylation buffer containing 100 mm Bicine-NaOH (pH 8.3), 10 mm MgCl2, and 20 mm NaHCO3 at 4 °C for 1 day, to bind the inhibitor (35). Approximately 10 mm 2-CABP (synthesized by KNC Laboratories Co., Ltd., Kobe, Japan) was added to the activated protein solution at 4 °C for 1 day. Crystallization attempts were carried out using the hanging-drop vapor diffusion method at 20 °C. Mixtures of the 1.5-μl protein solution and an equal volume of the reservoir solution were equilibrated against the 500-μl reservoir solution. Crystals with approximate dimensions of 0.6 × 0.2 × 0.05 mm were obtained within a week using a reservoir solution containing 100 mm acetate buffer (pH 6.0), 80–100 mm CaCl2, 5–6% (w/v) polyethylene glycol 6,000, and 10% (v/v) 2-methylpentane-2,4-diol.
X-ray Data Collection
Crystals were soaked briefly in a cryoprotectant solution, which had been prepared by the addition of 20% (v/v) ethylene glycol to the reservoir solution. They were mounted in a loop and flash-cooled in an N2 gas stream at −173 °C. Diffraction data were collected using synchrotron radiation at SPring-8 (BL41XU with an ADSC Quantum 210 CCD detector, an ADSC Quantum 315 CCD detector, and a Rayonix MX225HE CCD detector) and at the Photon Factory (BL5A with an ADSC Quantum 315 CCD detector, and AR-NW12 with an ADSC Quantum 210 CCD detector). All data sets were processed and scaled using the programs DENZO and SCALEPACK from the HKL2000 package (36).
Phasing, Model Building, and Refinement
The structures were determined by the molecular replacement method using the apoenzyme structure (PDB code 1GEH) (22) as a search model with the program MOLREP (37) in CCP4i (38, 39). The structural models were constructed using the program COOT (40). All structures were refined using the program REFMAC5 (41) in CCP4i. The atomic coordinates of Tk-Rubisco were deposited in the Protein Data Bank under accession numbers 3A12 (wild-type), 3KDN (SP4), and 3KDO (SP6). The protein-ligand interaction was investigated with the program LIGPLOT (42). Superposition of the structures was performed using the program COOT, on the basis of secondary-structure matching. The figures of structures were prepared with PyMOL.
B-factor Comparison among Three Determined Structures
The averaged B-factor of each Cα atom among 10 subunits in the asymmetric unit was calculated for the wild-type enzyme, and the mutants SP4 and SP6, respectively. The averaged B-factor distributions against the residue numbers for mutants were adjusted to that for the wild-type enzyme with the least squares method. C-terminal regions (residues 386–444) were excluded from the adjusting calculation, because the differences between the wild-type enzyme and SP6 mutant values were exclusively high (over 10).
Kinetic Examination
The conditions for activity measurements in the kinetic examination of the wild-type enzyme and the SP5-V330T mutant were the same as those applied in our previous study (29).
Effects of Temperatures on Enzyme Stabilities
Thermostabilities of the wild-type enzyme and the SP5-V330T mutant were analyzed by measuring the residual carboxylase activities of the proteins after incubation at 90 or 100 °C in 100 mm Bicine-NaOH (pH 8.3) and 10 mm MgCl2. Denatured proteins were removed by centrifugation (20 min, 15,000 × g, 4 °C) and the supernatant was used for activity measurements. Residual activities were measured with the enzymatic assay methods described in our previous study (29).
RESULTS AND DISCUSSION
Structure Determination of Tk-Rubisco
We have determined the crystal structures of the wild-type Tk-Rubisco, and the mutants SP4 and SP6 in complex with 2-CABP at 2.30, 2.09, and 2.36 Å, respectively. The resolutions are improved compared with that of the wild-type apoenzyme (2.8 Å) (22). Final statistics for all data sets are shown in Table 2. Approximately 90% of the residues in all structures were in the most favored region in the Ramachandran plot (43). Although the crystals of the wild-type enzyme and the mutants belong to different space groups (P21212 and P21, respectively: Table 2), their overall structures are virtually identical. Superpositions of the SP4 and SP6 mutants on the wild-type enzyme result in a root mean square distance of 0.27 Å for 437 Cα atoms and 0.38 Å for 436 Cα atoms out of a total 444 residues in a subunit, respectively. The active sites in all 10 subunits are distant (more than 20 Å) from the adjacent physiological decamers in the crystal. Thus, we do not take the space group difference into consideration in the further discussion.
TABLE 2.
Crystallographic statistics
| Data set | Wild-type | SP4 | SP6 |
|---|---|---|---|
| Resolution (Å)a | 50-2.30 (2.34-2.30) | 50-2.09 (2.13-2.09) | 50-2.36 (2.40-2.36) |
| Completeness (%)a | 99.9 (100.0) | 96.5 (70.3) | 99.6 (99.2) |
| I/σ (I)a | 15.2 (3.9) | 15.4 (2.3) | 13.1 (2.5) |
| Rmerge (%)a,b | 10.5 (37.9) | 7.1 (38.4) | 8.2 (38.9) |
| Unit cell parameters | |||
| Space group | P21212 | P21 | P21 |
| a (Å) | 173.29 | 97.47 | 97.48 |
| b (Å) | 246.38 | 246.24 | 246.57 |
| c (Å) | 144.59 | 133.08 | 134.83 |
| β (°) | 104.10 | 104.73 | |
| Refinement | |||
| Resolution limits (Å) | 50–2.30 | 50–2.09 | 50–2.36 |
| Number of reflections | 271,962 | 345,208 | 249,468 |
| R/Rfree (%)c | 19.9/24.4 | 21.3/25.3 | 21.9/26.4 |
| Root mean square deviation for bond distances (Å) | 0.006 | 0.004 | 0.002 |
| Root mean square deviation for bond angles (°) | 0.995 | 0.768 | 0.589 |
| Model | 36,651 atoms | 36,999 atoms | 36,065 atoms |
| Proteind | 4,366 residues | 4,371 residues | 4,379 residues |
| Substrate | 10 CABP | 10 CABP | 10 CABP |
| Solvent molecules | 2,306 waters | 2,652 waters | 1,811 waters |
| 10 Mg2+ | 10 Mg2+ | 10 Mg2+ | |
| PDB entry | 3A12 | 3KDN | 3KDO |
a Values for the outermost resolution shell are shown in parentheses.
b Rmerge = ∑|Ii − 〈Ii〉|/∑<Ii>, where Ii is the observed intensity and 〈Ii〉 is the average intensity over symmetry equivalent measurements.
c R = ∑‖Fobs| − |Fcalc‖/∑|Fobs|. Rfree is the same as R, but for a 5% subset of all reflections that were never used in crystallographic refinement.
d They contain 10 carbamylated lysine residues (3 letter code: KCX).
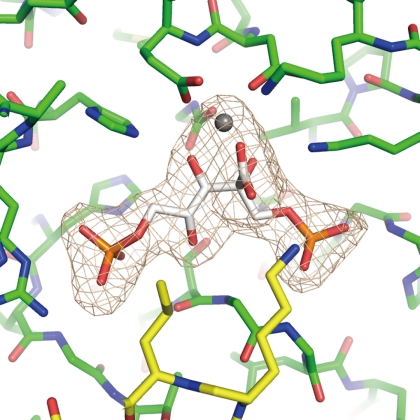
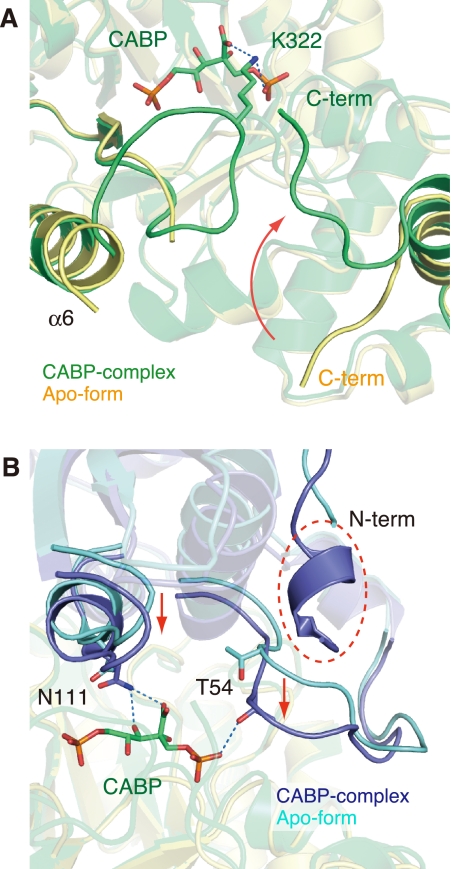
The inhibitor (2-CABP) and the metal (Mg2+) ion are clearly observed in the electron density maps at each of the 10 active sites in all three structures (Fig. 2). The active-site loop 6, which had been disordered in the apo form previously determined by our group (22), can also be observed. Superposition of the inhibitor complex (wild-type) on the apo form shows induced fit movements at the C-terminal region (from Gly-439 to Val-444), the two loops containing the active site residues (Thr-54 and Asn-111), and the N-terminal region (from Phe-5 to Tyr-11) to close the entrance of the active site (Fig. 3).
FIGURE 2.
Active site of Tk-Rubisco. A close-up view of the wild-type Tk-Rubisco is shown. Tk-Rubisco and a 2-CABP molecule are shown in green and white, respectively. An Mg2+ ion is shown as a gray sphere. Loop 6, which cannot be observed in the apo form, is shown in yellow. The electron density shown in light gray is contoured at 3.0 σ.
FIGURE 3.
Comparison between the inhibitor complex and the apo form. Both forms are the wild-type enzymes. A, the environment around the active site. The complex with 2-CABP and the apo form (PDB code 1GEH) are shown in green and yellow, respectively. A 2-CABP molecule is shown as a stick model, and hydrogen bonds are indicated by blue dashed lines. The movement of the C-terminal loop is indicated by a red arrow. B, the molecule opposite (A) in the functional dimer. The complex with 2-CABP and the apo form are shown in dark and light blues, respectively. The movements of the loops are indicated by red arrows, and the N-terminal region, which cannot be observed in the apo form, is indicated by a red dashed circle.
Comparison of Wild-type Tk-Rubisco with the Mutants
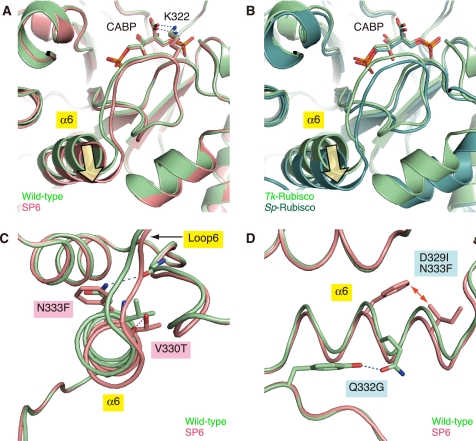
As described above, the overall structure of the wild-type Tk-Rubisco, and the mutants SP4 (with equivalent activity to the wild-type enzyme at ambient temperatures) and SP6 (with enhanced activity) can be superposed well on each other. However, the position of the α-helix 6 region in the SP6 mutant is different from that of the wild-type enzyme (Fig. 4A). The N-terminal side of the helix appears to be tilted toward the solvent region compared with that of the wild-type enzyme (yellow arrow in Fig. 4A). This mutant was created by replacement of the α-helix 6 of Tk-Rubisco with the corresponding region of Sp-Rubisco (PDB code 8RUC) (44), and the tilted conformation of the helix is also seen in Sp-Rubisco (yellow arrow in Fig. 4B). The helix in the SP4 mutant is less dislocated than that in the SP6 mutant (supplemental Fig. S1). These results imply that the tilted conformation is related to the higher activity at ambient temperatures.
FIGURE 4.
Superposition of wild-type Tk-Rubisco on the SP6 mutant (A, C, and D) or on Sp-Rubisco (B). Light green, red, and dark green schematics represent the structures of the wild-type Tk-Rubisco, the SP6 mutant, and the wild-type Sp-Rubisco, respectively. A and B, environment around the active site. Hydrogen bonds are indicated by blue (wild-type) and red (SP6) dashed lines, respectively. Yellow arrows indicate the tilt of the α-helix 6. C, close-up view from the N-terminal side of α-helix 6. D, close-up view from the side of α-helix 6. The red arrow indicates the unfavored steric interaction.
This helix tilt can be explained by the local flexibility introduced by the following four factors. The first factor is the loss of a hydrogen bond by the mutation N333F (Fig. 4C). The hydrogen bond between the side chain of Asn-333 and the main chain O atom of Val-315 in the wild-type Tk-Rubisco is broken by the N333F replacement in the SP6 mutant. The α-helix 6 in the SP6 mutant loses an anchor to its neighboring secondary structure and becomes flexible. Moreover, this factor cooperates with the bulky group of Phe-333 pushing the α-helix 6 to shift from its original position, and makes this helix more unstable. Sp-Rubisco, where a phenylalanine residue is located at this position (Phe-345), does not have the hydrogen bond (44). The N333F replacement in the SP4 mutant (SP4-N333F, Fig. 1A) showed an 88% higher carboxylase activity (kcat(C2)) and a 20% higher kcat(CO2)/KCO2 than the original SP4 mutant (29), which also supports the notion that this residue contributes to an increase in the activity.
The second factor is the loss of a hydrogen bond by the mutation Q332G (Fig. 4D). The hydrogen bond between the amido oxygen atom of Gln-332 and the hydroxy group on Tyr-342 in the wild-type Tk-Rubisco is broken by Q332G replacement in the SP6 mutant. The absence of this bond can also be presumed to result in an increase in α-helix 6 flexibility.
The third factor is the steric hindrance caused by the double mutation D329I/N333F (Fig. 4D). The side chains of Ile-329 and Phe-333 in the SP6 mutant are in unfavorable interatomic contact, whereas Asp-329 and Asn-333 in the wild-type enzyme are well accommodated. The averaged distance between Ile-329 and Phe-333 among the 10 molecules in the asymmetric unit is ∼3.2 Å. This contact may bend the helix toward the solvent region. This factor does not appear in Sp-Rubisco but is present in the SP6 mutant (supplemental Fig. S2).
The fourth factor is hydrogen bond formation by the mutation V330T (Fig. 4C). A new hydrogen bond is introduced between the hydroxy group of Thr-330 and the main chain O atom of Glu-326 in the SP6 mutant. This bond connects the α-helix 6 with loop 6, and allows loop 6 to move relative to that in the wild-type enzyme, as shown in Fig. 4C. The corresponding hydrogen bond is conserved in Sp-Rubisco (44). The introduction of a threonine residue into this helix may also be destabilizing because this residue is constrained to essentially one conformation in an α-helix at an entropic cost. The importance of this position is also supported by the findings for the T342V mutant in a cyanobacterial type I Rubisco (45) (corresponding to Val-330 in Tk-Rubisco), which exhibits decreased carboxylase activity. The other three factors described above promote the flexibility of the α-helix 6, whereas this fourth factor functions to transfer the flexibility to the active-site loop 6. It is suggested that the flexibility, mainly occurring by breaking the anchors between this helix and the core of Tk-Rubisco, contributes to the mobility of loop 6 and α-helix 6, and this mobility is favorable for increasing the CO2 incorporation rate and the kcat(CO2) value at ambient temperatures.
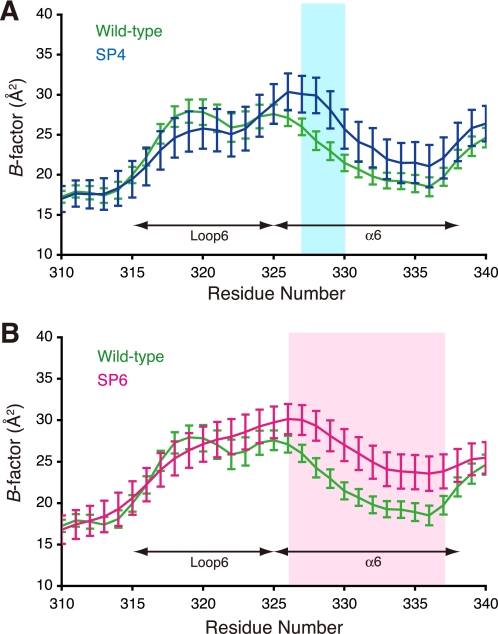
The flexibility of protein structure affects the crystallographic temperature factors (B-factors), which are related with thermal vibrations. A comparison of the B-factors among the three structures reveals that mutated residues in both the SP4 and SP6 mutants show significantly higher B-factors than the corresponding residues in the wild-type enzyme (Fig. 5; the results for all residues are shown in supplemental Fig. S3). These larger B-factor values also indicate the local flexibility of the mutated residues. Because more residues were mutated in the SP6 mutant than in the SP4 mutant, the α-helix 6 in the SP6 mutant can be considered to be more flexible than that in the SP4 mutant. This difference may be one of the reasons why the activity was not enhanced by the SP4 mutant but was enhanced by the SP6 mutant relative to the wild-type enzyme. The B-factors of loop 6 (residues 315–325) are almost equivalent between the wild-type Tk-Rubisco and the SP6 mutant. This can be explained by the hydrogen-bond fixation between Lys-322 on this loop and the inhibitor (Fig. 4A). The loop 6 motion of the SP6 mutant is probably larger than that of the wild-type enzyme when they are not bound with the inhibitor.
FIGURE 5.
Comparison of the B-factors. Each data point represents the adjusted B-factor of each Cα atom ± S.E. The B-factor values for the wild-type Tk-Rubisco as well as the mutants SP4 (A) and SP6 (B) are shown in green, blue, and red, respectively. The apparent differences between the wild-type data and the data for each mutant are shown in the colored regions (SP4, light blue; SP6, pink).
The tilt of α-helix 6 has also been observed in the V331A/G344S mutant of Rubisco from the green alga Chlamydomonas reinhardtii (46). This tilt supports the formation of a hydrophobic patch between residues 331 and 342 (corresponding to Ala-318 and Val-330 in Tk-Rubisco, respectively) (supplemental Fig. S4). Disruption of this hydrophobic patch is known to result in the loss of enzymatic activity based on various mutagenesis studies (46–48). Ala-318 in Tk-Rubisco is an insertion residue in loop 6 compared with other Rubiscos (supplemental Fig. S4A), and Ala-318 is close to Val-330 (supplemental Fig. S4B). A mutant with single amino acid deletion in this loop completely lost activity (reported as an SP1 mutant in our previous study (29)), supporting that the hydrophobic patch is vital for the activity. However, as both the wild-type enzyme and the SP6 mutant maintain the hydrophobic patch, this patch can be presumed to have little effect on the difference of enzymatic activity between these two enzymes.
Creating a More Active Mutant Based on Structural Analyses
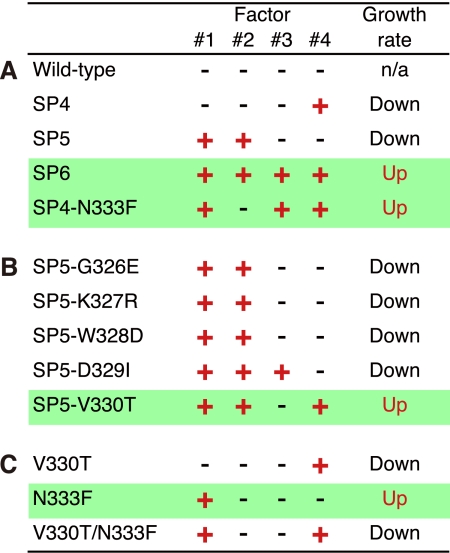
The four factors, related with the flexibility and the higher activity of Tk-Rubisco at ambient temperatures, are summarized in Fig. 6 (the factors introduced by the mutations N333F, Q332G, D329I/N333F, and V330T are named 1, 2, 3, and 4, respectively). In the SP6 mutant with enhanced activity, the entire α-helix 6 (residues 326–336, Fig. 1A) is replaced with the corresponding region of Sp-Rubisco, which satisfies all four factors (Fig. 6A). In contrast, the SP4 mutant, which is deficient compared with the wild-type enzyme, is replaced with only the N-terminal half of the α-helix 6 (residues 326–330), and satisfies only factor number 4 (by the mutation V330T). In a previous study on the mutagenesis of the SP4 mutant (29), the SP4-N333F mutant, which has a higher activity than the wild-type Tk-Rubisco, satisfies factors 1 (by the mutation N333F) and 3 (by the mutations D329I, originally contained in the SP4 mutant, and N333F) in addition to factor 4 (Fig. 6A). In this study, we have therefore attempted to optimize the SP5 mutant to create a Rubisco with further enhancement of the activity.
FIGURE 6.
The four factors playing the dominant role in the Tk-Rubisco activity. Factors 1, 2, 3, and 4 are the loss of a hydrogen bond by the mutation N333F, the loss of a hydrogen bond by the mutation Q332G, the steric hindrance by the double mutation D329I/N333F, and a new hydrogen bond by the mutation V330T, respectively. The growth rate was measured using Δ3c strains harboring each mutant. The red “+” satisfies the corresponding factor and the black “−” does not. Green lines highlight the mutants improving the growth rates. A, the wild-type Tk-Rubisco and mutants previously reported (25). B, SP5-mutant derivatives. C, the mutants used to investigate Val-330 and Asn-333.
The SP5 mutant, which also is inferior to the wild-type Tk-Rubisco in terms of the enzymatic activity, is replaced with the C-terminal half of the α-helix 6 (residues 331–336, Fig. 1A), and satisfies factors 1 (by the mutation N333F) and 2 (by the mutation Q332G) (Fig. 6A). These two factors are common in the mutants SP5 and SP6, the latter of which has higher activity than the wild-type enzyme. To investigate the contribution of the other two factors toward the enzymatic activity, we created mutants SP5-D329I (satisfying factor 3 by the mutations D329I and N333F, originally contained in the SP5 mutant, in addition to factors 1 and 2) and SP5-V330T (satisfying factor 4 by the mutation V330T in addition to factors 1 and 2) (Fig. 6B). These two mutants were expected to show improved enzymatic activities at ambient temperatures. Additionally, three mutants (SP5-G326E, SP5-K327R, and SP5-W328D) were also constructed to cover the other three residues, which in the SP5 mutant are different from those in the SP6 mutant (Fig. 1B), for comparison. All of these novel mutants were introduced into the R. palustris Δ3 strain to check their activities in vivo.
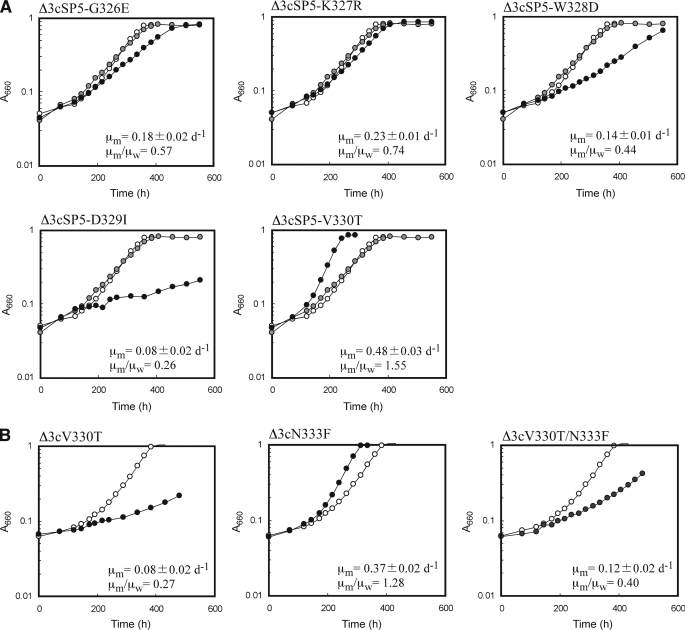
R. palustris Δ3cSP5-V330T cells, which harbored the SP5-V330T gene on the chromosome, exhibited a higher specific growth rate (0.48 day−1) under photoautotrophic conditions than the wild-type Tk-Rubisco (0.31 day−1) or any of the mutants reported in the present and previous papers (29) (Fig. 7A). The kinetic analyses of the SP5-V330T mutant in vitro at ambient temperatures also revealed that its kcat(CO2) and kcat(CO2)/KCO2 values were 71 and 57% higher than those of the wild-type enzyme, respectively (Table 3). This kcat(CO2)/KCO2 value was the highest among all the mutants we have measured to date (29). These enhanced activities both in vivo and in vitro must be related with an increase of flexibility around α-helix 6. The significant decrease of the thermostability of the SP5-V330T mutant at both 90 and 100 °C compared with the wild-type enzyme (Table 4) also supports an increase of flexibility in the SP5-V330T mutant. Intriguingly, on the other hand, the cells harboring the SP5-D329I gene (Δ3cSP5-D329I) grew more slowly than those with the wild-type Tk-Rubisco gene (Δ3cTk-Rubisco) (Fig. 7A). As expected, the other three mutants, SP5-G326E, SP5-K327R, and SP5-W328D, also did not show an increase in the specific growth rates of the R. palustris Δ3 strains. In these three mutants as well as the SP5-D329I mutant, the flexibilities around α-helices 6 are most likely insufficient to increase the enzymatic activities. Furthermore, as a strong and reliable correlation exists between cell growth rate and carboxylase activity (25), they probably also show lower carboxylase activities in vitro.
FIGURE 7.
Representative photoheterotrophic growth curves of the Δ3c strains. Open, gray, and closed circles indicate the growth of the cells harboring the wild-type Tk-Rubisco, the SP5 mutant, and targeted mutants, respectively. The average specific growth rate of each strain harboring the Tk-Rubisco mutant (μm) (3 to 5 measurements; n = 3 to 5) was compared with that harboring the wild-type enzyme (μw). A, SP5 mutant derivatives. B, the mutants used to investigate Val-330 and Asn-333. The averaged μw values were 0.31 ± 0.02 day−1 (n = 4) (A) and 0.29 ± 0.02 day−1 (n = 3) (B). The corresponding value for Δ3cSP5 was 0.23 ± 0.02 day−1 (n = 4).
TABLE 3.
Kinetic properties of purified Tk-Rubisco and its mutant
| Kinetic constant | Wild-type | SP5-V330T | Spinach (30) |
|---|---|---|---|
| μ = 0.30 day−1a | μ = 0.48 day−1a | ||
| VCO2 (μmol/min/mg) | 0.37 ± 0.02 | 0.64 ± 0.03 | 2.3 |
| kcat(CO2) (s−1·site−1) | 0.31 | 0.53 | 2.6 |
| KCO2 (μm) | 60 ± 7 | 67 ± 6 | 14.3 |
| kcat(CO2)/KCO2 | 0.0051 | 0.0080 | 0.182 |
| VO2 (μmol/min/mg) | 0.28 | 0.25 | |
| kcat(O2) (s−1·site−1) | 0.23 | 0.21 | |
| KO2 (μm) | 267 ± 0 | 123 ± 0 | |
| kcat(O2)/KO2 | 0.00085 | 0.00169 | |
| τ (VCO2KO2/VO2KCO2) | 6.0 ± 0.4 | 4.7 ± 0.3 | 93.8 ± 0.8 |
| KRuBP (μm) | < 0.5 | < 0.5 |
a μ represents specific growth rates of recombinant strains that harbor the indicated proteins noted.
TABLE 4.
Thermostability of wild-type Tk-Rubisco and its mutant
| Temperature | Half-lives of protein activities |
|
|---|---|---|
| Wild-type | SP5-V330T | |
| min | ||
| 90 °C | 220 ± 10 | 150 ± 3 |
| 100 °C | 48 ± 2 | 2.9 ± 0.2 |
All of the mutants with enhanced activities so far (SP6, SP4-N333F, and SP5-V330T) satisfy factors 1 and 4 (25) (green lines in Fig. 6, A and B). To analyze the importance of these two factors for the improvement of the enzymatic activities, three additional mutants, V330T (satisfying factor 4), N333F (satisfying factor 1), and V330T/N333F (satisfying both factors 1 and 4) were constructed (Figs. 1C and 6C), and introduced into the R. palustris Δ3 strain (Δ3cV330T, Δ3cN333F, and Δ3cV330T/N333F, respectively). As a result, the specific growth rates of Δ3cV330T and Δ3cV330T/N333F were slower (0.08 and 0.12 day−1, respectively) than that of Δ3cTk-Rubisco (0.29 day−1), whereas that of Δ3cN333F was higher (0.37 day−1) (Fig. 7B). The results for the N333F mutant indicate that the mutation N333F alone improves functional performance at ambient temperatures, and more investigations will be needed to completely clarify the mechanism. The results for the V330T mutant are as expected because this mutant lacks the essential factor 1. Although the double mutant V330T/N333F satisfies both factors 1 and 4, Δ3cV330T/N333F displayed a much lower specific growth rate compared with Δ3cTk-Rubisco. This suggests that these two factors alone are insufficient to increase activity. The three mutants with enhanced activities satisfy, in addition to factors 1 and 4, factors 2 and 3 (SP6), factor 3 (SP4-N333F), and factor 2 (SP5-V330T), respectively (Fig. 6, A and B). These results indicate that either one of factors 2 or 3 is also required, together with factors 1 and 4, to obtain an increase in activity. The slower growth rate in the SP5-D329I mutant described above can be explained by the lack of the essential factor 4.
In summary, we determined the crystal structures of the wild-type Tk-Rubisco, and the mutants SP4 and SP6. Analyses of the differences among these structures revealed that the four factors represented in Fig. 6 play a major role in determining levels of enzymatic activity at ambient temperatures. This finding led us to create the mutant SP5-V330T, the most active examined thus far. The R. palustris Δ3 strain harboring this mutant reached the specific growth rates that are comparable (∼80%) with those of cells with the native Rubiscos from R. palustris (25). Although lower than the wild-type enzyme, the SP5-V330T mutant still exhibits remarkable thermostability (Table 4). It indicates that this mutant can surely tolerate additional mutations without losing its basic structure, which is promising for further engineering of the enzyme to obtain a Rubisco with activity to exceed the performance of the current enzyme, and eventually, native Rubiscos from mesophilic sources.
Supplementary Material
Acknowledgments
We thank the beamline scientists and technical staff at the Photon Factory and SPring-8.
This work was supported by the Targeted Proteins Research Program (B-29) from the Ministry of Education, Culture, Sports, Science and Technology, Japan. The synchrotron radiation experiments were performed at the Photon Factory and SPring-8 with the approval of the Photon Factory Advisory Committee (proposals 2008S2-001 and 2008G149) and the Japan Synchrotron Radiation Research Institute (JASRI) (proposals 2008A1511, 2008A1973, 2008A2001, 2008A2048, 2009A1012, and 2009A1013).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The atomic coordinates and structure factors (codes 3A12, 3KDN, and 3KDO) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CBB
- Calvin-Benson-Bassham
- 2-CABP
- 2-carboxy-d-arabinitol 1,5-bisphosphate
- Rubisco
- ribulose-1,5-bisphosphate carboxylase/oxygenase
- Tk-Rubisco
- Rubisco from Thermococcus kodakarensis
- Sp-Rubisco
- Rubisco from spinach
- PDB
- Protein data bank
- Bicine
- N,N-bis(2-hydroxyethyl)glycine.
REFERENCES
- 1.Watson G. M., Tabita F. R. (1997) FEMS Microbiol. Lett. 146, 13–22 [DOI] [PubMed] [Google Scholar]
- 2.Shively J. M., van Keulen G., Meijer W. G. (1998) Annu. Rev. Microbiol. 52, 191–230 [DOI] [PubMed] [Google Scholar]
- 3.Andrews T. J., Lorimer G. H. (1987) in The Biochemistry of Plants (Hatch M. D., Boardman N. K. eds) pp. 131–218, Academic Press, San Diego [Google Scholar]
- 4.Harpel M. R., Serpersu E. H., Hartman F. C. (1995) in Techniques in Protein Chemistry (Crabb J. W. ed) pp. 357–364, Academic Press, San Diego [Google Scholar]
- 5.Andersson I., Backlund A. (2008) Plant Physiol. Biochem. 46, 275–291 [DOI] [PubMed] [Google Scholar]
- 6.Knight S., Andersson I., Brändén C. I. (1990) J. Mol. Biol. 215, 113–160 [DOI] [PubMed] [Google Scholar]
- 7.Schneider G., Lindqvist Y., Brändén C. I., Lorimer G. (1986) EMBO J. 5, 3409–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider G., Lindqvist Y., Lundqvist T. (1990) J. Mol. Biol. 211, 989–1008 [DOI] [PubMed] [Google Scholar]
- 9.Hartman F. C., Harpel M. R. (1994) Annu. Rev. Biochem. 63, 197–234 [DOI] [PubMed] [Google Scholar]
- 10.Schneider G., Lindqvist Y., Brändén C. I. (1992) Annu. Rev. Biophys. Biomol. Struct. 21, 119–143 [DOI] [PubMed] [Google Scholar]
- 11.Gatenby A. A. (1988) Photosynth. Res. 17, 145–157 [DOI] [PubMed] [Google Scholar]
- 12.van der Vies S. M., Bradley D., Gatenby A. A. (1986) EMBO J. 5, 2439–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gatenby A. A., van der Vies S. M., Bradley D. (1985) Nature 314, 617–620 [Google Scholar]
- 14.Gutteridge S., Sigal I., Thomas B., Arentzen R., Cordova A., Lorimer G. (1984) EMBO J. 3, 2737–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somerville C. R., Somerville S. C. (1984) Mol. Gen. Genet. 193, 214–219 [Google Scholar]
- 16.Parry M. A., Andralojc P. J., Mitchell R. A., Madgwick P. J., Keys A. J. (2003) J. Exp. Bot. 54, 1321–1333 [DOI] [PubMed] [Google Scholar]
- 17.Hanson T. E., Tabita F. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4397–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashida H., Saito Y., Kojima C., Kobayashi K., Ogasawara N., Yokota A. (2003) Science 302, 286–290 [DOI] [PubMed] [Google Scholar]
- 19.Ezaki S., Maeda N., Kishimoto T., Atomi H., Imanaka T. (1999) J. Biol. Chem. 274, 5078–5082 [DOI] [PubMed] [Google Scholar]
- 20.Watson G. M., Yu J. P., Tabita F. R. (1999) J. Bacteriol. 181, 1569–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finn M. W., Tabita F. R. (2003) J. Bacteriol. 185, 3049–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitano K., Maeda N., Fukui T., Atomi H., Imanaka T., Miki K. (2001) Structure 9, 473–481 [DOI] [PubMed] [Google Scholar]
- 23.Sato T., Atomi H., Imanaka T. (2007) Science 315, 1003–1006 [DOI] [PubMed] [Google Scholar]
- 24.Maeda N., Kitano K., Fukui T., Ezaki S., Atomi H., Miki K., Imanaka T. (1999) J. Mol. Biol. 293, 57–66 [DOI] [PubMed] [Google Scholar]
- 25.Yoshida S., Inui M., Yukawa H., Kanao T., Tomizawa K., Atomi H., Imanaka T. (2006) J. Biotechnol. 124, 532–544 [DOI] [PubMed] [Google Scholar]
- 26.Maeda N., Kanai T., Atomi H., Imanaka T. (2002) J. Biol. Chem. 277, 31656–31662 [DOI] [PubMed] [Google Scholar]
- 27.Schreuder H. A., Knight S., Curmi P. M., Andersson I., Cascio D., Brändén C. I., Eisenberg D. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9968–9972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor T. C., Andersson I. (1996) Nat. Struct. Biol. 3, 95–101 [DOI] [PubMed] [Google Scholar]
- 29.Yoshida S., Atomi H., Imanaka T. (2007) Appl. Environ. Microbiol. 73, 6254–6261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uemura K., Anwaruzzaman, Miyachi S., Yokota A. (1997) Biochem. Biophys. Res. Commun. 233, 568–571 [DOI] [PubMed] [Google Scholar]
- 31.Atomi H., Fukui T., Kanai T., Morikawa M., Imanaka T. (2004) Archaea 1, 263–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morikawa M., Izawa Y., Rashid N., Hoaki T., Imanaka T. (1994) Appl. Environ. Microbiol. 60, 4559–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujii T., Nakazawa A., Sumi N., Tani H., Ando A., Yabuki M. (1983) Agric. Biol. Chem. 47, 2747–2753 [Google Scholar]
- 34.Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 35.Lorimer G. H., Miziorko H. M. (1980) Biochemistry 19, 5321–5328 [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 37.Vagin A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 38.Collaborative Computational Project N. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 39.Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 40.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 41.Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 42.Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 43.Laskowski R. A., Moss D. S., Thornton J. M. (1993) J. Mol. Biol. 231, 1049–1067 [DOI] [PubMed] [Google Scholar]
- 44.Andersson I. (1996) J. Mol. Biol. 259, 160–174 [DOI] [PubMed] [Google Scholar]
- 45.Read B. A., Tabita F. R. (1994) Arch. Biochem. Biophys. 312, 210–218 [DOI] [PubMed] [Google Scholar]
- 46.Karkehabadi S., Satagopan S., Taylor T. C., Spreitzer R. J., Andersson I. (2007) Biochemistry 46, 11080–11089 [DOI] [PubMed] [Google Scholar]
- 47.Chen Z. X., Yu W. Z., Lee J. H., Diao R., Spreitzer R. J. (1991) Biochemistry 30, 8846–8850 [DOI] [PubMed] [Google Scholar]
- 48.Chen Z. X., Spreitzer R. J. (1989) J. Biol. Chem. 264, 3051–3053 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.