Abstract
Fredericamycin (FDM) A is a pentadecaketide natural product that features an amide linkage. Analysis of the fdm cluster from Streptomyces griseus ATCC 43944, however, failed to reveal genes encoding the types of amide synthetases commonly seen in natural product biosynthesis. Here, we report in vivo and in vitro characterizations of FdmV, an asparagine synthetase (AS) B-like protein, as an amide synthetase that catalyzes the amide bond formation in FDM A biosynthesis. This is supported by the findings that (i) inactivation of fdmV in vivo afforded the ΔfdmV mutant strain SB4027 that abolished FDM A and FDM E production but accumulated FDM C, a biosynthetic intermediate devoid of the characteristic amide linkage; (ii) FdmV in vitro catalyzes conversion of FDM C to FDM B, a known intermediate for FDM A biosynthesis (apparent Km = 162 ± 67 μm and kcat = 0.11 ± 0.02 min−1); and (iii) FdmV also catalyzes the amidation of FDM M-3, a structural analog of FDM C, to afford amide FDM M-6 in vitro, albeit at significantly reduced efficiency. Preliminary enzymatic studies revealed that, in addition to the common nitrogen sources (l-Gln and free amine) of class II glutamine amidotransferases (to which AS B belongs), FdmV can also utilize l-Asn as a nitrogen donor. The amide bond formation in FDM A biosynthesis is proposed to occur after C-8 hydroxylation but before the carbaspirocycle formation.
Keywords: Antibiotics, Bacterial Genetics, Bacterial Metabolism, Enzyme Catalysis, Enzyme Mechanisms, Enzymes, Asparagine Synthetase, Natural Product Biosynthesis
Introduction
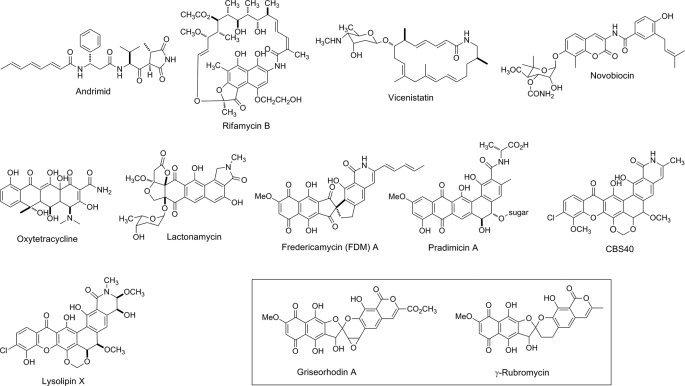
Amide bonds are common in natural products and represent fundamental linkages for numerous ribosomal and nonribosomal peptide-derived molecules; they are formed by the ribosome or by the condensation domain of nonribosomal peptide synthetases, respectively. Amide bonds can also be introduced into diverse natural products via different manifolds (Fig. 1). For instance, AdmF, a transglutaminase homolog, catalyzes amide bond formation in andrimid, highlighting a novel condensation strategy (1, 2). In the ansamycin-type antibiotics, the macrolactam ring in rifamycin is formed by an amide synthetase, RifF (3); whereas in some other macrolactams, like vicenistatin, the amide is proposed to result from thioesterase domain docking at the C terminus of the polyketide synthase (4). Alternative scenarios invoke amide bond formation as a tailoring step following construction of the core scaffold. For instance, in the aminocoumarin antibiotics, the amide bonds linking the 3-amino-4,7-dihydroxycoumarin moiety and related acyl moieties are formed by amide synthetases such as NovL (for novobiocin) and its homologs CloL, CouL, and SimL (5, 6). Novel novobiocin analogs can be envisioned by substituting NovL with the other three amide synthetases with significant substrate specificity discrepancies.
FIGURE 1.
Structures of selected amide bond-containing natural products with γ-rubromycin and griseorhodin A (boxed) as exceptions whose biosynthetic gene clusters feature FdmV homologs that apparently do not function as amide synthetases.
As a major class of natural products, aromatic polyketides display an amazingly creative occupancy of chemical space translating often to their profound bioactivities. Despite the diversity of chemical structures displayed by aromatic polyketides, very few of them contain amide moieties. Several amide-containing aromatic polyketides are shown in Fig. 1. Among them, the amide moiety in oxytetracycline is proposed to originate from a specific malonamyl starter unit (7, 8). In the case of lactonamycin, labeling studies suggest that the amino group originates from glycine or a glycine-derived starter unit (9). Fredericamycin (FDM)2 A (10), pradimicin A (11), CBS40 (12), and lysolipin X (13) are all aromatic polyketides originating from polyketide chains of at least 24 carbons in length. Amide construction in these natural products has not yet received significant attention, and thus the mechanisms associated with these transformations remain obscure.
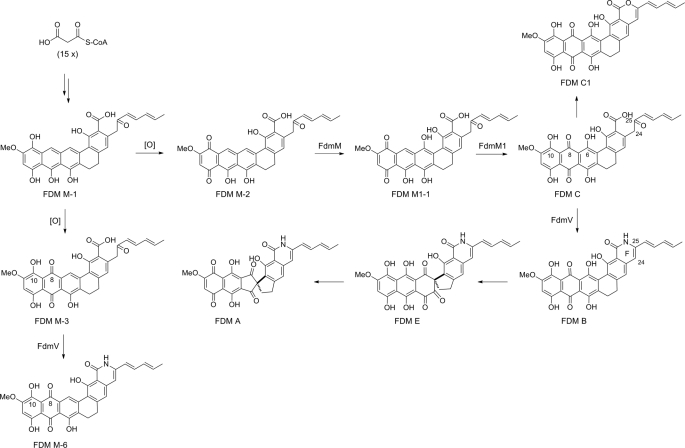
FDM A is a pentadecaketide-derived aromatic compound isolated from Streptomyces griseus ATCC 49344 in 1981 (10, 14). It is notable for its remarkable antitumor potential and its unique carbaspirocyclic structure (15, 16). We previously cloned and sequenced the fdm gene cluster, setting the stage to study the biosynthetic pathway responsible for FDM A production (17). Reexamination of S. griseus wild-type fermentation recently resulted in the isolation of three FDM analogs FDM B, FDM C, and FDM E (18, 19) (Fig. 2). FDM B and FDM E both contain the lactam ring F, and FDM C has a carboxylic acid group comparably situated. These facts support the proposal that the amide nitrogen of FDM A is incorporated following installation of the polycyclic ring system. The isolation of four new FDM analogs, FDM M-1, FDM M-2, FDM M-3, and FDM M1-1, all sharing the same FDM C scaffold, from the ΔfdmM or ΔfdmM1 mutant strains further supports the idea of amide formation as a postpolyketide synthase tailoring step (20) (Fig. 2). Finally, the lactam ring F can be found in both FDM A and FDM E, indicating that the spirocyclic moiety is installed following amide formation (19). Bioinformatics analysis, however, failed to reveal any typical amide synthetase gene within the cloned fdm biosynthetic gene cluster. FdmV, an asparagine synthetase (AS) B homolog, was thus postulated to catalyze amide bond formation en route to FDM B (17).
FIGURE 2.
Biosynthetic pathway for FDM A supported by FDM intermediates isolated from S. griseus wild-type and ΔfdmM, ΔfdmM1, and ΔfdmV mutant strains.
Here, we report in vivo and in vitro characterizations of FdmV as an amide synthetase that catalyzes the conversion of FDM C to FDM B in FDM A biosynthesis. FdmV can also convert FDM M-3 to the corresponding amide FDM M-6 and can utilize l-Asn, in addition to l-Gln and free amine, as a nitrogen source. FdmV represents a subgroup of AS B-like amide synthetases that are responsible for amide bond incorporation into several different kinds of secondary metabolites.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
Escherichia coli DH5α (21), BL21 (21), and ET12567/pUZ8002 (22) were grown in Luria-Bertani broth (21). S. griseus ATCC 49344 wild-type and recombinant strains were cultured in R2YE medium (23) for seed cultures and in Antibiotic Producing Medium for FDM production (17). For protoplast preparation, Streptomyces lividans K4-114 (24) and S. griseus strains were grown in YEME medium (23). MYME was used for genomic DNA isolation (23). Plasmids pGEM-3Zf(+) (Promega), pSPORT1 (Invitrogen), and pET29a(+) (Novagen) were from commercial sources. Plasmids pBS4028 (17), pBS4045 (25), pSET151 (23), pSET152 (23), pUWL201PW (26), and pWHM1250 (27) were described previously. Ampicillin at 100 μg/ml, apramycin at 50 μg/ml, kanamycin at 50 μg/ml, and thiostrepton at 10 μg/ml were used, respectively, in liquid media for selection (21, 23).
DNA Manipulation and Sequence Analyses
Restriction enzymes, T4 DNA ligase, and Pfx DNA polymerase were purchased from Invitrogen. DNA restriction and ligation reactions were performed according to the manufacturer's recommendations. Competent E. coli cells were prepared and transformed according to standard procedures (21). Streptomyces genomic DNA isolation, protoplast preparation, and transformation were performed as described previously (23). Southern hybridization was carried out with DIG Easy Hybridization kit (Roche Applied Sciences) following the manufacturer's instructions. Homologous sequence database searching and multiple alignments were executed with BLASTP and ClustalX, respectively.
Construction and Complementation of the ΔfdmV Mutant
To construct the ΔfdmV mutant strain SB4027, a 5.5-kb PstI fragment containing the fdmV gene was first excised from pBS4028 and inserted into the same site of pGEM-3Zf(+) to yield pBS4057. A 0.8-kb BamHI fragment of the aac(3)IV apramycin resistance gene was then cloned from pSET152 (23) and inserted into pBS4057 to replace a 1.2-kb BglII fragment, internal to fdmV, to afford pBS4058. The mutated fdmV gene, with the aac(3)IV gene replacing an internal fragment of fdmV, was subsequently recovered as a 5.1-kb PstI fragment from pBS4058 and cloned into the same site of pSET151 to afford pBS4059. pBS4059 was introduced into the S. griseus wild-type strain via E. coli-S. griseus conjugation. Exconjugants that were apramycin-resistant and thiostrepton-sensitive were selected as the desired ΔfdmV mutant strain SB4027, the genotype of which was confirmed by Southern blot analysis using the 0.8-kb SphI–BglII fragment containing the fdmU gene as a probe (supplemental Fig. S1). Finally, the previously reported plasmid pBS4045, which overproduces the fdm activator FdmR1 (25), was introduced into SB4027 via protoplast transformation, affording the ΔfdmV variant SB4028.
To construct the fdmV expression plasmid pBS4060, the fdmV gene was first moved as a 2.5-kb SrfI–NotI fragment from pBS4028 into the Ecl136II–NotI sites of pSPORT1, then recovered as a 2.5-kb PstI–HindIII fragment, and finally inserted into the same sites of pWHM1250 to afford pBS4060. Introduction of pBS4060 into SB4027 via protoplast transformation yielded the recombinant strain SB4029, in which the ΔfdmV mutation was complemented by the expression of a functional copy of fdmV in trans.
Production and Isolation of FDM C
The ΔfdmV mutant strain SB4028 was fermented under similar conditions as the S. griseus wild-type (25) for the production of FDM C. Briefly, after 6 days at 28 °C, 250 rpm, the cultures were acidified to pH 2.0 with 2 m HCl. The pellet was harvested by centrifugation (3,000 × g, 10 min) and extracted three times with acetone. Following solvent removal in vacuo, the residue was extracted with EtOAc three times. Solvent removal in vacuo was followed by chromatography over a polyamide 6 column (Fluka, Steinheim, Germany) using CHCl3-CH3OH in a gradient from 100:0 to 70:30. FDM C was eluted mainly with a 70:30 ratio of CHCl3-CH3OH. These fractions containing FDM C were then concentrated and subjected to silica gel chromatography (230–400 mesh; Natland Inc., Morrisville, NC) eluted with CHCl3-CH3OH-CH3COOH (200:0:0.1, 190:10:0.1, 180:20:0.1, 170:30:0.1, and 150:50:0.1). FDM C was eluted by CHCl3-CH3OH-CH3COOH 170:30:0.1. The FDM C-containing fractions were finally concentrated and purified by semipreparative HPLC on an Alltima C-18 column (5 μm, 250 × 10 mm; Alltech., Deerfield, IL) as described previously (19).
Overproduction and Purification of FdmV in S. lividans
To construct the fdmV overexpression plasmid pBS4061, the fdmV gene was first cloned by PCR using 5′-GTAATTCCATATGTGCGGCATCGCAGGCTG-3′ (NdeI underlined) as an upstream primer and 5′-GATTCTCGAGCAGCTCGACGTCGCCC-3′ (XhoI underlined) as a downstream primer. The resultant product was sequenced to confirm PCR fidelity and cloned as a 1.9-kb NdeI–XhoI fragment into the same sites of pET29a(+) to afford pBS4062. It was then moved as a 2.5-kb NdeI–ApoI fragment, containing the fdmV gene with the C-His6 tag sequence from pBS4062, and inserted into the NdeI–EcoRI sites of pUWL201PW to yield pBS4061, in which the expression of fdmV is under the control of the constitutive ErmE* promoter (23). To overproduce FdmV, pBS4061 was transformed into S. lividans K4-114 to afford SB4030. Spores of S. lividans SB4030 were inoculated into R2YE with thiostrepton and grown at 28 °C, 250 rpm overnight. The resultant seed culture was used to inoculate the same medium and fermented under the same conditions for 3 days. Cells were harvested by centrifugation, resuspended in 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, treated with lysozyme (1 mg/ml) for 1 h at 30 °C, and lysed by sonication. The lysis mixture was centrifuged at 12,000 × g for 30 min, and the resulting supernatant was loaded onto a nickel-nitrilotriacetic acid-agarose column (Qiagen, Valencia, CA) preequilibrated with binding buffer (300 mm NaCl, 20 mm Tris-HCl, pH 8.0, and 5 mm imidazole). The column was washed with 10 volumes of binding buffer followed by 6 volumes of washing buffer (300 mm NaCl, 20 mm Tris-HCl, pH 8.0, and 20 mm imidazole). FdmV was eluted with 6 volumes of elution buffer (300 mm NaCl, 20 mm Tris-HCl, pH 8.0, and 100 mm imidazole), and the samples were concentrated with ultrafiltration centrifugation tubes (Ultracel series 30-kDa; Millipore) and desalted with PD-10 desalting columns from GE Healthcare. The eluted FdmV was subjected to chromatography on a HiTrapTM HP 5-ml anion-exchange column (GE Healthcare). A 20–70 mm NaCl gradient in 20 mm Tris-HCl, pH 8.0, was applied, and the C-His6-tagged FdmV was eluted with 37 mm NaCl. The purified FdmV was found to be stable in 30% glycerol at -70 °C for at least 1 month.
Biochemical Characterizations of FdmV
Standard assay (at 30 °C) mixtures (100 μl) were composed of 100 mm Tris-HCl, pH 8.5, 10 mm l-glutamine, 10 mm MgCl2, 2 mm ATP, 0.1 mm FDM C, and 10% dimethyl sulfoxide (DMSO). FDM C was added as a 2 mm DMSO solution. The reactions were started by adding FdmV to a final concentration of 1 μm and terminated by adding 4 μl of 1 n HCl. Identical assays with boiled FdmV were carried out as negative controls.
The optimized reaction conditions for FdmV were determined by varying the DMSO concentration (5, 10, 15, and 20%) or the assay pH (6.5–9.0). Kinetic studies of the enzyme were performed by changing the FDM C concentrations from 50 to 200 μm under otherwise optimized conditions. Experiments were done in triplicate to arrive at each data point, and data were fit to the Michaelis-Menten equation to determine Km and kcat.
Chemical Synthesis of FDM B
FDM C (40 mg) was mixed with 1 g of CH3CO2NH4 in 3 ml of acetic acid and heated at 60 °C for 12 h to afford crude FDM B. FDM B was then purified by semipreparative HPLC and confirmed by spectroscopic analyses as described previously (19).
Analytical and Spectroscopic Procedures
Electrospray ionization-mass spectroscopy (ESI-MS) or high-resolution electrospray ionization-MS was performed on an Agilent 1100 LC/MSD SL quadrupole mass spectrometer (Santa Clara, CA). Atmospheric pressure chemical ionization mass spectroscopy (APCI-MS) was performed on an Agilent 1100 LC/MSD VL quadrupole mass spectrometer. High-resolution matrix-assisted laser desorption ionization-Fourier transform-mass spectroscopy (MALDI-FT-MS) was measured on an IonSpec HiResMALDI-FT mass spectrometer (Lake Forest, CA). NMR data were obtained using a Varian Unity Inova 500 MHz NMR spectrometer. Analytical or semipreparative HPLC was carried out on a Varian HPLC system with in-line Prostar 330 PDA detector. Analytical HPLC was performed on a Microsorb-MV 100-5 C-18 column (5 μm, 250 × 4.6 mm, Varian), and the semipreparative HPLC was on an Alltima C-18 column (5 μm, 250 × 10 mm, Alltech). The HPLC conditions have been described previously (19).
RESULTS
Prediction of FdmV as an AS B-like Protein
Bioinformatics analysis revealed that FdmV shows high sequence similarity to proteins involved in the biosynthesis of diverse natural products. Several of these enzymes are involved in aromatic polyketide biosynthetic pathways including LlpA from the lysolipin X pathway (50.7% identity) (13), PdmN from the pradimicin A pathway (48.9% identity) (28), RubR from the γ-rubromycin pathway (46.4% identity) (29), GrhP from the griseorhodin A pathway (44.7% identity) (30), OxyD from the oxytetracycline pathway (48.9% identity) (7, 8), and TcsG from the chlortetracycline pathway (50.1% identity) (31). Additionally, Ant-Orf1 from the anthramycin biosynthetic pathway (32) and PhzH from the phenazine-1-carboxamide pathway (33) show 48.6% and 40.9% identity to FdmV, respectively (supplemental Fig. S2). None of these enzymes, however, has been characterized functionally.
A conserved domain search of FdmV shows that it contains both the N-terminal glutaminase domain and the C-terminal amide synthetase domain present in AS B (34). FdmV displays 21.5% identity with the typical AS B from E. coli, which belongs to the class II glutamine amidotransferases (supplemental Fig. S2) (35). The AS B glutaminase domain hydrolyzes l-Gln, supplying a free ammonia group for the conversion of aspartic acid to asparagines, catalyzed by its amide synthetase domain (36). The highly conserved residues in the AS B active site can also be found in FdmV, such as Cys2, Arg50, Glu77, and Asp99 in the N-terminal glutaminase domain and Leu233 and Gly348 involved in ATP binding in the C-terminal domain of AS B (36) (supplemental Fig. S2). FdmV therefore was predicted to catalyze a reaction similar to that of AS B.
Inactivation of fdmV in S. griseus
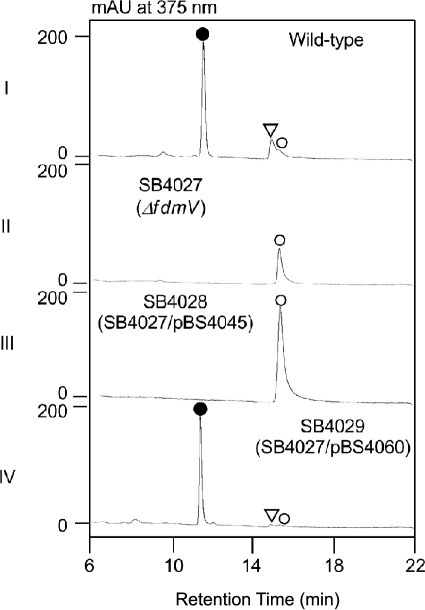
To probe the function of FdmV in vivo, we first inactivated fdmV in S. griseus by substituting a 1.2-kb fragment inside fdmV with the aac(3)IV apramycin resistance gene to afford the ΔfdmV mutant SB4027; the desired genotype of SB4027 was confirmed by Southern hybridization (supplemental Fig. S1). FDM A and FDM E production was abolished in SB4027, which instead accumulated FDM C with a titer of ∼2 mg/liter. To enhance FDM C production, an additional copy of the well studied fdm activator FdmR1 was added to SB4027 in trans, affording the FDM C-overproducing strain SB4028 (25). FDM C production was dramatically improved with a titer of ∼190 mg/liter in SB4028, greatly facilitating its isolation and characterization (Fig. 3).
FIGURE 3.
Metabolite profiles from selected S. griseus wild-type and recombinant strains as analyzed by HPLC: I, wild-type; II, ΔfdmV mutant SB4027; III, SB4028, SB4027 with overproduced FdmR1; IV, SB4029, SB4027 complemented by expressing fdmV in trans. ●, FDM A; ▿, FDM E; ○, FDM C.
Introduction of the fdmV-expressing plasmid pBS4060 into SB4027 afforded SB4029, in which the ΔfdmV mutation was complemented by the expression of a functional copy of fdmV in trans. FDM A production was fully restored in SB4029 (Fig. 3), eliminating any concern of potential polar effects of the ΔfdmV mutation in SB4027 on genes downstream of fdmV within the fdm cluster.
Isolation and Characterization of FDM C
FDM C produced by SB4028 was brought to analytical purity using sequential chromatographic steps. Its identity as FDM C was established via HPLC, MS, and NMR analyses. APCI-MS revealed the purified compound to have a mass of 572 Da (573.2 for [M+H]+ ion and 571.2 for [M−H]− ion), and high resolution electrospray ionization-MS of the compound (571.1231 for [M−H]− ion) enabled us to deduce its molecular formula as C31H24O11 (calculated 571.1240 for [M−H]− ion), which matches exactly that of FDM C. The purified compound displayed the same HPLC retention time and UV-visible absorption spectrum as FDM C reported previously and was finally confirmed to be FDM C by 1H NMR and 1H-1H correlation spectroscopy experiments (supplemental Table S1) (18).
Heterologous Expression of fdmV and Purification of Recombinant FdmV
All attempts to produce functional FdmV in E. coli failed. In E. coli BL21, the N-His6-tagged FdmV was well overproduced but completely insoluble, whereas no production of C-His6-tagged FdmV was observed. Soluble C-His6-tagged FdmV was subsequently overproduced in S. lividans K4-114. Recombinant FdmV was purified to homogeneity by sequential affinity and anion-exchange chromatography, and the purified FdmV migrated as a single band with the predicted size of 70.1 kDa upon SDS-PAGE analysis (Fig. 4).
FIGURE 4.
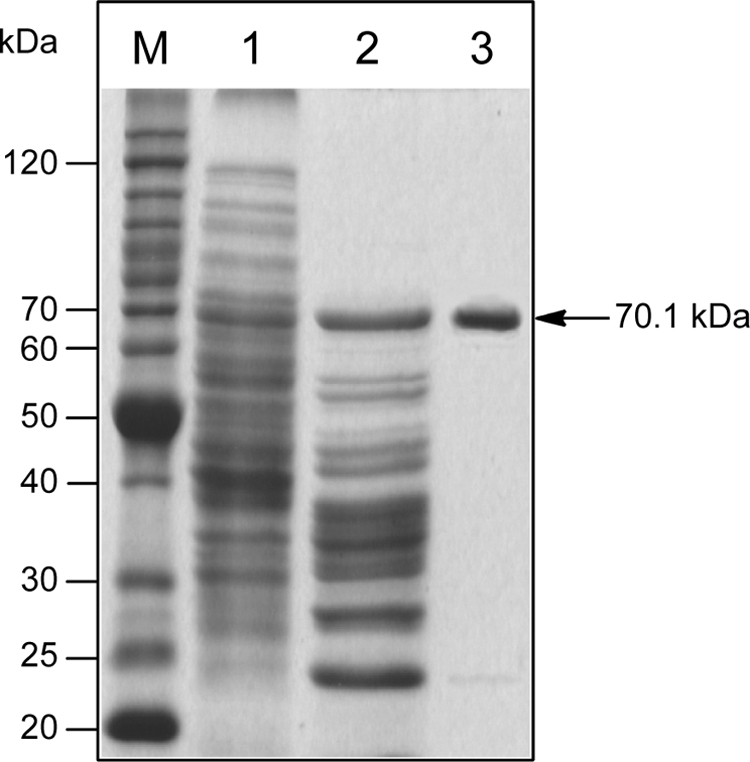
Purification of recombinant C-His6-tagged FdmV from SB4030 as monitored by SDS-PAGE. Lane M, BenchmarkTM protein ladder (Invitrogen); lane 1, total soluble proteins; lane 2, partially purified FdmV after nickel-nitrilotriacetic acid affinity chromatography; lane 3, purified FdmV after HiTrapTM HP anion-exchange chromatography.
FdmV Is an Amide synthetase
Enzyme assays of FdmV activity were designed according to known procedures for AS B, in which l-Gln serves as the nitrogen source (Fig. 5B). Because FDM C is not readily water-soluble, appropriate amounts of DMSO were added to the reaction. The initial FdmV assay was set as follows: 100 mm Tris-HCl, pH 8.0, 10 mm l-Gln, 10 mm MgCl2, 2 mm ATP, 0.1 mm FDM C, and 5% DMSO. After incubation at 30 °C for 9 h, ∼70% of FDM C was converted to a new compound, whose HPLC retention time and MS and UV-visible spectra were identical to authentic FDM B (18).
FIGURE 5.
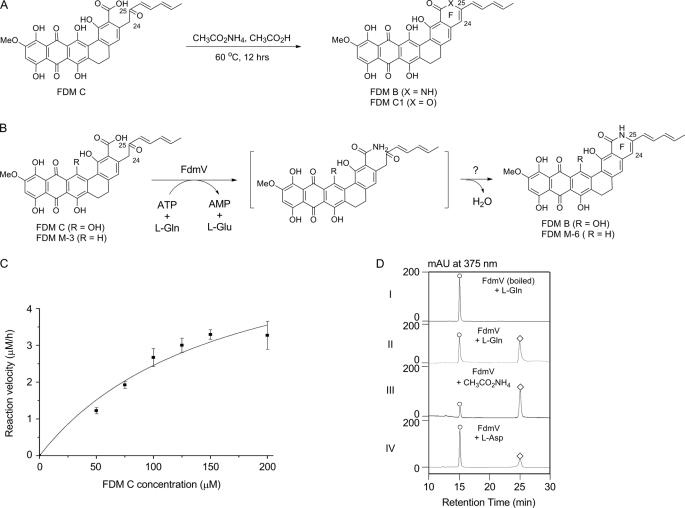
In vitro characterization of FdmV as an amide synthetase that catalyzes FDM C and FDM M-3 amidation to afford FDM B and FDM M-6, respectively. A, chemical synthesis of FDM B from FDM C. B, FdmV-catalyzed amidation of FDM C and FDM M-3 to FDM B and FDM M-6, respectively. C, kinetic analysis of FdmV-catalyzed amidation of FDM C with variable FDM C and saturating l-Gln and ATP. D, FdmV-catalyzed amidation of FDM C utilizing l-Gln, free NH3, or l-Asn as an ammonia source: completed assay with boiled FdmV as a control (I) and identical assays with l-Gln (II), CH3CO2NH4 (III), or l-Asp (IV) as an ammonia source. ○, FDM C; ♢, FDM B.
Authentic FDM B was chemically synthesized by mixing FDM C and CH3CO2NH4 in a 1:25 ratio in acetic acid and heating to 60 °C for 12 h. Under these conditions, ∼90% of FDM C was converted to FDM C1 and FDM B in about a 1:1 ratio (Fig. 5A). FDM C1 was confirmed by HPLC co-injection with an authentic standard and analysis of MS data (APCI-MS, 553.1 for [M+H]+ ion and 551.2 for [M−H]− ion). FDM B was identified based on its molecular mass of 553 Da (APCI-MS, 554.1 for [M+H]+ ion and 552.1 for [M−H]− ion); its molecular formula was determined to be C31H23NO9 by high resolution MALDI-FT-MS (554.1463 for [M+H]+ ion and calculated 554.1451), and it showed the same UV-visible spectrum as reported previously for FDM B (18). Although its poor solubility prohibited acquisition of high quality 1H NMR spectra for FDM B (18), the characteristic signal normally observed at 4.25 ppm corresponding to the C24 proton of FDM C was clearly absent in the spectra of FDM B. The absence of this signal is consistent with double bond formation between C24 and C25 in FDM B (Fig. 5A).
Enzymatic Characterizations of FdmV
Assay conditions for evaluating FdmV as an amide synthase were then studied at varying DMSO concentrations (5, 10, 15, and 20%) and pH values (ranging from 6.5 to 9.0 in 0.5 steps). The optimized conditions were found to be at pH 8.5 with 10% DMSO (supplemental Fig. S3). Under these conditions, a time course of FdmV showed the conversion of FDM C to FDM B was time-dependent, and the product formation was linear with respect to the time until 2 h. The conversion of FDM C to FDM B with constant l-Gln (10 mm) and ATP (2 mm) followed the Michaelis-Menten kinetics with an apparent Km of 162 ± 67 μm and a kcat of 0.11 ± 0.02 min−1 (Fig. 5C). The solubility of FDM C is extremely poor; the highest FDM C concentration attainable was 200 μm. Saturating concentrations of FDM C could therefore not be attained under the assay conditions. This limitation prohibited further kinetics studies on the other two substrates, l-Gln and ATP.
In addition to l-Gln, ammonium acetate and l-Asn can also be utilized as nitrogen donors in the FdmV-catalyzed amidation. With all the other variables held constant, the specific activity of FdmV toward 100 μm FDM C was determined to be 0.21 ± 0.03 × 10−3 μm min−1 mg−1 when using 10 mm l-Asn as amino source, which was clearly lower than l-Gln (specific activity is 0.53 ± 0.07 × 10−3 μm min−1 mg−1). The specific activity was determined to be 1.02 ± 0.03 × 10−3 μm min−1 mg−1 with 10 mm ammonium acetate representing the free amine as a nitrogen donor (Fig. 5D).
Several FDM analogs (FDM M-2, FDM M-3, and FDM M1-1) (Fig. 2) (20) and selected monocyclic benzoic acid derivatives (Fig. S4) were next tested as potential substrates of FdmV. All reactions were carried out using standard assay conditions. Among the eight substrate analogs tested, only FDM M-3 was amidated by FdmV (Fig. 5B and supplemental Fig. S5). The product was determined to be the corresponding lactam FDM M-6 on the basis of molecular mass as determined by APCI-MS (538.2 for [M+H]+ ion) and its molecular formula of C31H23NO8 based on the high resolution MALDI-FT-MS (538.1498 for [M+H]+ ion and calculated 538.1502). The rate of FDM M-6 formation from FDM M-3 was found to be very slow with less than 10% of FDM M-3 converted after an overnight incubation.
DISCUSSION
FdmV, an AS B-like protein and the only viable amide synthetase candidate identified within the fdm cluster, was envisaged to catalyze the conversion of FDM C to FDM B (17). Indeed, the accumulation of FDM C in the ΔfdmV mutant SB4027 in vivo supported this postulate (Fig. 3). This was further confirmed by in vitro conversion of FDM C to FDM B by recombinant FdmV (Fig. 5). At this stage, we have no evidence to exclude the dehydration-forming lactam ring F in FDM B as a spontaneous reaction. Based on the fact that FdmV cannot recognize the FDM intermediates FDM M-2 and FDM M1-1, amide formation in FDM A biosynthesis was determined to occur after the multiple hydroxylation steps of the polycyclic ring (Figs. 2 and 5).
AS B is a class II glutamine amidotransferase, and, like other members of the class (e.g. glutamine phosphoribosyl pyrophosphate amidotransferase, glucosamine-6-phosphate synthase, and glutamate synthase), has been well studied, as these enzymes play a central role in primary metabolism and human disease (34, 37). The class II glutamine amidotransferases all possess a conserved Cys2 in their N-terminal glutaminase domains and can utilize both l-Gln and free NH3 as nitrogen sources. Glucosamine-6-phosphate synthase differs in this respect in that it can only utilize l-Gln (34). FdmV should be a member of the class II glutamine amidotransferase based on its similarity to AS B, the conserved Cys2, and its ability to use both l-Gln and free NH3 (Fig. 5 and supplemental Fig. S2). Significantly, it was determined that FdmV is also capable of using l-Asn as a nitrogen source, indicating relaxed substrate specificity of its N-terminal glutaminase domain. In contrast, the FdmV C-terminal amide synthetase domain showed a high degree of substrate specificity as reflected by the inability of FdmV to amidate all tested substrate analogs with the exception of FDM M-3 (Fig. 5 and supplemental Fig. S4). Inspection of the substrates of FdmV revealed that both FDM C and FDM M-3 contain C-8 keto and C-10 hydroxyl groups that are absent in FDM M-2 and FDM M1-1, indicating that the oxidation states of FDM congeners at ring A and B are critical for FdmV activity (Fig. 2).
Database searching showed that four FdmV homologs (LlpA, PdmN, RubR, and GrhP) are involved in the biosynthesis of aromatic polyketides sharing similar biosynthetic pathways with FDM A (all derived from polyketide chains at least 24 C in length) (Fig. 1) (20). Among these aromatic polyketides, lysolipin X (LlpA) (13) possesses a lactam ring like FDM A. Pradimicin A (PdmN) (28) contains an amide bond formed putatively using d-Ala as the amino donor. However, neither γ-rubromycin (RubR) (29) nor griseorhodin A (GrhP) (30) contains an amide linkage (Fig. 1), although genes encoding FdmV homologs can be found in their biosynthetic gene clusters (supplemental Fig. S2). Multiple alignments of FdmV homologs reveal that the Cys2 indispensable for glutaminase activity is mutated to Ser in PdmN, RubR and GrhP (supplemental Fig. S2), implying that their ability to supply amino groups by hydrolyzing l-Gln is lost. In the case of PdmN, the amide synthetase activity is retained, and the enzyme uses d-Ala, instead of free NH3, as the nitrogen donor. This is reminiscent of the β-lactam synthetase involved in clavulanic acid biosynthesis (38) as well as the carbapenam synthetase CarA (39). Both enzymes are AS B homologs lacking the N-terminal Cys2 and drive intramolecular amide bond formation to afford each respective β-lactam. In the case of RubR and GrhP, it seems that not only is glutaminase activity lost, but so too is amide synthetase activity. It was reported recently that inactivation of grhP abolishes griseorhodin A production while also leading to accumulation of two new griseorhodin analogs, indicating that GrhP serves a role in griseorhodin biosynthesis not related to amide installation (40). Given the high degree of similarity between FdmV and these four enzymes and the significant diversity of catalytic activities (supplemental Fig. S2), these enzymes provide an excellent opportunity for combinatorial engineering to afford novel aromatic compounds.
Besides the aforementioned homologs, there are several other proteins involved in secondary metabolite production that show significant similarity to FdmV (supplemental Fig. S2). OxyD and TcsG are indispensable enzymes for oxytetracycline (7, 8) and chlorotetracycline (31) biosynthesis, respectively. In vivo studies have indicated that OxyD is an amide synthetase responsible for formation of a critical malonamyl starter unit during polyketide biosynthesis (7, 8). PhzH has been proposed to be a catalyst responsible for the conversion of phenazine-1-carboxylic acid to phenazine-1-carboxamide on the basis of in vivo data (33). Ant-Orf1 from the anthramycin biosynthetic pathway has been suggested to be an amide synthetase responsible for formation of the dehydroproline acrylamide moiety (32). Sequence alignment reveals that the N-terminal Cys2 and other active site residues are conserved in all four of these proteins (supplemental Fig. S2).
Conclusively, the AS B-like enzyme FdmV was clearly assigned as an amide synthetase catalyzing the conversion of FDM C to FDM B. Unveiling of FdmV homologs from different kinds of natural product biosynthetic clusters suggests that the AS B-like amide synthetases are widely distributed in secondary metabolite pathway, which adds another optional mechanism for the amidation of secondary metabolites.
Supplementary Material
Acknowledgments
We thank the Analytical Instrumentation Center of the School of Pharmacy, University of Wisconsin-Madison, for support in obtaining MS and NMR data.
This work was supported, in whole or in part, by National Institutes of Health Grant CA113297.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S5.
- FDM
- fredericamycin
- APCI-MS
- atmospheric pressure chemical ionization-mass spectroscopy
- AS
- asparagine synthetase
- DMSO
- dimethyl sulfoxide
- MALDI-FT-MS
- matrix-assisted laser desorption ionization-Fourier transform-mass spectroscopy.
REFERENCES
- 1.Fortin P. D., Walsh C. T., Magarvey N. A. (2007) Nature 448, 824–827 [DOI] [PubMed] [Google Scholar]
- 2.Magarvey N. A., Fortin P. D., Thomas P. M., Kelleher N. L., Walsh C. T. (2008) ACS Chem. Biol. 3, 542–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pompeo F., Mushtaq A., Sim E. (2002) Protein Expr. Purif. 24, 138–151 [DOI] [PubMed] [Google Scholar]
- 4.Ogasawara Y., Katayama K., Minami A., Otsuka M., Eguchi T., Kakinuma K. (2004) Chem. Biol. 11, 79–86 [DOI] [PubMed] [Google Scholar]
- 5.Luft T., Li S. M., Scheible H., Kammerer B., Heide L. (2005) Arch. Microbiol. 183, 277–285 [DOI] [PubMed] [Google Scholar]
- 6.Anderle C., Hennig S., Kammerer B., Li S. M., Wessjohann L., Gust B., Heide L. (2007) Chem. Biol. 14, 955–967 [DOI] [PubMed] [Google Scholar]
- 7.Zhang W., Ames B. D., Tsai S. C., Tang Y. (2006) Appl. Environ. Microbiol. 72, 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W., Watanabe K., Wang C. C., Tang Y. (2006) J. Nat. Prod. 69, 1633–1636 [DOI] [PubMed] [Google Scholar]
- 9.Zhang X., Alemany L. B., Fiedler H. P., Goodfellow M., Parry R. J. (2008) Antimicrob. Agents Chemother. 52, 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandey R. C., Toussaint M. W., Stroshane R. M., Kalita C. C., Aszalos A. A., Garretson A. L., Wei T. T., Byrne K. M., Geoghegan R. F., Jr., White R. J. (1981) J. Antibiot. 34, 1389–1401 [DOI] [PubMed] [Google Scholar]
- 11.Tomita K., Nishio M., Saitoh K., Yamamoto H., Hoshino Y., Ohkuma H., Konishi M., Miyaki T., Oki T. (1990) J. Antibiot. 43, 755–762 [DOI] [PubMed] [Google Scholar]
- 12.Hornung A., Bertazzo M., Dziarnowski A., Schneider K., Welzel K., Wohlert S. E., Holzenkämpfer M., Nicholson G. J., Bechthold A., Süssmuth R. D., Vente A., Pelzer S. (2007) ChemBioChem. 8, 757–766 [DOI] [PubMed] [Google Scholar]
- 13.Lopez P., Hornung A., Welzel K., Unsin C., Wohlleben W., Weber T., Pelzer S. (2010) Gene 461, 5–14 [DOI] [PubMed] [Google Scholar]
- 14.Misra R., Pandey R. C., Silverton J. V. (1982) J. Am. Chem. Soc. 104, 4478–4479 [Google Scholar]
- 15.Byrne K. M., Hilton B. D., White R. J., Misra R., Pandey R. C. (1985) Biochemistry 24, 478–486 [DOI] [PubMed] [Google Scholar]
- 16.Misra R., Pandey R. C., Hilton B. D., Roller P. P., Silverton J. V. (1987) J. Antibiot. 40, 786–802 [DOI] [PubMed] [Google Scholar]
- 17.Wendt-Pienkowski E., Huang Y., Zhang J., Li B., Jiang H., Kwon H., Hutchinson C. R., Shen B. (2005) J. Am. Chem. Soc. 127, 16442–16452 [DOI] [PubMed] [Google Scholar]
- 18.Sontag B., Müller J. G., Hansske F. G. (2004) J. Antibiot. 57, 823–828 [DOI] [PubMed] [Google Scholar]
- 19.Chen Y., Luo Y., Ju J., Wendt-Pienkowski E., Rajski S. R., Shen B. (2008) J. Nat. Prod. 71, 431–437 [DOI] [PubMed] [Google Scholar]
- 20.Chen Y., Wendt-Pienkowski E., Rajski S. R., Shen B. (2009) J. Biol. Chem. 284, 24735–24743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J., Fritsch E. F., Maniatis T. (2000) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 22.MacNeil D. J., Gewain K. M., Ruby C. L., Dezeny G., Gibbons P. H., MacNeil T. (1992) Gene 111, 61–68 [DOI] [PubMed] [Google Scholar]
- 23.Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000) Practical Streptomyces Genetics, The John Innes Foundation, Norwich, UK [Google Scholar]
- 24.Ziermann R., Betlach M. C. (1999) BioTechniques 26, 106–110 [DOI] [PubMed] [Google Scholar]
- 25.Chen Y., Wendt-Pienkowski E., Shen B. (2008) J. Bacteriol. 190, 5587–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doumith M., Weingarten P., Wehmeier U. F., Salah-Bey K., Benhamou B., Capdevila C., Michel J. M., Piepersberg W., Raynal M. C. (2000) Mol. Gen. Genet. 264, 477–485 [DOI] [PubMed] [Google Scholar]
- 27.Madduri K., Kennedy J., Rivola G., Inventi-Solari A., Filippini S., Zanuso G., Colombo A. L., Gewain K. M., Occi J. L., MacNeil D. J., Hutchinson C. R. (1998) Nat. Biotechnol. 16, 69–74 [DOI] [PubMed] [Google Scholar]
- 28.Kim B. C., Lee J. M., Ahn J. S., Kim B. S. (2007) J. Microbiol. Biotechnol. 17, 830–839 [PubMed] [Google Scholar]
- 29.Martin R., Sterner O., Alvarez M. A., de Clercq E., Bailey J. E., Minas W. (2001) J. Antibiot. 54, 239–249 [DOI] [PubMed] [Google Scholar]
- 30.Li A., Piel J. (2002) Chem. Biol. 9, 1017–1026 [DOI] [PubMed] [Google Scholar]
- 31.Nakano T., Miyake K., Endo H., Dairi T., Mizukami T., Katsumata R. (2004) Biosci. Biotechnol. Biochem. 68, 1345–1352 [DOI] [PubMed] [Google Scholar]
- 32.Hu Y., Phelan V., Ntai I., Farnet C. M., Zazopoulos E., Bachmann B. O. (2007) Chem. Biol. 14, 691–701 [DOI] [PubMed] [Google Scholar]
- 33.Mavrodi D. V., Bonsall R. F., Delaney S. M., Soule M. J., Phillips G., Thomashow L. S. (2001) J. Bacteriol. 183, 6454–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards N. G., Schuster S. M. (1998) Adv. Enzymol. 72, 145–198 [DOI] [PubMed] [Google Scholar]
- 35.Zalkin H. (1993) Adv. Enzymol. 66, 203–309 [DOI] [PubMed] [Google Scholar]
- 36.Larsen T. M., Boehlein S. K., Schuster S. M., Richards N. G.., Thoden J. B., Holden H. M., Rayment I. (1999) Biochemistry 38, 16146–16157 [DOI] [PubMed] [Google Scholar]
- 37.Richards N. G., Kilberg M. S. (2006) Annu. Rev. Biochem. 75, 629–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller M. T., Bachmann B. O., Townsend C. A., Rosenzweig A. C. (2001) Nat. Struct. Biol. 8, 684–689 [DOI] [PubMed] [Google Scholar]
- 39.Miller M. T., Gerratana B., Stapon A., Townsend C. A., Rosenzweig A. C. (2003) J. Biol. Chem. 278, 40996–41002 [DOI] [PubMed] [Google Scholar]
- 40.Yunt Z., Reinhardt K., Li A., Engeser M., Dahse H. M., Gütschow M., Bruhn T., Bringmann G., Piel J. (2009) J. Am. Chem. Soc. 131, 2297–2305 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.