Abstract
Oligogalacturonate lyases (OGLs; now also classified as pectate lyase family 22) are cytoplasmic enzymes found in pectinolytic members of Enterobacteriaceae, such as the enteropathogen Yersinia enterocolitica. OGLs utilize a β-elimination mechanism to preferentially catalyze the conversion of saturated and unsaturated digalacturonate into monogalacturonate and the 4,5-unsaturated monogalacturonate-like molecule, 5-keto-4-deoxyuronate. To provide mechanistic insights into the specificity of this enzyme activity, we have characterized the OGL from Y. enterocolitica, YeOGL, on oligogalacturonides and determined its three-dimensional x-ray structure to 1.65 Å. The model contains a Mn2+ atom in the active site, which is coordinated by three histidines, one glutamine, and an acetate ion. The acetate mimics the binding of the uronate group of galactourono-configured substrates. These findings, in combination with enzyme kinetics and metal supplementation assays, provide a framework for modeling the active site architecture of OGL. This enzyme appears to contain a histidine for the abstraction of the α-proton in the −1 subsite, a residue that is highly conserved throughout the OGL family and represents a unique catalytic base among pectic active lyases. In addition, we present a hypothesis for an emerging relationship observed between the cellular distribution of pectate lyase folding and the distinct metal coordination chemistries of pectate lyases.
Keywords: Bacteria; Carbohydrate Metabolism; Intestine; Manganese; Protein Structure; Yersinia enterocolitica; Oligogalacturonate; Pectin Metabolism; Pectin, Pectate Lyase, Carbohydrate Active Enzyme, Beta-elimination
Introduction
The distal gut microbiota of monogastric animals is a bioreactor consisting of 10–100 trillion bacteria from up to a thousand different species (1, 2). This diverse community, referred to as the “forgotten organ” (3), unlocks the rich source of chemical energy present within the glycosidic bonds of plant cell wall polysaccharides ingested as part of the host diet. These potential substrates are indigestible by the full repertoire of glycoside hydrolases and lyases encoded within the human genome. Consistent with this biological role, analysis of the gut metagenome has revealed an enormous arsenal of enzymes active in the modification and utilization of plant cell wall polysaccharides (2, 4). Recently, using transcriptomic technology, some of the catabolic strategies employed by the model gut microbe Bacteroides thetaiotaomicron have illuminated the dynamic relationship that exists between dietary regime, host glycan assemblies, and the composition of the microbiota (2, 5, 6). However, how these induced enzymes operate at the biochemical level and deconstruct complex target substrates still remains poorly understood.
By comparison with the natural flora, the processes by which enteropathogenic bacterial populations interact within this dynamic microecosystem has received significantly less attention in the literature. In particular, the mechanisms by which pathogens compete for nutrients during infection, colonization, and disease progression remain to be determined. At the turn of this century, hexuronide transport and utilization systems responsive to KdgR (2-keto-3-deoxygluconate repressor protein) were identified in the genomes of several gut pathogens (7, 8). These observations have provided a platform for exploring the capability of infectious agents, such as Yersinia enterocolitica, to metabolize pectic plant cell wall polysaccharides.
Pectin is chemically the most complex carbohydrate found in nature. It is located within the primary cell walls and middle lamella of most terrestrial plants, indicating that it is involved in intercellular interactions (9). Pectin is a negatively charged and heavily hydrated polysaccharide that cross-links the load-bearing cellulosic components of the cell wall. This class of structural carbohydrate is defined by the presence of a galacturonic acid (GalA)4 component within its backbone, interconnected through α-1,4 linkages. The abundance and composition of its decorations, however, vary significantly, which forms the basis for its classification into three major subtypes: homogalacturonan, rhamnogalacturonan I, and rhamnogalacturonan II (9). The identity of these side chains and the inherent charge status of uronate groups in homogalacturonan are critical factors for regulating the function of distinct pectic polysaccharides during various phytophysiological events. For example, methylesterification of homogalacturonan, which neutralizes the GalA subunits, regulates dynamic and reversible packing of fibers in a process called “gellation.” (10)
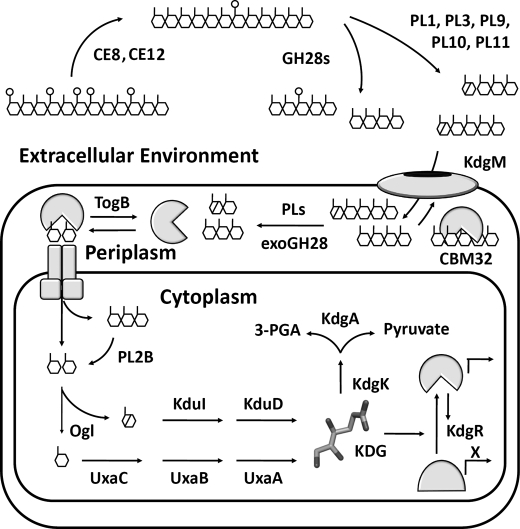
The pectic network is an energy-rich substrate targeted for catabolism by pectinolytic microorganisms. This complex carbohydrate is an amenable substrate, when compared with crystalline structural polysaccharides, due to its polarity, hydration level, and flexibility. Extracellular depolymerization of pectin is associated with the activity of secreted pectinases from dedicated phytopathogens and saprophytes. Conversely, the intracellular stages of pectic utilization, involving reactions that convert pectic fragments released by upstream extracellular processes into smaller metabolites, are common to a wider variety of pectinolytic bacteria. Recently, we reported several advances toward understanding the molecular basis for polygalacturonate depolymerization and transport using the enteropathogen Y. enterocolitica as a model organism (see Ref. 11 for a recent review) (Fig. 1). The mechanisms governing the intracellular conversion of oligogalacturonides into downstream metabolites remain less understood.
FIGURE 1.
General pectin degradation pathway found in pectinolytic bacteria from Enterobacteraciae (11). Extracellular pectin depolymerization is primarily restricted to phytopathogens and involves a consortium of enzymes that liberate fragments from the highly polymerized pectic network within the plant cell wall. Pectinases involved in these processes include PLs, polygalacturonases (GH28s), and carbohydrate esterases (CE8). Products diffuse into the periplasmic space through anionic porins belonging to the KdgM family, where a specialized collection of proteins operates to retain and concentrate (CBM32) and process pectic substrates into mono-, di-, and trisaccharides (PLs and exoGH28). Intracellular transport of oligogalacturonides is facilitated by integral membrane systems, including the ATP-dependent transporter TogMNAB. Within the cell, pectin catabolism remains poorly understood at the structural level; however, a pathway has been established based upon genetics, biochemistry, and comparative genomics (7, 12). There are two branches of metabolism dedicated to saturated monogalacturonate (GalA; UxaA, UxaB, and UxaC), and unsaturated monogalacturonate (DKI; KduI and KduD), the product of pectate lyases. These pathways converge to produce KDG, which is the inducer of KdgR, a repressor protein that regulates the expression of virtually every pectin utilization gene known. KDG is converted into the metabolites 3-phosphoglyceraldehyde and pyruvate by a two-step phosphorylation and hydrolysis reaction path catalyzed by KdgK and KdgA, respectively.
The highly conserved cytosolic pathway was first documented in Erwina chrysanthemi (now renamed Dickeya dadantii Ech586) using biochemical and genetic approaches in the 1980s and 1990s (see Ref. 12 for an extensive review) (Fig. 1). In the initial stages, intracellular di- and trigalacturonides are processed into saturated monogalacturonate (GalA) and a 4,5-unsaturated GalA-like monosaccharide (DKI; 5-keto-4-deoxyuronate) by the combined activities of an exo-acting family 2 pectate lyase (referred to as YePL2B in Y. enterocolitica (13, 14)) and oligogalacturonate lyase (YeOGL/YePL22 in Y. enterocolitica), a novel lyase family preferentially active on digalacturonides (14) (see Fig. 2 for a generalized reaction mechanism). GalA and DKI then undergo a short parallel series of chemical modifications culminating in the production of 2-keto-3-deoxygluconate (KDG), the central metabolite in the regulation of pectin-inducible genes. Despite this understanding of the pathway, the molecular processes that define substrate recognition and catalytic mechanism within each stage of the intracellular catabolism of pectic fragments still remain to be defined.
FIGURE 2.
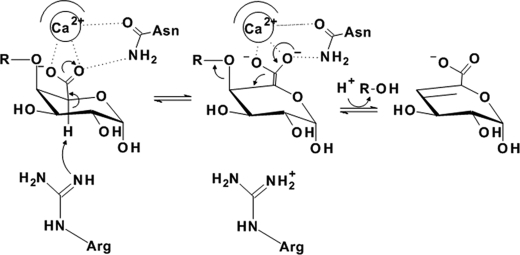
General β-elimination reaction mechanism for catalysis by pectate lyases. α1,4-Linked galacturonate substrates adopting the relaxed 4C1 chair conformation present the ideal geometry for an anti periplanar transelimination with the C5 hydrogen and C4 hydroxyl group both orientated in axial configurations. The rate-limiting step is the acidification and deprotonation of C5, a process that is facilitated by the effects of the electrophilic uronate group, catalytic divalent metals, and localized basic residues within the active site, which draw charge from the C5 carbon and increase the reactivity of its lone hydrogen atom. Abstraction occurs via general base attack by an arginine (family 1, 2, 3, and 10) or lysine (family 9) (43), the high pKa of these residues accounting for the basic pH optima observed across the PL landscape. Recently, the contributions of an ancillary lysine in family 1 PLs and asparagines in family 9 have been proposed to influence the catalytic rate by stabilizing the enolate-enolate intermediate (45). These residues have been implicated in hydrogen donation and/or bonding to the nascent oxyanion of the uronate group following electron transfer to the transient double bond between C5 and C6. Decomposition of the intermediate occurs by protonation of the scissile glycosidic oxygen and elimination of the axial O4 by electron shuttling from the C5–C6 bond to the C4–C5 bond of the sugar ring. This unsaturation distorts the relaxed chair structure of the pyranose because the trigonal planar geometry of C4 and C5 results in migration of C4 into the plane of C2, C3, and C5. The substrate is shown is a disaccharide, with the R-group depicting a second GalA or DKI molecule (note the Asn residue shown may be equivalent to Gln350).
In this study, we have explored in further detail the mechanism of oligogalacturonate lyase (OGL) activity by using the oligogalacturonate lyase from Y. enterocolitica, YeOGL. These findings illuminate several features of oligogalacturonide β-elimination, which is distinct from polysaccharide lyases, define a new family of cytoplasmic pectate lyase within the Carbohydrate-Active Enzymes Database (CAZy; available on the World Wide Web) (15), and present a hypothesis to explain the relationship between cellular localization of pectate lyases and the coordination of distinct metal cofactors.
EXPERIMENTAL PROCEDURES
Molecular Cloning, Gene Expression, Protein Production, and Purification
Recombinant YeOGL was prepared according to techniques described previously (16). Briefly, the full-length gene locus YE1876 (ogl), which encodes a 388-amino acid 44.2-kDa protein, was amplified by polymerase chain reaction from Y. enterocolitica subsp. enterocolitica 8081 genomic DNA (ATCC number 23715TM) using forward (ATATGCTAGCATGGCTAAAGGTAAACAAATCCCCCTGA) and reverse (TATACTCGAGCTATTTCCAGACTGATTCAGGCAATGTGGCT) oligonucleotide primers. The gene fragment was restricted using engineered NheI 5′ and XhoI 3′ sites and ligated into pET28a digested with complementary restriction enzymes to create the expression vector pET28-YeOGL. YeOGL encoded by pET28-YeOGL contains a thrombin-cleavable N-terminal His6 tag. The construct was transformed into chemically competent BL21-pLysS (DE3) cells, and 1.0-liter cultures of LB were grown supplemented with 50 μg/ml kanamycin. When cultures reached an optical density of ∼0.6 (at 600 nm), the temperature was decreased to 16 °C, and recombinant gene expression was induced by the addition of isopropylthiogalactopyranoside to a final concentration of 0.2 mm. Induced cultures were incubated overnight with shaking. Cells were then pelleted, resuspended in binding buffer (500 mm NaCl, 20 mm Tris-HCl, pH 8.0), and lysed by French press. The lysate was clarified by centrifugation at 17,000 × g for 45 min, and the supernatant was loaded directly onto a nickel-charged immobilized metal affinity chromatography column. After extensive washing with binding buffer, bound polypeptides were eluted by a stepwise 5–500 mm imidazole gradient in binding buffer. Collected protein eluate was analyzed by SDS-PAGE for purity, pooled, and used for biochemical assays following buffer dialysis to remove imidazole. Protein dedicated for crystallization trials was further processed by buffer exchanging into 20 mm Tris-HCl, pH 8.0, 2 mm CaCl2 and treatment with 1.5 units of thrombin at room temperature overnight. The extent of protein cleavage was evaluated by SDS-PAGE, and then YeOGL was concentrated in a stirred ultrafiltration device at 4 °C under nitrogen pressure. Protein was loaded onto an S-200 gel filtration column in 20 mm Tris-HCl to further purify the protein. The major protein peak was detected by UV absorbance, pooled, and concentrated to 12.5 mg/ml for crystallization trials.
Crystallization and Structure Solution
YeOGL was screened for crystallization conditions using the hanging drop vapor diffusion method by mixing with an equal volume of mother liquor and incubating at 18 °C. Crystals formed in 0.2 m CaCl2, 0.1 m NaOAc, pH 4.8, 20% PEG 3350 and were cryoprotected with 15% ethylene glycol. Diffraction data were collected at the Canadian Light Source on beamline CMCF1 and processed using iMOSFLM and SCALA (17) (see Table 1 for a list of data collection and refinement statistics). The structure was solved with PHASER (18), using the Vibrio parahemeolyticus homolog of YeOGL (VpOGL; Protein Data Bank entry 3C5M, 68% identity) as the search model. The YeOGL model was corrected and completed by successive rounds of manual building in COOT (19) and refinement with REFMAC (20), all within the CCP4 suite of programs (21). Waters were added using the ARP/WARP option in REFMAC and then visually inspected prior to structure deposition. The final coordinates were validated with SFCHECK (22) and PROCHECK (23). The modeled GalA complex was constructed by exporting a monogalacturonate in an energy-minimized state from the Glycam Biomolecule Builder (24) and superimposing the uronate group with the ligated acetate ion. The monosaccharide was modeled with the axial O4 projecting toward solvent and H5 toward His242, the closest basic amino acid in the active site with appropriate geometry for proton abstraction.
TABLE 1.
Crystallographic statistics for the structure of YeOGL
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a (Å) | 66.65 |
| b (Å) | 75.51 |
| c (Å) | 78.51 |
| α (degrees) | 90.00 |
| β (degrees) | 90.00 |
| γ (degrees) | 90.00 |
| Resolution (Å) | 30.00-1.65 |
| Rmerge | 0.085 (0.35) |
| I/σI | 14.9 (4.0) |
| Completeness (%) | 97.7 (85.5) |
| Multiplicity | 6.8 (4.4) |
| Average mosaicity | 0.44 |
| Refinement | |
| Resolution (Å) | 35.42-1.65 (1.69-1.65) |
| No. of reflections | 44,824 |
| Rwork/Rfree | 0.18/0.20 |
| No. of atoms | 3527 |
| Protein | 3019 |
| Acetate/ion | 4/3 |
| Water | 497 |
| B-Factors (Å2) | |
| Protein | 15.4 |
| Acetate/ion | 19.0/12.0 |
| Water | 30.8 |
| r.m.s deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (degrees) | 1.554 |
| Ramachandran statistics | |
| Favored (%) | 98.9 |
| Allowed (%) | 0.5 |
| Outlier (%) | 0.5 (Gly373) |
Kinetic Assays
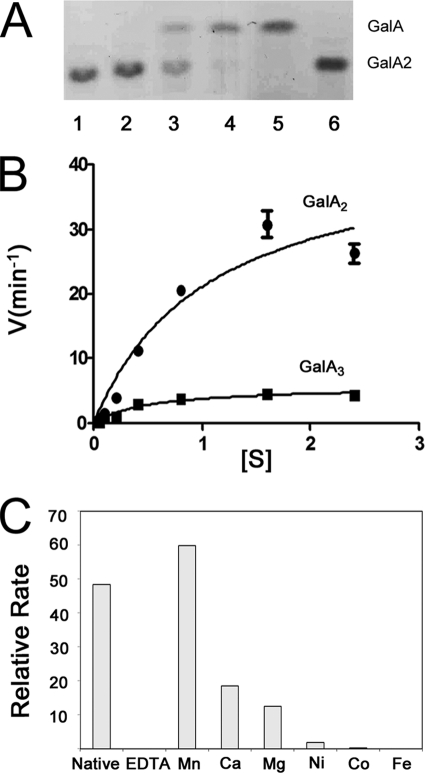
Activity of YeOGL on digalacturonate was first demonstrated using thin layer chromatography. Enzyme digests were set up with 0.1 μm enzyme and 1.0 mm substrate in Tris-HCl, pH 7.0 (a previously reported condition for EcOGL (14)) and incubated for 10 min at 37 °C. At 0, 1, and 5 min, aliquots were removed and assayed for the formation of monosaccharide products by loading TLC silica gel plates with aluminum backing (EMD; 55553-7) and run in 1-butanol/acetic acid/water (2:1:1, v/v/v) solvent for 4 h. Plates were dried and developed using orcinol sulfuric acid reagent (sulfuric acid/ethanol, 3:70 (v/v), orcinol 1%). Sugars were visualized by heating. Activity of YeOGL was quantified using wild-type enzyme at 0.5 μm and substrate concentrations ranging from 0.025 to 6.4 mm. The formation of product was monitored by absorbance at 230 nm. The highest concentration points had an inhibitory effect on activity, an effect commonly observed for pectate lyases (25), and were excluded from the Michaelis-Menten calculations. All assays for wild-type and catalytic mutants were performed in 0.1 mm Tris-HCl, pH 7.0, supplemented with 1.0 mm MnCl2 at 25 °C. The molar extinction coefficient used to quantify DKI production (the unsaturated reaction product) was 4,600 m−1 (26). Reactions were performed in triplicate for statistical significance, and the data were processed using GraphPad Prism® version 5 software.
Metal supplementation assays were performed as described for YePL2A (13). Each reaction series was run in triplicate for statistical accuracy. Control reactions were set up using 0.5 μm YeOGL and 0.2 mm GalA2 in 0.1 mm Tris-HCl, pH 7.0. Activities were ablated with 2 mm EDTA and recovered by the addition of a 10 mm divalent cation-chloride salt, which saturates the chelating agent on a molar basis (EDTA/metal, 5:2). In the presence of MnCl2, the reaction rate was increased above that of the wild-type enzyme activity.
RESULTS AND DISCUSSION
Structure of YeOGL
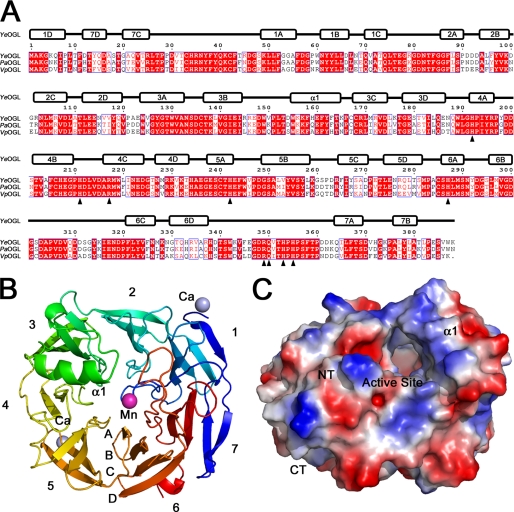
Although the existence of OGLs has been known for some time, no representative structure has been reported in the literature, and thus the structural features that contribute to the function of these enzymes remained unknown. To explore the structural basis of OGL activity, we determined the high resolution x-ray crystal structure of YeOGL. The entire 388 amino acids of OGL plus 572 water molecules, one manganese ion, two calcium ions, and one acetate ion (a component of the crystallization buffer) in bidentate coordination with the manganese ion are visible within the electron density (to be deposited in proof). The metal was determined to be Mn2+ based upon high resolution structural data supporting appropriate covalent coordination bond lengths, ligated nitrogen chemistry, coordination sphere symmetry, and anomalous signal (see below). YeOGL adopts a seven-bladed β-propeller fold (Fig. 3) that has structural similarity to the WD40 family of proteins (27, 28). A DALI-Lite version 3 search (29) revealed the highest similarity to OGL from V. parahemeolyticus (VpOGL), solved by the Northeast Structure Genomics initiative (Protein Data Bank entry 3C5M, Z value = 59.1, r.m.s. deviation = 1.1 Å, with 68% identity over 373 aligned amino acids), which was used as the search model to solve the structure of YeOGL. Other than these two bacterial lyases, however, the closest structural homologs are proteins that display very little sequence and functional similarity. The next two closest hits belong to components of eukaryotic enzymes, including the Bα subunit of the protein phosphatase 2A holoenzyme (28) (Protein Data Bank entry 3DW8, Z value = 22.1, r.m.s. deviation = 3.2 Å, with 10% identity over 286 aligned amino acids) and the histone-binding protein RBBP7 (27) (Protein Data Bank code 3CFS, Z value = 22.1, r.m.s. deviation = 3.1 Å, with 10% identity over 290 aligned amino acids). These observations suggest that the seven-bladed β-propeller is a plastic fold capable of accommodating diverse functionalities.
FIGURE 3.
β-Propeller fold of YeOGL. A, structural alignment of OGLs from Y. enterocolitica (YeOGL), E. chrysanthemi (EcOGL), and V. parahemeolyticus (VpOGL). The secondary structure elements of YeOGL are displayed above the aligned primary structure. Amino acids targeted for mutagenesis are indicated with black triangles below the sequences. B, schematic representation of YeOGL structure colored from the N terminus (blue) to C terminus (red). The seven β-propellers are labeled (1–7) with each strand (A–D) packing from the core toward the surface. The three bound metal ions are displayed as spheres with the catalytic manganese colored in purple and the surface-bound calciums in light blue. C, electrostatic surface potential of YeOGL. The protein is centered to display the entrance to the active site that is lined with several basic patches.
Each propeller in YeOGL consists of a repeating antiparallel four-stranded β-substructure (labeled A–D, using nomenclature described for other structural homologs) that flows from the core of the protein toward the outer surface of the protein (Fig. 3, A and B). The innermost strands (strand A) are connected to the outermost strand (strand D) of the preceding propeller by loops that vary greatly in length and structure and congregate on the upper surface of the protein, consistent with other seven-bladed propeller proteins (27, 28). One of these loop segments, a stretch of 11 amino acids connecting strands B and C of propeller 6 (Pro305–Glu316), could not be modeled because the density in this region was too poor. This is also observed in the structure of VpOGL, suggesting that the absence of density is due to flexibility in this loop region rather than crystallographic packing effects. The “pinwheel-like” fold is capped at the C terminus by two N-terminal antiparallel strands (7C and 7D) that deviate from the contiguous repeating β-structures to form the two outer strands of the seventh propeller. The C-terminal sequence exits through the bottom of the protein and projects into solvent. The only noticeable α-helical structure (α1) is a 12-amino acid sequence (Asp154–Lys165) within the third propeller that lies on the protein surface.
The solvent-accessible electrostatic surface model of YeOGL reveals that there is a prominent tunnel lined with basic amino acids burrowing from the surface toward a core metal binding site (Fig. 3C). Metal coordination pockets are a signature substructure within pectate lyases and a key feature of the β-elimination chemistry, highlighting that this is the location of the active site in YeOGL. The walls of the tunnel are formed by several loop regions, an observation consistent with the active sites of other WD40 enzymes (27, 28), and capped by the α-helix (α1). Surprisingly, the tunnel appears to exit into solvent on both sides of the protein, reminiscent of the possessive cellobiohydrolase (30), as opposed to displaying a “pocket-like” structure more common within exo-acting enzymes (31). In the full-length YeOGL, however, it is likely that the rear of the active site is occluded by the missing loop segment because the loop truncation occurs near this region. One entrance to the active site would require the substrate and product to access and depart the active site through the same portal.
The Unique Metal Binding Pocket of a Cytoplasmic Pectate Lyase
All described pectate lyase families utilize a catalytic metal during β-elimination of GalA. Mechanistically, uronate coordination contributes to electron delocalization and acidification of the α-proton at C5 of the aglycone sugar (Fig. 2). Calcium is the only metal that has been found in the active sites of extracellular lyases; interestingly, however, this metal also plays several other significant roles in the modification of pectic fragments and regulation of pectin ultrastructure. These include 1) the “egg box” model (32), a dynamic and physiologic process by which divalent cations cross-link the uronate groups, enabling the reversible gellation and solubilization of homogalacturonate chains, and 2) bridging substrate uronate groups and protein contact points at distal subsites within pectate lyases, interactions that contribute to the binding energy and substrate specificity. These reactions tend to be transient, however, and do not require an intact metal binding pocket in the absence of enzyme-substrate complexes. The prominent and diverse roles of calcium in pectinase activity and pectin structure may be linked to the prevalence of this metal cofactor within the plant cell wall (∼1 mm).
By comparison with extracellular lyases, the more specialized role of catalytic metals in pectate lyases that operate within the confines of bacterial membranes has only just begun to be defined. The initial two cases revealed that two E. chrysanthemi (D. dadantii Ech586) enzymes, PelW (a family 2 PL) and EcOGL (a family 22 PL), could harness novel metal cofactors, including manganese, nickel, and cobalt (14). In contrast to calcium, coordination of these transition metals requires signature nitrogen ligands presented by histidine residues (33). Recently, we provided a structure-function explanation for manganese coordination by two homologs of PelW from Y. enterocolitica, YePL2A and YePL2B (13). Within the coordination pocket of these enzymes, two histidine residues were identified that contribute to the octahedral geometry at distances consistent with the binding of manganese.
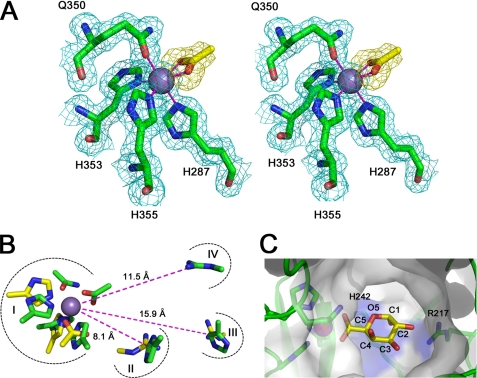
To further explore the emerging relationship between pectate lyase location and differential metal coordination chemistry, we have dissected the manganese binding pocket within YeOGL. The coordination sphere is composed of three histidines (His287, His353, and His355) and a glutamine (Gln350) residue that display coordinate bond distances ranging from 2.1 to 2.2 Å for all six ligands, consistent with compositions and distances described for other manganese complexes (33). Gln350 was modeled with Oϵ as the donor atom and Nϵ hydrogen bond contact with an ordered water. This is the most stable tautomeric form because Nϵ lacks a free electron pair for covalent coordinate bond formation. The remaining two atoms that complete the octahedral geometry were determined to be the oxygen atoms of an acetate carboxylate group, which is supported by B-factor and difference map analysis (see Table 2 and Fig. 4A). Acetate is a component of the crystallization buffer and mimics the chemistry of the GalA uronate group.
TABLE 2.
Atomic distances and temperature factors for the manganese coordination pocket of YeOGL
| Ligand | Distance | B-Factor | Occupancy |
|---|---|---|---|
| Å | Å2 | ||
| Mn | 7.5 | 1.0 | |
| Nϵ-His287 | 2.1 | 9.5 | 1.0 |
| Oϵ-Gln350 | 2.2 | 10.2 | 1.0 |
| Nδ-His353 | 2.2 | 8.7 | 1.0 |
| Nδ-His355 | 2.1 | 8.3 | 1.0 |
| OA-ACT | 2.1 | 17.0 | 1.0 |
| OB-ACT | 2.3 | 16.2 | 1.0 |
FIGURE 4.
The active site of YeOGL. A, the octahedral metal binding site is displayed in wall-eyed stereo format with the weighted maximum likelihood (20)/σA (53) 2Fo − Fc map (cyan) contoured to 1.0 σ (0.43 e−/Å3). The coordinated acetate ion (yellow sticks) is shown with an Fo − Fc omit map, produced from phases generated during refinements with the acetate molecule excluded from the model, contoured at 2.5 σ (0.18 e−/Å3). This map demonstrates the high quality of unbiased electron density for this molecule. The three histidines (His287, His353, and His355) and glutamine (Gln350) are shown in stick representation, and the Mn2+ ion is modeled as a sphere. B, superimposition of catalytic substructures within the active site of YePL2A (yellow, Protein Data Bank entry 2V8J) and YeOGL (green). Four aligned regions are shown: metal coordination pocket (I), catalytic base (II), and potential residues involved in substrate stabilization (III and IV). Distances of the residues from the metal are labeled. C, cut-away surface representation of the active site with a GalA modeled in the +1 subsite. The GalA uronate group of the aglycone is superimposed with the coordinated acetate ion with the axial H5 pointing toward the candidate Brønstead base His242 and O2 pointing toward the stabilizing Arg217, and O4 is presented for polymerization.
This analysis defines the third pectate lyase that coordinates Mn2+, two of which are cytoplasmic (PL2B/OGL) and one of which is periplasmic (PL2A) (13, 14) (Fig. 1). These select enzymes contrast with the more abundant examples of extracellular secreted pectate lyases that exclusively utilize calcium. The molecular basis for the relationship between cellular distribution and metal selectivity had yet to be elucidated. Although the possibility of metals contributing different mechanistic effects to the reaction must be considered because calcium and transition metals display distinct electron orbital structures, it appears likely that this relationship may reflect the distribution of trace elements within prokaryotes. Although the roles of intracellular calcium in bacteria are still poorly understood, emerging evidence has revealed that, similar to eukaryotes, calcium is a signaling molecule in many bacterial processes, such as transport, cell differentiation, and the maintenance of cell structure (34, 35). As a result, calcium levels within the cell are limiting and tightly regulated (100–300 nm) (36, 37). Therefore, intracellular pectate lyases may have evolved unique chemistries tailored for the coordination of metal cofactors (Mn2+ as opposed to Ca2+) that are available in sufficient concentrations within the microenvironment in which they function. The observation that PL2A utilizes manganese and is operational within the periplasm, however, provides an interesting twist on this possibility because the calcium-utilizing pectate lyase, PelX (PL9), has also been determined to be functional within the periplasm (7, 38). Analysis of the primary structure of the intact YePL2A polypeptide reveals the presence of a signature Lys-Arg dipeptide in its secretion signal. This sequence marker indicates that YePL2A is folded within the cytoplasm and subsequently transported intact through a TAT pathway across the inner membrane (39, 40). Y. enterocolitica lacks a PelX homolog and therefore may have diversified its collection of pectinases by evolving to secrete a PL2 homolog that would be active in the upstream stages of the pathway. These enzymes are observed in only a few pectinolytic microbes; however, in almost every case, they are found in tandem PL2A and PL2B pairs (see the CAZy Web site) (7). Typically secreted pectinases transit through a SEC-mediated pathway in an unfolded state and therefore would incorporate a catalytic metal within a folding microenvironment where calcium is prevalent. This observation is reminiscent of the targeted folding and selective metal acquisition of the Synechocystis metalloproteins MncA and CucA (41).
Structural Analysis of the YeOGL β-Elimination Machinery
Evolutionary convergence of the catalytic machinery in pectate lyases has been observed in families 1, 2, and 10 (13, 42). Although these enzymes have no sequence relatedness and adopt unique folds, they display a constellation of functional residues within their active site that is structurally conserved. These include the components of the metal binding pocket involved in α-proton acidification; a basic residue, typically an arginine, which operates as the Brønstead base during H5 abstraction; and a secondary arginine that flanks the substrate by binding to its C2 and C3 hydroxyl groups. Therefore, based upon this premise and in order to dissect the molecular basis of β-elimination within the active site of YeOGL, we have explored the evidence for structural convergence between YeOGL and YePL2A. These two enzymes, unrelated at the sequence level, represent the only two characterized pectate lyases available in the data base in complex with a Mn2+ ion. Comparing the active sites of these two lyases is complicated by the presence of the two alternate conformations of YePL2A that have been described, which represent distinct structural snapshots across the catalytic landscape of the enzyme. The Mn2+ complex, or “open” form, is poised for substrate binding; whereas the GalA3-Michaelis complex, or “closed” form, has locked its two catalytic arms around the substrate, resulting in a rearrangement of the core β-elimination machinery (13). The lack of a metal in the YePL2A-GalA3 complex results in a shift of the substrate orientation within the active site because the uronate group is not anchored by coordination. Preliminary rounds of analysis determined that the metal-bound complex of YeOGL and YePL2A (with both enzymes lacking sugar substrate) display the greatest level of structural similarity in the alignment of their catalytic residues. Therefore, in the following discussion, we will describe the superimposition of the two active sites using the metal as a central reference point.
Four main substructures were identified (I–IV) (Fig. 4B). In I, the metal binding site shares several structurally conserved donor atoms within both enzymes, including two histidines. Interestingly, the Gln350 of YeOGL did not align with any candidate residues from YePL2A. This glutamine, which also binds an ordered water molecule, may be functionally similar to the catalytic amides from family PL9 and PL10 lyases. It has been proposed that amide groups proximal to the uronate may stabilize the enolate-enolate intermediate through hydrogen bonding (42, 43) (Fig. 2). In II, the Brønstead base from YePL2A (Arg171) structurally aligns with His242 in YeOGL, the only basic residue with suitable proximity and geometry for abstracting the H5 of a bound GalA substrate. Histidine-catalyzed β-elimination has been proposed for the family PL8 Streptcoccus pneumoniae hyaluronate lyase (SpPL8) (44); however, to the best of our knowledge, this is the first description of a similar role for this residue in pectate lyases. The remaining two substructures, III and IV, represent two potential substrate stabilizing residues in YeOGL. In III, the His211 in YeOGL superimposes well with the Arg272 of YePL2A in its open conformation; however, this position is distal to the metal binding site (∼15.9 Å) and in an unfavorable position to interact directly with the substrate. In YePL2A, this is explained through a change in enzyme conformation; following GalA3 binding, Arg272 undergoes a positional shift of 4.4 Å toward the aglycone sugar. Further analysis of the YeOGL structure reveals that the arginine displayed in IV (Arg217) most likely interacts with GalA in subsite +1 (bond cleavage occurs between the sugars at subsites −1 and +1) based upon its configuration because both Nη of the guanidine group are pointing toward the metal binding pocket, consistent with other pectate lyase structures, functional group chemistry, and proximity. In YeOGL, the two Arg217 nitrogen atoms are 11.2 and 11.5 Å, respectively from the coordinated metal, which is very similar to the distances between the guanidine group of Arg272 (10.9 and 11.0 Å) and a bound water molecule in the GalA3-Michaelis complex of YePL2A (not shown). This observation suggests that there is more conformational flexibility within the active site of YePL2A than YeOGL and that the active site of YeOGL presents another strong example of the structural convergence of the β-elimination machinery in pectate lyases. The potential role of this highly conserved arginine in catalytic proton transfer to the glycosidic oxygen of the scissile bond, in YeOGL and other pectate lyases, remains to be established (42, 45).
Crystallographic investigations into the molecular basis of YeOGL recognition of oligogalacturonides have proven unsuccessful. Fortuitously, however, the coordination of an acetate molecule to the Mn2+ ion provides a framework for predicting the position of a GalA molecule within the active site (Fig. 4C). A YeOGL-GalA model was constructed by superposing the uronate group of GalA with the bound acetate group. To select between the two possible orientations, the monosaccharide was positioned such that the axial hydrogen is pointing toward the candidate Brønstead base, His242, as opposed to toward the top of the active site, a region lacking any definable basic residue suitable for potential proton abstraction. In this configuration, the O2 hydroxyl of GalA is directed toward the guanidine group of Arg217. Although the model does not account for subtle changes in conformation and orientation that may be induced by binding energy, the two molecules are already approaching hydrogen bond distance. O1 is pointing toward the core of the enzyme and the rear exit of the solvent tunnel, and the axial O4 is positioned toward the tunnel entrance to accommodate polymerization of another GalA. Based upon these observations, it is likely that in the native complex, GalA undergoes a small rotation around its planar axis so that O2 and O3 both interact with Arg217, and O4 extends closer toward the center of the solvent tunnel to alleviate any steric clash with the wall of the enzyme.
Transition Metal Utilization by YeOGL
to test the viability of our model and interrogate the role of Mn2+ in the elimination of oligogalacturonate, we have characterized the active site by enzyme kinetics and metal supplementation assays. Preliminary assays revealed that YeOGL is active on GalA2 (Fig. 5A). Calculated kinetic values (see Fig. 5B and Table 3) suggest that YeOGL has an activity identical to that of its EcOGL homolog (14), a result that is not surprising when considering the high level of amino acid conservation between the two proteins. The ∼8-fold higher maximal velocity of YeOGL on GalA2 (43.67 ± 5.16 min−1) compared with GalA3 (5.70 ± 0.49 min−1), underpins the complementary role of YePL2B/PelW. YePL2B/PelW is dedicated to the conversion of cytoplasmic trisaccharides into substrates for OGL, a process that will ultimately increase the efficiency of the upstream stages of cytoplasmic oligogalacturonide catabolism (see Fig. 1). Previously, a cluster of transition metals, ranging from the seventh to the tenth row of the fourth period, were previously shown to be capable of assisting in the catalytic activity of EcOGL to varying degrees (Mn2+ > Co2+ > Fe2+ > Ni2+) (14). This finding indicated that the metal-coordinating machinery of this lyase family is tailored to utilizing transition metals. Similar recovery assays performed for YeOGL also revealed a preferential relationship with Mn2+ because other cofactors failed to restore similar levels of enzyme activity (Fig. 5C). These assays support the structural observations for tailored metal binding within the OGL family discussed above.
FIGURE 5.
Activity of YeOGL. A, TLC gel of GalA22 digestion. Lanes are loaded as follows: no enzyme control (lane 1), increasing reaction time points (0–5 min) (lanes 2–4), GalA standard (lane 5), and GalA2 standard (lane 6). B, reaction rates are plotted with velocity (min−1) against substrate concentration (mm). C, metal supplementation assays. Rates shown represent quantified changes in UV absorbance. Error bars, S.D.
TABLE 3.
Kinetic values of YeOGL
| Substrate | Km | Kcat | Kcat/Km |
|---|---|---|---|
| mm | min−1 | min−1mm−1 | |
| GalA2 | 1.07 ± 0.28a | 43.67 ± 5.16 | 40.81 |
| GalA3 | 0.52 ± 1.2 | 5.70 ± 0.49 | 10.96 |
a S.D. values are based upon the rate calculations for three independently performed experiments.
Evolution of the OGL Family
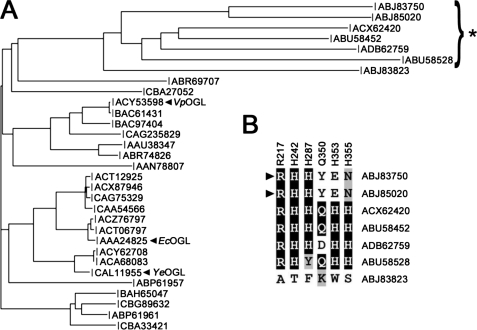
EcOGL was first discovered in 1968 (originally referred to as oligogalacturonide transeliminase (46)) and known to be found in diverse species of Enterobacteriaceae, including Y. enterocolitica, for over a decade (8); however, a detailed analysis of the relatedness of orthologs and its classification as a novel polysaccharide lyase family in the CAZy data base has been lacking. Therefore, to provide insight into the functional relatedness of the family, we did a BLAST search using the YeOGL as a query sequence. Initial searches identified 46 family members (with a low end cut-off value of ≥10 for several outlying sequences as determined in the Carbohydrate-Active Enzymes Database)5 that partition into two clusters, one of which contains ∼85% of the entries (relatedness value of 53–97%), including the characterized enzymes from E. chrysanthemi (CAA43990) and Y. enterocolitica (CAL11955) and the uncharacterized structural homolog from V. parahemeolyticus (BAC61431). The smaller seven-member out-group (relatedness threshold value of 10–52%) consists of uncharacterized putative enzymes from Candidatus Solibacter usitatus (ABJ83750, ABJ85020, and ABJ83823), Roseiflexus castenholizii (ABU58452 and ABU58528), and single entries from Geobacillus sp. and Haloterrigena turkmenica. The presence of OGL in each of these species indicates that these microorganisms have a propensity to utilize pectin as an energy source.
To compare the functional residues across the OGL family, a ClustalW alignment (47) was performed on sequences from each representative species; redundant entries from individual strains were omitted for clarity (Fig. 6). There is high level sequence conservation across the entire family. Inspection of the putative catalytic histidine (H242) and stabilizing arginine (R217) reveals that they are virtually invariant. Only one sequence (ABJ83823) from S. usitatus displays unique residues at these positions, which in addition contains a completely remodeled metal binding site and raises the question of whether it is even a functional lyase. Closer analysis of the other two entries from S. usitatus (ABJ83750 and ABJ85020) may provide insight into the potential relationship between metal selectivity and cellular localization we have described above. Both of these putative enzymes contain bona fide signal peptides. The lack of a consensus TAT sequence (not shown) suggests that they are targeted for a general secretion pathway and therefore are folded outside the cytoplasm, where Ca2+ is abundant. Closer investigation of catalytic machinery in these homologs reveals that they contain the putative catalytic histidine; however, noticeable substitutions are present within the metal binding site. The His353 and His355 from YeOGL are replaced with a glutamate and asparagine residue, respectively. These functional groups contribute oxygen donor atoms during coordinate bond formation, indicating a potential loss of Mn2+ and gain of Ca2+ binding specificity. In addition, although His287 is conserved in the primary structure, it is located in a stretch of amino acids displaying very little similarity in the S. usitatus proteins, and therefore, it is difficult to justify a spatial and functional conservation of the histidine at this site. These two predicted enzymes from the OGL family, therefore, provide further support for the hypothesis that pectate lyase catalytic metal selectivity is based upon the location of enzyme folding rather than mechanistic effects.
FIGURE 6.
Pylogenetic tree of the OGL family. Related sequence targets are shown as a phylogram. The out-group is indicated with an asterisk. The inset displays the level of conservation of functional residues for the seven out-group sequences, and the putative secreted pectate lyases are indicated with black triangles.
The observation that Yersinia pestis, the causative agent of the bubonic plague, possesses an OGL homolog is intriguing. In addition to this cytoplasmic lyase, the genome of every known Y. pestis strain was shown to contain genetic evidence of a well developed pectin utilization pathway (7) (see the CAZy Web site). These observations appear inconsistent with traditional life cycle models of the pathogen because Y. pestis is widely considered an enzootic disease and is not known to colonize the digestive tract of its mammalian host. It is difficult to rationalize therefore where the microbe would have access to dietary pectin and concomitantly why it would contain a pathway tailored to its catabolism. Recently, however, it has been reported that Y. pestis can persist within soil (48–50), a process that may be facilitated by scavenging dietary carbohydrates, such as pectin. Utilizing plant cell wall polysaccharides as a transient energy source may explain the mechanistic basis of how Y. pestis lies dormant within and passages through the environment as it waits for an opportunity to colonize new host reservoirs. This possibility awaits further investigation, however, because the presence of pectin utilization genes within Y. pestis may more simply reflect the evolutionary history of this genus, and this pathway may in fact be a decommissioned remnant containing non-functional enzymes.
CONCLUSION
Early structural analysis suggested that PLs harness a structurally conserved β-helix fold to modify and dismantle pectin (51, 52). Structural biology within the past decade, however, has illuminated the great structural diversity in protein folds that actually exists within nature for this class of carbohydrate-active enzymes (this work and see Refs. 13 and 42). Analysis of the catalytic substructures within diverse PL families has established a new paradigm for our understanding of PL structure-function relationships: the remarkable structural convergence of active site chemistry. Tailoring distinct protein scaffolds with highly similar catalytic apparatus strongly suggests that the β-elimination machinery and the stereochemical identity of their target substrate place heavy selective pressure on the evolutionary processes driving the structural convergence of these enzyme families. Beyond the core features of the PL active site, however, several variations on this theme have begun to surface with the increase of available structural information. First, it appears that the OGL family utilizes a histidine as a Brønstead base during H5 abstraction. Outside of family 9 pectate lyases, which utilize a lysine, all other described examples exclusively display catalytic arginines. The significance of this chemical substitution probably reflects the localized OGL environment. Indeed, the optimal pH of YeOGL is between 7.3 and 7.7 (EcOGL (14); YeOGL not shown), consistent with the buffered solution of the bacterial cell. Additionally, this pH optimum is more alkaline than the pKa of histidine (pKa = 6.1), an observation that contrasts with that of extracellular lyases. Secreted enzymes display a pH optimum around 9.0 (25), which requires localized pKa effects perturbing the catalytic arginine (pKa = 12.5) to display a more acidic character. Second, the metal binding site of YeOGL possesses three histidines and a glutamine, tailored for the coordination of Mn2+. This is the second lyase, representing two different PL families with unique protein folds, that harnesses a metal cofactor other than calcium. What distinguishes these two enzyme families from previous complexes is that YeOGL and YePL2B are cytoplasmic lyases and therefore fold and operate in an environment that limits the availability of calcium ions. The periplasmic homolog of YePL2B, YePL2A, contains a TAT secretion signal in its signal peptide, indicating that it is folded within the cell and secreted into the periplasm intact. This observation suggests that it is the environment in which the enzyme is folded that dictates the metal cofactor selection within the active site of pectate lyases and not the mechanistic influences.
As the field continues to advance, it will be exciting to discover whether the vast arsenal of uncharacterized pectate lyases encoded within the distal gut microbiome also display structural convergence in the architecture of their catalytic site and a metal selectivity dependent on cellular localization or if this dynamic and densely populated community has evolved novel platforms for the β-elimination of pectate.
Acknowledgments
We thank B. Henrissat and P. M. Coutinho for assistance in defining the cut-off parameters for the OGL family.
This work was supported by grants from the Georgia Research Alliance (to H. J. G.) and the Natural Sciences and Engineering Research Council of Canada (to A. B. B.).
The atomic coordinates and structure factors (code 3PE7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
B. Henrissat and P. M. Coutinho, personal communication.
- GalA
- galacturonic acid
- GalA2
- digalacturonate
- GalA3
- trigalacturonate
- PL
- pectate lyase
- OGL
- oligogalacturonate lyase
- KDG
- 2-keto-3-deoxygluconate
- r.m.s.
- root mean square
- DKI
- 5-keto-4-deoxyuronate.
REFERENCES
- 1.Bäckhed F., Ley R. E., Sonnenburg J. L., Peterson D. A., Gordon J. I. (2005) Science 307, 1915–1920 [DOI] [PubMed] [Google Scholar]
- 2.Martens E. C., Koropatkin N. M., Smith T. J., Gordon J. I. (2009) J. Biol. Chem. 284, 24673–24677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Hara A. M., Shanahan F. (2006) EMBO Rep. 7, 688–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonnenburg J. L., Xu J., Leip D. D., Chen C. H., Westover B. P., Weatherford J., Buhler J. D., Gordon J. I. (2005) Science 307, 1955–1959 [DOI] [PubMed] [Google Scholar]
- 5.Martens E. C., Roth R., Heuser J. E., Gordon J. I. (2009) J. Biol. Chem. 284, 18445–18457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martens E. C., Chiang H. C., Gordon J. I. (2008) Cell Host Microbe 4, 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodionov D. A., Gelfand M. S., Hugouvieux-Cotte-Pattat N. (2004) Microbiology 150, 3571–3590 [DOI] [PubMed] [Google Scholar]
- 8.Rodionov D. A., Mironov A. A., Rakhmaninova A. B., Gelfand M. S. (2000) Mol. Microbiol. 38, 673–683 [DOI] [PubMed] [Google Scholar]
- 9.Caffall K. H., Mohnen D. (2009) Carbohydr. Res. 344, 1879–1900 [DOI] [PubMed] [Google Scholar]
- 10.Mohnen D. (2008) Curr. Opin. Plant Biol. 11, 266–277 [DOI] [PubMed] [Google Scholar]
- 11.Abbott D. W., Boraston A. B. (2008) Microbiol. Mol. Biol. Rev. 72, 301–316, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hugouvieux-Cotte-Pattat N., Condemine G., Nasser W., Reverchon S. (1996) Annu. Rev. Microbiol. 50, 213–257 [DOI] [PubMed] [Google Scholar]
- 13.Abbott D. W., Boraston A. B. (2007) J. Biol. Chem. 282, 35328–35336 [DOI] [PubMed] [Google Scholar]
- 14.Shevchik V. E., Condemine G., Robert-Baudouy J., Hugouvieux-Cotte-Pattat N. (1999) J. Bacteriol. 181, 3912–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boraston A. B., Creagh A. L., Alam M. M., Kormos J. M., Tomme P., Haynes C. A., Warren R. A., Kilburn D. G. (2001) Biochemistry 40, 6240–6247 [DOI] [PubMed] [Google Scholar]
- 17.Evans P. (2006) Acta Crystallogr. D Biol Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 18.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 20.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 21.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 22.Vaguine A. A., Richelle J., Wodak S. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 191–205 [DOI] [PubMed] [Google Scholar]
- 23.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 24.Kirschner K. N., Yongye A. B., Tschampel S. M., González-Outeiriño J., Daniels C. R., Foley B. L., Woods R. J. (2008) J. Comput. Chem. 29, 622–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tardy F., Nasser W., Robert-Baudouy J., Hugouvieux-Cotte-Pattat N. (1997) J. Bacteriol. 179, 2503–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scavetta R. D., Herron S. R., Hotchkiss A. T., Kita N., Keen N. T., Benen J. A., Kester H. C., Visser J., Jurnak F. (1999) Plant Cell 11, 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murzina N. V., Pei X. Y., Zhang W., Sparkes M., Vicente-Garcia J., Pratap J. V., McLaughlin S. H., Ben-Shahar T. R., Verreault A., Luisi B. F., Laue E. D. (2008) Structure 16, 1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y., Chen Y., Zhang P., Jeffrey P. D., Shi Y. (2008) Mol. Cell 31, 873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rouvinen J., Bergfors T., Teeri T., Knowles J. K., Jones T. A. (1990) Science 249, 380–386 [DOI] [PubMed] [Google Scholar]
- 31.Davies G., Henrissat B. (1995) Structure 3, 853–859 [DOI] [PubMed] [Google Scholar]
- 32.Liners F., Letesson J. J., Didembourg C., Van Cutsem P. (1989) Plant Physiol. 91, 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harding M. M. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 401–411 [DOI] [PubMed] [Google Scholar]
- 34.Dominguez D. C. (2004) Mol. Microbiol. 54, 291–297 [DOI] [PubMed] [Google Scholar]
- 35.Michiels J., Xi C., Verhaert J., Vanderleyden J. (2002) Trends Microbiol. 10, 87–93 [DOI] [PubMed] [Google Scholar]
- 36.Gangola P., Rosen B. P. (1987) J. Biol. Chem. 262, 12570–12574 [PubMed] [Google Scholar]
- 37.Gilroy S., Fricker M. D., Read N. D., Trewavas A. J. (1991) Plant Cell 3, 333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shevchik V. E., Kester H. C., Benen J. A., Visser J., Robert-Baudouy J., Hugouvieux-Cotte-Pattat N. (1999) J. Bacteriol. 181, 1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeLisa M. P., Samuelson P., Palmer T., Georgiou G. (2002) J. Biol. Chem. 277, 29825–29831 [DOI] [PubMed] [Google Scholar]
- 40.De Buck E., Lammertyn E., Anné J. (2008) Trends Microbiol. 16, 442–453 [DOI] [PubMed] [Google Scholar]
- 41.Tottey S., Waldron K. J., Firbank S. J., Reale B., Bessant C., Sato K., Cheek T. R., Gray J., Banfield M. J., Dennison C., Robinson N. J. (2008) Nature 455, 1138–1142 [DOI] [PubMed] [Google Scholar]
- 42.Charnock S. J., Brown I. E., Turkenburg J. P., Black G. W., Davies G. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jenkins J., Shevchik V. E., Hugouvieux-Cotte-Pattat N., Pickersgill R. W. (2004) J. Biol. Chem. 279, 9139–9145 [DOI] [PubMed] [Google Scholar]
- 44.Li S., Kelly S. J., Lamani E., Ferraroni M., Jedrzejas M. J. (2000) EMBO J. 19, 1228–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seyedarabi A., To T. T., Ali S., Hussain S., Fries M., Madsen R., Clausen M. H., Teixteira S., Brocklehurst K., Pickersgill R. W. (2010) Biochemistry 49, 539–546 [DOI] [PubMed] [Google Scholar]
- 46.Moran F., Nasuno S., Starr M. P. (1968) Arch. Biochem. Biophys. 125, 734–741 [DOI] [PubMed] [Google Scholar]
- 47.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ayyadurai S., Houhamdi L., Lepidi H., Nappez C., Raoult D., Drancourt M. (2008) Microbiology 154, 2865–2871 [DOI] [PubMed] [Google Scholar]
- 49.Drancourt M., Houhamdi L., Raoult D. (2006) Lancet Infect. Dis. 6, 234–241 [DOI] [PubMed] [Google Scholar]
- 50.Eisen R. J., Petersen J. M., Higgins C. L., Wong D., Levy C. E., Mead P. S., Schriefer M. E., Griffith K. S., Gage K. L., Beard C. B. (2008) Emerg. Infect. Dis. 14, 941–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenkins J., Pickersgill R. (2001) Prog. Biophys. Mol. Biol. 77, 111–175 [DOI] [PubMed] [Google Scholar]
- 52.Jenkins J., Mayans O., Pickersgill R. (1998) J. Struct. Biol. 122, 236–246 [DOI] [PubMed] [Google Scholar]
- 53.Read R. J. (1986) Acta Crystallogr. A 42, 140–149 [Google Scholar]