Abstract
Cellular stress in early mitosis activates the antephase checkpoint, resulting in the decondensation of chromosomes and delayed mitotic progression. Checkpoint with forkhead-associated and RING domains (CHFR) is central to this checkpoint, and its activity is ablated in many tumors and cancer cell lines through promoter hypermethylation or mutation. The interaction between the PAR-binding zinc finger (PBZ) of CHFR and poly(ADP-ribose) (PAR) is crucial for a functional antephase checkpoint. We determined the crystal structure of the cysteine-rich region of human CHFR (amino acids 425–664) to 1.9 Å resolution, which revealed a multizinc binding domain of elaborate topology within which the PBZ is embedded. The PBZ of CHFR closely resembles the analogous motifs from aprataxin-like factor and CG1218-PA, which lie within unstructured regions of their respective proteins. Based on co-crystal structures of CHFR bound to several different PAR-like ligands (adenosine 5′-diphosphoribose, adenosine monophosphate, and P1P2-diadenosine 5′-pyrophosphate), we made a model of the CHFR-PAR interaction, which we validated using site-specific mutagenesis and surface plasmon resonance. The PBZ motif of CHFR recognizes two adenine-containing subunits of PAR and the phosphate backbone that connects them. More generally, PBZ motifs may recognize different numbers of PAR subunits as required to carry out their functions.
Keywords: ADP-ribosylation, Crystal Structure, Ligand-binding Protein, Ligand Complex, Mitosis, Zinc Finger, Antephase Checkpoint
Introduction
Progress through the eukaryotic cell division cycle is regulated by mechanisms, or checkpoints, that detect potentially disastrous defects, such as DNA damage (1). Cells are particularly sensitive to defects during mitosis, and several checkpoints regulate the passage into mitosis/M phase from interphase/G2 and progress through mitosis to ensure the accurate segregation of chromosomes. The early prophase or antephase checkpoint responds to cellular stresses, such as microtubule poisons and UV irradiation, encountered in the period before late prophase and leads to decondensation of chromosomes and a delay in mitotic progression (reviewed in Ref. 2) (3, 4). Once the nuclear envelope is broken down, cells are committed to mitosis, although completion of mitosis is subject to the correct assembly of the mitotic spindle and satisfaction of the spindle checkpoint. The CHFR (checkpoint with forkhead-associated and really interesting new gene (RING) finger domains) protein is a key component of the antephase checkpoint (2, 5–7). CHFR is inactivated in many tumors and cancer cell lines through promoter hypermethylation or mutation (5, 7). Under conditions of mitotic stress, cancer cell lines lacking CHFR function, such as HeLa, enter metaphase without delay and demonstrate higher mitotic indices compared with cell lines that express CHFR. The antephase checkpoint can be rescued in these cell lines by the re-expression of CHFR (5). The molecular pathway that links cellular stress to chromosome decondensation through CHFR has yet to be elucidated, although several activities of CHFR have been described, and many interaction partners of CHFR have been identified. The RING finger of CHFR has E3 ubiquitin ligase activity, which is essential for the antephase checkpoint (8). CHFR is autoubiquitinated and mediates the ubiquitination of substrate proteins Aurora-A and Plk1, two mitotic kinases that regulate the G2/M transition and mitotic progression (9, 10). CHFR works together with E2 enzymes, such as Ubc4 and Ubc5, to synthesize Lys48-linked chains that target substrates for proteasomal destruction, and Ubc13/Uev1a, to synthesize Lys63-linked chains to mediate signaling (8, 11). The precise function of CHFR-mediated ubiquitination remains unclear; there is evidence for and against the proteolytic degradation of ubiquitinated substrates, and in addition the poly-ubiquitin chains recruit factors, such as p38 stress kinases, to mediate downstream signaling (9, 12). CHFR knock-out mice exhibit increased tumor susceptibility, which together with the absence of CHFR expression in many tumors suggests that it is a tumor suppressor (5, 9). The mechanisms through which the absence of CHFR could drive tumor progression are not known. Candidate mechanisms include increased genomic instability through an aberrant antephase checkpoint or through down-regulation of histone deacetylase 1 (13, 14).
Poly(ADP-ribose) (PAR)2 is a polymeric post-translational modification of proteins associated with the regulation of DNA damage repair and mitosis (15). There are at least 17 PAR polymerases (PARPs) in the human genome and also a number of enzymes that modify proteins singly with mono(ADP-ribose) (mADPr). PARPs catalyze the displacement of the nicotinamide group of NAD+ by the growing PAR chain to form a 1–2-O-glycosidic linkage with the 2′-OH group of either the adenine ribose or the nicotinamide ribose, resulting in a branched and heterogeneous polymer. Several PARPs have mitotic functions; e.g. tankyrase-1 modifies the spindle-associated protein NuMA, and this activity is required for the proper assembly and maintenance of bipolar spindles (16, 17). PAR synthesis is essential for a functional antephase checkpoint, and CHFR interacts with PAR through a 20-amino acid PAR binding zinc finger motif (PBZ) at the C-terminal end of its cysteine-rich region (see Fig. 1A) (18). CHFR lacking the PBZ does not co-localize with nuclear PAR foci in interphase HEK293T cells and cannot rescue antephase checkpoint function in HeLa cells despite retaining autoubiquitination activity (18). The loss of checkpoint function might, however, be due to some other defect of CHFR lacking the PBZ that was not controlled for, such as binding and ubiquitination of substrates (e.g. Aurora-A and HDAC1), which requires the cysteine-rich region (9, 14). Nevertheless, it seems likely that the CHFR-PAR interaction is an important part of the antephase checkpoint and could form part of the checkpoint sensor for cellular stress and microtubule poisons or be required for proper localization of CHFR.
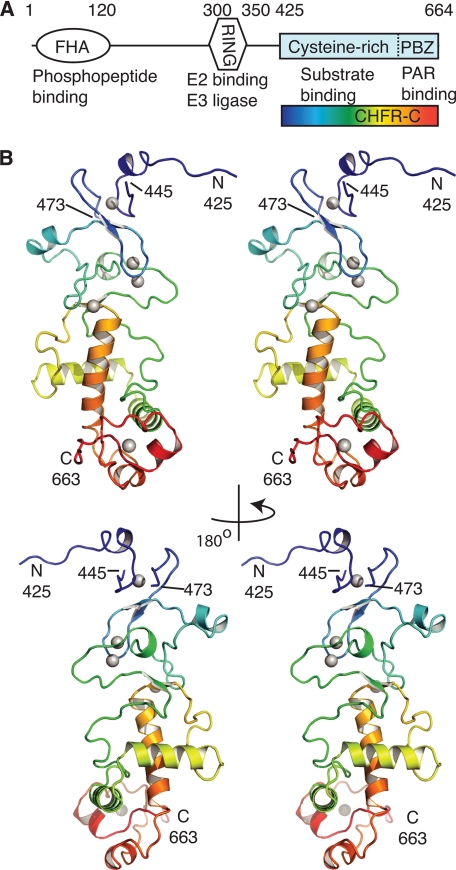
FIGURE 1.
Architecture of the cysteine-rich domain of CHFR. A, the domain structure of human CHFR is shown with residue numbering (above), and functional annotation (below). The CHFR-C region that was crystallized is indicated with a rainbow-colored box. B, the overall architecture of CHFR-C is shown as two pairs of stereo images related by a 180° rotation about the y axis. The main chain of CHFR-C is shown in a schematic representation colored blue to red from N to C terminus. Zinc ions are shown as gray spheres. See also supplemental Movie S1.
The structure of the CHFR PBZ and the basis of molecular recognition of PAR are unknown. NMR structures of PBZs from the DNA damage factor APLF (aprataxin and PNK-like factor) and an uncharacterized Drosophila protein CG1218-PA have been determined (19–21). The heterogeneity of PAR has frustrated attempts to derive high resolution structures of the PAR-PBZ interaction. Nevertheless, studies with APLF and ligands that resemble small PAR fragments have identified a single adenine binding site within a hydrophobic pocket that is crucial for PAR binding (20, 21). NMR chemical shift experiments using PAR and mADPr suggest that this pocket has a conserved function in CG1218-PA and CHFR (19). The binding site for PAR extends over more of the PBZ surface than just this pocket, although it is not known which other PAR features are recognized. The binding site of PAR on CHFR appears to be more extensive than on other PBZs and is greater than that of mADPr, although this might be an artifact of the isolated PBZ motif removed from the context of the cysteine-rich region (19). Many of the key details of PAR recognition by PBZs remain to be discovered. For example, it is not clear whether individual PBZs recognize more than one subunit of PAR, which is presumably important for discrimination between PAR and mADPr.
The forkhead-associated domain is the only region of CHFR for which a structure has been determined (22). Because there are no structures of the other domains of CHFR or details of its interactions with molecular partners, we investigated the purification and crystallization of the human CHFR protein. Herein, we report the crystal structure of the C-terminal region of human CHFR and the details of its interaction with PAR.
EXPERIMENTAL PROCEDURES
Cloning, Protein Expression, and Purification
CHFR cysteine-rich domain constructs 407–664 (CHFR-C1) and 394–664 (CHFR-C2) were cloned into the pETM6T1 vector (derived from pET44 (Novagen)) with an N-terminal, tobacco etch virus-cleavable His6-NusA tag for expression in Escherichia coli BL21-CodonPlus (DE3)-RIL cells (Stratagene). Cells were grown in lysogeny broth medium at 37 °C to an optical density of 0.4, induced by the addition of 0.4 mm isopropyl β-d-thiogalactopyranoside and incubated overnight at 21 °C. 0.4 mm ZnCl2 was added to the medium before induction. Cells were lysed in a buffer containing 100 mm NaCl, 50 mm Tris, pH 8.0, 5% glycerol, 10 mm 2-mercaptoethanol, and EDTA-free protease inhibitor tablet (Roche Applied Science). Proteins were purified by anion exchange using an anion exchange-Sepharose 4 fast flow column (GE Healthcare) run with an increasing salt gradient from 0.1 to 1 m NaCl over 20 column volumes. The tag was cleaved overnight with tobacco etch virus protease at a ratio of ∼1:20 to eluted protein. The proteins were reloaded onto the anion exchange-Sepharose column to separate the cleaved proteins from the tag and then further purified using a Superdex 200 16/60 gel filtration column (GE Healthcare), which was equilibrated in 150 mm NaCl, 25 mm Tris, pH 8.5, and 2% (v/v) glycerol. Proteins were concentrated in gel filtration buffer to 8 mg/ml.
Full-length CHFR, prepared for thermal denaturation and PAR-binding assays, was cloned into a modified version of the pRSF vector (Novagen) with an N-terminal cleavable His6-FLAG-double streptavidin tag. The full-length protein was expressed using the same method as for the CHFR cysteine-rich domain constructs. Cells were lysed in a buffer containing 200 mm NaCl, 50 mm Tris, pH 8.0, 5% (v/v) glycerol, 10 mm 2-mercaptoethanol, and EDTA-free protease inhibitor tablet. The protein was purified using Strep-Tactin resin (Fisher) and eluted by cleaving the tag with 3C protease in lysis buffer (without protease inhibitors). The protein was further purified using a Superdex 200 16/60 gel filtration column as described for the CHFR cysteine-rich domain constructs. Mutations were introduced into full-length CHFR by the QuikChange site-directed mutagenesis method (Stratagene), and mutant proteins were expressed and purified in the same way as the wild type full-length protein.
Crystallization and Structure Determination
The initial crystallization hit for the CHFR 407–664 construct was obtained from the Qiagen Classics Suite HT screen, which was set up using an Art Robbins Phoenix liquid handling system. Crystals of dimension less than 50 μm initially appeared after 3 weeks in 12% (w/v) PEG 20,000, 0.1 m MES, pH 6.5, buffer. An additive screen was carried out using additive screen HT (Hampton research) to optimize this condition further. Larger crystals (100 μm) were observed within 7 days when 0.1 m KCl was added to the original crystallization buffer.
All subsequent CHFR-C1 and CHFR-C2 crystals were grown by hanging drop vapor diffusion at 18 °C by mixing 1 μl of protein with an equal volume of well buffer containing 12% (w/v) PEG 20,000, 0.1 m KCl, and 0.1 m MES, pH 6.5, buffer. For the co-crystallization of CHFR 407–664 with adenosine 5′-diphosphoribose (mADPR) and AMP, the protein was incubated on ice with 5 mm mADPR (Sigma) and 10 mm AMP (Sigma), respectively, for 1 h, before adding 1 μl of this mixture to 1 μl of well buffer. All crystals were soaked for 5 min in well buffer (plus 5 or 10 mm of the ligand), supplemented with 20% (w/v) PEG 20,000, and then flash-frozen in liquid nitrogen. Nucleotide-free CHFR 394–664 crystals were used for the ligand-soaking experiments. The synthesis of P1P2-diadenosine 5′-pyrophosphate (AMP2) was carried out essentially as previously described (23) (see supplemental material for further details). Crystals were transferred to a fresh drop-containing well buffer supplemented with 10 mm AMP2 for 8 h and then transferred to a fresh drop-containing well buffer (plus 10 mm AMP2) supplemented with 20% (w/v) PEG 20,000 and flash frozen.
Data were collected at the Diamond light source, UK (beamlines I02, I03, and I04). Fluorescence scans of these crystals showed the presence of bound zinc, which was resolved into 10 sites. One crystal of the CHFR 407–664 mADPR complex was used to collect data sets at two wavelengths, one close to the zinc edge and a high resolution data set far from the edge (Table 1). Data were processed using Mosflm and Scala (24). Single-wavelength anomalous dispersion experimental phases were calculated using data set 1 (figure of merit was 0.35, which was improved to 0.69 after density modification), and the initial model was built using the Autosol and Autobuild programs, part of the PHENIX software suite (25). The structure obtained in the presence of mADPR was used as a model to solve the structure of the nucleotide-free, AMP- and AMP2-bound protein by molecular replacement in PHASER (26). COOT (27) and PHENIX were used for subsequent model building and refinement. Structure figures were prepared using PyMOL (28).
TABLE 1.
Summary of crystallographic analysis
| mADPr | Apo | |
|---|---|---|
| Space group | C2 | C2 |
| Lattice constants | ||
| a (Å) | 161.5 | 163.0 |
| b (Å) | 51.7 | 52.0 |
| c (Å) | 82.4 | 82.5 |
| β (degrees) | 105.8 | 105.7 |
| 1 | 2 | ||
|---|---|---|---|
| Data collection | |||
| X-ray source | Diamond I02 | Diamond I02 | Diamond I04 |
| Wavelength (Å) | 1.2832 | 0.9795 | 0.9763 |
| Resolution range (Å)a | 65.05–2.49 (2.62–2.49) | 49.22–1.89 (1.99–1.89) | 49.51–1.98 (2.09–1.98) |
| Unique reflections | 23,184 (3333) | 52,431 (7631) | 46,604 (6829) |
| Completeness (%) | 99.5 (99.3) | 99.5 (99.9) | 99.5 (99.8) |
| Multiplicity | 7.1 (7.1) | 3.3 (3.3) | 3.5 (3.5) |
| Rmerge (%) | 7.1 (24.0) | 8.8 (44.8) | 10.2 (46.1) |
| Mn I/σ(I) | 17.6 (6.7) | 8.7 (2.5) | 9.1 (2.6) |
| Refinement | |||
| Resolution range (Å) | 32.5–1.89 | 39.7–1.98 | |
| No. of amino acids | 425 | 424 | |
| No. of waters | 710 | 591 | |
| R factor (%) | 16.8 | 16.1 | |
| Rfreeb (%) | 19.4 | 20.2 | |
| Bond deviation (Å) | 0.003 | 0.007 | |
| Angle deviation (degrees) | 0.845 | 1.028 | |
| Molprobity output scores | |||
| All-atom Clashscore | 5.95 | 3.62 | |
| Rotamer outliers (%) | 2.93 | 2.93 | |
| Ramachandran outliers (%) | 0.00 | 0.00 | |
| Ramachandran favored (%) | 98.8 | 98.6 | |
| Molprobity score | 1.68 | 1.51 |
| AMP | AMP2 | |
|---|---|---|
| Space group | C2 | C2 |
| Lattice constants | ||
| a (Å) | 162.8 | 163.1 |
| b (Å) | 51.6 | 52.1 |
| c (Å) | 82.5 | 83.6 |
| β (degrees) | 105.9 | 105.3 |
| Data collection | ||
| X-ray source | Diamond I03 | Diamond I03 |
| Wavelength (Å) | 0.9763 | 0.9763 |
| Resolution range (Å)a | 65.41–2.37 (2.50–2.37) | 65.63–2.60 (2.74–2.60) |
| Unique reflections | 26,821 (3880) | 21,061 (3068) |
| Completeness (%) | 99.6 (99.5) | 99.5 (99.8) |
| Multiplicity | 3.5 (3.5) | 3.4 (3.5) |
| Rmerge (%) | 12.4 (40.9) | 9.3 (43.1) |
| Mn I/σ(I) | 8.2 (3.1) | 8.6 (2.6) |
| Refinement | ||
| Resolution range (Å) | 39.69–2.37 | 36.26–2.60 |
| No. of amino acids | 427 | 427 |
| No. of waters | 213 | 95 |
| R factor (%) | 17.9 | 19.5 |
| Rfreeb (%) | 23.1 | 24.6 |
| Bond deviation (Å) | 0.008 | 0.009 |
| Angle deviation (degrees) | 1.043 | 1.140 |
| Molprobity output scores | ||
| All-atom Clashscore | 1.46 | 2.33 |
| Rotamer outliers (%) | 3.70 | 3.49 |
| Ramachandran outliers (%) | 0.00 | 0.00 |
| Ramachandran favored (%) | 98.3 | 96.7 |
| Molprobity score | 1.34 | 1.64 |
a Values in parentheses are for the highest resolution shell.
b Free R factor was computed using 8% of the data assigned randomly.
Thermal Denaturation Assays
Thermal denaturation assays using Sypro Orange dye (Sigma) were used to measure the relative thermal stability of CHFR proteins. Sypro Orange fluoresces when bound to hydrophobic regions exposed in proteins. As the proteins are heated, they unfold, and there is an increase in fluorescence. 100-μl samples of 5 μm CHFR were prepared on ice in 50 mm NaCl, 25 mm Tris 8.5, 2% (v/v) glycerol, and 5× Sypro Orange. 25-μl aliquots of each protein sample were then added to three wells of a 96-well PCR plate (Bio-Rad), and each protein was analyzed in triplicate. The thermal denaturation assay was conducted in an iCycler, iQ5 Real Time PCR Detection system (Bio-Rad). Samples were prechilled in the machine to 4 °C for 10 min, and then the plate was heated up from 4 to 95 °C at 2 °C/min. Fluorescence intensity of the Sypro Orange was measured at excitation/emission wavelengths of 470/600 nm. The fluorescence data were fitted to the equation, y = (anx + bn) + ((adx + bd) − (anx + bn))/(1 + e(Tm − x)/m), using Prism 5 (GraphPad Software Inc.), where an and ad represent the slopes of the native and denatured base lines, bn and bd are the y intercepts of the native and denatured baselines, Tm is the apparent melting temperature, and m describes the slope of the denaturation.
Affinity Measurements Using Surface Plasmon Resonance (SPR)
SPR assays were performed on a BIAcore 3000 instrument (GE Healthcare) at 25 °C using running buffer (25 mm Tris/HCl, pH 8.5, 150 mm NaCl, 2% (v/v) glycerol, 0.5 mm TCEP, 0.005% (v/v) Tween 20). 35 RU of biotinyl-PAR (see supplemental material) was immobilized on flow cells 2 and 4 of BIAcore SA sensor chips (GE Healthcare), 1 and 3 being left blank. All flow cells were then blocked with 5 mm biotin. CHFR proteins were dialyzed against running buffer and injected at 40 μl/min for 375 s over pairs of blank and PAR-containing flow cells. Individual sensorgrams were recorded for each CHFR protein at each of >10 concentrations over a range from 0.1 nm to 2 μm (supplemental Fig. S7). The sensor chip surface was regenerated between injections by perfusion of 35 μl of 20 mm glycine/HCl, pH 2.0, at 10 μl/min.
Sensorgrams were processed using BIAevaluation 3.0 software (Biacore AB). Sensorgrams recorded from flow cells 2 and 4 were corrected for passive refractive index changes and nonspecific interactions by subtraction of the corresponding sensorgram recorded from flow cells 1 and 3. With 35 RU of immobilized biotinyl-PAR, maximum binding of CHFR was ∼110 RU. The association phase of sensorgrams (t = 0–360 s) were fitted to a biphasic exponential equation, r = Req1(1 − e−Ka1t) + Req2(1 − e−Ka2t), using Prism 5, to derive values for response at equilibrium (Req), where Req = Req1 + Req2. Req was plotted against sample concentration and fitted by non-linear regression to a binding isotherm described by the equation Req = ([CHFR]n × 110 RU)/([CHFR]n + KDn).
Generation of the CHFR-PAR Model
The PAR model Protein Data Bank file and refinement parameters (.cif) were initially produced using the PRODRG server and edited to correct the obvious stereochemical errors. One adenine group and one AMP group were automatically fitted into the electron density maps (of the AMP and AMP2 ligand structures, respectively) using COOT, which was also used for several rounds of manual fitting (to remove any clashes) and geometry refinement. The final structure was energy-minimized as described in the supplemental material.
RESULTS
Overall Architecture of the CHFR Cysteine-rich Region
CHFR consists of an N-terminal FHA domain, a central RING domain, and C-terminal cysteine-rich region (CHFR-C, residues 407–664) (Fig. 1A). It is not known whether these domains are independent or whether they interact. Searches of the SWISSPROT, SMART, and Pfam databases suggested that this putative zinc binding region is unrelated to any domains of known three-dimensional structure (29, 30) with the exception of the PBZ motif (residues 620–644) (18, 19). We therefore set out to determine the structure of the cysteine-rich region using x-ray crystallography. Two different fragments of CHFR that included the entire cysteine-rich region were crystallized in space group C2: residues 407–664 in the presence of mono-ADP-ribose (CHFR-C1/mADPR) and residues 394–664 in apo-form (CHFR-C2). The structure of CHFR-C1/mADPR was solved by single-wavelength anomalous dispersion phasing, which revealed two protein chains having five zinc sites each, and was refined to a final resolution of 1.9 Å (Table 1 and Fig. 1B). The apo-form was solved by molecular replacement and refined to 2.5 Å. The structures were highly similar both within (Cα root mean square deviation = 1.13 Å) and between (Cα root mean square deviation = 0.22 Å) crystals except for the last three residues, which diverge by 3–11 Å, indicating some flexibility in this region. In both structures, residues 425–445 and 473–663 are modeled, whereas no ordered electron density was observed for residues 446–472.
The cysteine-rich region forms a single continuous structural unit comprising an N-terminal region that has only a few short secondary structure elements, a central α-helical region, and a C-terminal PBZ motif (Fig. 2). The secondary structure of CHFR-C comprises eight α-helices and four β-strands. The strands are each made up of three residues and formed into two short β-sheets. No structures that significantly resemble the whole CHFR-C domain were found from a search of the Protein Data Bank using the DALI server (31). Fig. 1B shows how the protein chain traces a meandering path between the first four zinc binding sites (colored blue to green) past the α-helical region (colored green, yellow, and orange) and down to contact the PBZ (colored red). The chain returns back through two α-helices (colored green and yellow) and connects to the N-terminal region via the second half of zinc motif 4. A long helix (colored orange in Fig. 1B) then leads the chain finally into the PBZ (colored red). The long loops between β3-α4 and α5-β4 are ordered through a network of hydrogen bonds and through packing of conserved hydrophobic side chains against the hydrophobic residues of α4, α5, and α6, as shown for Phe536 in supplemental Fig. S1.
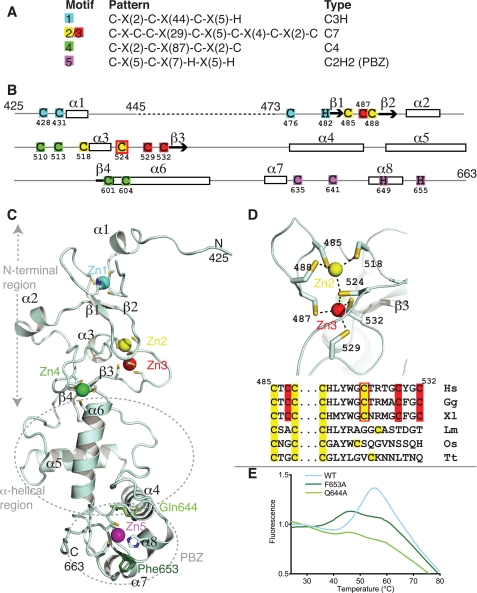
FIGURE 2.
Details of the five zinc binding sites of CHFR-C. A, the five zinc binding sites are arranged as four motifs described by their ProSite pattern and the type of coordinating residues (C, cysteine; H, histidine). B, the amino acids included in the model are shown with a solid line, onto which are mapped the pattern of zinc-coordinating residues and the secondary structure. The dashed line represents a region that was not ordered. C, schematic representation of the CHFR-C structure, with the bound zinc ions shown as colored spheres, as defined in A. Two residues within the PBZ that are crucial for protein stability are shown as green sticks. D, magnified view of zinc binding motif 2/3 (top) and amino acid sequence alignment of human (Hs), chicken (Gg), Xenopus (Xl), Leishmania (Lm), rice (Os), and Tetrahymena (Tt) equivalent motifs color-coded as in B. E, thermal denaturation data showing the loss of stability caused by mutation of PBZ residues Phe653 and Gln644. See also supplemental Fig. S1.
The Cysteine-rich Region Has Four Different Zinc-binding Motifs
There are four zinc-binding motifs within CHFR-C, which bind five zinc ions (Fig. 2 and supplemental Movie S1). It is usually the case that tandem zinc binding domains are modular, with distinct motifs within the sequence in which the four residues that coordinate each zinc ion are clustered (see the Protein of the Month: Zinc Fingers InterPro Web site). This is not the case in CHFR, in which zinc binding sites 2, 3, and 4 are interleaved (colored yellow, red, and green, respectively, in Fig. 2). Motifs 1, 2/3, and 4 have extraordinarily large gaps between some of the coordinating residues. We were not able to find any structures that matched the first two motifs using DALI, and to our knowledge, the coordination geometry found in motif 2/3 has not previously been observed. These deviations from the norm explain why it was not possible to confidently predict the type of motifs found in CHFR-C, and they raise the issue that such extremely large gaps between coordinating residues might frustrate the prediction of other zinc binding domains. With one exception, which is explained below, the zinc binding sites of CHFR are conserved throughout evolution, and we are confident that the structure of human CHFR-C region serves as an archetype for other homologues.
Zinc binding motif 1, which is holding the first ordered region onto the rest of the domain, is a C3H type zinc finger (colored cyan in Fig. 2). Only one of the zinc binding residues in this motif (His482) is located in a regular secondary structural element, whereas the others (cysteines 428, 431, and 476) are in loop regions (Fig. 2, B and C). There is a 44-residue gap between the second and third zinc-coordinating cysteine residues in motif 1, longer than is found in typical C3H zinc fingers. The loop has expanded dramatically in vertebrates compared with other eukaryotes, perhaps indicating an altered function of this region of the protein.
The two zinc ions of zinc-binding motif 2/3 stabilize the β1-β2 loop region together with the short helix α3 (Fig. 2, B–D). This is, to our knowledge, a novel zinc binding motif that we have termed C7, in which a bridging cysteine residue coordinates two different zinc ions. The first zinc binding site in this motif is formed by cysteines 485, 488, 518, and 524, and Cys524 again together with cysteines 487, 529, and 532 form the second zinc binding site (Fig. 2D). Interestingly, this double zinc binding motif is only a feature of CHFR proteins found in vertebrates, suggesting that this motif has evolved from a more conventional C4 motif (Fig. 2D).
Zinc binding motif 4 links the N-terminal region of CHFR-C to the central, α-helical region through two cysteine residues interleaved with the residues of motif 2/3 (cysteines 510 and 513), one located in α6 (Cys604), and one located at the end of β4 (Cys601), coordinating a single zinc ion (Fig. 2, B and C). The arrangement of cysteine residues within zinc binding motif 4 is structurally similar to the GATA type zinc finger fold (the structure found to have the highest DALI Z score (3.2) was that of the nitrogen metabolite repression regulator area zinc finger (Protein Data Bank entry 2VUT)). However, there are 87 residues between the second and third cysteine residues that coordinate the zinc in motif 4, whereas most GATA-type zinc fingers have a much shorter loop of 17–20 residues (see the Protein of the Month: Zinc Fingers InterPro Web site).
Crystal Structures of CHFR·Ligand Complexes Identify Two Adenine Binding Sites on the PBZ Motif
Zinc binding motif 5 is a PBZ motif, which has been previously characterized as the site of interaction between CHFR and PAR (18). Structures of PBZ motifs from human APLF and Drosophila CG1218-PA have been recently determined by NMR spectroscopy, and our high resolution x-ray structure confirms that the arrangement of the residues that coordinate the zinc ion (Cys635, Cys641, His649, and His655) is similar in CHFR (19–21). As predicted by Isogai et al. (19), residues 647–654 of the CHFR PBZ (which correspond to CHFR residues 635–642 in their work because they have used CHFR isoform 3 numbering) form an α-helical structure similar to that found in CG1218-PA. They also found that the C-terminal residues of CHFR (residues 652–664 in our structure) were significantly shifted upon PAR binding, unlike the corresponding residues in CG1218-PA, suggesting the presence of a more extensive PAR binding site in CHFR. The PBZ motif is an integral part of CHFR-C, in contrast to the PBZ motifs of APLF and CG1218-PA, which reside in otherwise unstructured regions of the proteins. This has important consequences for functional studies because some conserved residues in CHFR that have been implicated in PAR binding fulfill a structural role. For example, residues Arg632 and Gln644 are in fact buried in the protein structure, form part of the contact region between the PBZ motif and the rest of the CHFR-C domain, and are unlikely to interact with binding partners (18, 19). (Fig. 2C and supplemental Fig. S1). Phe653 is partly buried under one of the zinc-coordinating residues (His649), and this residue is potentially important for the structural integrity of the PBZ itself (Fig. 2C). We found that mutation of Gln644 or Phe653 destabilized the protein, and we predict that mutation of Arg632 would also be deleterious (Fig. 2E). The reduced thermal stability of these mutations confirms that the PBZ motif has a structural role in addition to binding PAR. These results also highlight the need for a structural model of the interaction between PAR and CHFR. One of the cancer-associated mutations in CHFR (F536S) presumably inactivates the protein through structural destabilization because Phe536 holds three structural elements together (32) (supplemental Fig. S1).
The CHFR PBZ motif is the most conserved part of CHFR-C in vertebrates, suggesting that it has the most conserved function (supplemental Fig. S2). We therefore investigated the structures of CHFR·ligand complexes to obtain further details of the binding of PAR to CHFR. Our attempts to determine the structure of a CHFR-C·PAR complex were unsuccessful, resulting in poorly diffracting crystals or the absence of any additional electron density. This is probably because PAR is a heterogeneous, branched polymer of high molecular weight. An ideal ligand for x-ray co-crystal structure determination would be the minimal subunit of PAR containing two adenine moieties; however, access to this subunit by total synthesis has not yet been reported. Instead, we were able to obtain crystal structures of CHFR-C with various ligands that are structurally similar to different regions of PAR, from which we could obtain more information about the specific residues that are important for binding. The structures of the ligands selected, mADPR, AMP, and AMP2, and the region of PAR they represent are shown in Fig. 3A. The co-crystal structures of AMP and AMP2 with CHFR-C were solved by molecular replacement using the CHFR-C1·mADPR structure as a model. The CHFR-C·AMP and CHFR-C·AMP2 structures were refined to 2.37 and 2.60 Å, respectively. In all structures, one of the two CHFR-C chains was involved in crystal contacts that blocked the ligand binding sites (supplemental Fig. S3), and the following discussion refers only to the ligand binding sites in the other chain.
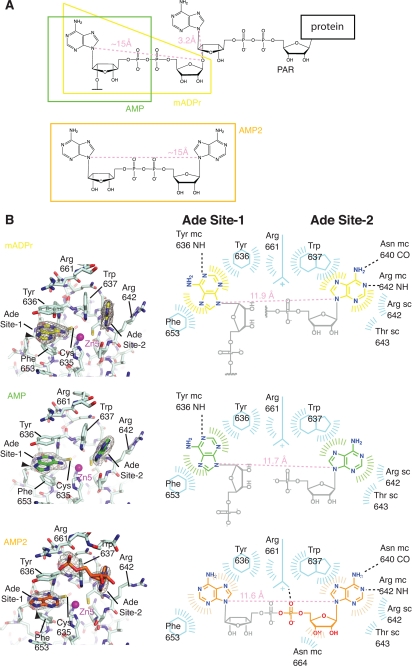
FIGURE 3.
The PBZ domain of CHFR has two adenine binding sites. A, chemical structures of PAR attached to a substrate protein and the AMP2 ligand. The ligands for which co-crystal structures with CHFR-C were determined are marked. Maximum theoretical distances within the linker motifs are shown in light pink. B, structures of mADPr-bound, AMP-bound, and AMP2-bound PBZ domains (left-hand images) and schematic diagrams of the interactions (right-hand images). The 2mFo − DFc electron density maps shown around the ligands in the left-hand images are contoured at 1 σ. The black arrowheads indicate the position and orientation of the bond between the modeled adenine groups and the unmodeled ribose groups in site 1. In the corresponding schematic diagrams, the portions of the ligands that were modeled are colored as in the left-hand image, and the unmodeled portions are shaded gray. Dashed lines indicate putative hydrogen bonds, and flashes around ligands or protein residues indicate Van der Waals contacts. The number of flashes gives a rough indication of the extent of the contacts, and the direction points toward the other component in contact. Actual distances within linker motifs are shown in light pink.
The structure of CHFR-C bound to mADPR revealed two binding sites for adenine groups within the CHFR PBZ, labeled site-1 and site-2 in Fig. 3B. The electron density was consistent with a higher degree of ligand mobility in site 2 than site 1, and the mean B-factors of the modeled ligands support that interpretation (52 Å2 in site 2, 35 Å2 in site 1, S.D. 5 Å2 in both). Similar electron density was observed in the AMP-bound structure, although again the site 1 density was clearer. There was no apparent density that could be interpreted to represent the rest of the ligand in both cases (supplemental Fig. S4). In both structures, the adenine group in site 1 stacks between the aromatic rings of Tyr636 and Phe653. The adenine group in site 2 is stabilized by stacking interactions with Trp637 and contact with the side chains of Arg642 and Thr643 (Fig. 3B). The adenine groups in sites 1 and 2 are suitably positioned to make hydrogen bond contacts with the main chain of Tyr636 and Asn640/Arg642, respectively. In the apo-structure, these residues make contacts to ordered water molecules that occupy the adenine binding sites (supplemental Fig. S4). The apo-structure also shows that binding of the ligands does not alter the protein conformation or nucleotide-binding site, with the exception of side chain motions of Tyr636 and Arg642.
In the structure of CHFR-C·AMP2, electron density is observed for two adenine groups and part of the phosphoribose linker. AMP2 was modeled as one adenine (site 1) and one AMP molecule (site 2) (Fig. 3B). Arg661 and the C-terminal residues of CHFR-C are more ordered in the AMP2-bound structure compared with the other structures and form additional contacts with the ordered phosphate and ribose groups. The orientation of the adenine in site 1 appears to be different from that found in the AMP and mADPr structures (Fig. 3B and supplemental Fig. S5). The obvious consequence of this is that the point of attachment of the ribose is in a similar position in all three structures but that the orientation of the attached linker would be rotated through ∼90° in the AMP2 structure, as shown by the black arrowheads in Fig. 3B. To explain this difference, we modeled the binding of AMP2 to CHFR-C and found that the geometry of the linker is incompatible with the orientation of the site 1 adenine in the AMP or mADPr structures. So although the linker would appear to be of the correct length, the geometry of the two adenine sites is such that a longer linker or a linker with a different geometry is required to connect the two sites as observed in the mADPr or AMP structures. This incompatibility and the observation that the C terminus of CHFR and the phosphate and ribose groups of the ligand are only ordered in the CHFR-C·AMP2 structure suggest that one molecule of the AMP2 is bound to the protein, with the caveat that we were not able to model the entire ligand linker motif, presumably due to its flexibility.
A Structural Model of the CHFR-PAR Interaction and Its Validation by Surface Plasmon Resonance
We devised a model for binding of a minimal fragment of PAR to the CHFR PBZ based on analysis of the three ligand-bound structures of CHFR-C (Fig. 4, A and B, and supplemental Movie S2). The position and orientation of the adenine in site 1 were based on the AMP- and mADPR-bound structures. We assumed that the structures of monomeric ligands reflect the highest affinity positions, perhaps because of the additional hydrogen bond formed with the main chain of Tyr636. We assumed that the AMP2-bound structure provided the optimal model for the adenine in site 2 due to the additional hydrogen bonds and increased contact area (Fig. 3B). In contrast to the AMP2-bound model, the additional ribose group that makes a longer linker between adenine groups in PAR permits the correct geometry so that the two adenine groups fit in their optimum orientation. The surface of CHFR that binds PAR has an overall positive charge to complement the negatively charged PAR (supplemental Fig. S2). The positive charge extends from the PBZ motif up through the helical region, which might imply that this face of CHFR is oriented toward the PAR. This could influence the binding of other proteins, which might be more likely on the opposite more acidic face, when CHFR is bound to PAR.
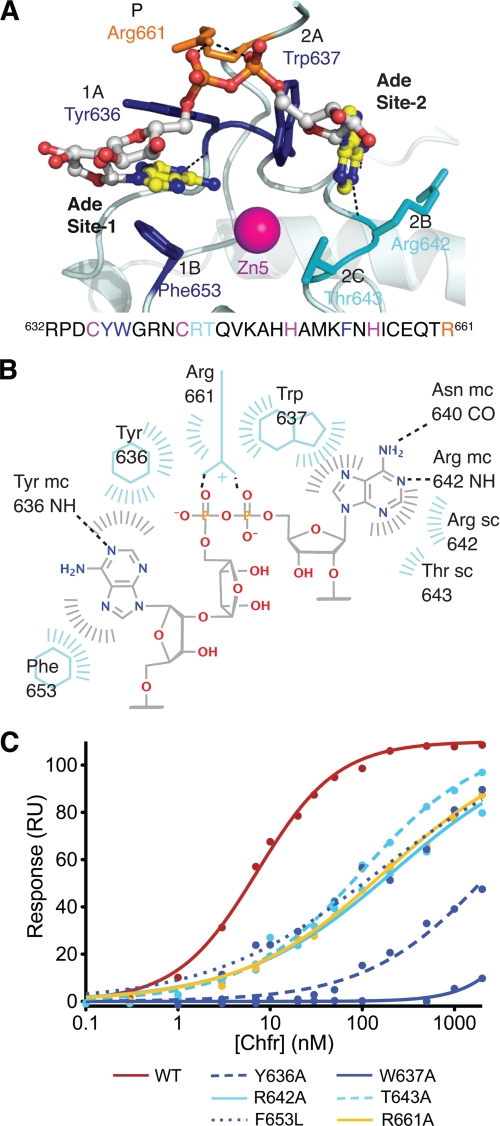
FIGURE 4.
A structural model for PAR binding to CHFR. A, model of the binding of a two-adenine-containing fragment of PAR bound to the PBZ motif of CHFR (light blue). The PAR is shown as a ball-and-stick model and is colored as follows: yellow, carbon in adenine groups; white, carbon in ribose groups; blue, nitrogen; red, oxygen; orange, phosphorus. The side chains of the residues that contact PAR are labeled according to the part of PAR they contact (i.e. Ade site 1 or 2 or phosphate (P)), shown as sticks and colored as follows: dark blue, core aromatic residues; cyan, other residues that contact adenine in site 2; orange, residue that contacts phosphate linker. See also supplemental Movie S2. B, schematic diagram to illustrate the contacts between the PAR model and the PBZ of CHFR. C, binding curves derived from surface plasmon resonance data show the reduced binding of CHFR mutants relative to WT.
To validate the binding mode of ligands we observed in our crystal structures and PAR model, we mutated several residues predicted to be important, shown in Fig. 4A. Tyr636, Trp637, Arg642, Thr643, and Arg661 were individually mutated to alanine in the full-length CHFR protein, and all of these mutants have stability equivalent to that of wild-type protein by thermal denaturation (Table 2 and supplemental Fig. S6). However, when Phe653 was mutated to alanine, the protein was unfolded (Fig. 2E), but mutation of this residue to leucine did not affect the overall fold of the protein, so this less dramatic mutant was used in further experiments instead. The binding properties of the wild type CHFR and the mutants were analyzed by SPR using immobilized PAR (Table 2, Fig. 4C, and supplemental Fig. S7). We found that full-length wild-type CHFR binds PAR with high affinity (7 nm), and all of the CHFR mutants tested had at least a 10-fold weaker binding (Table 2). As expected, mutation of the aromatic residues had the greatest effect, and the replacement of Tyr636 or Trp637 with alanine decreased binding affinity by a factor of over 100. The substantial decrease in PAR binding upon mutation of these residues provides support for two adenine-binding sites in CHFR and our proposed model of PAR binding. We cannot rule out the possibility that the model is imperfect in some details, such as the orientation of the adenine ring in site 1. This will only be clarified when PAR that is suitable for structural studies becomes available.
TABLE 2.
Summary of thermal denaturation and PAR binding data
Values shown are ±S.E.
| Construct | Tm | KD |
|---|---|---|
| °C | nm | |
| WT | 50.6 ± 0.1 | 7.0 ± 0.3 |
| Y636A | 52.7 ± 0.3 | 2500 ± 400 |
| W637A | 51.1 ± 0.2 | >5000 |
| R642A | 52.7 ± 0.1 | 220 ± 20 |
| T643A | 53 ± 1 | 100 ± 7 |
| F653L | 47.5 ± 0.2 | 150 ± 20 |
| R661A | 52.8 ± 0.1 | 180 ± 20 |
DISCUSSION
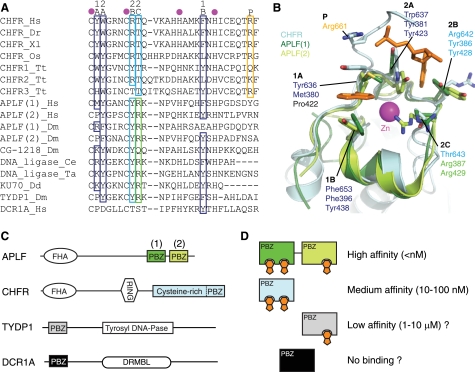
The residues that form the PAR-binding interface are highly conserved across CHFR homologues, and therefore the binding mode that we have elucidated is likely to be conserved (Fig. 5A). With some notable exceptions, as explained below, these residues are also conserved in most other PBZ motifs. In general, three of the four conserved residues that form the adenine binding sites are aromatic or hydrophobic, and one is basic (labeled 1A, 1B, 2A, and 2B in Fig. 5). The basic residue could contribute to the interaction in three ways: in forming part of the hydrophobic pockets; as part of the overall positive charge of the PBZ surface; and in direct recognition of the phosphates. The fifth residue that contacts the adenines, at position 2C, is not conserved, although it tends to be a threonine in CHFR homologues and an arginine in other PBZ motifs.
FIGURE 5.
Conservation of PAR-binding features within PBZ motifs. A, multiple alignment of protein sequences obtained from the PBZ motifs of CHFR homologues and other proteins. The residues that coordinate the zinc ion are marked with magenta circles, and the residues that contact PAR in human CHFR are indicated as defined in Fig. 4A. Conserved PAR-binding residues are boxed and colored using the scheme defined in Fig. 4. The green box marks a non-conserved residue that is predicted to contact PAR in other PBZ motifs. B, superposition of human CHFR and APLF motifs (light blue, CHFR; dark green, APLF motif 1; light green, APLF motif 2) with PAR-interacting residues on CHFR and their structural equivalents on APLF shown as sticks. Only the ordered cores of APLF motifs are shown (APLF motif 1, residues 373–405; APLF motif 2, residues 417–442) because the N and C termini have poorly defined conformations. C, domain structures of selected proteins that contain PBZ motifs. Other domains are as follows: forkhead-associated (FHA); really interesting new gene (RING); tyrosyl-DNA phosphodiesterase (Tyrosyl DNA-Pase); DNA repair metallo-β-lactamase (DRMBL). D, a schematic model to illustrate that the binding affinities for PAR can be predicted to vary over a wide range, dependent on the number of adenine binding sites (orange) within single or tandem PBZ motifs. The PBZ domains of TYDP1 and DCR1A are predicted to exhibit low affinity for PAR and no PAR binding, respectively.
Strikingly, site 1 appears to be absent in some other PBZ motifs because a proline residue is present at position 1A, which is unable to form a hydrogen bond (e.g. APLF PBZ motif 2; Fig. 5). Comparison of the binding affinities of the PBZ-containing proteins CHFR (this study) and APLF (18, 21) for immobilized PAR, measured using SPR, supports our hypothesis. Both CHFR and the first PBZ motif of APLF have high affinity for PAR (KD of 7 and 50 nm, respectively). The second PBZ motif of APLF, in which Ade site 1 is blocked by a proline at position 1A, has a similar affinity as the Y636A point mutant of CHFR in which this site is disrupted (KD of 8 and 2.5 μm, respectively). These data are consistent with the model in which the single PBZ motif of CHFR and the first PBZ motif of APLF bind two subunits of PAR and the second PBZ motif of APLF binds a single subunit. NMR studies on the two PBZ motifs of APLF identified an adenine binding site identical to the site 2 that we have characterized on CHFR (20, 21). These studies, and further NMR studies on the Drosophila CG-1218 protein and on the isolated PBZ motif of CHFR provide some evidence to corroborate the existence of site 1 in some PBZ motifs. Notably, chemical shift data on the interaction between the first PBZ motif of APLF and mADPR show a large shift for Met380, and a single NOE was detected between the adenine ring and Phe396. Chemical shifts at both sites and in the phosphate-binding C terminus of the isolated CHFR PBZ motif were identified upon binding of PAR or mADPR to CHFR (19).
PBZ motifs are combined with other types of domain in multiple contexts in several proteins (Fig. 5C). For example, CHFR has a single PBZ motif embedded within a larger structured domain, whereas APLF has two PBZ motifs that are located within an unstructured region of the protein and separated by a flexible linker. The coupling of two PBZ motifs results in very high affinity for PAR; e.g. the full-length APLF or GST-tagged (and hence potentially dimeric) CHFR have KD values of less than 1 nm. In addition, we predict that a subset of PBZ motifs have two adenine binding sites and thus recognize two subunits of PAR, whereas other PBZ motifs recognize only one PAR subunit. This will result in different levels of affinity for PAR, dependent on the number of PBZ motifs and the number of PAR subunits each motif can bind to. KD values could range from less than 1 nm (e.g. APLF tandem domains), through 10–100 nm (CHFR), to possibly a 1–10 μm range (TYDP1) (Fig. 5D). The PBZ motif of CHFR also serves a structural role, unlike the PBZ motifs of APLF. The predicted PBZ of DCR1A lacks all of the PAR-binding residues, except for the aromatic at position 1B that is important for stability, and might serve a primarily structural role.
Because PARP inhibitors have shown promise as cancer therapeutics, it has been suggested that the interaction of PBZ motifs with PAR could be a potential drug target (20). The predicted wide range of affinities of PBZ motif proteins for PAR suggests that targeting specific PAR-PBZ interactions might be a considerable challenge. This study describes several advances that are crucial for this endeavor, such as a soakable system for the study of PBZ-ligand interactions, high resolution crystal structures of the CHFR PBZ motif bound to ligands, and the identification of the crucial residues required for the interaction. These resources should stimulate the field to produce tool compounds with which to investigate the biological functions of protein-PAR interactions and the therapeutic potential of this new class of target.
Supplementary Material
Acknowledgments
We thank Junjie Chen for CHFR cDNA; the ISMB Biophysics Centre at Birkbeck (University of London) for use of the SPR instrument; the staff of DIAMOND beamlines I02, I03, and I04 for assistance; Isaac Westwood for contributions to data collection; Antony Oliver and Ammar Ali for assistance in preparing PARP1 protein; and Jon Wilson and Charlotte Dodson for critical comments on the manuscript.
This work was supported by Cancer Research UK Project Grant C24461/A9549 (to R. B.) and Cancer Research UK Centre Grant C309/A8274.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Movies S1 and S2.
The atomic coordinates and structure factors (codes 2XP0, 2XOC, 2XOZ, and 2XOY) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PAR
- poly(ADP-ribose)
- PBZ
- PAR-binding zinc finger
- PARP
- poly(ADP-ribose) polymerase
- mADPR
- adenosine 5′-diphosphoribose
- AMP2
- P1P2-diadenosine 5′-pyrophosphate
- SPR
- surface plasmon resonance
- RU
- resonance units.
REFERENCES
- 1.Morgan D. O. (2007) The Cell Cycle, Principles of Control, Oxford University Press [Google Scholar]
- 2.Chin C. F., Yeong F. M. (2010) Mol. Cell. Biol. 30, 22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rieder C. L., Cole R. W. (1998) J. Cell Biol. 142, 1013–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rieder C. L., Cole R. (2000) Curr. Biol. 10, 1067–1070 [DOI] [PubMed] [Google Scholar]
- 5.Scolnick D. M., Halazonetis T. D. (2000) Nature 406, 430–435 [DOI] [PubMed] [Google Scholar]
- 6.Toyota M., Sasaki Y., Satoh A., Ogi K., Kikuchi T., Suzuki H., Mita H., Tanaka N., Itoh F., Issa J. P., Jair K. W., Schuebel K. E., Imai K., Tokino T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7818–7823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Privette L. M., Petty E. M. (2008) Transl. Oncol. 1, 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaturvedi P., Sudakin V., Bobiak M. L., Fisher P. W., Mattern M. R., Jablonski S. A., Hurle M. R., Zhu Y., Yen T. J., Zhou B. B. (2002) Cancer Res. 62, 1797–1801 [PubMed] [Google Scholar]
- 9.Yu X., Minter-Dykhouse K., Malureanu L., Zhao W. M., Zhang D., Merkle C. J., Ward I. M., Saya H., Fang G., van Deursen J., Chen J. (2005) Nat. Genet. 37, 401–406 [DOI] [PubMed] [Google Scholar]
- 10.Kang D., Chen J., Wong J., Fang G. (2002) J. Cell Biol. 156, 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bothos J., Summers M. K., Venere M., Scolnick D. M., Halazonetis T. D. (2003) Oncogene 22, 7101–7107 [DOI] [PubMed] [Google Scholar]
- 12.Matsusaka T., Pines J. (2004) J. Cell Biol. 166, 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maddika S., Sy S. M., Chen J. (2009) J. Biol. Chem. 284, 12998–13003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh Y. M., Kwon Y. E., Kim J. M., Bae S. J., Lee B. K., Yoo S. J., Chung C. H., Deshaies R. J., Seol J. H. (2009) Nat. Cell Biol. 11, 295–302 [DOI] [PubMed] [Google Scholar]
- 15.Schreiber V., Dantzer F., Ame J. C., de Murcia G. (2006) Nat. Rev. Mol. Cell Biol. 7, 517–528 [DOI] [PubMed] [Google Scholar]
- 16.Chang P., Coughlin M., Mitchison T. J. (2005) Nat. Cell Biol. 7, 1133–1139 [DOI] [PubMed] [Google Scholar]
- 17.Chang W., Dynek J. N., Smith S. (2005) Biochem. J. 391, 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahel I., Ahel D., Matsusaka T., Clark A. J., Pines J., Boulton S. J., West S. C. (2008) Nature 451, 81–85 [DOI] [PubMed] [Google Scholar]
- 19.Isogai S., Kanno S., Ariyoshi M., Tochio H., Ito Y., Yasui A., Shirakawa M. (2010) Genes Cells 15, 101–110 [DOI] [PubMed] [Google Scholar]
- 20.Eustermann S., Brockmann C., Mehrotra P. V., Yang J. C., Loakes D., West S. C., Ahel I., Neuhaus D. (2010) Nat. Struct Mol. Biol. 17, 241–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li G. Y., McCulloch R. D., Fenton A. L., Cheung M., Meng L., Ikura M., Koch C. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 9129–9134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stavridi E. S., Huyen Y., Loreto I. R., Scolnick D. M., Halazonetis T. D., Pavletich N. P., Jeffrey P. D. (2002) Structure 10, 891–899 [DOI] [PubMed] [Google Scholar]
- 23.Millo E., Zocchi E., Galatini A., Benatti U., Damonte G. (2008) Synth. Commun. 38, 3260–3269 [Google Scholar]
- 24.Collaborative Computational Project, Number 4. (1994) Acta Crystallogr. D 50, 760–76315299374 [Google Scholar]
- 25.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P., Cowtan K. (2004) Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 29.Schultz J., Milpetz F., Bork P., Ponting C. P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonnhammer E. L., Eddy S. R., Durbin R. (1997) Proteins 28, 405–420 [DOI] [PubMed] [Google Scholar]
- 31.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mariatos G., Bothos J., Zacharatos P., Summers M. K., Scolnick D. M., Kittas C., Halazonetis T. D., Gorgoulis V. G. (2003) Cancer Res. 63, 7185–7189 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.