Abstract
Sphingosine kinase 1 (SK1) is an enzyme that catalyzes the phosphorylation of sphingosine to produce the bioactive lipid sphingosine 1-phosphate (S1P). We demonstrate here that the SK1 inhibitor, SKi (2-(p-hydroxyanilino)-4-(p-chlorophenyl)thiazole) induces the proteasomal degradation of SK1 in human pulmonary artery smooth muscle cells, androgen-sensitive LNCaP prostate cancer cells, MCF-7 and MCF-7 HER2 breast cancer cells and that this is likely mediated by ceramide as a consequence of catalytic inhibition of SK1 by SKi. Moreover, SK1 is polyubiquitinated under basal conditions, and SKi appears to increase the degradation of SK1 by activating the proteasome. In addition, the proteasomal degradation of SK1a and SK1b in androgen-sensitive LNCaP cells is associated with the induction of apoptosis. However, SK1b in LNCaP-AI cells (androgen-independent) is less sensitive to SKi-induced proteasomal degradation and these cells are resistant to SKi-induced apoptosis, thereby implicating the ubiquitin-proteasomal degradation of SK1 as an important mechanism controlling cell survival.
Keywords: Apoptosis, Enzyme Degradation, Proteasome, Sphingolipid, Ubiquitination
Introduction
Sphingosine 1-phosphate (S1P)5 is a bioactive lipid that has an important role in regulating the growth, survival, and migration of mammalian cells. S1P binds to a family of five GPCR termed S1Pn (where n = 1–5) that regulate various effectors, such as MAP kinase (1). S1P is produced by the enzyme sphingosine kinase (SK1 and SK2 isoforms), which catalyzes the phosphorylation of sphingosine to produce S1P (2, 3). There are three N-terminal variants of SK1. SK1a (GenBankTM number: NM_001142601) is a 42.5 kDa protein, while SK1b (GenBankTM number: NM_182965) is a 51 kDa protein identical to SK1a, but with an 86 amino acid N-terminal extension. The third form has a molecular mass of 43.9 kDa and is identical to SK1a except for a 14 amino acid N-terminal extension (GenBankTM number: NM_021972) and migrates with similar mobility as SK1a on SDS-PAGE. The SK1a annotation used here therefore includes SK1a and possibly SK1a+14.
SK1 has been demonstrated to have an important role in cancer (4). For instance, enforced overexpression of SK1 increases V12-Ras-dependent transformation of cancer cells (5), S1P levels, estrogen-dependent tumorigenesis, and blocks apoptosis of MCF-7 cells induced by anti-cancer drugs (6). SK1/S1P is also required for EGF-induced MCF-7 cell migration, proliferation and survival (7) and breast cancer cell growth (8). High SK1 expression is also correlated with poor prognosis in ER+ breast cancer, and SK1 induces a migratory phenotype in response to S1P in MCF-7 cells, via SK1-dependent changes in S1P3 expression and PAK1/ERK-1/2 regulation (9). There is no evidence that mutations occur in the SK1 gene linked to cancer and therefore, the term non-oncogene addiction has been used to describe its role in cancer progression (10). The S1P signaling pathway has also been implicated in promoting the proliferation of androgen-independent prostate cancer PC-3 cells (11). Moreover, irradiation of a radiation-sensitive cancer cell line, TSU-Pr1 results in a decrease in SK1 activity and a concomitant increase in ceramide (the precursor of sphingosine), which induces apoptosis of these cells. In addition, radiation-resistant LNCaP cells can be forced to undergo irradiation-induced apoptosis when treated with SK1 inhibitors (12). Indeed, the loss of cell viability induced by chemotherapeutic agents (e.g. etoposide) in vitro and in vivo, and their efficacy, is correlated with the extent to which these agents reduce SK1 activity (13) while overexpression of recombinant SK1 renders prostate cancer cells more resistant to chemotherapeutic agents (14). LNCaP cells forced to overexpress recombinant SK1 do not undergo growth inhibition after androgen deprivation (15). Taken together, these findings support the hypothesis that prostate cancer cells acquire the ability to survive (in the presence of chemotherapeutic agents) and proliferate in the absence of androgens via a mechanism that involves SK1. We have also shown that hypoxia induces an increase in the expression of SK1 in human pulmonary smooth muscle cells (16), and that this may have an important role in arterial smooth muscle remodeling (increased proliferation and survival) in pulmonary hypertension. Anelli et al. (17) also reported that hypoxia increases SK1 transcriptional regulation leading to increased SK1 protein, intracellular S1P production and S1P release from U87MG glioma cells. Indeed, siRNA knockdown of HIF-2α abolishes the induction of SK1 and the production of extracellular S1P after treatment of cells with CoCl2 (a hypoxia-mimicking agent).
SK inhibitors including N,N-dimethylsphingosine, SKi (2-(p-hydroxyanilino)-4-(p-chlorophenyl)thiazole (18), SK1-I ((2R,3S,4E)-N-methyl-5-(4′-pentylphenyl)-2-aminopent-4-ene-1,3-diol or BML-258) (19), B-5354c (20), F-12509A (21), S-15183a,b (22) have been tested in in vitro cancer lines. In the current study, we have used SKi and N,N-dimethylsphingosine to interrogate the role of SK1 in human pulmonary smooth muscle cells, prostate cancer cells and breast cancer cells. In this regard, we have shown that SK1 inhibitors induce a novel proteasomal degradation of SK1. These findings suggest that SKi and N,N-dimethylsphingosine may be more clinically efficacious than is predicted from simple inhibitor-SK1 activity relationships because of their ability to potentially create SK1-null states in diseased cells.
EXPERIMENTAL PROCEDURES
Materials
All general biochemicals, anti-HA antibody, and anti-actin antibody were from Sigma. High glucose Dulbecco's modified Eagle's medium (DMEM), Mimimum Essential medium (MEM), RPMI 1640, European Fetal Calf Serum (EFCS), penicillin-streptomycin (10,000 units/ml penicillin and 10,000 μg/ml streptomycin), l-glutamine, and Lipofectamine TM2000 were from Invitrogen (Paisley, UK). Charcoal-filtered fetal bovine serum was from Lonza. Human pulmonary aortic smooth muscle cells, human smooth muscle cell growth medium and passaging solutions were from TCS Cellworks (Buckingham, UK). MCF-7 Neo and MCF-7 HER218 cells were gifted from R. Schiff (Baylor College). LNCaP and LNCaP-AI cells were gifted from H. Leung (Beatson Institute, Glasgow). Oligo(dT)12–18 was synthesized by Eurofins MWG Operon (Ebersberg, Germany). dNTP mix was from GE Healthcare. BioscriptTM reverse transcriptase enzyme was from Bioline (London, UK). Taq polymerase was from Thermo Scientific (Abgene/Thermo Fisher Scientific, Epsom, UK). SK1 PCR primers were synthesized by Biomers.net. Anti-phosphorylated ERK1/2 and anti-cyclin D1 antibodies were from Santa Cruz Biotechnology. Anti-ERK2 antibody was from BD Transduction Laboratories (Oxford, UK), Anti-SK1a and anti-SK1b antibodies were a kind gift from A. Huwiler (University of Bern, Switzerland (23)). Anti-FLAG M2 antibody was from Stratagene (La Jolla, CA). Anti-PARP and anti-cleaved caspase-3 antibodies were from Cell Signaling Technology (supplied by New England Biolabs (UK) Ltd., Hitchin, UK). The HA-tagged ubiquitin plasmid construct was obtained from Addgene (Cambridge). pCMV(HA) was from Clontech (Basingstoke, UK). FLAG-tagged SK1 was generated by one of us (24). Sphingosine, MG132, lactacystin, PD098059, Ac-DEVD-CHO, recombinant SK1a, Fumonisin B1, C2-dihydroceramide, C2-ceramide, and dihydrosphingosine were from Enzo Life Sciences (Exeter, UK). DharmaFECTTM 2 reagent and SK1 siRNA were from Dharmacon (Dharmacon, Cromlington, UK). Scrambled siRNA: was from Qiagen (Crawley, UK). SKi, cycloheximide, and CA074Me were from Merck Biosciences (Nottingham, UK). N,N-Dimethylsphingosine was from Avanti Polar Lipids (Alabaster, AL) as were lipids for mass spectroscopy analysis including sphingosine, dihydrosphingosine, a 17-carbon analog of Sph (C17-Sph), S1P, a 17-carbon analog of S1P (C17-S1P), N-myristoyl (14:0), N-palmitoyl (16:0), N-oleoyl (18:1), N-stearoyl (18:0), N-arachidoyl (20:0), N-nervonoyl (24:1), N-lignoceroyl (24:0) sphingosines (ceramides, Cer), N-palmitoyl (16:0), N-oleoyl (18:1), N-stearoyl (18:0), N-arachidoyl (20:0), N-behenoyl (22:0), N-nervonoyl (24:1), N-lignoceroyl (24:0) DHSph (dihydroceramides, DHCer), and N-heptadecanoyl-sphingosine (17:0-Cer).
Cell Culture
Human pulmonary aortic smooth muscle cells (hPASMC) were grown in human smooth muscle cell growth medium. MCF-7 breast cancer cells (either stably expressing vector (Neo) or human epidermal growth factor receptor 2 (HER2)) were grown in DMEM with 10% EFCS and 1% penicillin-streptomycin, 0.4% geneticin, and 15 μg/ml insulin. Human prostate cancer LNCaP and LNCaP-AI cell lines were maintained in RPMI 1640 medium supplemented with 10% EFCS or 10% delipidated serum, respectively, 1% penicillin-streptomycin and 1% l-glutamine. HEK293 cells were maintained in MEM with 10% EFCS and 1% penicillin-streptomycin. All cells were maintained in a humidified atmosphere at 37 °C with 5% CO2. Cells were treated with inhibitors or vehicle for 24 (MCF-7 and hPASMC) or 48 h (LNCaP cells). For LNCaP/LNCaP-AI cells, SKi, MG132, PD098059, CA074Me, and AC-DEVD-CHO were replenished after 24 h.
Transfection
HEK293 cells were transfected with FLAG-tagged hSK1 and HA-tagged-ubiquitin plasmid construct, using the LipofectamineTM2000 reagent according to the manufacturer's instruction. Transfection was performed with serum starvation for 24 h before treatments and harvesting.
siRNA Transfection
These were carried out using Dharmafect Reagent 2 according to the protocol provided by the manufacturer. siRNA final concentration was 200 nm, and cells were treated for 48 h before stimulation.
Generation of Cell Lines with Inducible SK1 Expression
HEK293 cells with doxycycline-inducible expression of FLAG-tagged human SK1 were generated as previously described (25). The construct for inducible expression of non-phosphorylatable SK1 (SK1S225A) cDNA containing a C-terminal FLAG epitope tag (24) was subcloned into pcDNA5/FRT/TO-SK1(FLAG)-AU following digestion with BamHI and NotI. Similarly, cDNAs for wild-type and non-phosphorylatable SK1 variants that constitutively localize to the plasma membrane (Lck-SK1 and Lck-SK1S225A (26)) were produced by digestion with HindIII, blunted with Pfu, then digested with NotI and subcloned into pcDNA5/FRT/TO-AU previously digested with EcoRV and NotI. Flp-In T-Rex HEK293 cells (Invitrogen) with doxycycline-inducible expression of these constructs were then generated as previously described using (26).
Expression of wild-type SK1 and variants in Flp-In T-Rex HEK293 cells were induced with low concentrations of doxycycline hyclate (50–200 ng/ml) that resulted in ∼10-fold increases in SK1 activity above basal levels. After 24 h, cells were treated with 10 μm SKi, 10 μm MG132, or both. DMSO was used as the vehicle control. After a further 24 h, cells were harvested, lysed, and subjected to SDS-PAGE and immunoblotting with anti-FLAG (Sigma), anti-ERK1/2 (Promega), or anti-α-tubulin (Abcam) antibodies.
RNA Isolation
RNase-free equipment was used at all times. After stimulation, adherent cells were washed with phosphate-buffered saline (PBS) before total RNA was extracted using the NucleoSpin® RNA II kit (Abgene/Thermo Fisher Scientific, Epsom, UK), according to the manufacturer's instructions. RNA was stored at −80 °C.
Reverse Transcription Reaction
1500 ng of total RNA was treated with 1 unit amplification grade DNase (Sigma) for 15 min at room temperature, followed by DNase 1 inactivation by heating (70 °C for 10 min) in the presence of Stop solution. First strand synthesis was carried out in each reaction using 1500 ng total RNA catalyzed by the enzyme BioscriptTM reverse transcriptase (200 units). The reaction was primed using 500 ng of Oligo(dT)18 in a final volume of 20 μl. The reverse transcription mixture also contained 500 μm of each dNTP, 1× first-strand buffer (50 mm Tris-HCl (pH 8.6), 40 mm KCl, 1 mm MnSO4, 1 mm DTT). First-strand synthesis was carried out for 50 min at 42 °C and inactivated for 15 min at 70 °C. Each time first-strand synthesis was performed, a separate reaction was carried out minus reverse transcriptase (−RT), to establish a lack of genomic DNA. 2 μl of cDNA was used as a template in subsequent PCR to determine the expression of SK1 or GAPDH.
Polymerase Chain Reaction (PCR)
RT-PCR was performed to identify SK1 and GAPDH mRNA transcripts. The primers used for the reactions were as follows: SK1: FWD: CTGTCACCCATGAACCTGCTGTC, REV: CATGGCCAGGAAGAGGCGCAGCA; GAPDH: FWD: TGAAGGTCGGTGTCAACGGATTTGGC, REV: CATGTAGGCCATGAGGTCCACCAC.
The PCR reaction conditions were: 1 cycle initial denaturation at 94 °C for 2 min, 25 (GAPDH)/35 (SK1) cycles amplification at 94 °C for 1 min 30 s, 55 °C for 30 s and 72 °C for 1 min 40 s, followed by a final extension at 72 °C for 5 min.
Real Time PCR
Total RNA extraction of LNCaP cells grown in T-25 cell culture flasks was carried out using a GenElute mammalian Total RNA Miniprep Kit (Sigma-Aldrich). The procedure included an on-column DNase treatment of the RNA. The extracted total RNA was quantified and assessed by a NanoDrop-2000C spectrophotometer and 5 μg of each RNA sample was used in a first-strand Superscript III cDNA synthesis reaction (Invitrogen) and primed with a NV-clamped Oligo d(T)18 primer (Eurofins, Germany). cDNA synthesis reactions lacking Superscript III were also set up in parallel to act as RT controls for genomic DNA contamination assessment. 100 ng of cDNA was used in each 25 μl of Solaris qPCR reaction (Thermo Scientific) with the exception of the no-template control (NTC; H20) reactions. The supplied Solaris ROX was used in each reaction as a passive reference dye. The qPCR reactions were run on a Bio-Rad DNA Engine with Chromo4 Real-Time PCR Detection System (Bio-Rad) under the required Solaris cycling conditions. The data were collected using Opticon Monitor 3.1 (Bio-Rad). The primers were: SK1-For: ACCATTATGCTGGCTATGAGC, Rev: CAGCAATAGCGTGCAGTT; Probe: TGAAGACCTCCTGACCA; Cyclin D1: For: ACGCTTCCTCTCCAGAGTGAT, Rev: TTGACTCCAGCAGGGCTT; Probe: TGCCAGGAGCAGATCGGAAG; GAPDH-For: GCCTCAAGATCATCAGCAATG, Rev: CTTCCACGATACCAAAGTTGTC; Probe: GCCAAGGTCATCCATGA. Experiments were performed in triplicate and results expressed as the mean C(t) ratio of SK1/GAPDH or CD1/GAPDH ± S.D.
Preparation of Whole Cell Extracts
hPASMC, LNCaP, and LNCaP-AI cell extracts for SDS-PAGE and Western blot analysis or immunoprecipitation analysis were prepared by washing treated cells with 5 ml PBS and then resuspending cell pellets in whole cell lysis buffer (137 mm NaCl, 2.7 mm KCl, 1 mm MgCl2, 1 mm CaCl2, 1% v/v Nonidet P-40, 10% v/v glycerol, 20 mm Tris, pH 8, containing 0.2 mm PMSF, 0.2 mm leupeptin, 0.2 mm aprotinin, 0.5 mm Na3VO4, 100 μm NaF, 10 mm β-glycerophosphate). Samples were repeatedly (×6) passed through a 23-gauge needle using a syringe and rotated for 30 min at 4 °C to allow for efficient lysis. Cell debris was pelleted by centrifugation at 22,000 × g for 10 min at 4 °C and the supernatant (whole cell extract) subsequently collected. The protein content was measured using the Pierce BCA Assay Kit (Fisher Scientific UK, Loughborough). For each sample, 10–20 μg of protein, combined with Laemmli buffer (0.5 m Tris, 2 mm Na4P2O7, 5 mm EDTA, 2% w/v SDS, pH 6.7 containing 5% v/v glycerol, 0.25% w/v bromphenol blue, 10% (v/v) β-mercaptoethanol) were used for SDS-PAGE and Western blotting. MCF-7 cell lysates for SDS-PAGE and Western blot analysis were prepared by adding boiling 1× sample buffer to adherent cells and harvesting the lysate, which was repeatedly (×6) passed through a 23-gauge needle and syringe.
Immunoprecipitation
HEK293 cell extracts for immunoprecipitation were prepared as outlined above. Lysate (equivalent to ∼30 μg protein) was precleared for 1 h with protein G-Sepharose beads (Sigma) and FLAG-tagged SK1 immunoprecipitated overnight at 4 °C in whole cell lysis buffer using fresh G-Sepharose beads and 5 μg of the anti-FLAG antibody or the equivalent volume of whole cell lysis buffer as a control. Beads were washed once with 1 ml of wash buffer (10 mm HEPES, 100 mm NaCl, pH 7.0) containing 0.5% (v/v) Nonidet P-40, and once with 1 ml wash buffer without detergent before boiling in 20 μl of Laemmli buffer. Recovered complexes were resolved by SDS-PAGE and ubiquitinated SK1-FLAG was detected by Western blot analysis using anti-HA antibody.
Western Blotting
Analysis of proteins by SDS-PAGE and Western blotting was performed as previously described by us (27) using anti-phosphorylated ERK1/2, anti-ERK2, anti-PARP, anti-caspase-3, anti-SK1a, anti-SK1b, anti-actin, anti-cyclin D, anti-HA, and anti-FLAG M2 antibodies.
Proteasome Activity Assays
Proteasome activity was measured in cells using a Proteasome Glo Chymotrypsin-Like Cell-based assay kit (Promega) per the manufacturer's instructions. Results are presented as 100% of the basal luminogenic proteasome activity. All results shown are the mean of triplicate assays with S.E.
Analysis of Sphingoid Bases, Sphingoid Base-1-Phosphates, and Ceramides
Analyses of the sphingolipids were performed by combined LC/MS/MS. The instrumentation employed was an API4000 Q-trap hybrid triple quadrupole linear ion-trap mass spectrometer (Applied Biosystems, Foster City, CA) equipped with a turboionspray ionization source interfaced with an automated Agilent 1100 series liquid chromatograph and autosampler (Agilent Technologies, Wilmington, DE). The sphingolipids were ionized via electrospray ionization (ESI) with detection via multiple reaction monitoring (MRM). Analysis of sphingoid bases and the molecular species of ceramides employed ESI in positive ions with MRM analysis using a minor modification of published methods (28, 29). Briefly, resolution of sphingoid bases was achieved with a Discovery C18 column (2.1 × 50 mm, 5 μm particle size, Supelco, Bellefonte, PA) and a gradient from methanol/water/formic acid (61:38:1, v/v) with 5 mm ammonium formate to methanol/acetonitrile/formic acid (39:60:1, v/v) with 5 mm ammonium formate at a flow rate of 0.5 ml/min. The MRM transitions employed for detection of sphingoid bases were as follows: m/z 286 > 268 (C17-Sph, internal standard); m/z 300 > 282 (Sph); and m/z 302 > 284 (DHSph).
Ceramide molecular species were resolved using a 3 × 100 mm X-Terra XDB-C8 column (3.5 μm particle size, Waters, Milford, MA) and a gradient from methanol/water/formic acid (61:39:0.5, v/v) with 5 mm ammonium formate to acetonitrile/chloroform/water/formic acid (90:10:0.5:0.5, v/v) with 5 mm ammonium formate at a flow rate of 0.5 ml/min. MRM transitions monitored for the elution of ceramide molecular species were as follows: m/z 510 > 264, 14:0-Cer; m/z 538 > 264, 16:0-Cer; m/z 540 > 284, 16:0-DHCer; m/z 552 > 264, 17:0-Cer (internal standard); m/z 564 > 264, 18:1-Cer; m/z 566 > 284, 18:1-DHCer; m/z 566 > 264, 18:0-Cer; m/z 568 > 284, 18:0-DHCer; m/z 594 > 264, 20:0-Cer; m/z 596 > 284, 20:0-DHCer; m/z 624 > 284, 22:0-DHCer; m/z 650 > 264, C22:0 Cer; m/z 622 > 264, 24:1-Cer; m/z 652 > 284, 24:1-DHCer; m/z 652 > 264, 24:0-Cer; m/z 654 > 284, 24:0-DHCer; m/z 680 > 264. S1P was quantified as bis-acetylated derivatives with C17-S1P as the internal standard employing reverse-phase HPLC separation, negative ion ESI, and MRM analysis. Details of this approach are described in Ref. 30.
Densitometry
Densitometric quantification of Western blots was performed using the Molecular Analyst Software (Bio-Rad). Statistical analysis was performed using unpaired Student's t test.
RESULTS
Effect of SK1 Inhibitors in Human Pulmonary Artery Smooth Muscle Cells (hPASMC)
We have used SK1 inhibitors to interrogate the role of SK1 in a number of cell types, including hPASMC, LNCaP prostate cancer cells and MCF-7 breast cancer cells. hPASMC express SK1a (Mr = 42kDa, Fig. 1, a and b). Remarkably, the chronic treatment of hPASMC with SKi (24 h) reduced SK1a expression (Fig. 1, a and b), and this reduction was detectable within 16 h of treatment (Fig. 1c). Thus, SKi not only inhibits SK1 catalytic activity (18) but also reduces the expression of SK1a in hPASMC. Similar results were obtained with a structurally dissimilar SK1 inhibitor, N,N-dimethylsphingosine (Fig. 1d).
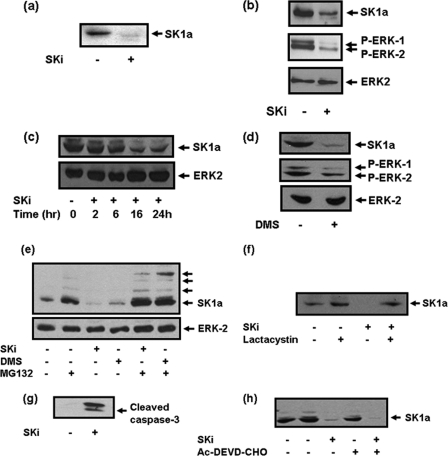
FIGURE 1.
Effect of SKi and N,N-dimethylsphingosine on SK1a in hPASMC. Western blots showing the effect of SKi (10 μm, 48 h) on (a) the expression of SK1a in hPASMC; (b) SK1a and phosphorylated ERK-1/2 levels in hPASMC; (c) time course for removal of SK1a from hPASMC. Also presented are Western blots showing the effect of (d) N,N-dimethylsphingosine (10 μm, 24 h) on SK1a and phosphorylated ERK-1/2 levels in hPASMC; (e) reversal by MG132 (10 μm, 30-min pretreatment) of the SKi and N,N-dimethylsphingosine (all 10 μm, 24 h) induced degradation of SK1a in hPASMC; (f) reversal by lactacystin (10 μm, 30-min pretreatment) of the SKi (10 μm, 24 h) induced degradation of SK1a in hPASMC (g) SKi (10 μm, 48 h) on caspase-3 activation in hPASMC; (h) lack of effect of AC-DEVD-CHO (50 μm, 30-min pretreatment) on the SKi (10 μm, 24 h)-induced down-regulation of SK1a in hPASMC. The Western blots were reprobed with anti-ERK-2 antibody to ensure comparable protein loading. Results are representative of at least 3–5 separate experiments.
We next investigated the mechanism by which these SK1 inhibitors induce down-regulation of SK1. In this regard, the treatment of hPASMC cells with the proteasomal inhibitor, MG132 inhibited the SKi- and N,N-dimethylsphingosine-induced down-regulation of SK1a (Fig. 1e). Treatment of cells with MG132 restored SK1a to a level above that in untreated cells and induced formation of additional SK1a immunoreactive proteins with high molecular mass compared with the native enzyme, suggesting that SK1a might be post-translationally modified, e.g. polyubiquitinated before it is degraded by the proteasome (Fig. 1e). Moreover, MG132 alone induced the formation of high molecular mass forms of SK1a suggesting that SK1a is degraded by the proteasome under basal conditions (Fig. 1e). It is therefore possible that SKi converts SK1 to a conformation that is then targeted to the ubiquitin-proteasomal pathway and/or that it induces activation of the proteasome to accelerate degradation of SK1a (see later). An alternative proteasomal inhibitor, lactacystin also blocked the effect of SKi on SK1a expression levels (Fig. 1f).
We next investigated the functional significance of the SKi-induced proteasomal degradation of SK1a in hPASMC. In this regard, treatment of cells with SKi induced the onset of apoptosis as evidenced by increased caspase-3 activation (Fig. 1g, indicated by formation of p19 and p17 caspase 3 auto-activation cleavage products). The treatment of cells with the caspase-3/7 inhibitor Ac-DEVD-CHO failed to prevent the SKi-induced degradation of SK1a, thereby excluding a role for caspase 3/7 in its removal from these cells (Fig. 1h). In addition, SKi and N,N-dimethylsphingosine also reduced the phosphorylation state of ERK-1/2 (Fig. 1, b and d).
Effect of SK1 Inhibitors in Human Prostate Cancer Cells (LNCaP cells)
We determined whether the effect of SKi on the proteasomal degradation of SK1 is a general mechanism that occurs in other cellular systems. For this purpose we used androgen-sensitive LNCaP and androgen-independent LNCaP-AI cells. LNCaP-AI cells have been selected by culturing LNCaP cells in androgen-deprivation conditions for prolonged periods of time. This mimics androgen ablation therapy used for the treatment of prostate cancer (31). LNCaP-AI cells are not dependent upon androgen for proliferation as they can grow in hormone-free medium. However, these cells do express androgen receptor (AR) and respond to androgens, being able to express androgen-regulated genes such as the PSA (prostatic specific antigen) gene (31). Therefore, LNCaP-AI cells are considered as a good in vitro model for hormone-refractory prostate cancer.
LNCaP and LNCaP-AI cells express both SK1a and SK1b (supplemental Fig. S1, a and b). Interestingly, the expression of SK1a and SK1b in LNCaP-AI is up-regulated compared with LNCaP cells. Messenger RNA transcript for SK1a/b (supplemental Fig. S1a, product size of amplicon: 480 bp and which will potentially include SK1a, SK1a+14, and SK1b mRNA transcript as primers are designed to a common region) and SK1a (Mr = 42kDa) and SK1b (Mr = 51kDa) protein expression (supplemental Fig. S1, detected by Western blotting with anti-SK1a and anti-SK1b specific antibodies) were increased in LNCaP-AI cells compared with LNCaP cells, suggesting that androgen escape might be associated with increased transcriptional up-regulation of SK1a/b.
We have also identified significant differences in the regulation of SK1a and SK1b in LNCaP and LNCaP-AI cells, which may contribute to the development of chemotherapeutic resistance in LNCaP-AI cells. In this regard, chronic treatment of LNCaP cells with SKi induced the down-regulation of SK1a and SK1b (Fig. 2a, supplemental Fig. S2). In common with SKi, N,N-dimethylsphingosine also induced down-regulation of SK1b (Fig. 2a) in LNCaP cells. The effect of SKi is post-transcriptional as treatment of LNCaP cells with cycloheximide, to inhibit de novo protein synthesis, did not block the effect of SKi on SK1b levels in LNCaP cells (Fig. 2a, supplemental Fig. S2). In addition, SKi treatment of LNCaP cells for 24 h, which induces SK1a (supplemental Fig. S3) and SK1b (Fig. 2a) degradation, did not decrease SK1 mRNA expression (supplemental Fig. S3). Moreover, the treatment of LNCaP cells with the proteasomal inhibitor, MG132 reduced the effect of SKi on SK1a and SK1b (Fig. 2, b and c, supplemental Fig. S2). In addition to this, others have reported, using the cathepsin B inhibitor, CA074Me, that SKi induces lysosomal degradation of SK1 in human podocytes (32). To investigate the role for lysosomal degradation of SK1a/b in response to SKi, we pretreated LNCaP cells with CA074Me. However, CA074Me had no effect on the SKi-induced degradation of SK1a and SK1b (supplemental Figs. S2 and S4). Thus, the lysosomal route of degradation does not appear to be functional in LNCaP cells. Moreover, lactacystin, which has no activity against cathepsin B reversed the effect of SKi on SK1a in hPASMC (Fig. 1f), thereby suggesting that the lysosomal route of degradation is also not operative in these cells.
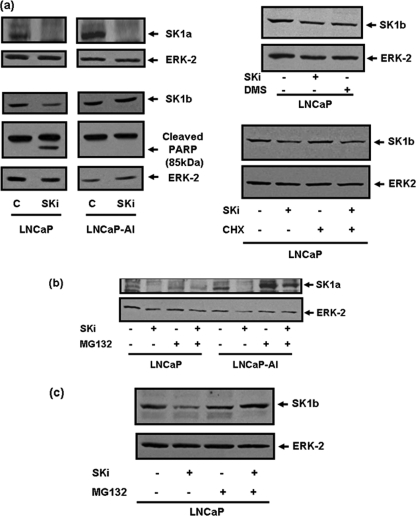
FIGURE 2.
SK1a and SK1b regulation and modulation by SKi in LNCaP cells. Western blots comparing (a) the effect of the SK1 inhibitor, SKi (10 μm, 48 h) or N,N-dimethylsphingosine (10 μm, 48 h) on expression levels of SK1a, SK1b, and cleaved p85 PARP levels. Also shown is the lack of effect of cycloheximide (5 μg/ml, 30-min pretreatment) on the SKi (10 μm, 24 h)-induced degradation of SK1b levels in LNCaP cells; (b, c) effect of MG132 (10 μm, 30-min pretreatment) on SKi (10 μm, 48 h)-induced down-regulation of (b) SK1a in LNCaP and LNCaP-AI cells and (c) SK1b in LNCaP cells. The Western blots were reprobed with anti-ERK-2 antibody to ensure comparable protein loading. In each case, results are representative of at least three separate experiments.
Significantly, the treatment of LNCaP-AI cells with SKi induced MG132-sensitive degradation of SK1a (Fig. 2b, supplemental Fig. S2). However, SK1b was less sensitive to SKi (Fig. 2a). SKi stimulated PARP cleavage in LNCaP cells indicating that these cells are forced to undergo the onset of apoptosis when SK1a and SK1b are degraded (Figs. 2a and 3a). However, SKi-treated LNCaP-AI cells do not undergo apoptosis (there was no PARP cleavage (Figs. 2a and 3a)) and this is associated with reduced sensitivity of SK1b to SKi in terms of its proteasomal degradation. In contrast with hPASMC, the treatment of LNCaP cells with SKi induced a substantial increase in the phosphorylation state of ERK-1/2 (Fig. 3b) which appears to be a rebound mechanism or alarm signal that affords protection against or places a break on apoptosis. Thus, pretreatment of LNCaP cells with PD098059, an inhibitor of MEK1 activation (an upstream regulator of ERK-1/2), which blocks the phosphorylation of ERK-1/2 in response to SKi (Fig. 3b) was associated with enhanced PARP cleavage (Fig. 3a). In contrast, SKi failed to increase ERK-1/2 phosphorylation in LNCaP-AI cells (Fig. 3b), where the onset of apoptosis was not induced.
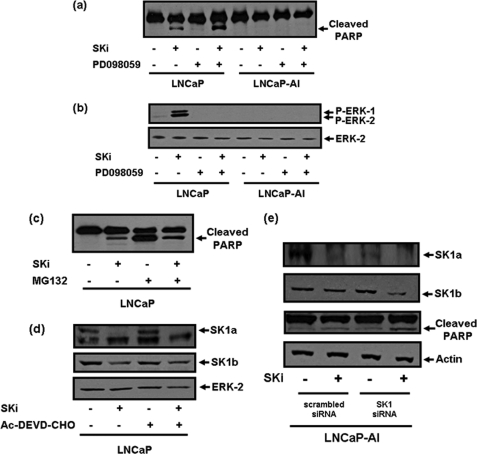
FIGURE 3.
SKi-induced apoptosis of LNCaP cells. Western blots with (a) anti-PARP antibody showing that SKi (10 μm, 48 h) induces PARP cleavage in LNCaP cells but not LNCaP-AI cells, and that this is increased by PD098059 (10 μm, 30-min pretreatment); (b) anti-phospho ERK-1/2 antibody showing the SKi-induced rebound activation of ERK-1/2 in LNCaP but not LNCaP-AI cells, and that this is blocked by PD098059 (10 μm, 30 min pretreatment); (c) anti-PARP antibody showing that SKi (10 μm, 48 h) protects against MG132 (10 μm, 48 h)-induced PARP cleavage in LNCaP cells; (d) anti-SK1a or anti-SK1b antibody showing that AC-DEVD-CHO (100 μm, 1-h pretreatment) does not block SKi (10 μm, 48 h)-induced degradation of SK1a or SK1b in LNCaP cells; (e) anti-SK1a or anti-SK1b or anti-PARP antibodies to show the effect of SK1 siRNA (200 nm, 72 h) (compared with scrambled siRNA) with and without SKi (10 μm, 24 h) on SK1a and SK1b expression and on PARP cleavage in LNCaP-AI cells. The Western blots were reprobed with anti-ERK-2 or anti-actin antibodies to ensure comparable protein loading. Results are representative of three separate experiments.
MG132 alone also induced the onset of apoptosis of LNCaP cells as evidenced by substantial PARP cleavage (Fig. 3c), while the treatment of these cells with SKi reduced the MG132-induced cleavage of PARP (Fig. 3c). These findings are consistent with the possibility that SKi can counter (but does not entirely overcome) inhibition of the proteasome by MG132, e.g. SKi activates the proteasome.
The SKi-induced proteasomal degradation of SK1a or SK1b was not blocked by the caspase-3/7 inhibitor Ac-DEVD-CHO, suggesting that apoptotic caspase-3 and caspase-7 are not involved in the SKi-induced degradation of SK1a or SK1b in these cells (Fig. 3d).
Effect of SK1 siRNA on SK1 Signaling in LNCaP and LNCaP-AI Cells
To further investigate the role of SK1a and SK1b in the regulation of prostate cancer cell survival, we used siRNA to knock down the expression of these enzymes. The siRNA used is targeted at a common region in SK1a and SK1b mRNA, and will therefore theoretically knock down expression of both isoforms. However, while SK1 siRNA was effective at knocking down the expression of SK1a (Fig. 3e) in LNCaP-AI cells, it failed to reduce SK1b expression (Fig. 3e, supplemental Fig. S2). This is also the case in LNCAP cells (data not shown). This is likely due to a slow turnover rate of SK1b. Indeed, SK1b is very stable as 24.5 h treatment with cycloheximide had no effect on its expression (Fig. 2a, supplemental Fig. S2). We reasoned therefore, that the treatment of LNCaP-AI cells with SK1 siRNA should sensitize these cells to SKi (24 h) because degradation in response to SKi will be unopposed by de novo synthesis of the enzyme, and this is indeed the case (Fig. 3e, supplemental Fig. S2). Thus, in contrast with scrambled siRNA-treated LNCaP-AI cells, where the sensitivity of SKi to proteosomal degradation of SK1b is reduced compared with LNCaP cells (Figs. 2a and 3e), the degradation of SK1b was increased by combined treatment of LNCaP-AI cells with SK1 siRNA and SKi (Fig. 3e, supplemental Fig. S2).
We therefore next addressed the effect of the combined treatment of LNCaP-AI cells with SK1 siRNA and SKi on the apoptotic status of these cells. Evidence for a key role for SK1b in regulating cell survival and resistance to SKi in LNCaP-AI cells was obtained by results showing that the combined treatment of these cells with SK1 siRNA and SKi induced PARP cleavage compared with scrambled siRNA-treated cells (Fig. 3e). The treatment of LNCaP-AI cells with SK1 siRNA alone did not substantially increase PARP cleavage (Fig. 3e). We conclude that LNCaP-AI cells are forced to undergo apoptosis when both SK1a and SK1b expression is reduced.
Effect of SK1 Inhibitors in Human Breast Cancer Cells (MCF-7 Neo and MCF-7 HER2 Cells)
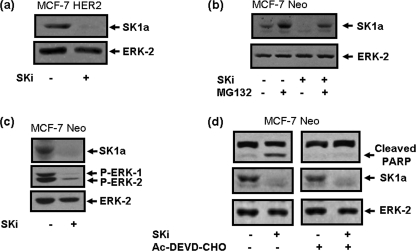
To investigate whether SK1 is susceptible to targeting for proteasomal degradation in breast cancer cells, we used MCF-7 Neo cells (stably transfected with Neo vector) and MCF-7 HER2 cells (MCF-7 HER218 clone, stably transfected with a HER2 plasmid construct). Treatment of MCF-7 Neo and MCF-7 HER2 cells with SKi induced the degradation of SK1a (Mr = 42kDa) (Fig. 4, a and b) and this was blocked by treatment with MG132 (Fig. 4b). Moreover, the treatment of MCF-7 Neo cells with SKi induced a reduction in basal ERK-1/2 phosphorylation and promoted PARP cleavage (Fig. 4, c and d), indicating that SKi induces the onset of apoptosis of these cells. Pretreatment of MCF-7 Neo cells with Ac-DEVD-CHO reduced PARP cleavage in response to SKi, but had no effect on proteasomal degradation of SK1a (Fig. 4d), thereby confirming that apoptotic caspase-3/7 do not participate in the degradation of SK1a in these cells.
FIGURE 4.
Effect of SKi on SK1a in MCF-7 cells. Western blots showing (a) effect of SKi (10 μm, 24 h) on SK1a in MCF-7 HER2 cells; (b) reversal by MG132 (10 μm, 30-min pretreatment) of the SKi (10 μm, 24 h)-induced degradation of SK1a in MCF-7 Neo cells; (c) effect of SKi (10 μm, 24 h) on SK1a and phosphorylated ERK-1/2 levels in MCF-7 Neo cells; (d) the lack of effect of AC-DEVD-CHO (100 μm, 30-min pretreatment) on SKi (10 μm, 24 h)-induced degradation of SK1a and concomitant inhibition of SKi-induced PARP cleavage in MCF-7 Neo cells. Images are from the same Western blot. The Western blots were reprobed with anti-ERK-2 antibody to ensure comparable protein loading. Results are representative of at least three separate experiments.
The Effect of SKi on Sphingolipids and Fumonisin B1 on Proteasomal Degradation of SK1
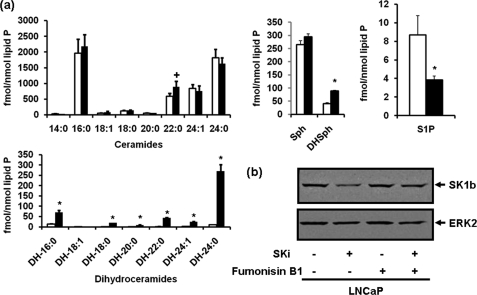
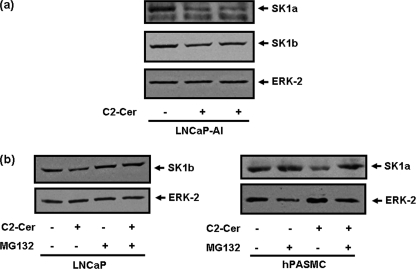
The treatment of LNCaP cells with SKi decreased S1P levels and increased C22:0 ceramide, dihydroceramide, and dihydrosphingosine levels in LNCaP cells (Fig. 5a). Therefore, to test a possible role for ceramide/dihydroceramide in the proteasomal degradation of SK1, we treated cells with fumonisin B1 (to inhibit ceramide synthase and thus conversion of sphingosine to ceramide and dihydrosphingosine to dihydroceramide), which reduced the effect of SKi on the down-regulation of SK1b (Fig. 5b, supplemental Fig. S2). Treatment of LNCaP cells with dihydrosphingosine or C2-dihydroceramide had no effect on SK1a or SK1b expression, respectively (supplemental Fig. S5). These findings suggest that ceramide is the most likely sphingolipid metabolite responsible for inducing the proteasomal degradation of SK1, as a consequence of catalytic inhibition of SK1 by SKi. To further test for a possible role of ceramide in the proteasomal degradation of SK1, we used the short chain cell permeable ceramide, C2-ceramide. Treatment of LNCaP-AI cells with C2-ceramide induced the down-regulation of SK1a and SK1b (Fig. 6a). Similarly, SK1a was down-regulated by C2-ceramide in hPASMC and this was inhibited by MG132 as was the response for SK1b in LNCaP cells (Fig. 6b, supplemental Fig. S2).
FIGURE 5.
Effect of SKi on sphingolipid levels and of fumonisin B1 on SKi-induced SK1b degradation. a, bar charts showing the effect of SKi (10 μm, 24 h) on sphingolipid levels in LNCaP cells. *, p < 0.05 for control versus SKi-treated cells for all dihydroceramide species, S1P and dihydrosphingosine and p = 0.05 for C22:0 ceramide for n = 3 cell samples; (b) Western blot showing the effect of fumonisin B1 (100 μm, 1-h pretreatment) on SKi (10 μm, 24 h)-induced proteasomal degradation of SK1b in LNCaP cells. The Western blots were reprobed with anti-ERK-2 antibody to ensure comparable protein loading. Results are representative of three separate experiments.
FIGURE 6.
Effect of C2-ceramide on SK1a and SK1b expression. Western blot showing the effect of (a) C2 ceramide (50 μm, 24 h) on SK1a and SK1b levels in LNCaP-AI cells; (b) reversal by MG132 (10 μm, 30 min pretreatment) of the C2-ceramide (50 μm, 24 h)-induced reduction in SK1b levels in LNCaP cells and SK1a in hPASMC. The Western blots were reprobed with anti-ERK-2 antibody to ensure comparable protein loading. Results are representative of three separate experiments.
Effect of SKi on Polyubiquitination and Proteasomal Degradation of SK1
It is well established that in order for proteins to be targeted to the proteasome for degradation, the proteins have first to be modified by polyubiquitination (a modification that instructs the cell to remove the protein). We therefore tested whether SK1 is regulated by the ubiquitin-proteasomal pathway. For this purpose, we used the HEK293 cell expression system in which we co-expressed FLAG-tagged SK1a and HA-ubiquitin. This expression system exhibits high efficiency transfection, thereby improving sensitivity for detection of poylubiquitinated SK1a. HEK293 cells express endogenous SK1a (Mr = 42 kD, Fig. 7a). In common with LNCaP, MCF-7 cells and hPASMC, the treatment of HEK293 cells with SKi reduced the expression of endogenous SK1a (Fig. 7a).
FIGURE 7.
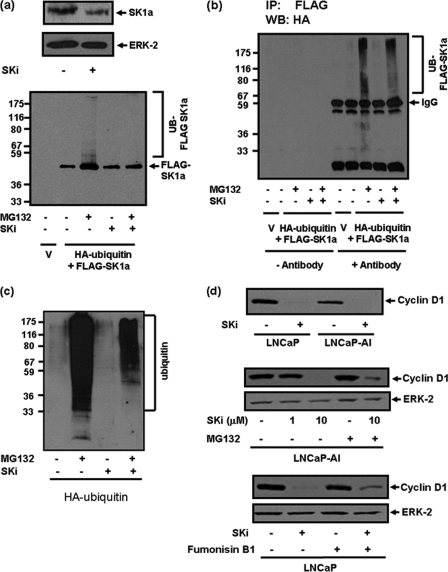
Polyubiquitination of SK1a. Western blots showing: (a) overexpression of FLAG tagged SK1a in HEK293 cells, and effect of SKi (10 μm, 24 h) on endogenous SK1a (detected with anti-SK1 antibody) and FLAG-tagged SK1a (detected with anti-FLAG tag antibody); (b) anti-FLAG SK1a immunoprecipitates immunostained with anti-HA tag antibody to demonstrate the effect of MG132 (10 μm, 30-min pretreatment) and SKi (10 μm, 24 h) or both on polyubiquitination of FLAG-SK1a in FLAG-tagged SK1a/HA-tagged ubiquitin overexpressing HEK293 cells. Results are representative of three separate experiments; (c) polyubiquitination of total cellular proteins in lysates of HA-tagged ubiquitin overexpressing HEK293 cells treated with MG132 (10 μm, 30 min pretreatment) and SKi (10 μm, 6 h) or both. Results are representative of two separate experiments; (d) effect of SKi (10 μm, 48 h) on cyclin D1 expression in LNCaP and LNCaP-AI cells, and partial reversal with MG132 (10 μm, 30 min pretreatment) in LNCaP-AI cells. Also shown is a Western blot of the effect of fumonisin B1 (100 μm, 30-min pretreatment) on SKi (10 μm, 24 h)-induced degradation of cyclin D1 in LNCaP cells. The Western blots were reprobed with anti-ERK-2 antibody to ensure comparable protein loading.
We were able to enforce expression of FLAG SK1a in HEK293 cells (Fig. 7a). The treatment of these cells with MG132 induced an increase in the amount of native FLAG-tagged SK1a (Mr = 42 kDa), and promoted the accumulation of high molecular mass forms of FLAG-tagged SK1a (Fig. 7a). Moreover, HA-tagged polyubiquitinated FLAG tagged SK1 could be detected in anti-FLAG SK1 immunoprecipitates of MG132-treated cells on Western blots immunostained with anti-HA tag antibody (Fig. 7b). These findings demonstrate that FLAG-tagged SK1 is regulated by the ubiquitin-proteasomal pathway in HEK293 cells.
The treatment of HEK293 cells with SKi failed to reduce the amount of FLAG-tagged SK1a expressed (Fig. 7a). This is perhaps to be expected given that the effect of SKi is against a background of continual enforced ectopic expression of FLAG-tagged SK1a from the plasmid. However, an effect of SKi on FLAG-tagged SK1a is revealed when HEK293 cells are treated with SKi and MG132 together. Under these conditions, SKi reversed the increase in native FLAG-tagged SK1a (Mr = 42kDa) levels in response to MG132 and also reduced the amount of polyubiquitinated SK1a forms (Fig. 7, a and b). These findings suggest that SKi can counter (but not entirely overcome) the inhibition of the proteasome by MG132 in HEK293 cells to induce degradation of polyubiquitinated FLAG-tagged SK1a, thereby suggesting that SKi might also activate the proteasome in these cells. Moreover, SKi appears to partially counter inhibition of the proteasome by MG132 in LNCaP, LNCaP-AI, and MCF-7 cells. Thus, SK1a expression is recovered in cells treated with SKi/MG132, but the amount recovered is lower than that observed in cells treated with MG132 alone (Figs. 2b and 4b). However, there is full recovery in hPASMC (Fig. 1e). These findings likely reflect differences in the relative flux of ubiquitinated proteins through the proteasomal pathway between cell types.
We also examined whether SKi might alter the ubiquitination of cellular proteins in HA-ubiquitin overexpressing HEK293 cells. Indeed, there is considerable flux of endogenous ubiquitinated proteins through the proteasome, as demonstrated by the finding that treatment of HA-ubiquitin overexpressing HEK293 cells with MG132 increased the accumulation of several ubiquitinated cellular proteins (Fig. 7c). Moreover, treatment of these cells with SKi reduced the accumulation of ubiquitinated proteins, which is again consistent with the possibility that SKi activates the proteasome to reduce the block by MG132. Moreover, there is low level basal ubiquitination of proteins in cells that have not been treated with MG132 (Fig. 7c). SKi had no effect on this basal ubiquitination (Fig. 7c), suggesting that SKi does not inhibit ubiquitination per se.
We also assessed the effect of SKi on the expression of cyclin D1, which regulates cell cycle progression. In this regard, we demonstrated that treatment of both LNCaP and LNCaP-AI cells with SKi induced a reduction in cyclin D1 expression, and this could be partially reversed by MG132 (Fig. 7d). In addition, SKi treatment of LNCaP cells did not decrease cyclin D1 mRNA expression (supplemental Fig. S3). Moreover, treatment of cells with fumonisin B1 also partially reversed the SKi-induced degradation of cyclin D1 (Fig. 7d).
Spatial Context of the Proteasomal Degradation of SK1
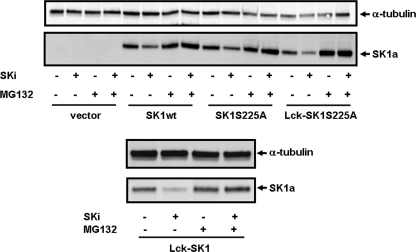
To evaluate whether the subcellular localization of SK1 has any effect on the ability of SK1 to be proteasomally degraded in response to SKi, we used a FlpIn system in HEK293 cells, to induce overexpression of WT, S225A mutant, WT-Lck (plasma membrane localization), or Lck-S225A mutant (plasma membrane localization) SK1a. This expression system produces low level overexpression of SK1, which we deemed necessary to increase the sensitivity of SK1 to be proteasomally degraded in response to SKi in HEK293 cells. The treatment of HEK293 cells overexpressing these SK1a forms with SKi resulted in their MG132-sensitive degradation (Fig. 8), suggesting that subcellular localization does not alter the ability of SK1a to be regulated by the ubiquitin-proteasomal pathway. Moreover, the ability of the ERK-2 phosphorylation-deficient S225A SK1a mutant to undergo proteasomal degradation in response to SKi suggests that the potential phosphorylation of SK1 by ERK-2 is likely to have no effect on this process.
FIGURE 8.
Effect of SKi on spatially localized SK1. Western blots showing the effect of SKi (10 μm, 24 h) and MG132 (10 μm, 24 h) on FLAG-WT, S225A, Lck WT and Lck-S225A mutant SK1a levels in HEK293 cells. Cell lysates were also immunostained with anti-tubulin antibody to ensure comparable protein loading. Results are representative of 2–3 separate experiments.
Effect of SKi on Proteasomal Activity
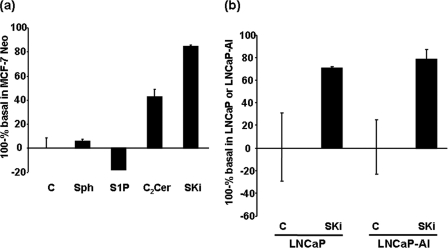
To assess whether SKi and C2-ceramide activate and/or S1P inhibits the proteasome, we used the Proteasome-GloTM Cell-Based assay (Promega), which is optimized for cell permeabilization, proteasome activity, and luciferase activity. The assay contains a luminogenic proteasome substrate, Suc-LLVY-aminoluciferin (succinyl-leucine-leucine-valine-tyrosine aminoluciferin), which is in competition with endogenous ubiquitinated protein substrates for the proteasome. Thus, results of the luminogenic assay are presented as proteasomal activity against endogenous substrates (100% of the basal luminogenic proteasome activity). In this regard, we demonstrate here that the treatment of MCF-7 Neo cells with C2-ceramide or SKi significantly increased proteasomal activity against endogenous substrates in MCF-7 cells (Fig. 9a). Treatment of these cells with S1P had a slight inhibitory effect on proteasomal activity against endogenous substrates (Fig. 9a). The degree of activation induced by SKi was reduced by increasing the concentration of luminogenic substrate in the assay (data not shown), thereby confirming that the assay measures competition between luminogenic and endogenous substrates. The treatment of LNCaP and LNCaP-AI cells with SKi increased proteasomal activity against endogenous substrates to similar extents (Fig. 9b).
FIGURE 9.
Effect of SKi, S1P, and C2-ceramide on proteasomal activity. Proteasomal luminogenic assays demonstrating the effect of (a) treating MCF-7 Neo cells with C2-ceramide (50 μm, 6 h) or SKi (10 μm, 6 h) or S1P (5 μm, 6 h) on proteasomal activity (100-% of basal luminogenic proteasome activity) against endogenous substrates (n = 3 assays per treatment); (b) treating LNCaP and LNCaP-AI cells with SKi (10 μm, 24 h) on proteasomal activity (100-% of basal luminogenic proteasome activity) in each cell type against endogenous substrates (n = 3 assays per treatment).
DISCUSSION
The major finding of this study is that SK1 inhibitors can target SK1 to the ubiquitin-proteasomal degradation pathway. Targeting SK1 to the ubiquitin-proteasomal degradation pathway might be due to changes induced by SKi in the amount of ceramide (a consequence of catalytic inhibition of SK1 by SKi) and subsequent proteasomal activation. Indeed, others have demonstrated that C2-ceramide activates the ubiquitin-mediated proteolytic pathway in astrocytes (33). Moreover, this possibility is supported by our findings that the effect of SKi was partially reversed by the ceramide synthase inhibitor, fumonisin B1 and that C2-ceramide induced proteasomal degradation of SK1 while C2-dihydroceramide failed to do so. These results are also in line with studies showing that the same concentration of fumonisin B1 partially blocked the increase of ceramide and the induction of apoptosis in response to phorbol ester (PMA) or PMA plus radiation in LNCaP cells (34).
Treatment of LNCaP cells with SKi increased C22:0 ceramide levels, while decreasing S1P levels. However, SKi also increased the levels of various dihydroceramide species. Indeed others have reported that SKi increases dihydroceramide formation in ovarian cancer cells (35). These findings suggest that SKi (in addition to inhibiting SK1 activity) might also inhibit dihyroceramide desaturase activity. Interestingly, SKi failed to increase C24:0 ceramide levels. This finding suggests that C24:0 ceramide might be synthesized predominantly through the de novo pathway as SKi (presumably via inhibition of dihydroceramide desaturase) substantially increased C24:0 dihydroceramide mass. In contrast, C22:0 ceramide might be predominantly formed from the sphingosine salvage pathway, supported by our finding that SKi causes only a minor increase in C22:0 dihydroceramide mass. Thus, only minor amounts of C22:0 ceramide are likely formed by the de novo pathway. In addition, fumonisin B1 partially reversed the SKi-induced proteasomal degradation of SK1 suggesting that fumonisin B1 might target the ceramide synthase-catalyzed conversion of sphingosine to C22:0 ceramide. Fumonisin B1 is unlikely to manifest its effects by inhibition of the CerS in the de novo ceramide synthesis pathway as neither dihydrosphingosine nor C2-dihydroceramide induced the proteasomal degradation of SK1. The synthesis of various ceramide species might also be defined as suggested by Hannun and co-workers (36, 37), by subcellular localization or even localization to different lipid microdomains within the same subcellular compartment of the various enzymes involved in the synthesis of ceramides. This possibility would provide an explanation for the differential effect of SKi on C22:0 and C24:0 ceramide levels e.g. SK1 may have preferential access to the C22:0 ceramide/sphingosine pool.
There is supporting evidence to demonstrate that SK1 is polyubiquitinated in mammalian cells. In this regard, Kihara et al. (38) have previously reported that SK1a and SK1b (a smaller 34 kDa species than the SK1b referred to in the present study) are regulated by the ubiquitin-proteasomal degradation pathway and that both forms are polyubiquitinated, with SK1b appearing to be more susceptible to polyubiquitination. We did not detect the 34 kDa SK1b in MCF-7, LNCaP cells, or hPASMC.
We also found that the treatment of HEK293 cells with MG132 increased the amount of polyubiquitinated SK1a, whereas SKi reduced this effect, without affecting basal ubiquitination. These findings are consistent with the possibility that SKi activates the proteasome to increase removal of polyubiquitinated SK1a. This was confirmed by measurement of the effect of SKi on proteasomal activity in MCF-7, LNCaP, and LNCaP-AI cells. The activation of the proteasome by SKi is also in line with results demonstrating that SKi reduces MG132-induced PARP cleavage of LNCaP cells. The stimulation of PARP cleavage by MG132 is very substantial indeed, and this is significantly lowered by SKi, which by itself induces a lower level of PARP cleavage, which is likely independent of the unfolded protein response (UPR), and which is contingent on proteasomal degradation of SK1. In this regard, it is well established that MG132 induces apoptosis of cells by inhibiting the proteasome. This results in the accumulation of unfolded proteins, which results in the UPR and induction of a caspase-12, caspase-7, and caspase-3 activation cascade and resulting in apoptosis. This can therefore be partially reversed by SKi-induced activation of the proteasome.
Based on the ability of SKi to induce proteasomal degradation of SK1, we have used this inhibitor to interrogate the role of SK1 in various mammalian cell systems. Our conclusions from these studies are described herein.
hPASMC
We have established that SKi induces the proteasomal degradation of SK1a in hPASMC. This is correlated with a reduction in the phosphorylation state of ERK-1/2 (an enzyme that is required for maintenance of cell survival) and increased caspase 3 auto-activation, indicating that the reduction in SK1a expression is linked with the induction of apoptosis and that SK1a is protective upstream of caspase-3 activation and apoptosis. Similar data were obtained using an alternative SK inhibitor, N,N-dimethylsphingosine.
Breast Cancer Cells
We have shown that SKi induces proteasomal degradation of SK1a in MCF-7 cells and this is associated with the onset of apoptosis. Pretreatment of MCF-7 cells with Ac-DEVD-CHO reduced the SKi-induced PARP cleavage but had no effect on the proteasomal degradation of SK1a. These findings are important as they clearly eliminate caspase-3/7 as being responsible for the SKi-induced degradation of SK1a.
Prostate Cancer Cells
We have demonstrated that SKi induces the apoptosis of androgen-sensitive LNCaP prostate cancer cells. This appears to be a consequence of lowering SK1a/b levels below a threshold required for survival. However, SKi fails to induce apoptosis in androgen-independent LNCaP-AI cells where the reduction in total SK1 level might not fall below this threshold. This might be due to the fact that the expression levels of SK1a and SK1b in LNCaP-AI cells are elevated above those in LNCaP cells. Thus, reduced sensitivity to SKi in LNCaP-AI cells might be a matter of titration of SK1b above a threshold level. This possibility is supported by the finding that the sensitivity of SK1b to SKi in terms of proteasomal degradation is enhanced by combined treatment of LNCaP-AI cells with SK1 siRNA and SKi. However, it should also be noted that SKi activates the proteasome to similar extents in LNCaP and LNCaP-AI cells, suggesting that other additional mechanisms might be operative such as modification of SK1b that reduce its sensitivity to SKi-induced proteasomal degradation. This might occur at a site up-stream of ceramide production based on our finding that C2-ceramide, which bypasses inhibition of SK1b, induces proteasomal degradation of SK1b to a similar extent in LNCaP and LNCaP-AI cells.
Finally, while siRNA treatment of LNCaP-AI cells with SK1 siRNA eliminates SK1a, this does not result in apoptosis. However, apoptosis can be induced in LNCaP-AI cells when both SK1a and SK1b are degraded by the proteasome in response to combined treatment with SKi and SK1 siRNA, thereby highlighting ubiquitin-proteasomal degradation of SK1 as an important mechanism controlling cell survival.
Summary
In conclusion, we demonstrate here that SK1a and SK1b are regulated by the ubiquitin-proteasomal degradation pathway. We also demonstrate that SK1 inhibitors activate this pathway to remove SK1 from cells. The removal of SK1 through the ubiquitin-proteasomal route is linked to down-stream activation of caspase-3/7, which are well established regulators of apoptosis. While, we demonstrate that SKi appears to induce the proteasomal degradation of polyubiquitinated proteins, we can link the specific degradation of SK1 with the onset of apoptosis. This is based on two findings. These are: (i) combined treatment of LNCaP-AI cells (which are refractory to SKi-induced degradation of SK1b, and normally do not undergo apoptosis in response to SKi) with SK1 siRNA/SKi reduces both SK1a and SK1b expression and this is then associated with the onset of apoptosis, and (ii) SKi activates the proteasome to equal extents in both LNCaP and LNCaP-AI cell, suggesting that the reduced sensitivity of SK1b to SKi-induced proteasomal degradation in LNCaP-AI cells is at a point up-stream of the action of ceramide at the proteasome. Thus, SK1b may maintain S1P levels above a threshold, below which LNCaP-AI cells undergo apoptosis.
Our findings with SKi and apoptosis are supported by studies from Taha et al. (39), who demonstrated that siRNA knock down of SK1 in MCF-7 breast cancer cells induces apoptosis. This involves caspase activation and is associated with increased ceramide formation and Bax oligomerization. Moreover, caspase activation and Bax oligomerization was significantly attenuated by inhibition of ceramide synthesis using the serine palmitolyl transferase inhibitor, myriocin. In addition, French et al. (18) have demonstrated that SKi induces apoptosis of various cancer cells. Our findings therefore extend these studies by demonstrating that inhibition of SK1 catalytic activity by SKi may induce apoptosis by removal of SK1 via the ubiquitin-proteasomal degradation pathway. In addition, the finding that SK1b in LNCaP-AI cells is less sensitive to SKi compared with androgen sensitive LNCaP cells might provide an explanation as to why androgen-independent prostate cancer cells acquire resistance to SK1 inhibitors.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL079396 (to V. N.). This work was also supported by Grants 23158/A7536 from CRUK, PG/06/137/21825 from the British Heart Foundation (to S. P. and N. J. P.), and by the Fay Fuller Foundation (to S. M. P. and T. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- S1P
- sphingosine 1-phosphate
- SKi
- 2-(p-hydroxyanilino)-4-(p-chlorophenyl)thiazole
- SK
- sphingosine kinase
- MRM
- multiple reaction monitoring
- hPASMC
- human pulmonary aortic smooth muscle cells.
REFERENCES
- 1.Pyne S., Pyne N. J. (2000) Biochem. J. 349, 385–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pyne S., Lee S. C., Long J., Pyne N. J. (2009) Cell. Signal. 21, 14–21 [DOI] [PubMed] [Google Scholar]
- 3.Taha T. A., Hannun Y. A., Obeid L. M. (2006) J. Biochem. Mol. Biol. 39, 113–131 [DOI] [PubMed] [Google Scholar]
- 4.Pyne N. J., Pyne S. (2010) Nat. Rev. Cancer 10, 489–503 [DOI] [PubMed] [Google Scholar]
- 5.Xia P., Gamble J. R., Wang L., Pitson S. M., Moretti P. A., Wattenberg B. W., D'Andrea R. J., Vadas M. A. (2000) Curr. Biol. 10, 1527–1530 [DOI] [PubMed] [Google Scholar]
- 6.Nava V. E., Hobson J. P., Murthy S., Milstien S., Spiegel S. (2002) Exp. Cell Res. 281, 115–127 [DOI] [PubMed] [Google Scholar]
- 7.Sarkar S., Maceyka M., Hait N. C., Paugh S. W., Senkala H., Milstien S., Spiegel S. (2005) FEBS Lett. 579, 5313–5317 [DOI] [PubMed] [Google Scholar]
- 8.Goetzl E. J., Dolezalova H., Kong Y., Zeng L. (1999) Cancer Res. 59, 4732–4737 [PubMed] [Google Scholar]
- 9.Long J. S., Edwards J., Watson C., Tovey S., Mair K. M., Schiff R., Natarajan V., Pyne N. J., Pyne S. (2010) Mol. Cell. Biol. 30, 3827–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vadas M., Xia P., McCaughan G., Gamble J. (2008) Biochim. Biophys. Acta 1781, 442–447 [DOI] [PubMed] [Google Scholar]
- 11.Akao Y., Banno Y., Nakagawa Y., Hasegawa N., Kim T., Murate T., Igarashi Y., Nozawa Y. (2006) Biochem. Biophys. Res. Commun. 342, 1284–1290 [DOI] [PubMed] [Google Scholar]
- 12.Nava V. E., Cuvillier O., Edsall L. C., Kimura K., Milstien S., Gelmann E. P., Spiegel S. (2000) Cancer Res. 60, 4468–4474 [PubMed] [Google Scholar]
- 13.Pchejetski D., Golzio M., Bonhoure E., Calvet C., Doumerc N., Garcia V., Mazerolles C., Rischmann P., Teissié J., Malavaud B., Cuvillier O. (2005) Cancer Res. 65, 11667–11675 [DOI] [PubMed] [Google Scholar]
- 14.Pchejetski D., Doumerc N., Golzio M., Naymark M., Teissié J., Kohama T., Waxman J., Malavaud B., Cuvillier O. (2008) Mol. Cancer Ther. 7, 1836–1845 [DOI] [PubMed] [Google Scholar]
- 15.Dayon A., Brizuela L., Martin C., Mazerolles C., Pirot N., Doumerc N., Nogueira L., Golzio M., Teissie J., Serre G., Rischmann P., Malavaud B., Cuvilllier O. (2008) PLoS One 4, e8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad M., Long J. S., Pyne N. J., Pyne S. (2006) Prostaglandins Lipid Mediators 79, 278–286 [DOI] [PubMed] [Google Scholar]
- 17.Anelli V., Gault C. R., Cheng A. B., Obeid L. M. (2008) J. Biol. Chem. 283, 3365–3375 [DOI] [PubMed] [Google Scholar]
- 18.French K. J., Upson J. J., Keller S. N., Zhuang Y., Yun J. K., Smith C. D. (2006) J. Pharm. Exp. Therap. 318, 596–603 [DOI] [PubMed] [Google Scholar]
- 19.Paugh S. W., Paugh B. S., Rahmani M., Kapitonov D., Almenara J. A., Kordula T., Milstien S., Adams J. K., Zipkin R. E., Grant S., Spiegel S. (2008) Blood 112, 1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kono K., Tanaka M., Ogita T., Kohama T. (2000) J Antibiot. 53, 759–764 [DOI] [PubMed] [Google Scholar]
- 21.Kono K., Tanaka M., Ogita T., Hosoya T., Kohama T. (2000) J Antibiot. 53, 459–466 [DOI] [PubMed] [Google Scholar]
- 22.Kono K., Tanaka M., Ono Y., Hosoya T., Ogita T., Kohama T. (2000) J Antibiot. 54, 415–420 [DOI] [PubMed] [Google Scholar]
- 23.Huwiler A., Döll F., Ren S., Klawitter S., Greening A., Römer I., Bubnova S., Reinsberg L., Pfeilschifter J. (2006) Biochim. Biophys. Acta 1761, 367–376 [DOI] [PubMed] [Google Scholar]
- 24.Pitson S. M., Moretti P. A., Zebol J. R., Lynn H. E., Xia P., Vadas M. A., Wattenberg B. W. (2003) EMBO J. 22, 5491–5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pham D. H., Moretti P. A., Goodall G. J., Pitson S. M. (2008) BioTechniques 45, 155–156 [DOI] [PubMed] [Google Scholar]
- 26.Pitson S. M., Xia P., Leclercq T. M., Moretti P. A., Zebol J. R., Lynn H. E., Wattenberg B. W., Vadas M. A. (2005) J. Exp. Med. 201, 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alderton F., Rakhit S., Kong K. C., Palmer T., Sambi B., Pyne S., Pyne N. J. (2001) J. Biol. Chem. 276, 28578–28585 [DOI] [PubMed] [Google Scholar]
- 28.Murphy R. C., Fiedler J., Hevko J. (2001) Chem. Rev. 101, 479–526 [DOI] [PubMed] [Google Scholar]
- 29.Merrill A. H., Caligan T. B., Wang E., Peters K., Ou J. (2000) Methods Enzymol. 312, 3–9 [DOI] [PubMed] [Google Scholar]
- 30.Berdyshev E. V., Gorshkova I. A., Garcia J. G., Natarajan V., Hubbard W. C. (2005) Anal. Biochem. 339, 129–136 [DOI] [PubMed] [Google Scholar]
- 31.Halkidou K., Gnanapragasam V. J., Mehta P. B., Logan I. R., Brady M. E., Cook S., Leung H. Y., Neal D. E., Robson C. N. (2003) Oncogene 22, 2466–2477 [DOI] [PubMed] [Google Scholar]
- 32.Ren S., Xin C., Pfeilschifter J., Huwiler A. (2010) Cell Physiol. Biochem. 26, 97–104 [DOI] [PubMed] [Google Scholar]
- 33.Calatayud C. A., Pasquini L. A., Pasquini J. M., Soto E. F. (2005) Dev. Neurosci. 27, 397–407 [DOI] [PubMed] [Google Scholar]
- 34.Garzotto M., Haimovitz-Friedman A., Liao W. C., White-Jones M., Huryk R., Heston W. D., Cardon-Cardo C., Kolesnick R., Fuks Z. (1999) Cancer Res. 59, 5194–5201 [PubMed] [Google Scholar]
- 35.Illuzzi G., Bernacchioni C., Aureli M., Prioni S., Frera G., Donati C., Valsecchi M., Chigorno V., Bruni P., Sonnino S., Prinetti A. (2010) J. Biol. Chem. 285, 18594–18602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hannun Y. A., Obeid L. M. (2008) Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 37.Canals D., Jenkins R. W., Roddy P., Hernández-Corbacho M. J., Obeid L. M., Hannun Y. A. (2010). J. Biol. Chem. 285, 32476–32485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kihara A., Anada Y., Igarashi Y. (2006) J. Biol. Chem. 281, 4532–4539 [DOI] [PubMed] [Google Scholar]
- 39.Taha T. A., Kitatani K., El-Alwani M., Bielawski J., Hannun Y. A., Obeid L. M. (2006) FASEB J. 20, 482–484 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.