Abstract
The differentiation, maintenance, and repair of skeletal muscle is controlled by interactions between genetically determined transcriptional programs regulated by myogenic transcription factors and environmental cues activated by growth factors and hormones. Signaling through the insulin-like growth factor 1 (IGF1) receptor by locally produced IGF2 defines one such pathway that is critical for normal muscle growth and for regeneration after injury. IGF2 gene and protein expression are induced as early events in muscle differentiation, but the responsible molecular mechanisms are unknown. Here we characterize a distal DNA element within the imprinted mouse Igf2-H19 locus with properties of a muscle transcriptional enhancer. We find that this region undergoes a transition to open chromatin during differentiation, whereas adjacent chromatin remains closed, and that it interacts in differentiating muscle nuclei but not in mesenchymal precursor cells with the Igf2 gene found more than 100 kb away, suggesting that chromatin looping or sliding to bring the enhancer in proximity to Igf2 promoters is also an early event in muscle differentiation. Because this element directly stimulates the transcriptional activity of an Igf2 promoter-reporter gene in differentiating myoblasts, our results indicate that we have identified a bona fide distal transcriptional enhancer that supports Igf2 gene activation in skeletal muscle cells. Because this DNA element is conserved in the human IGF2-H19 locus, our results further suggest that its muscle enhancer function also is conserved among different mammalian species.
Keywords: Chromatin Remodeling, Differentiation, Gene Regulation, Gene Transcription, Insulin-like Growth Factor (IGF), Muscle Development
Introduction
The differentiation, maintenance, regeneration, and repair of skeletal muscle requires ongoing interactions between signaling pathways activated by hormones and growth factors and an intrinsic regulatory program controlled by myogenic transcription factors (1–4). Among growth factors with major actions on muscle are the insulin-like growth factors IGF1 and IGF2 (5, 6),3 two closely related single-chain secreted proteins that bind with high affinity to the IGF1 receptor (7), leading to activation of several intracellular signal transduction pathways that act downstream of this membrane-spanning protein-tyrosine kinase (7). Much experimental evidence supports the importance of IGF actions in muscle. In mice, targeted IGF1 receptor deficiency caused marked muscle hypoplasia and neonatal death secondary to respiratory failure from severe muscle weakness (8). In contrast, targeted overexpression of IGF1 stimulated an increase in muscle mass throughout life (9), enhanced anabolic responses to exercise (10), and slowed the development of experimental muscular dystrophy (11). Moreover, analysis of quantitative trait loci for muscle in the pig identified a single-nucleotide polymorphism in an IGF2 intron that influenced IGF2 gene expression, with one variant being associated with a 3-fold greater abundance of IGF2 mRNA in muscle and a 3–4% increase in total muscle mass (12).
The IGF2 gene resides on human chromosome 11p15.5 and on a syntenic segment of mouse chromosome 7 and is part of an imprinted cluster with the adjacent upstream insulin gene (Ins2 in mice (13)) and downstream H19 (13). In mice, the Igf2 gene is composed of six exons (14), and gene expression is regulated by three adjacent promoters, termed P1–P3, each with its own unique leader exon, whereas exons 4–6 encode the IGF2 precursor protein (14). The human IGF2 gene is more complicated, because it has an additional upstream promoter (15). In both species, IGF2 is transcribed from the paternally derived chromosome in most tissues, and H19 is expressed from the maternal chromosome by regulation through an imprinting control region (ICR) located between the two genes (13, 16). The ICR contains binding sites for the nuclear factor, CCTC binding factor (CTCF) (16, 17), which when bound to DNA in chromatin on the maternally derived chromosome facilitates H19 transcription by directing distal enhancers to the H19 promoter (16, 17). On the paternal chromosome, DNA in the ICR is methylated, and CTCF cannot bind, and the enhancers have access to the IGF2 promoters (16, 17).
IGF2 gene transcription, mRNA production, and protein biosynthesis are induced as early events during muscle differentiation in culture (18, 19), and the secreted IGF2 functions as an autocrine differentiation-promoting factor (20, 21), as evidenced by impaired differentiation when IGF2 synthesis or access to the IGF1 receptor is blocked (19, 21) and by accelerated and enhanced differentiation when IGF2 is overexpressed (20, 22). The molecular mechanisms responsible for IGF2 gene activation during muscle differentiation are unknown, although the single nucleotide porcine IGF2 polymorphism associated with increased muscle mass appears to prevent binding of a putative transcriptional repressor to the IGF2 gene (12, 23, 24). Here we characterize a conserved distal enhancer that interacts with the mouse Igf2 gene in differentiating myoblasts but not in mesenchymal progenitors and that can promote Igf2 gene transcription in muscle.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
DMEM, Superscript III first strand synthesis kit, TRIzol reagent, trypsin/EDTA solution, and horse serum were from Invitrogen, and FBS and newborn calf serum were from Hyclone (Logan, UT). Restriction enzymes, buffers, ligases, and polymerases were from New England Biolabs (Beverly, MA), BD Biosciences (Clontech), and Fermentas (Hanover, MD). Protease inhibitor tablets were purchased from Roche Applied Sciences; okadaic acid was from Alexis Biochemicals (San Diego, CA), sodium orthovanadate was from Sigma, and proteinase K was from Roche Applied Sciences. TransIT-LT-1 was from Mirus Corp. (Madison, WI), and Hoechst 33258 nuclear dye was from Polysciences (Warrington, PA). The BCA protein assay kit was from Pierce, Immobilon-FL was from Millipore Corporation (Billerico, MA), and AquaBlock tm/EIA/WIB solution was from East Coast Biologicals (North Berwick, ME). DNA purification kits were from Qiagen, and luciferase assay reagents were from Promega (Madison, WI). The following antibodies were from the Developmental Studies Hybridoma Bank: F5D (anti-myogenin, from W. E. Wright) and CT3 (anti-troponin T, from J. J-C. Lin). The polyclonal antibody to Akt was from Cell Signaling Technology (Beverly, MA). AlexaFluor 680-conjugated goat anti-mouse IgG was from Invitrogen, and IR800-conjugated goat anti-rabbit IgG was from Rockland (Gilbertsville, PA). All other chemicals were reagent grade and were purchased from commercial suppliers.
Cell Culture
The cells were incubated at 37 °C in humidified air with 5% CO2. C2 myoblasts (passages 4–10) were grown on gelatin-coated tissue culture dishes in DMEM with 10% heat-inactivated FBS and 10% newborn calf serum. At confluent density, the cells were washed, and low serum differentiation medium was added (differentiation medium (DM) was DMEM with 2% horse serum). C3H 10T1/2 mouse embryonic fibroblasts (catalogue number CCL226; ATCC, Manassas, VA) were incubated on gelatin-coated tissue culture dishes in growth medium (DMEM with 10% heat-inactivated FBS) at 37 °C in humidified air with 5% CO2. They were converted to myoblasts by infection at ∼50% of confluent density with a recombinant adenovirus for mouse MyoD (Ad-MyoD), as described (25), followed by incubation in DM after reaching confluent density as above.
Animal Studies
Male and female C57Bl6 mice were housed at the Oregon Health & Science University Animal Care Facility on a 12-h light/dark schedule with free access to food and water and received care according to institutional and National Institutes of Health guidelines. Pregnant mice were euthanized by exposure to CO2; the pups were isolated by Caesarean section and euthanized after exposure to CO2. Three-month-old male mice were euthanized by cervical dislocation. Freshly isolated tissues were flash-frozen in liquid nitrogen and pulverized prior to RNA extraction. The Oregon Health & Science University Animal Care and Use Committee approved all animal studies.
Gene Transfer with Recombinant Adenoviruses
Recombinant adenoviruses for MyoD (Ad-MyoD) and β-galactosidase (Ad-β-Gal) were purified on discontinuous cesium chloride gradients and titered by optical density, as described (25). Prior to infection, the viruses were diluted in DMEM plus 2% fetal calf serum, filtered through a Gelman syringe filter (0.45 μm), and then were added to cells at 37 °C for 120 min. After the addition of an equal volume of DMEM with 20% fetal bovine serum, the cells were incubated for a further 24 h. The cells then were washed twice with phosphate-buffered saline and incubated in DM.
Analysis of Igf2 Gene Expression
Whole cell and nuclear RNA were isolated as described (21). RNA concentrations were determined spectrophotometrically at 260 nm and quality assessed by agarose gel electrophoresis. RNA (2.5 μg) was reverse transcribed in a final volume of 20 μl, with either oligo(dT) primers (for total RNA) or random hexamers (for nuclear RNA), and PCR was performed with 0.1 μl of cDNA and the primer pairs in Table 1. The linear range of product amplification was established in pilot studies for each primer pair, and the cycle number that reflected the approximate midpoint was used in final experiments. This varied from 20 to 27 cycles for total RNA and from 25 to 30 cycles for nuclear RNA. The results were visualized after electrophoresis through 1.0% agarose gels.
TABLE 1.
Primers used for RT-PCR
| Gene | Location | Primer sequence | Product |
|---|---|---|---|
| bp | |||
| Nuclear RNA | |||
| Igf2 | Exon 3 | 5′-GCAAACTGGACATTAGCTTCT-3′ | 597 |
| Intron 3–4 | 5′-CCCTTGGGTAACTAAAATCATCTT-3′ | ||
| Myogenin | Intron 2–3 | 5′-GGGATCACTCAGTCAGTGTTGTAA-3′ | 537 |
| Exon 3 | 5′-TCTCTGCTTTAAGGAGTCAGCTAAA-3′ | ||
| S17 | Exon 2 | 5′-ATCCCCAGCAAGAAGCTTCGGAACA-3′ | 439 |
| Intron 2–3 | 5′-GAACCGACTTTGTCTCTACATCAAG-3′ | ||
| Total RNA | |||
| Igf2 | Exon 1 | 5′-CAGCAGCTCCCACTTCATCCG-3′ | 400 |
| Exon5 | 5′-TGGCACGGCTTGAAGGCCTGC-3 | ||
| Igf2 | Exon 2 | 5′-CGGCCTCTGCGACTCGGGCAG-3′ | 485 |
| Exon 5 | 5′-TGGCACGGCTTGAAGGCCTGC-3 | ||
| Igf2 | Exon 3 | 5′-CCTGTGAGAACCTTCCAGCCT-3′ | 396 |
| Exon 5 | 5′-TGGCACGGCTTGAAGGCCTGC-3 | ||
| Myogenin | Exon 1 | 5′-GGGGACCCCTGAGCATTGTCC-3′ | 512 |
| Exon 3 | 5′-CAGCCTGACAGACAATCTCAGTT-3′ | ||
Construction of Igf2 Promoter-Reporter Plasmids
Mouse myogenin and mouse Igf2 promoter-luciferase plasmids have been described (18). For these experiments, Igf2 P3 was inserted in plasmid pGL3 (Promega). DNA fragments pictured in Fig. 2 were isolated from mouse genomic DNA after PCR by standard methods, except for 4.3-kb element D, which was obtained from Dr. Jie Chen (University of Illinois, Urbana, IL). Region 1, CS6, and CS9 were cloned via 5′ SalI and 3′ XbaI linkers into the corresponding sites in the Igf2 P3 pGL3 plasmid (see supplemental Table S1 for details). All of the subfragments were generated by restriction enzyme digestion or PCR and were purified after preparative agarose gel electrophoresis by ion exchange chromatography (Qiaex II gel extraction kit; Qiagen) and subcloned into Igf2 promoter-luciferase plasmids. All of the DNA manipulations were confirmed by sequencing at the Oregon Health & Science University DNA Services Core.
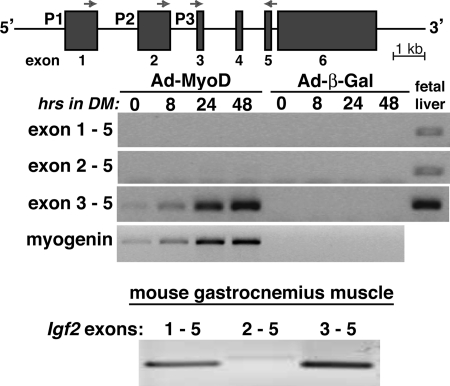
FIGURE 2.
Promoter 3-specific induction of Igf2 gene expression during muscle differentiation. Top panel, schematic of the mouse Igf2 gene. The three promoters and six exons are indicated, as are the locations of exon-specific PCR primers. Middle panel, results of time course gene expression experiments by RT-PCR for Igf2 and myogenin using 10T1/2 mesenchymal stem cells infected with Ad-MyoD or Ad-β-Gal and incubated in DM for up to 48 h. For Igf2, only transcripts containing exons 3 and 5 gave a positive result in differentiating myoblasts, demonstrating that only Igf2 promoter 3 is activated in this muscle differentiation model. Mouse day 18 fetal liver RNA serves as a positive control for active transcription from each Igf2 promoter (14). Bottom panel, results of RT-PCR experiments measuring promoter-specific Igf2 transcripts using gastrocnemius muscle from 8-week-old male mice. Transcripts directed by promoter 3 and containing exons 3 and 5 are more abundant than mRNAs derived from promoters 1 or 2.
Transient Transfections and Luciferase Reporter Gene Assays
C2 and C3H 10T1/2 cells were plated onto gelatin-coated 12-well plates and were transfected at 50 or 25% of confluent density, respectively, with individual Igf2 promoter-luciferase reporter plasmids or with mouse myogenin promoter-luciferase (0.4 μg of plasmid DNA/well for C2 cells and 0.2 μg for 10T1/2 cells). C2 cell extracts were harvested 1 day later (undifferentiated), or DM was added, and cellular proteins were isolated after an additional 48 h (differentiated). For 10T1/2 cells, 1 day after transfection the cells were infected with Ad-MyoD or adenoviruses for β-galactosidase, and after an additional day in growth medium, the extracts were harvested (undifferentiated), or DM was added, and the cells were incubated for an additional 24 h before protein isolation (differentiated). Cell extracts from an individual experiment were stored at −80 °C until luciferase assay, and the results were normalized to cellular protein concentrations (21). At least three experiments were performed for each promoter-reporter plasmid using duplicate transfections per experiment.
Protein Extraction and Immunoblotting
Whole cell protein lysates were prepared as described (19) and stored in aliquots at −80 °C until use. Protein concentrations were determined with the BCA protein assay kit, and aliquots (25 μg/lane) were separated by SDS-PAGE, transferred to Immobilon-FL, blocked in AquaBlock, and incubated with primary and secondary antibodies, as described (21). The membranes were washed according to a protocol from LiCoR and scanned on an Odyssey Infrared Imaging System using v3.0 analysis software (LiCoR Biosciences, Lincoln, NE). Primary antibodies were used at the following dilutions: anti-myogenin (1:100), anti-troponin T (1:1000), and anti-Akt (1:2000).
Immunocytochemistry
The cells were fixed, permeabilized, blocked, and incubated with antibodies as described (21). The primary antibodies were added in blocking buffer for 16 h at 4 °C (anti-troponin T, 1:1000 dilution; anti-myogenin, 1:50; and secondary antibodies at 1:1000). The images were captured with a Roper Scientific Cool Snap FX CCD camera attached to a Nikon Eclipse T300 fluorescent microscope using IP Labs 3.5 software. Hoechst staining was performed as described (21).
Analysis of Chromatin Structure by Restriction Endonuclease Accessibility
Isolated nuclei from 1 × 107 cells were incubated overnight at 37 °C in 100 μl of AluI buffer (New England Biolabs; 50 mm NaCl, 10 mm Tris-Cl, 10 mm MgCl2, 1 mm DTT, pH 7.9) with 1 unit/μl of AluI. The nuclei incubated in the same buffer without AluI served as a negative control. Following the addition of 100 μl of 2× proteinase K digestion buffer (100 mm TrisCl, 200 mm NaCl, 2 mm Na2EDTA, 1% SDS, pH 7.5) for 2 h at 55 °C, 100 μl of AluI buffer plus 100 μl of 2× proteinase K buffer containing 100 μg of proteinase K were added and incubated overnight at 37 °C. DNA was isolated by extraction with a phenol-chloroform-isoamyl alcohol solution and ethanol precipitation and dissolved in 500 μl of 10 mm TrisCl, 1 mm Na2EDTA, pH 7.9. PCR was performed using 25–50 ng of this DNA per reaction (see Table 2 for primers). The linear range of product amplification was established for each primer pair in pilot studies, and the cycle number that reflected the approximate midpoint was used in the final experiments. This varied from 30 to 35 cycles. The results were visualized after electrophoresis through 1.0% agarose gels.
TABLE 2.
Primers for restriction accessibility assay
| DNA fragment | DNA strand | Primer sequence | Product |
|---|---|---|---|
| bp | |||
| Fragment 1 | Top | 5′-CTTCCAGACTCATCAAGAATA-3′ | 299 |
| Bottom | 5′-GAACAACTGTGGGGACCAAAG-3′ | ||
| Fragment 2 | Top | 5′-ATTGCAGGCAGTGGGTGGA-3′ | 300 |
| Bottom | 5′-ATAGAAATGCCTCTTAAGAGT-3′ | ||
| Fragment 3 | Top | 5′-GGCTTCCCGCCATCTCGA-3′ | 285 |
| Bottom | 5′-TGGGGTTAGGAGCAGCTGT-3′ | ||
| Fragment 4 | Top | 5′-AAGGAGGATTTAGCTCGGGAG-3′ | 399 |
| Bottom | 5′-CTGGGGTCCGGCTCACAT-3′ | ||
| Fragment 5 | Top | 5′-ATGTGACCCGGACCCCAGGCC-3′ | 291 |
| Bottom | 5′-GACAGGCCTTGTGTTCTTGCA-3′ |
Chromatin Conformation and Capture Assays
These studies followed published protocols (26–28). The nuclei from 1 × 107 cells were fixed by the addition of 0.5 ml of lysis buffer (10 mm Hepes, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DTT, 1% Triton X-100, pH 7.9) plus protease inhibitors and 2% formaldehyde for 2–3 min at 15 °C and quenched by the addition of glycine to 0.125 m for 5 min on ice. Following two washes with ice-cold lysis buffer, the fixed nuclei were resuspended in 0.5 ml of 1.2× BglII restriction enzyme buffer (100 mm NaCl, 50 mm Tris-HCl, 10 mm MgCl2, 1 mm DTT, pH 7.9) containing 0.3% SDS and incubated at 37 °C for 1 h with shaking at 1000 rpm. Triton X-100 was added to a 1.8% final concentration, and samples were incubated for 1 h at 37 °C with shaking followed by the addition of 1500 units of BglII and incubation for 16 h at 37 °C and 1000 rpm. After the addition of SDS to 1.3% for 20 min at 65 °C, each sample was mixed with 7 ml of 1× DNA ligation buffer, Triton X-100 was added to 1%, and the nuclei were incubated for 1 h at 37 °C and 400 rpm. After equilibration at 16 °C, T4 DNA ligase was added (100 units), followed by incubation for 5 h at 16 °C with slow agitation. After sequential incubation with proteinase K (300 μg for 16 h at 65 °C) and RNase A (3 μl of a 100 mg/ml solution for 30 min at 37 °C), DNA was isolated by phenol-chloroform extraction and ethanol precipitation and dissolved in 1 ml of 10 mm TrisCl, 1 mm Na2EDTA, pH 7.9. PCRs were performed with 1 μl of DNA, and the primers are listed in Table 3. The linear range of product amplification was established for each primer pair in pilot studies using an artificial template that was generated by overlap extension PCR (29), and the cycle number that reflected the approximate midpoint was used in the final experiments. This varied from 30 to 35 cycles. The results were visualized after electrophoresis through 1.2% agarose gels.
TABLE 3.
Primers for chromatin conformation capture assay
| DNA fragment | Primer sequence |
|---|---|
| Enhancer 1 | 5′-AAACAGCATCCTTAGCCTATGATG-3′ |
| Enhancer 2 | 5′-ACCAGCCTGGCTACCACCTG-3′ |
| Promoter 1 | 5′-TAGAGGGTTACAAGGTAGG-3′ |
| Promoter 2 | 5′-AGAGAGGCCAAACGTCATCGT-3′ |
| Enhancer 3 | 5′-ATGTGACCCGGACCCCAGGCC-3′ |
| Enhancer 4 | 5′-GACAGGCCTTGTGTTCTTGCA-3′ |
Statistical Analysis
The data are presented as the means ± S.D. The statistical significance was determined using a paired Student's t test. The results were considered statistically significant when p < 0.05.
RESULTS
Promoter-specific Activation of Igf2 Gene Transcription during Skeletal Muscle Differentiation
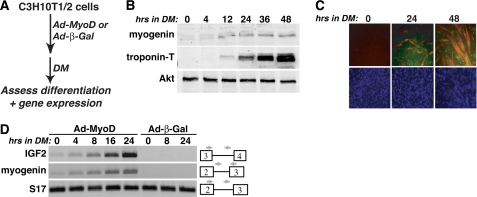
Previous studies have found that IGF2 gene expression and protein secretion were induced within early stages of differentiation of skeletal myoblasts in culture (18, 19, 21) and have shown that IGF2 actions, mediated by autocrine activation of the IGF1 receptor and the phosphatidylinositol 3-kinase-Akt pathway, are necessary to sustain muscle differentiation (19, 30). In C3H110T1/2 mouse mesenchymal stem cells, adenoviral-mediated expression of the myogenic transcription factor, MyoD, can potently convert these uncommitted progenitors to a myoblast fate (31), with rapid and robust up-regulation of muscle genes and proteins and myotube formation occurring once DM is added (19) (Fig. 1, A–C). Under these conditions we find that transcription of the endogenous Igf2 gene is also rapidly stimulated after the addition of DM, with kinetics of activation very similar to those of the early differentiation gene myogenin (Fig. 1D).
FIGURE 1.
Activation of Igf2 gene transcription during muscle differentiation. Shown are the results of time course experiments with 10T1/2 mesenchymal stem cells infected with Ad-MyoD or Ad-β-Gal and incubated in DM for up to 2 days. A, experimental scheme. B, induction of muscle proteins myogenin and troponin-T and constant expression of Akt during differentiation, as assessed by immunoblotting. C, myotube formation measured by immunocytochemistry for myogenin (green) and troponin-T (red), and nuclear staining by Hoechst dye (blue). D, time course of transcription for Igf2, myogenin, and S17 genes, measured by semi-quantitative RT-PCR. The locations of relevant exons and PCR primers are shown.
The Igf2 gene on mouse chromosome 7 is composed of six exons and five introns, and its transcription is governed by three tandem promoters, P1–P3, each of which regulate a unique leader exon (14). In mouse fetal tissues all three promoters are active (14). To examine promoter usage in Ad-MyoD-converted muscle cells, we developed a selective RT-PCR assay in which transcripts containing individual leader exons are assessed. In mouse fetal liver all three promoters are active, as evidenced by positive RT-PCR products with primers derived from each leader exon coupled to a shared coding exon primer, whereas in Ad-MyoD-converted 10T1/2 cells, only transcripts directed by P3 accumulate during muscle differentiation (Fig. 2). In addition, in mouse gastrocnemius muscle, mRNAs directed by P3 were more abundant than transcripts controlled by P1 or P2 (Fig. 2).
Identifying a DNA Element That Can Stimulate Igf2 Promoter Activity during Skeletal Muscle Differentiation
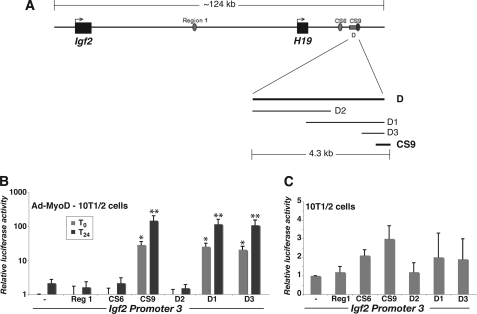
Although it has been established that IGF2 is highly expressed in skeletal muscle in vivo (32) and that its gene expression is induced during myoblast differentiation in culture (Figs. 1 and 2 and Refs. 18, 19, and 33), very little is known about the mechanisms of Igf2 gene regulation in muscle. No transcriptional response elements have been identified, and no transcription factors have been characterized. Studies using transgenic mice have been employed to investigate aspects of regulation of the Igf2-H19 locus and have defined several chromosomal segments that could direct gene activity to mesenchymal tissues, including muscle (34–37), but the identified domains have not been evaluated in any detail. We analyzed four previously mapped DNA elements from the mouse Igf2-H19 locus for activity during muscle differentiation by fusing each of them 5′ to a minimal Igf2 P3-luciferase reporter gene and transfecting them into Ad-MyoD 10T1/2 cells. Of these four DNA segments, three were described as having enhancer activity in mesenchymal tissues in transgenic mice (CS6, CS9, D (35–37)), and one had been identified as a putative repressor (Region 1 (34)) (Fig. 3A). Region D, a ∼4.3-kb DNA fragment located ∼22–27 kb 3′ to mouse H19, also was found to stimulate promoter activity in cultured muscle cells (33). When assessed in transient reporter gene assays in Ad-MyoD-converted 10T1/2 cells, Igf2 P3 alone was ineffective, and the addition of either Region 1 or CS6 did not stimulate its transcriptional activity (Fig. 3B). In contrast, CS9 and region D each dramatically enhanced the activity of Igf2 P3 in myoblasts, particularly after induction of differentiation, and dissection of the D region revealed that a fragment of only 756 bp, D3, located at its 3′ end, was just as active as the entire 4.3-kb segment (Fig. 3B). By contrast, none of the DNA fragments tested were able to stimulate Igf2 P3 in 10T1/2 mesenchymal stem cells (Fig. 3C).
FIGURE 3.
Identification of DNA elements that mediate muscle-specific induction of Igf2 gene transcription. A, schematic of Igf2-H19 locus on mouse chromosome 7. Region 1, CS6, CS9, and D sites are indicated, as is dissection of the D segment into D1–D3. B, results of luciferase reporter gene experiments with Ad-MyoD-infected 10T1/2 cells incubated in DM for 0 or 24 h, using mouse Igf2 promoter 3 ± Region 1 (Reg1), CS6, CS9, or D DNA segments (means ± S.D., n = 5–8 experiments; *, p < 0.001; **, p < 0.0002 versus IGF2 promoter 3). Note the log scale on the ordinate. C, results of luciferase reporter gene experiments with 10T1/2 cells, using the same recombinant plasmids as in B. Note the linear scale on the ordinate.
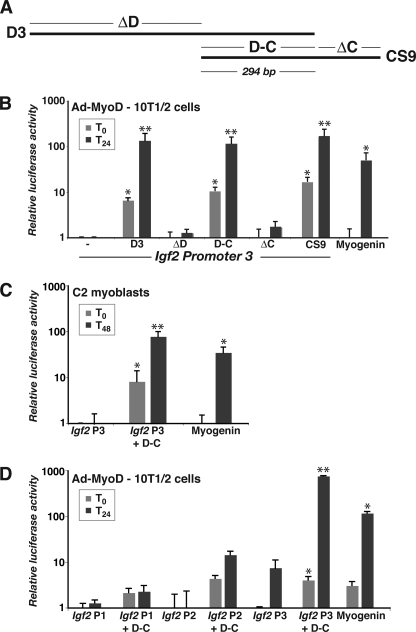
Because DNA sequence analysis revealed that 756-bp D3 overlapped 486-nucleotide CS9 by 294 bp, we next devised a series of experiments to functionally map the putative muscle enhancer (Fig. 4A). As pictured in Fig. 4B, the 294-bp overlapping DNA segment (fragment D-C) was as potent as D3 or CS9 in differentiating Ad-MyoD converted 10T1/2 cells and also stimulated Igf2 P3 activity in differentiating C2 myoblasts (Fig. 4C). Also, the D-C fragment did not enhance the activity of Igf2 P1 or P2 (Fig. 4D), suggesting selectivity for P3, the Igf2 promoter that becomes activated during muscle differentiation in culture (Fig. 2). Taken together, the results in Figs. 3 and 4 provide initial support for the hypothesis that this DNA element located 3′ to H19 is a distal muscle enhancer for mouse Igf2.
FIGURE 4.
Functional dissection of the putative Igf2 muscle enhancer. A, diagram of D3 and CS9 regions (see Fig. 3A for genomic context). B, results of luciferase reporter gene experiments with Ad-MyoD-infected 10T1/2 cells incubated in DM for 0 or 24 h, using mouse Igf2 promoter 3 (P3) ± D3, ΔD, D-C, ΔC, CS9, or the mouse myogenin promoter (mean ± S.D., n = 5 experiments; *, p < 0.01; **, p < 0.001 versus Igf2 P3). C, results of luciferase reporter gene experiments with C2 myoblasts incubated in DM for 0 or 48 h, using mouse Igf2 P3, Igf2 P3 + D-C, or the mouse myogenin promoter (mean ± S.D., n = 5 experiments; *, p < 0.01; **, p < 0.005 versus IGF2 P3). D, results of luciferase reporter gene experiments with Ad-MyoD-infected 10T1/2 cells incubated in DM for 0 or 24 h, using mouse Igf2 P1, P2, or P3 ± D-C, or the mouse myogenin promoter (mean ± S.D., n = 3 experiments; *, p < 0.01; **, p < 0.005 versus Igf2 P3). Note the log scales on the ordinates for B and C.
Chromatin Changes Accompany Muscle Differentiation at the Distal Igf2-H19 DNA Element
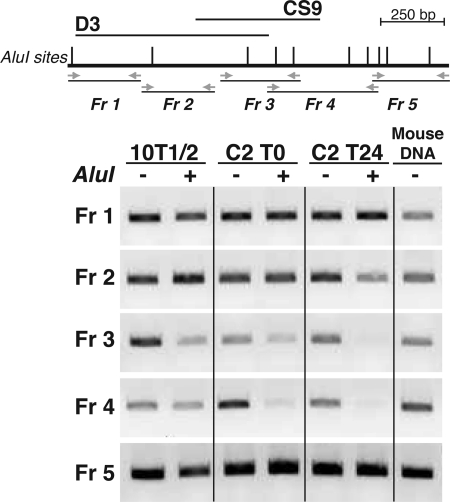
Previous studies have found that chromatin at muscle promoters undergoes dramatic changes triggered by transcription factor binding during differentiation, including histone modifications and recruitment of transcriptional co-factors (21, 38–41). We asked whether the distal Igf2-H19 DNA element was similarly plastic and used a restriction endonuclease accessibility assay to examine its chromatin environment. Nuclei, from mesenchymal precursor cells and from myoblasts before and after onset of differentiation, were incubated with AluI, and the isolated DNA was analyzed by PCR. As pictured in Fig. 5, DNA in chromatin from 10T1/2 cells is relatively impermeable to digestion by AluI throughout the D3-CS9 region, whereas in undifferentiated C2 myoblasts, limited cleavage is observed, which increases dramatically after 24 h in DM. Of particular note is the focused region of accessibility to AluI, which consists of the 294 bp shared between D3 and CS9 and the remaining 192 bp of CS9 and does not extend further in either the 5′ or 3′ directions (Fig. 5). These results show that onset of muscle differentiation is accompanied by discrete chromatin changes centered on the D3-CS9 element 3′ to H19.
FIGURE 5.
Mapping chromatin changes during muscle differentiation in the putative Igf2 muscle enhancer. Top panel, schematic of the D3 and CS9 region 3′ to H19 showing location of AluI sites (gray vertical lines) and PCR primers (horizontal arrows) defining fragments (Fr) 1–5. Bottom panel, results of restriction endonuclease accessibility assays in 10T1/2 cells, and in C2 myoblasts before (T0) and 24 h after onset of differentiation (T24). Mouse genomic DNA serves as a positive control for PCR. The results shown are representative of three independent experiments.
Identifying Long Range Interactions between the Distal Chromosomal DNA Element and Igf2 during Muscle Differentiation
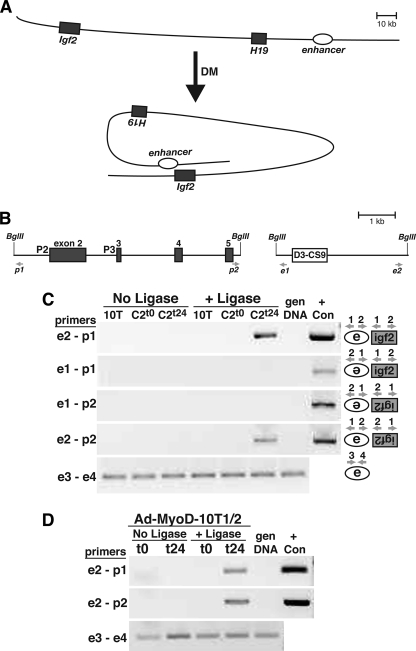
For a putative enhancer to regulate gene transcription at a distant promoter, their physical association must occur. We used a chromosomal conformation and capture assay to determine whether the distal DNA element 3′ to H19 interacted with the Igf2 gene during muscle differentiation, as outlined in the hypothetical diagram pictured in Fig. 6A. Chromosomal DNA, obtained from cross-linked and BglII-digested nuclei isolated from mesenchymal precursor cells and from myoblasts, was ligated and assessed by PCR using the primers depicted in Fig. 6B. The results show that there is an inducible physical interaction between the D3-CS9 region and Igf2 that only occurs after onset of muscle differentiation in both C2 myoblasts and Ad-MyoD-infected 10T1/2 cells (Fig. 6, C and D). Because the locations of the two BglII sites in the IGF2 gene were not near P3, we could not map the association between the distal enhancer and the active promoter to any greater precision than several kilobases. Nevertheless, these observations demonstrate that there is a dynamic rearrangement of the Igf2-H19 locus coincident with onset of muscle differentiation that results in the transfer of information from a site over 100 kb away to activate Igf2 gene transcription from promoter 3.
FIGURE 6.
The distal enhancer physically interacts with Igf2 promoter 3 during muscle differentiation. A, schematic of Igf2-H19 locus showing putative chromosomal looping during muscle differentiation and potential inducible association of the D-C enhancer region with Igf2 promoter 3. B, higher resolution view of mouse Igf2 exons 2–5 (left panel), and the putative distal enhancer (D3-CS9, right panel), showing locations of BglII sites and PCR primers used in the chromatin conformation and capture assays. C, results of chromatin conformation capture experiments using 10T1/2 mesenchymal stem cells (10T) or C2 myoblasts incubated in DM for 0 or 24 h. D, results of chromatin conformation and capture experiments using Ad-MyoD-infected 10T1/2 cells incubated in DM for 0 or 24 h. For C and D, the results are presented ± incubation of chromatin with DNA ligase. Also, PCR primer pairs for each group of experiments are indicated to the left of each panel, and in C the orientation of association between the enhancer and promoter being tested in each panel is diagrammed to the right. gen DNA, mouse genomic DNA (negative control); + Con, positive control for each primer pair, generated by overlap extension PCR (29). Primer pairs e3-e4 serve as positive controls for DNA quality and quantity. The results depicted are representative of three independent experiments for both C and D.
Mapping Studies Identify Multiple Functional Elements in the Distal Igf2 Muscle Enhancer
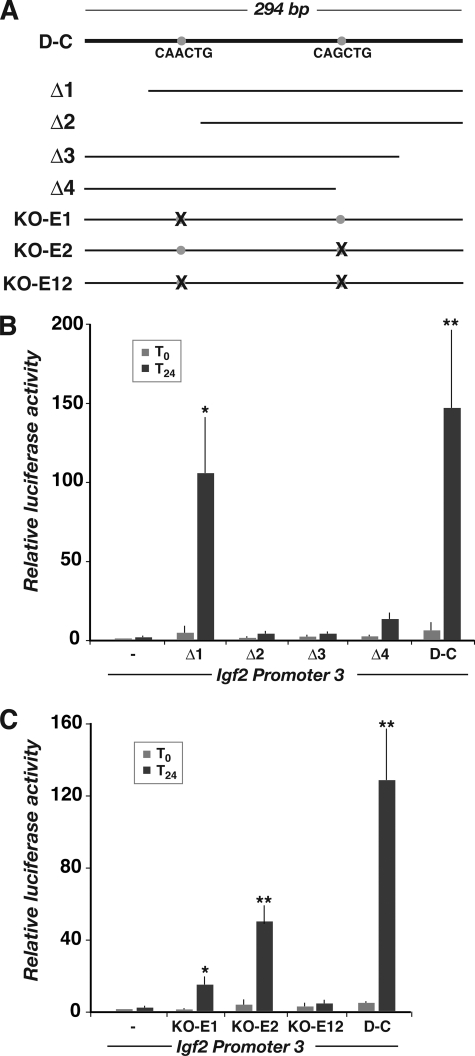
We performed additional promoter-reporter gene experiments to define the components of the 294-bp distal enhancer that were required to stimulate Igf2 gene transcription during muscle differentiation. Results with a series of 5′ deleted DNA fragments revealed that the initial 50 bp was dispensable for function (Δ1) but that removal of an additional 40 bp led to a ∼20-fold decline in transcriptional activity in differentiating myoblasts (Δ2; Fig. 7, A and B). Similarly, stepwise deletions at the 3′ end of the D-C element also caused a greater than 20-fold decrease in function (Δ3 and Δ4; Fig. 7, A and B). In addition, mutation of the first of two E-boxes found within the 294-bp segment reduced enhancer activity by nearly 90% (KO-E1), and mutation of both eliminated activation of Igf2 P3 during muscle differentiation (Fig. 7, A and C). Of note, comparative mapping revealed that a conserved DNA fragment was found in the human genome in a similar location 3′ to H19 on chromosome 11p15.5 and that its DNA sequence was ∼80% identical to the 294-nucleotide D-C region from bp 118–294 (data not shown).
FIGURE 7.
Functional dissection of the putative Igf2 muscle enhancer. A, diagram of the 294-base pair region of overlap between the D3 and CS9 segments. The locations of putative E-boxes are indicated by gray circles, and the DNA sequences are listed below. The serial deletions and mutations used in promoter-reporter experiments are diagramed below. B and C, results of luciferase reporter gene experiments using Ad-MyoD-infected 10T1/2 cells incubated in DM for 0 or 24 h. B, results with deletions Δ1–Δ4 (mean ± S.D., n = 5 experiments; *, p < 0.01; **, p < 0.005 versus Igf2 P3). C, results with the E-box mutations listed above (mean ± S.D., n = 3 experiments; *, p < 0.05; **, p < 0.02 versus Igf2 P3).
DISCUSSION
A role for IGF2 has been established in skeletal muscle based on evidence that its production has been linked temporally and functionally with muscle regeneration after injury (32) and on the association of a DNA polymorphism in the porcine IGF2 gene with increased muscle IGF2 mRNA expression and enhanced muscle mass (12). Despite these connections with muscle growth and repair and studies showing that IGF2 gene activation occurs as an early event during muscle differentiation (18, 19, 33), the mechanisms regulating IGF2 gene activity in muscle have proven to be elusive. Here we identify and characterize a distal DNA element within the imprinted mouse Igf2-H19 locus that has the properties of a muscle transcriptional enhancer. Our key findings include the demonstration that this region undergoes a transition to open chromatin during muscle differentiation, whereas adjacent chromatin remains closed, and more importantly, it physically interacts in differentiating muscle nuclei with the Igf2 gene found more than 100 kb away, suggesting that chromatin looping or sliding to bring the enhancer in proximity to Igf2 promoters is also an early event in skeletal muscle differentiation. Because additional studies show that a ∼250-bp component of this element can stimulate the transcriptional activity of Igf2 promoter 3 in differentiating myoblasts but not in mesenchymal progenitor cells, our results in aggregate indicate that we have identified a bona fide distal transcriptional enhancer that supports Igf2 gene activation in skeletal muscle cells. Because this segment of DNA is conserved in an analogous location in the human IGF2-H19 locus on chromosome 11p15.5, our results further suggest that its muscle enhancer function also is conserved among different mammalian species.
Studies revealing parent-of-origin-specific fetal growth impairments in heterozygous knock-out mice first identified Igf2 as an imprinted gene (42). Subsequent genetic manipulations led to the characterization and mapping of the larger imprinted Igf2-H19 locus (43–46), defined the existence of an ICR in the intergenic region between Igf2 and H19 (47, 48), and showed how binding of CTCF is able to establish a chromatin boundary to control the transcriptional activity of each gene (Refs. 47 and 48; also reviewed in Refs. 16 and 17). Other experiments, employing transgenic mice carrying parts of the Igf2-H19 locus or using chromatin conformation and capture assays, were able to identify the existence of distal enhancers with tissue-limited properties (28, 35–37), but these studies have not progressed beyond initial descriptions. For example, the ∼4.3-kb region D first identified in transgenic mice (Fig. 3 and Ref. 36) was found by chromatin conformation and capture assay to associate with the Igf2 gene in mouse muscle tissue (28), and was shown to stimulate activity of the H19 promoter in cultured muscle cells (33), but was not studied in any detail with Igf2 promoters, and was not recognized to overlap in DNA sequence with CS9 (Fig. 3), which itself was first identified through analyses in transgenic mice (35).
The three Igf2 promoters all appear to function in the embryo and fetus (14), although the contribution of each to IGF2 mRNA production in different developing tissues has not been established. In muscle cells in culture, only promoter 3 appears to be active, with significant induction occurring during differentiation, and the mechanisms by which the distal enhancer selectively targets just this promoter remain to be elucidated. It also is unknown whether the enhancer plays a role in Igf2 gene activation during muscle regeneration and whether it primarily targets promoter 3, because in postnatal gastrocnemius muscle in vivo under normal growth conditions, promoter 1 also is functional (Fig. 2). Similarly, the individual components of the enhancer remain to be dissected, and the critical transcription factors need to be identified. Our initial studies using reporter gene experiments suggest that at least one E-box within the ∼250-bp minimal element is necessary for full activity of the enhancer but also show that other regions appear to be required to stimulate promoter function during muscle differentiation (Fig. 7).
Recent whole genome analyses of the E-box-binding transcription factor, MyoD, which plays a central role in specifying myoblast fate, in initiating muscle differentiation, and in facilitating muscle regeneration (reviewed in Refs. 49 and 50) have found that it can bind in chromatin to many E-box sequences (CANNTG, where N = G, A, T, or C; preferred sites are CAGCTG and CACCTG (51)), including those that are far from muscle genes (51). Within the D3-CS9 segment of mouse chromatin downstream of H19 that encompasses the muscle enhancer (Fig. 5), there are at least six E-boxes, including two that match a preferred sequence, CAGCTG. Both of these latter DNA segments, plus another E-box within the D3-CS9 overlapping segment that when mutated reduces functional enhancer activity (Fig. 7C), bind MyoD in chromatin in confluent and differentiated C2C12 myoblasts and in differentiated myotubes derived from mouse myoblasts in primary culture, as seen in results found in the NCBI sequence read archive (accession number SRP001761). Thus, based on these data, MyoD may be one of the transcription factors that can regulate the distal muscle enhancer.
Other critical transcriptional and chromatin modifying factors that participate in the actions of the distal enhancer to stimulate Igf2 gene transcription during muscle differentiation remain to be identified. Of particular interest will be the chromatin-associated proteins that mediate the long distance transfer of information from the enhancer to promote Igf2 gene activation in differentiating myoblasts. Because the distal enhancer characterized here as a tissue-limited activator of Igf2 gene transcription also has been found to associate in muscle with the H19 promoter located ∼27 kb away (28) and because the entire Igf2-H19 locus is subject to parental imprinting (reviewed in Refs. 16 and 17), of equivalent interest will be the detailed biochemical mechanisms by which the ICR and CTCF differentially direct specific chromatin loops containing the enhancer toward the H19 or Igf2 gene promoters.
Excessive signaling through the IGF1 receptor has been associated with several negative outcomes, including increased cancer risk (52) and accelerated aging and tissue senescence (53, 54). At the same time, IGF therapy has been proposed for muscle disorders and to counteract sarcopenia associated with aging and chronic disease (5, 55). It is clear that better understanding of the regulation and actions of this potent growth factor system in muscle or in other tissues is needed to ensure optimal therapeutic benefit.
Supplementary Material
Acknowledgments
We thank members of our laboratory for advice during the course of these studies.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK42748-21 (to P. R.) and Training Grants T32 DK007680 and T32 DK007674 (to D. T. A.). This work was also supported by a predoctoral fellowship from the American Heart Association (to S. F. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- IGF
- insulin-like growth factor
- ICR
- imprinting control region
- Ad-MyoD
- recombinant adenovirus for MyoD
- Ad-β-Gal
- recombinant adenovirus for β-galactosidase
- DM
- differentiation medium.
REFERENCES
- 1.Lassar A., Münsterberg A. (1994) Curr. Opin. Cell Biol. 6, 432–442 [DOI] [PubMed] [Google Scholar]
- 2.Naya F. J., Olson E. (1999) Curr. Opin Cell Biol. 11, 683–688 [DOI] [PubMed] [Google Scholar]
- 3.McKinsey T. A., Zhang C. L., Olson E. N. (2002) Trends Biochem. Sci 27, 40–47 [DOI] [PubMed] [Google Scholar]
- 4.Palacios D., Puri P. L. (2006) J. Cell. Physiol. 207, 1–11 [DOI] [PubMed] [Google Scholar]
- 5.Glass D. J. (2003) Nat. Cell Biol. 5, 87–90 [DOI] [PubMed] [Google Scholar]
- 6.Rotwein P. (2003) Growth Horm. IGF Res. 13, 303–305 [DOI] [PubMed] [Google Scholar]
- 7.Nakae J., Kido Y., Accili D. (2001) Endocr. Rev. 22, 818–835 [DOI] [PubMed] [Google Scholar]
- 8.Liu J. P., Baker J., Perkins A. S., Robertson E. J., Efstratiadis A. (1993) Cell 75, 59–72 [PubMed] [Google Scholar]
- 9.Barton-Davis E. R., Shoturma D. I., Musaro A., Rosenthal N., Sweeney H. L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15603–15607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paul A. C., Rosenthal N. (2002) J. Cell Biol. 156, 751–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barton E. R., Morris L., Musaro A., Rosenthal N., Sweeney H. L. (2002) J. Cell Biol. 157, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Laere A. S., Nguyen M., Braunschweig M., Nezer C., Collette C., Moreau L., Archibald A. L., Haley C. S., Buys N., Tally M., Andersson G., Georges M., Andersson L. (2003) Nature 425, 832–836 [DOI] [PubMed] [Google Scholar]
- 13.Edwards C. A., Ferguson-Smith A. C. (2007) Curr. Opin. Cell Biol. 19, 281–289 [DOI] [PubMed] [Google Scholar]
- 14.Rotwein P., Hall L. J. (1990) DNA Cell Biol. 9, 725–735 [DOI] [PubMed] [Google Scholar]
- 15.Rotwein P. (1999) in The IGF System (Rosenfeld R. G., Roberts C. T., Jr., eds) pp. 19–35, Humana Press, Totowa, NJ [Google Scholar]
- 16.Wallace J. A., Felsenfeld G. (2007) Curr. Opin. Genet. Dev. 17, 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips J. E., Corces V. G. (2009) Cell 137, 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kou K., Rotwein P. (1993) Mol. Endocrinol. 7, 291–302 [DOI] [PubMed] [Google Scholar]
- 19.Wilson E. M., Hsieh M. M., Rotwein P. (2003) J. Biol. Chem. 278, 41109–41113 [DOI] [PubMed] [Google Scholar]
- 20.Florini J. R., Magri K. A., Ewton D. Z., James P. L., Grindstaff K., Rotwein P. S. (1991) J. Biol. Chem. 266, 15917–15923 [PubMed] [Google Scholar]
- 21.Wilson E. M., Rotwein P. (2006) J. Biol. Chem. 281, 29962–29971 [DOI] [PubMed] [Google Scholar]
- 22.Stewart C. E., James P. L., Fant M. E., Rotwein P. (1996) J. Cell. Physiol. 169, 23–32 [DOI] [PubMed] [Google Scholar]
- 23.Markljung E., Jiang L., Jaffe J. D., Mikkelsen T. S., Wallerman O., Larhammar M., Zhang X., Wang L., Saenz-Vash V., Gnirke A., Lindroth A. M., Barrés R., Yan J., Strömberg S., De S., Pontén F., Lander E. S., Carr S. A., Zierath J. R., Kullander K., Wadelius C., Lindblad-Toh K., Andersson G., Hjälm G., Andersson L. (2009) PLoS Biol. 7, e1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butter F., Kappei D., Buchholz F., Vermeulen M., Mann M. (2010) EMBO Rep. 11, 305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson E. M., Tureckova J., Rotwein P. (2004) Mol. Biol. Cell 15, 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling J. Q., Li T., Hu J. F., Vu T. H., Chen H. L., Qiu X. W., Cherry A. M., Hoffman A. R. (2006) Science 312, 269–272 [DOI] [PubMed] [Google Scholar]
- 27.Kurukuti S., Tiwari V. K., Tavoosidana G., Pugacheva E., Murrell A., Zhao Z., Lobanenkov V., Reik W., Ohlsson R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10684–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon Y. S., Jeong S., Rong Q., Park K. Y., Chung J. H., Pfeifer K. (2007) Mol. Cell. Biol. 27, 3499–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 30.Wilson E. M., Rotwein P. (2007) J. Biol. Chem. 282, 5106–5110 [DOI] [PubMed] [Google Scholar]
- 31.Weintraub H. (1993) Cell 75, 1241–1244 [DOI] [PubMed] [Google Scholar]
- 32.DeVol D. L., Rotwein P., Sadow J. L., Novakofski J., Bechtel P. J. (1990) Am. J. Physiol. 259, E89–E95 [DOI] [PubMed] [Google Scholar]
- 33.Erbay E., Park I. H., Nuzzi P. D., Schoenherr C. J., Chen J. (2003) J. Cell Biol. 163, 931–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ainscough J. F., John R. M., Barton S. C., Surani M. A. (2000) Development 127, 3923–3930 [DOI] [PubMed] [Google Scholar]
- 35.Ishihara K., Hatano N., Furuumi H., Kato R., Iwaki T., Miura K., Jinno Y., Sasaki H. (2000) Genome Res. 10, 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaffer C. R., Srivastava M., Park K. Y., Ives E., Hsieh S., Batlle J., Grinberg A., Huang S. P., Pfeifer K. (2000) Genes Dev. 14, 1908–1919 [PMC free article] [PubMed] [Google Scholar]
- 37.Kaffer C. R., Grinberg A., Pfeifer K. (2001) Mol. Cell. Biol. 21, 8189–8196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bergstrom D. A., Penn B. H., Strand A., Perry R. L., Rudnicki M. A., Tapscott S. J. (2002) Mol. Cell 9, 587–600 [DOI] [PubMed] [Google Scholar]
- 39.Penn B. H., Bergstrom D. A., Dilworth F. J., Bengal E., Tapscott S. J. (2004) Genes Dev. 18, 2348–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simone C., Forcales S. V., Hill D. A., Imbalzano A. N., Latella L., Puri P. L. (2004) Nat. Genet. 36, 738–743 [DOI] [PubMed] [Google Scholar]
- 41.Serra C., Palacios D., Mozzetta C., Forcales S. V., Morantte I., Ripani M., Jones D. R., Du K., Jhala U. S., Simone C., Puri P. L. (2007) Mol. Cell 28, 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeChiara T. M., Robertson E. J., Efstratiadis A. (1991) Cell 64, 849–859 [DOI] [PubMed] [Google Scholar]
- 43.Bartolomei M. S., Zemel S., Tilghman S. M. (1991) Nature 351, 153–155 [DOI] [PubMed] [Google Scholar]
- 44.Leighton P. A., Ingram R. S., Eggenschwiler J., Efstratiadis A., Tilghman S. M. (1995) Nature 375, 34–39 [DOI] [PubMed] [Google Scholar]
- 45.Pfeifer K., Leighton P. A., Tilghman S. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13876–13883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webber A. L., Ingram R. S., Levorse J. M., Tilghman S. M. (1998) Nature 391, 711–715 [DOI] [PubMed] [Google Scholar]
- 47.Bell A. C., Felsenfeld G. (2000) Nature 405, 482–485 [DOI] [PubMed] [Google Scholar]
- 48.Hark A. T., Schoenherr C. J., Katz D. J., Ingram R. S., Levorse J. M., Tilghman S. M. (2000) Nature 405, 486–489 [DOI] [PubMed] [Google Scholar]
- 49.Berkes C. A., Tapscott S. J. (2005) Semin. Cell Dev. Biol. 16, 585–595 [DOI] [PubMed] [Google Scholar]
- 50.Tapscott S. J. (2005) Development 132, 2685–2695 [DOI] [PubMed] [Google Scholar]
- 51.Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G. J., Parker M. H., MacQuarrie K. L., Davison J., Morgan M. T., Ruzzo W. L., Gentleman R. C., Tapscott S. J. (2010) Dev. Cell 18, 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pollak M. (2008) Nat. Rev. Cancer 8, 915–928 [DOI] [PubMed] [Google Scholar]
- 53.Berryman D. E., Christiansen J. S., Johannsson G., Thorner M. O., Kopchick J. J. (2008) Growth Horm. IGF Res. 18, 455–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fontana L., Partridge L., Longo V. D. (2010) Science 328, 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoffman E. P., Nader G. A. (2004) Nat. Med. 10, 584–585 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.