Abstract
Thrombospondin-1 (TSP1) can inhibit angiogenic responses directly by interacting with VEGF and indirectly by engaging several endothelial cell TSP1 receptors. We now describe a more potent mechanism by which TSP1 inhibits VEGF receptor-2 (VEGFR2) activation through engaging its receptor CD47. CD47 ligation is known to inhibit downstream signaling targets of VEGFR2, including endothelial nitric-oxide synthase and soluble guanylate cyclase, but direct effects on VEGFR2 have not been examined. Based on FRET and co-immunoprecipitation, CD47 constitutively associated with VEGFR2. Ligation of CD47 by TSP1 abolished resonance energy transfer with VEGFR2 and inhibited phosphorylation of VEGFR2 and its downstream target Akt without inhibiting VEGF binding to VEGFR2. The inhibitory activity of TSP1 in large vessel and microvascular endothelial cells was replicated by a recombinant domain of the protein containing its CD47-binding site and by a CD47-binding peptide derived from this domain but not by the CD36-binding domain of TSP1. Inhibition of VEGFR2 phosphorylation was lost when CD47 expression was suppressed in human endothelial cells and in murine CD47-null cells. These results reveal that anti-angiogenic signaling through CD47 is highly redundant and extends beyond inhibition of nitric oxide signaling to global inhibition of VEGFR2 signaling.
Keywords: Cell-surface Receptor, Endothelium, Fluorescence Resonance Energy Transfer (FRET), Receptor Tyrosine Kinase, Thrombospondin, CD47, Angiogenesis
Introduction
VEGF-A is essential for developmental angiogenesis and plays important roles in adult animals to control vascular permeability and homeostasis, blood pressure, and pathological angiogenesis associated with wound healing and cancer (1–4). Based on its major role as an angiogenic factor and autocrine growth factor for many cancers, therapeutic angiogenesis inhibitors that sequester VEGF or inhibit signaling through VEGF receptor-2 (VEGFR2)2/KDR and extend survival of some cancer patients have been developed for clinical use (5, 6). VEGF binding to VEGFR2 and its coreceptor neuropilin-1 on endothelial cells activates the tyrosine kinase activity of VEGFR2 and initiates several signaling pathways that promote endothelial cell survival, motility, and proliferation and release of nitric oxide (2).
Physiological VEGF/VEGFR2 signaling is regulated by endogenous angiogenesis inhibitors (7). These are typically secreted proteins that can act by binding and sequestering VEGF or by engaging cell-surface receptors that convey inhibitory signals downstream of VEGFR2. The first identified endogenous angiogenesis inhibitor was thrombospondin-1 (TSP1) (8–10). TSP1 inhibits angiogenic responses stimulated by several factors, including FGF2, lysophosphatidic acid, and VEGF.
Several mechanisms have been proposed to account for inhibition of VEGF-mediated angiogenesis by TSP1 (11). The scavenger receptor CD36 is required for TSP1 to inhibit FGF2-stimulated endothelial migration in vitro and angiogenesis in the corneal assay (12, 13). CD36 ligation induces Fyn phosphorylation and p38 activity, leading to increased endothelial cell apoptosis (13). CD36 is also a fatty acid translocase, and TSP1 binding to CD36 inhibits uptake of myristate into endothelial cells and myristate-stimulated activation of Src kinases and cGMP signaling (14). This provides a mechanism for TSP1 to inhibit the previously reported activation of endothelial NOS (eNOS) by exposure of endothelial cells to extracellular myristate (15). Recently, CD36 was reported to co-immunoprecipitate with VEGFR2, suggesting direct cross-talk between these two receptors (16). Although a recombinant CD36-binding domain of TSP1 at micromolar concentrations inhibited VEGF-stimulated VEGFR2 phosphorylation, differential permeability responses to VEGF in TSP1-null versus wild-type mice implied that endogenous TSP1 at high picomolar concentrations stimulates rather than inhibits VEGF signaling via this pathway.
At nanomolar concentrations, TSP1 can also inhibit VEGF signaling by directly binding to the protein and by competing with VEGF for binding to cell-surface heparan sulfate proteoglycans (17). These activities are mediated by the type 1 and 3 repeats of TSP1 and its N-terminal heparin-binding domain, respectively. A subsequent study revealed that direct binding of VEGF to TSP1 can further inhibit VEGF signaling by internalization of the VEGF-TSP1 complex via the TSP1 receptor LRP1 (LDL-related receptor protein-1) (18).
TSP1 also inhibits the Akt/eNOS/NO/cGMP/cGMP-dependent protein kinase cascade stimulated by VEGF (11, 19). At nanomolar TSP1 concentrations, this inhibition can be mediated by CD36 (14), but at physiological circulating plasma TSP1 concentrations (100–200 pm), CD47 is the necessary TSP1 receptor for inhibiting activation of soluble guanylate cyclase (20) or cGMP-dependent protein kinase (21). CD47-null and TSP1-null mice show similarly enhanced survival of full thickness skin grafts and increased angiogenic responses in the wound bed (22).
CD47 is the only TSP1 receptor that is known to mediate inhibition of VEGF signaling at physiological circulating TSP1 concentrations, but this inhibition has been documented only downstream of NO. However, VEGFR2 stimulates angiogenesis through several additional signaling pathways (2). Because endogenous TSP1 also limits phosphorylation of Akt and Src (which are upstream of the known CD47 targets) in retina (23), we further examined the effects of CD47 on upstream signaling through VEGFR2. We show here that CD47 constitutively associates with VEGFR2 and that ligation of CD47 by TSP1 or other CD47 ligands inhibits this association and VEGFR2 phosphorylation.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Human umbilical vein endothelial cells (HUVEC; Lonza) and bovine aortic endothelial cells (BAEC) (24) were cultured at 37 °C in 5% CO2 using EGM2 medium (Thermo Fisher Scientific Inc., Waltham, MA). Human dermal microvascular endothelial cells (HDMVEC) were generously provided by Dr. Frank Cuttitta (Angiogenesis Core Facility, NCI, National Institutes of Health). The TSP1-derived CD47-binding peptide 7N3 (FIRVVMYEGKK) and inactive control peptide 604 (FIRGGMYEGKK) were synthesized by Peptides International (Louisville, KY) (25). Human TSP1 was purified from the supernatant of thrombin-activated platelets obtained from the National Institutes of Health Blood Bank as described (26). Recombinant proteins containing the type 1 domains (TSP1 type 1 repeats; 3TSR) or C-terminal signature domains of TSP1 (residues 648–1170; E3CaG) were prepared as described previously and provided by Dr. Deane Mosher (University of Wisconsin) (27) and Dr. Jack Lawler (Harvard Medical School) (28). VEGF-165 was obtained from Open Biosystems.
A VEGFR2-mCherry plasmid encoding mCherry fused to the C terminus of human VEGFR2 was purchased from GeneCopoeia (Germantown, MD). CD47-GFP plasmid encoding an in-frame fusion of enhanced GFP at the C terminus of full-length human CD47 isoform 2 was obtained from Dr. Dennis E. Discher (University of Pennsylvania, Philadelphia, PA) (29). Alexa Fluor 594-conjugated donkey anti-rabbit IgG and Alexa Fluor 488-conjugated goat anti-mouse IgG antibodies were purchased from Invitrogen. Anti-VEGFR2 and anti-phospho-Tyr1175 VEGFR2 antibodies were obtained from Cell Signaling (Danvers, MA) or Upstate. Anti-CD47 antibody B6H12 was from Abcam, and H-100 was from Santa Cruz Biotechnology. Rabbit polyclonal antibodies against phospho-Ser473 Akt, phospho-Thr308 Akt, and Akt were from Cell Signaling.
Immunoprecipitation and VEGFR2 Phosphorylation
HUVEC, BAEC, or HDMVEC were plated at 1 × 106 cells in 10-cm2 plates. After achieving 80% confluence, the cells were serum-starved overnight in the presence of 0.1% BSA (Sigma). The cells were incubated with TSP1, 3TSR, or E3CaG for 20 min and then treated with VEGF for 5 min under each condition. Single-treatment controls were used in each assay. Cell lysates were made using either radioimmune precipitation assay buffer (50 mm Tris (pH 7.4), 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mm EGTA, and 1 mm NaF) or immunoprecipitation buffer (50 mm Tris-HCl, 150 mm NaCl, and 1% Nonidet P-40) along with 1× Complete Mini protease inhibitor mixture (Roche Applied Science) and Na3VO4. Cell lysates were centrifuged at 13,000 rpm for 15 min. A BCA assay (Pierce) was used to quantify total protein. Dynabeads (Invitrogen) were used for immunoprecipitation. The Dynabeads were washed three times with activation buffer. The cell lysates were incubated in Dynabeads-protein G along with anti-VEGFR2 antibody (1:500) and incubated for 2 h at 4 °C on a shaker. The beads were washed three times with lysate buffer and boiled at 95 °C for 5 min. The immunoprecipitated cell lysates were loaded on 4–12% NuPAGE gels (Invitrogen), and Western blotting was performed. For immunoprecipitation, primary antibody against phospho-VEGFR2 (1:1000; Cell Signaling) was used. Normalization of protein lysates used for Western blotting was performed with anti-β-actin (1:3000) or anti-VEGFR2 (1:1000) antibody.
Western Blotting
HUVEC and BAEC were seeded in EGM (Lonza) at 2 × 105 cells/well on 6-well plates. When 60% confluent, HUVEC and BAEC were serum-deprived for 24 and 18 h, respectively, before the addition of VEGF (20 ng/ml) and TSP1 (1 μg/ml), 7N3 (1 μm), or control peptide 604 (1 μm). After a 15-min incubation at 37 °C and 5% CO2 in EBM2 medium (Lonza) plus 0.1% BSA, cells were lysed at 4 °C in 1× SDS sample buffer. Cell lysates were boiled for 5 min, electrophoretically separated on 4–12% BisTris (HUVEC) and 7% Tris acetate (BAEC) NuPAGE gels (Invitrogen) for 1.5 h at 150 V, and transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore) for 2 h at 100 V. Membranes were blocked in 5% BSA, 0.1% Tween 20, and PBS and incubated overnight at 4 °C with primary antibody (1:1000) against phospho-Ser473 Akt, phospho-Thr308 Akt, phospho-VEGFR2, or VEGFR2. SuperSignal® West Dura extended duration chemiluminescent substrate (Pierce) was used for detection. Stripped membranes were reprobed with rabbit anti-Akt polyclonal antibody (1:1000) to confirm protein loading levels.
Western blots were quantified using the Bio-Rad Quantity 1.1 program. β-Actin was used as a loading control for all blots. Results are presented as the mean ± S.D. of three independent experiments.
Endogenous Immunostaining
HUVEC and HDMVEC were plated 10,000 cells/well in Lab-Tek II glass chamber cover slips, grown overnight, and serum-starved for 2 h in EBM2 medium. The cells were washed three times with PBS. The endothelial cells were fixed with 4% paraformaldehyde for 20 min at room temperature. The cells were washed with PBS three times for 5 min each. Cells were permeabilized with 0.25% Triton X-100 for 5 min. The fixed cells were washed 3 times with PBS for 5 min each. Cells were blocked with 1% BSA in 0.1% Tween 20 and PBS for 1 h at room temperature. The cells were incubated in a mixture of two primary antibodies (mouse anti-human CD47 (B6H12) and rabbit anti-human VEGFR2 (Upstate)) overnight at 4 °C. Primary anti-CD47 and anti-VEGFR2 antibodies alone were used as single controls. For HDMVEC, a primary anti-VEGFR2 antibody (Cell Signaling) was used. Secondary antibodies alone were used as controls for background. The cells were washed with PBS three times for 5 min each. The cells were incubated with a mixture of two secondary antibodies (Alexa Fluor 488 anti-rabbit and Alexa Fluor 594 anti-mouse) in 1% BSA for 1 h at room temperature in the dark. The cells were washed with PBS three times for 5 min each. The slides were mounted using VECTASHIELD mounting medium with DAPI (Vector Laboratories).
Transfection
HUVEC and NIH3T3 cells were plated overnight in either 6-well 10-cm2 plates or Lab-Tek chambers. The endothelial cells were transfected with CD47-GFP or VEGFR2-mCherry by using EndoFectin (GeneCopoeia) or Plus reagent and Lipofectamine (Invitrogen). The medium was replaced 3–5 h post-transfection. After 24–36 h, the cells were used for experiments. CD47 morpholino was transfected using the Amaxa Nucleofection kit (Lonza) in HUVEC. Transfection was performed according to protocol instructions. At 24–36 h post-transfection, the cells were harvested and used for experiments.
Confocal Microscopy
HUVEC were grown and transfected on Lab-Tek II chambers with GFP-tagged human CD47 along with VEGFR2-mCherry. Single CD47-GFP and VEGFR2-mCherry transfections were used as controls for each experiment. Cells were serum-starved for 2 h in EBM2 medium. Cells were treated with TSP1 (20 min) in the presence or absence of VEGF (5 min) at 37 °C in 5% CO2. Confocal images were sequentially acquired with Zeiss AIM software on a Zeiss LSM 510 confocal system with a Zeiss Axiovert 100 M inverted microscope and a 50-milliwatt argon UV laser tuned to 364 nm, a 25-milliwatt argon visible laser tuned to 488 nm, and a 1-milliwatt HeNe laser tuned to 543 nm. A Plan-Neofluar 63×/1.4 numerical aperture oil immersion objective was used at various digital zoom settings. Emission signals after sequential excitation were collected with a BP 385–470, BP 505–550, or LP 560 filter, respectively, using individual photomultipliers. Co-localization analyses of CD47-GFP and VEGFR2-mCherry proteins were performed with Zeiss AIM software (Version 4.2) with thresholds set based on the single-stained control image of each fluorophore acquired with the same confocal parameters as the double-stained sample.
FRET
CD47-GFP, VEGFR2-mCherry, and CD47 + VEGFR2 were transfected into HUVEC. Cells were serum-starved for 2 h in EBM2 medium. Cells were treated similarly with 7N3 and TSP1 in the presence or absence of VEGF, and FRET was analyzed. FRET analysis was performed using the Zeiss AIM macro for sensitized emission, where controls for background and donor alone (GFP) or acceptor alone (mCherry) were acquired with the same confocal settings as the acceptor plus donor samples. Fc-FRET and N-FRET algorithms were used to analyze the FRET data.
Cell Adhesion Assay
Changes in cell adhesion were quantified using an ACEA Biosciences instrument. HUVEC (20,000 cells/well) were incubated in 50 μl of EGM2 medium in duplicate overnight at 37 °C in 5% CO2. After 24 h, the medium was changed to basal medium containing 0.1% BSA for 20 min, and the cells were then treated with either TSP1, E3CaG, or 3TSR in the presence or absence of VEGF for 0–6 h. The impedance signal was normalized to 1 before treatment. Changes in cell spreading and/or proximity to the substrate were measured as an increase or a decrease in impedance and expressed normalized to the initial signal for each well as the cell index after treatment with respect to “untreated.” Untreated was taken as 1 in each replicate.
Cell Proliferation Assay
NIH3T3 cells were plated (≈1500 cells/well) in 96-well plates. VEGFR2-mCherry plasmid was transiently transfected in NIH3T3 cells using Plus reagent and Lipofectamine. Untransfected cells were used as negative controls for each experiment. At 3 h post-transfection, the medium was replaced with 0.5% FBS and TSP1 (2 nm), VEGF (30 ng/ml), and TSP1 (2 nm) + VEGF (30 ng/ml). Duplicate transfected plates were used for 0-h background. After 24–36 h, cell proliferation was measured using a CellTiter 96® AQueous One Solution cell proliferation assay (MTS) kit (Promega). Absorbance was measured at 492 nm after a 6-h incubation with MTS solution. Net proliferation was calculated by subtracting the 0-h absorbance values and normalizing to untransfected cells (= 100%).
VEGF Cell Binding Assay
VEGF was iodinated using IODO-GEN (Pierce) and the protocol suggested by the manufacturer, resulting in a specific activity of 15–25 mCi/μmol. Endothelial cells (1 × 106) were then added to tubes containing 10–15 fmol of 125I-VEGF with or without purified TSP1 in Dulbecco's PBS with Ca2+ and Mg2+ (Invitrogen) and 0.1% BSA. After incubation for 2 h at room temperature on a gyrotory shaker at 160 cycles/min, the cells were separated from the soluble material by centrifugation for 60 s through NyoSil M25 oil (Nye Lubricants, Fairhaven, MA). Oil and medium were aspirated, and 125I in the cell pellets was quantified directly. A 200-fold excess of unlabeled VEGF ligand was used to access nonspecific binding in the assay.
VEGF Binding to Immobilized TSP1
Immulon 2HB Removawells (Thermo, Franklin, MA) were coated with 50 μl of TSP1 (10 μg/ml) for 16–20 h at 4 °C. Nonspecific binding was blocked by incubating the wells with Dulbecco's PBS with Ca2+ and Mg2+ and 1% BSA for 2 h at room temperature. 10–15 fmol of 125I-VEGF was added in the presence or absence of competing protein or peptide and incubated at room temperature for 2 h. The liquid was then aspirated, the wells were rinsed twice with Dulbecco's PBS/BSA, and individual wells were quantified directly. A 200-fold excess of unlabeled VEGF ligand was used to access nonspecific binding in the assay.
Statistical Analysis
Three independent experiments were performed, and Student's t test was used for p values. Values <0.05 were considered statistically significance.
RESULTS
TSP1 Inhibits VEGFR2 Phosphorylation Independently of VEGF Sequestration
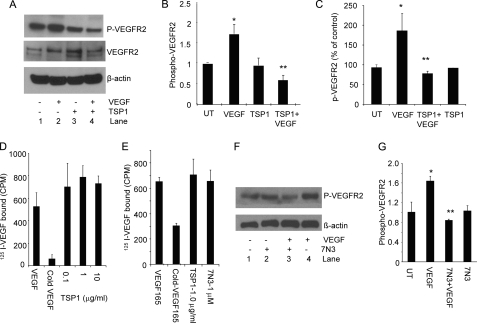
A preliminary time course using BAEC treated with 30 ng/ml VEGF demonstrated maximal VEGFR2 phosphorylation at 5 min (supplemental Fig. S1A), which was used for all subsequent experiments except where indicated. In addition to inhibiting VEGF signaling through its receptors CD36 and CD47, TSP1 is reported to directly sequester VEGF and thereby inhibit VEGFR2 phosphorylation (17, 18, 30). Consistent with these studies, VEGF-stimulated VEGFR2 phosphorylation at Tyr1175 in serum-starved BAEC was inhibited in a dose-dependent manner when the cells were pretreated for 20 min with 0.2–2 nm TSP1 (supplemental Fig. S1B). Based on three independent experiments, treatment with 1 μg/ml (2.2 nm) TSP1 inhibited VEGF-stimulated VEGFR2 phosphorylation by 3-fold relative to VEGF-stimulated cells (p = 0.0001) (Fig. 1, A and B). TSP1 alone did not significantly alter VEGFR2 phosphorylation at the same concentration. TSP1 at 2.2 nm similarly inhibited VEGF-induced VEGFR2 phosphorylation in HDMVEC (Fig. 1C).
FIGURE 1.
TSP1 and a CD47-binding peptide inhibit VEGFR2 signaling without inhibiting VEGF binding. A, TSP1 suppresses VEGFR2 phosphorylation. A representative Western blot is shown using anti-Tyr1175 VEGFR2 and anti-VEGFR2 antibodies with lysates from untreated BAEC (lane 1) and BAEC stimulated with VEGF (30 ng/ml) for 5 min (lane 2), treated with TSP1 (2 nm) alone (lane 3), and pretreated with TSP1 (2 nm) followed by VEGF (30 ng/ml) for 5 min (lane 4). B, quantification of VEGFR2 phosphorylation in three independent experiments normalized to loading controls. p values were as follows: VEGF versus untreated (UT), 0.00007 (*); and VEGF versus TSP1 + VEGF, 0.0001 (**). C, inhibition of VEGFR2 phosphorylation by TSP1 in HDMVEC (quantification of two independent experiments). p values were as follows: untreated versus VEGF, 0.09 (*); VEGF versus TSP1 + VEGF, 0.07 (**); VEGF versus TSP1, 0.08; and untreated versus TSP1, 0.7. D, iodinated VEGF binding to BAEC was assessed in the presence of the indicated concentrations of TSP1 or in the presence of a 200-fold excess of unlabeled VEGF. E, TSP1 and 7N3 inhibition of VEGF binding to BAEC. F, inhibition of VEGFR2 phosphorylation by peptide 7N3. Lane 1, untreated; lane 2, 7N3 (1 μm) alone; lane 3, 7N3 (1 μm) and VEGF (30 ng/ml); lane 4, VEGF (30 ng/ml)-stimulated. G, quantification of three independent experiments. p values were as follows: untreated versus VEGF; 0.007 (*); and VEGF versus 7N3 + VEGF, 0.012 (**).
However, the same concentrations of TSP1 failed to significantly inhibit 125I-VEGF binding to BAEC (Fig. 1D). Specific binding was confirmed by displacement with unlabeled VEGF (Fig. 1, D and E). However, TSP1 did not inhibit VEGF binding to these cells even at a 10-fold higher concentration than that required to optimally inhibit VEGFR2 phosphorylation. TSP1 also failed to inhibit 125I-VEGF binding to HUVEC (data not shown). Consistent with these data, the Kd for direct binding of 125I-VEGF to immobilized TSP1 was found to be ∼10 nm (data not shown). Therefore, ≤2.2 nm TSP1 inhibits VEGFR2 activation independently of inhibiting VEGF binding to the cell surface.
Ligation of CD47 Is Sufficient to Inhibit VEGFR2 Phosphorylation
TSP1 inhibits endothelial cell NO/cGMP signaling via CD47 at ≥10 pm concentrations (31). To explore the possibility that TSP1 also regulates VEGFR2 phosphorylation via CD47, we first used the TSP1-derived CD47-binding peptide 7N3 (FIRVVMYEGKK). VEGF-stimulated VEGFR2 phosphorylation in serum-starved BAEC was inhibited by pretreatment for 5 min with 1 μm 7N3 (Fig. 1, F and G), a concentration shown previously to maximally inhibit NO-mediated activation of soluble guanylate cyclase via CD47 (20). 7N3 alone did not exhibit any change in phosphorylation. The control peptide 604 (FIRGGMYEGKK) did not inhibit VEGF-stimulated phosphorylation (data not shown).
As was found for TSP1, peptide 7N3 at the concentration used did not inhibit VEGF binding to BAEC (Fig. 1E). Therefore, ligation of CD47 is sufficient to suppress VEGF-stimulated VEGFR2 phosphorylation on the time scale that we observed inhibition by TSP1 and without preventing VEGF binding to the cells.
TSP1 Inhibits VEGFR2 Phosphorylation via Its CD47-binding Domain but Not Its CD36-binding Domain
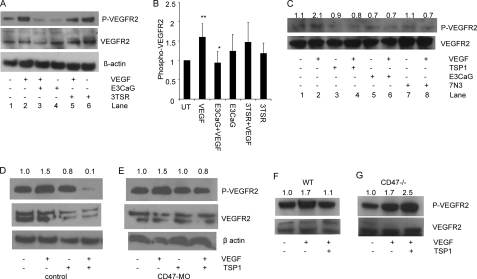
Although the binding and signaling activities of VVM peptides via CD47 are well documented, their role in the binding of intact TSP1 to CD47 remains unclear (32). To confirm that the CD47-binding domain of TSP1 is responsible for inhibition of VEGFR2 phosphorylation, we treated BAEC with a recombinant CD47-binding domain of TSP1 (E3CaG). After pretreatment with 2 nm E3CaG, VEGF-stimulated VEGFR2 phosphorylation was significantly reduced (p = 0.02) (Fig. 2, A and B). In contrast, pretreatment with a recombinant CD36-binding domain of TSP1 (3TSR) at the same concentration had no effect. We also increased the dose of 3TSR from 2 to 20 nm but did not observe a significant decrease in VEGFR2 phosphorylation (data not shown). From these data, we conclude that physiological concentrations of TSP1 regulate VEGFR2 phosphorylation via CD47 but not via CD36. Our results contrast with a previous report that ≥10 nm 3TSR inhibited VEGFR2 phosphorylation in HDMVEC (16). This may be due to the higher CD36 expression in microvascular cells. However, pretreatment of HDMVEC with TSP1 (2 nm), E3CaG (2 nm), or 7N3 (1 μm) similarly reduced VEGFR2 phosphorylation stimulated by VEGF to basal levels (Fig. 2C). Therefore, engagement of CD47 is sufficient to inhibit VEGFR2 phosphorylation even in microvascular endothelial cells that highly express the TSP1 receptor CD36.
FIGURE 2.
CD47 is necessary for inhibition of VEGFR2 signaling. A, TSP1 mediates suppression via its CD47-binding domain but not via its CD36-binding domain. Lane 1, untreated; lane 2, VEGF (30 ng/ml)-treated; lane 3, E3CaG (2 nm) + VEGF (30 ng/ml); lane 4, E3CaG (2 nm) alone; lane 5, 3TSR (2 nm) + VEGF (30 ng/ml); lane 6, 3TSR alone (2 nm). B, quantification of three independent experiments. p values were as follows: untreated (UT) versus VEGF; 0.01 (**); VEGF versus E3CaG + VEGF, 0.02 (*); and VEGF versus 3TSR + VEGF. 0.10. C, VEGFR2 Tyr1175 phosphorylation was assessed in unstimulated HDMVEC (lane 1) and in cells stimulated with VEGF (30 ng/ml) for 5 min (lane 2), treated with TSP1 (2 nm) alone (lane 3), pretreated with TSP1 (2 nm) and stimulated with VEGF (30 ng/ml) for 5 min (lane 4), treated with E3CaG (2 nm) alone (lane 5), pretreated with E3CaG and stimulated with VEGF for 5 min (lane 6), treated with peptide 7N3 (1 μm) alone (lane 7), and pretreated with peptide 7N3 and stimulated with VEGF for 5 min (lane 8). Numbers represent densitometry for phospho-VEGFR2/total VEGFR2 normalized to 1.0 for untreated cells. D and E, TSP1 does not suppress VEGFR2 phosphorylation after CD47 knockdown in HUVEC. D, control HUVEC (first lane) and HUVEC treated with VEGF (30 ng/ml) (second lane), TSP1 (2 nm) (third lane), TSP1 (2 nm) + VEGF (30 ng/ml) (fourth lane). E, CD47 morpholino (MO)-treated HUVEC left untreated (first lane) or treated with VEGF (30 ng/ml) (second lane), TSP1 (2 nm) (third lane), and TSP1 (2 nm) + VEGF (fourth lane). Numbers represent densitometric analysis of the phospho-VEGFR2 signal normalized to 1.0 for untreated cells. F and G, primary murine lung endothelial cells from wild-type and CD47-null mice were treated with VEGF in the absence or presence of TSP1 and analyzed by Western blotting for VEGFR2 phosphorylation. Numbers represent densitometric analysis of the phospho-VEGFR2 signal.
To confirm the role of CD47 in regulating VEGFR2 in endothelial cells, we decreased CD47 expression in HUVEC using a previously validated CD47 antisense morpholino (33). TSP1 inhibited VEGF-induced VEGFR2 phosphorylation in control morpholino-treated HUVEC (Fig. 2D), whereas inhibition of VEGF-induced phosphorylation was lost in cells treated with the CD47 morpholino (Fig. 2E). TSP1 also inhibited VEGF-stimulated phosphorylation of VEGFR2 in primary murine endothelial cells (Fig. 2F), but no inhibition was observed in equivalent primary cells from CD47-null mice (Fig. 2G). Therefore, CD47 is necessary for suppression of VEGFR2 phosphorylation by physiological levels of TSP1.
Akt Phosphorylation Downstream of VEGFR2 Is Inhibited by CD47 Ligation
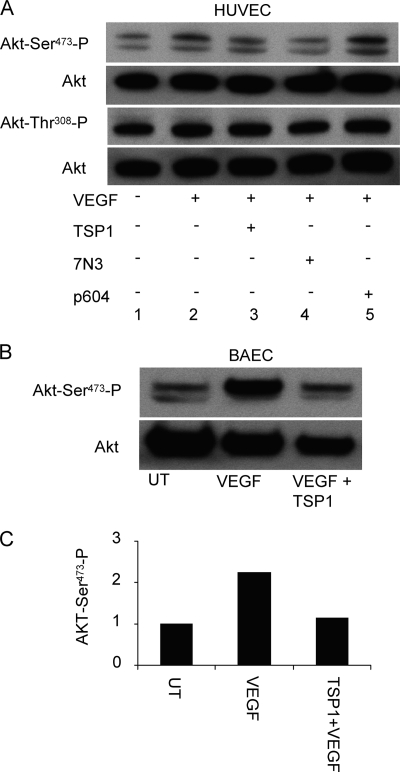
VEGF up-regulates NO synthesis in endothelial cells via Akt-mediated phosphorylation of eNOS at Ser1177 (34, 35). VEGF-induced migration of endothelial cells in turn depends upon phosphorylation of eNOS at Ser1177 (36). The effects of CD47 ligation on this upstream target of activated VEGFR2 were examined in BAEC and HUVEC. Endothelial cells treated with TSP1 (2 nm) or 7N3 (1 μm) in the presence or absence of VEGF (30 ng/ml) showed decreased Akt phosphorylation at Ser473 but not Thr308 in HUVEC (Fig. 3A). Similar inhibition of Ser473 phosphorylation by TSP1 was seen in BAEC (Fig. 3, B and C).
FIGURE 3.
CD47 ligation inhibits VEGF-induced activation of Akt. A, TSP1 regulates phosphorylation of Akt at Ser473 via CD47 in HUVEC. Lane 1, untreated cells; lane 2, VEGF-stimulated; lane 3, TSP1 (2 nm) + VEGF (30 ng/ml) treatment; lane 4, peptide 7N3 (1 μm) + VEGF (30 ng/ml) treatment; lane 5, control peptide 604 (1 μm) + VEGF (30 ng/ml) treatment. B, suppression of phosphorylation of Akt at Ser473 by TSP1 in BAEC. First lane 1, untreated (UT); second lane, VEGF-stimulated; third lane, TSP1/VEGF-treated. C, quantification of Akt phosphorylation in BAEC (one representative experiment).
Cell Adhesion Is Blocked by TSP1 via CD47
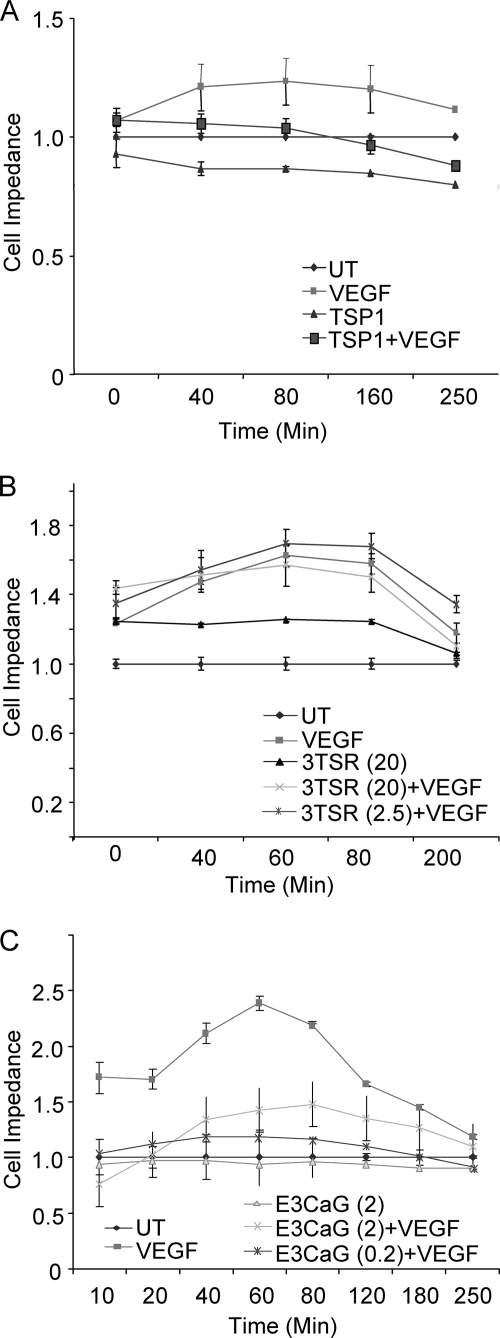
VEGF stimulates endothelial cell proliferation, migration, adhesion, and spreading (37–39). HUVEC transfected with GFP-actin and observed by confocal microscopy spread more rapidly over 20 min when treated with VEGF than when treated with 7N3 + VEGF (data not shown). To quantify the effect of TSP1 on VEGF-induced cell adhesion, we performed similar experiments using a quantitative real-time impedance-based assay. HUVEC were plated in duplicate in ACEA Biosciences plates and allowed to equilibrate for 24 h prior to treatment in the presence or absence of VEGF. In this assay, increased cell adhesion or spreading on the substrate increases impedance at this interface. VEGF treatment alone increased impedance across the cell layer as early as 5 min (Fig. 4A), presumably by increasing cell spreading on and/or proximity to the substrate. TSP1 suppressed the increase in impedance driven by VEGF. The VEGF-driven signal was also inhibited by E3CaG (0.2–2 nm) (Fig. 4C). In contrast, 3TSR (2.5–20 nm) did not inhibit the VEGF-stimulated response (Fig. 4B). Similarly, the CD47-binding peptide 7N3 (1 μm) was sufficient to inhibit VEGF-stimulated cell impedance (data not shown). Therefore, engagement of CD47 is sufficient to inhibit VEGF effects on endothelial cell adhesion.
FIGURE 4.
The CD47-binding domain of TSP1 inhibits VEGF-stimulated endothelial cell adhesion. A, changes in cell spreading and adhesive contact with the substrate were assessed by impedance and are presented as a normalized cell index. Cells were treated with VEGF (30 ng/ml) in the absence or presence of TSP1 (2 nm), and impedance was quantified for 4 h. B, cells were treated with VEGF in the presence of the indicated concentrations of the recombinant CD36-binding domain of TSP1 (3TSR; 2.5–20 nm). C, cells were stimulated with VEGF in the presence of the indicated concentrations of the recombinant CD47-binding domain of TSP1 (E3CaG; 0.2–2 nm). UT, untreated.
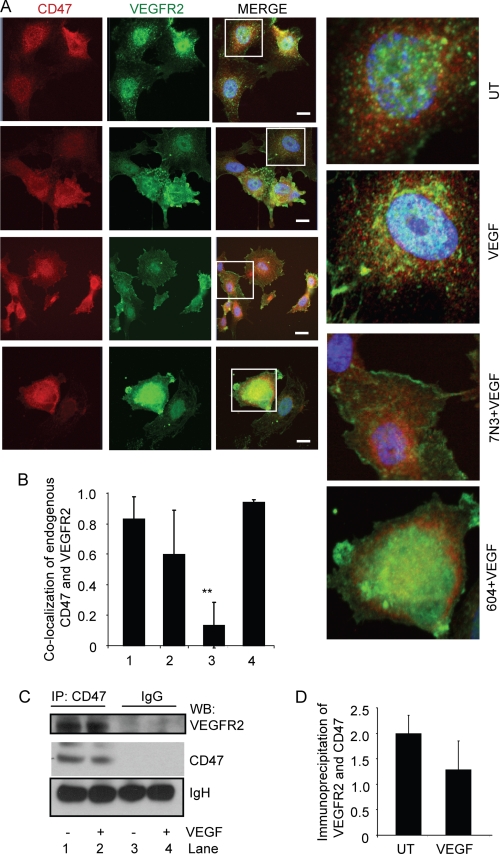
VEGFR2 and CD47 Interaction and Co-localization
To account for the suppression of VEGF-induced VEGFR2 phosphorylation, Akt phosphorylation, and cell adhesion, we hypothesized that CD47 may interact with VEGFR2. Based on immunostaining of endogenous CD47 and VEGFR2 in HUVEC, CD47 and VEGFR2 were partially co-localized in untreated cells (Fig. 5, A and B). The limited co-localization is consistent with the greater relative abundance of CD47 and its known lateral interactions with other membrane proteins such as integrins (40). Treating the cells with VEGF (30 ng/ml) did not significantly alter the co-localization of endogenous CD47 and VEGFR2. However, treatment with peptide 7N3 but not the control peptide 604 in the presence VEGF reduced the co-localization of CD47 and VEGFR2 (Fig. 5, A and B, and supplemental Fig. S2). We further addressed this potential interaction by immunoprecipitation of CD47. VEGFR2 was coprecipitated by an anti-CD47 antibody but not by an isotype-matched control IgG (Fig. 5C). Consistent with the results obtained for immunolocalization, treatment of cells with VEGF did not significantly inhibit co-immunoprecipitation of VEGFR2 with CD47 (Fig. 5, C and D). CD47 and VEGFR2 also co-localized in untreated HDMVEC (supplemental Fig. S3, A and B). As in large vessel endothelial cells, HDMVEC pretreated with TSP1 and stimulated with VEGF (30 ng/ml) showed a significant decrease in co-localization (p = 0.001) (supplemental Fig. S3). TSP1 and VEGF alone did not alter CD47 and VEGFR2 co-localization.
FIGURE 5.
Endogenous CD47 interacts with VEGFR2. A, untreated (UT) HUVEC or HUVEC treated for 5 min with 30 ng/ml VEGF alone or following a 5-min pretreatment with 1 μm CD47-binding peptide 7N3 or control peptide 604. The cells were fixed 5 min after the addition of VEGF and stained with antibodies to visualize endogenous CD47 (red) and VEGFR2 (green). Scale bars = 10 μm. B, quantitative analysis of co-localization. Bar 1, untreated HUVEC; bar 2, HUVEC stimulated with VEGF (30 ng/ml) for 5 min; bar 3, HUVEC simulated with peptide 7N3 (1 μm) + VEGF (30 ng/ml) p < 0.05 (**); bar 4, HUVEC simulated with VEGF (30 ng/ml) + control peptide 604 (1 μm). C, co-immunoprecipitation of endogenous VEGFR2 with CD47 in HUVEC. Lanes 1 and 3, untreated; lanes 2 and 4, VEGF-treated. Lanes 1 and 2 were immunoprecipitated (IP) using an anti-CD47 antibody, and lanes 3 and 4 were immunoprecipitated using control IgG. A representative blot is shown. D, quantification of three independent Western blots (WB).
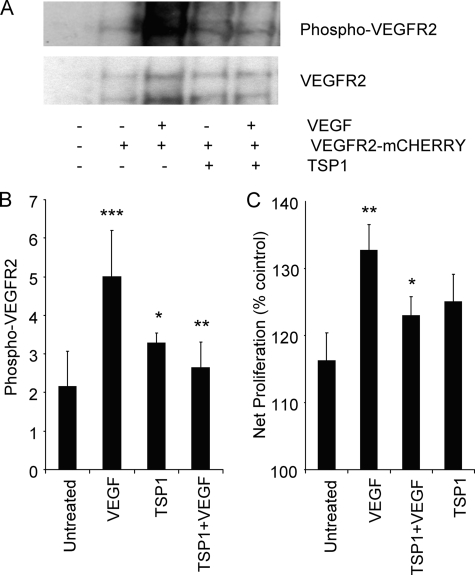
TSP1 Inhibits VEGF-induced Phosphorylation of VEGFR2-mCherry and Proliferation Responses
To further examine interactions between CD47 and VEGFR2, we utilized fusions of these receptors with fluorescent proteins. To validate that VEGFR2-mCherry replicates the signaling responses of endogenous VEGFR2, we transiently expressed the protein in NIH3T3 cells. After 24 h, the cells were serum-starved for 2 h and treated with VEGF (30 ng/ml) for 5 min alone or after pretreatment with TSP1 (2 nm). VEGF strongly stimulated phosphorylation of VEGFR2-mCherry, and this was blocked in cells treated with TSP1 (Fig. 6, A and B). 3T3 cells transfected with VEGFR2-mCherry showed enhanced proliferation to VEGF. In minimal medium with 0.5% FBS, transfected cells showed optimal proliferative responses to exogenous VEGF (data not shown). Under these conditions, the addition of TSP1 inhibited VEGF-stimulated proliferation of 3T3 cells transiently expressing VEGFR2-mCherry (Fig. 6C). Therefore, VEGF-mCherry is a functional Tyr kinase signaling receptor and responds to both VEGF and TSP1 in a manner similar to endogenous VEGFR2 in endothelial cells.
FIGURE 6.
Functional analysis of VEGFR2-mCherry expressed in NIH3T3 cells. A, phosphorylation of VEGFR2-mCherry in response to VEGF and inhibition by TSP1. A representative Western blot is shown for untransfected 3T3 cells (first lane); untreated 3T3 cells expressing VEGFR2-mCherry (second lane); and 3T3 cells expressing VEGFR2-mCherry treated with VEGF (30 ng/ml) (third lane), treated with TSP1 (2 nm) alone (fourth lane), and pretreated with TSP1 (2 nm) followed by VEGF (30 ng/ml) (fifth lane). B, quantification of Western blots from three independent experiments. p values were as follows: untransfected versus transfected and untreated; 0.1; untreated versus VEGF, 0.006 (***); VEGF versus TSP1 + VEGF, 0.02 (**); VEGF versus TSP1, 0.07 (*); and untreated versus TSP1 + VEGF, 0.07. C, TSP1 inhibits VEGF-induced cell proliferation of NIH3T3 cells expressing VEGFR2-mCherry p < 0.05 (*). Cell proliferation was quantified using MTS reagent (n = 3).
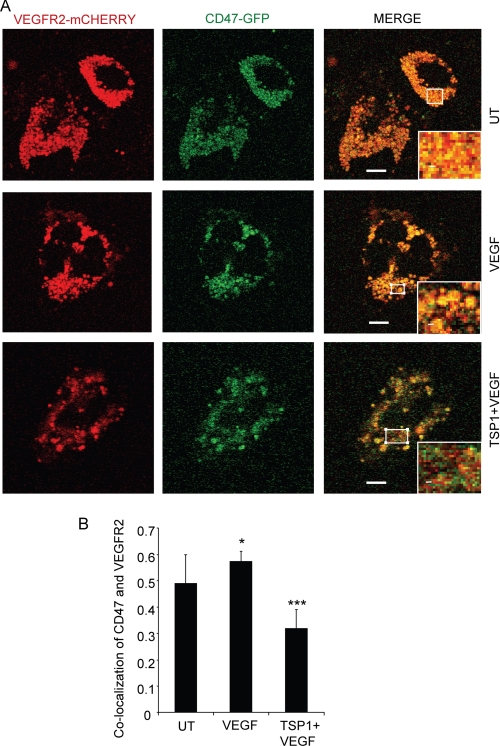
When transiently coexpressed in HUVEC, CD47-GFP and VEGFR2-mCherry fusion proteins were highly co-localized (Fig. 7A, upper panels). Treatment with VEGF for 5 min modestly increased the co-localization in live cells (Fig. 7A, middle panels, and B). However, pretreatment with TSP1 (2 nm) significantly reduced the co-localization of CD47 and VEGFR2 (Fig. 7A, lower panels, and B).
FIGURE 7.
TSP1 inhibits co-localization of CD47-GFP and VEGFR2-mCherry in live endothelial cells. A, CD47 and VEGFR2 co-localization is inhibited by TSP1. HUVEC were transiently transfected with plasmids encoding VEGFR2-mCherry and CD47-GFP and preincubated as indicated with 1 μg/ml TSP1 (2.2 nm) for 20 min before treatment for 5 min with 30 ng/ml VEGF. Images were acquired using live cells. Red, VEGFR2-mCherry; green, CD47-GFP. B, quantification of CD47 and VEGFR2 co-localization. p value were as follows: untreated (UT) versus VEGF, 0.1; untreated versus TSP1 + VEGF, 0.07 (*); and VEGF versus TSP1 + VEGF, 0.0001 (***). Scale bars = 10 μm.
FRET between CD47 and VEGFR2
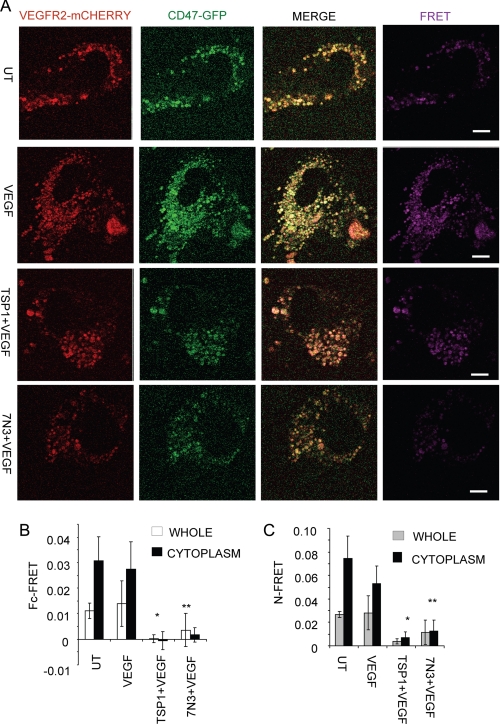
To assess a potential proximal interaction of CD47 with VEGFR2, HUVEC were transiently transfected with CD47-GFP and VEGFR2-mCherry and treated with TSP1 or 7N3 in the presence of VEGF, and FRET was performed by sensitized emission (Fig. 8A). Whole cell FRET analysis of untreated and VEGF-treated cells showed high FRET values (>0.01), but FRET was decreased or lost when cells were treated with TSP1 or 7N3 (0.0002 and 0.003, respectively).
FIGURE 8.
CD47 ligation inhibits FRET between CD47 and VEGFR2. A, representative FRET images between CD47 and VEGFR2. HUVEC were transiently transfected to express VEGFR2-mCherry (red) and CD47-GFP (green). Cells were pretreated with 1 μg/ml (2.2 nm) TSP1 as indicated for 20 min before the addition of VEGF. Live cells were imaged 5 min after the addition of VEGF. Right panels (purple), FRET. UT, untreated. B, quantification of whole cells and cytoplasm by Fc-FRET. C, quantification of whole cells and cytoplasm by N-FRET. p values for Fc-FRET were as follows: VEGF versus TSP1 + VEGF, 0.00050 (*); and VEGF versus peptide 7N3 + VEGF, 0.000013 (**). p values for N-FRET were as follows: VEGF versus TSP1 + VEGF, 0.00017 (*); and VEGF versus peptide 7N3 + VEGF, 0.000036 (**). Scale bars = 10 μm.
FRET quantification using either Fc-FRET (Fig. 8B) (41) or N-FRET (Fig. 8C) (42) over the entire cell or restricted to the cell cytoplasm showed similar differences. The cytoplasm of untreated cells showed slightly higher FRET (0.030) compared with VEGF-treated cells (0.027 by Fc-FRET). This is consistent with the immunostaining and co-immunoprecipitation experiments. However, treatment with TSP1 or 7N3 in the presence of VEGF dramatically decreased FRET (−0.0006 and 0.001, respectively, by Fc-FRET) (Fig. 8B). Analysis by N-FRET confirmed the conclusions from Fc-FRET (Fig. 8C). There was no decrease in FRET between no treatment and 7N3 peptide treatment alone (supplemental Fig. S4).
DISCUSSION
Although TSP1 has been recognized as an endogenous angiogenesis inhibitor for 20 years, the receptors and molecular mechanisms that mediate this activity remain controversial (11, 23, 43). Our data reveal a new mechanism by which TSP1 can regulate, through its receptor CD47, VEGF signaling in endothelial cells by controlling the activation of VEGFR2. Previously, the known mechanisms through which CD47 inhibits VEGFR2 signaling were limited to the downstream NO/cGMP cascade (11), which is only one branch of the VEGFR2 signaling network (2). The mechanism described here more globally inhibits this network by preventing VEGFR2 autophosphorylation at Tyr1175, which is critical for activation of downstream signaling through this receptor (44). We also identified CD47 as a novel VEGFR2-associated protein. The strong FRET signal between CD47-GFP and VEGFR2-mCherry suggests that CD47 binds directly to VEGFR2. Energy transfer by the Förster mechanism is generally limited to <10 nm, so even if another protein such as an integrin mediates the interaction, VEGFR2 and CD47 must be closely localized in this complex. Notably, ligation of CD47 by TSP1 or a CD47-binding peptide abolishes the close association of CD47 with VEGFR2. This is reminiscent of the dissociation of SHPS-1/SIRPα from its complex with CD47 in smooth muscle cells by comparable concentrations of TSP1 and by a related CD47-binding peptide (45).
Several receptors of TSP1 have been implicated in its anti-angiogenic activity (11, 31, 46), but their functions may be distinct. CD36 is necessary for TSP1 to inhibit FGF2-stimulated angiogenesis in rat cornea (13), but it is not clear that this requirement extends to VEGF-stimulated angiogenesis. Ligation of CD36 modulates VEGF signaling, and CD36 was recently found to co-immunoprecipitate in a complex with β1 integrins and VEGFR2 (16). TSP1 limits microvascular survival in retinal hyperoxia, which correlates with CD36 association with Src versus Fyn and antagonism of Akt survival signaling (23). β1 integrins were also implicated in the anti-angiogenic activity of TSP1 in conjunction with CD36 (47). However, engagement of α3β1 or α4β1 integrins by the N-terminal domain of TSP1 stimulates rather than inhibits angiogenesis in the chick chorioallantoic membrane assay (24, 48). CD36 can also activate Src kinases via its fatty acid translocase activity, and this is inhibited by TSP1 (14). However, none of these CD36-dependent activities are known to occur at physiological concentrations of TSP1, and CD36 is not sufficient for the effects of TSP1 on VEGFR2 signaling shown here because they are absent in CD47-null or knockdown endothelial cells that presumably express unchanged levels of CD36.
The present data indicate that inhibition of VEGF signaling through CD47 is highly redundant. Ligation of CD47 by physiological concentrations of TSP1 inhibits VEGF-induced cGMP synthesis in endothelial cells (19, 20) eNOS, soluble guanylate cyclase, and cGMP-dependent protein kinase are known targets of this CD47 signal (49, 50). Direct inhibition of VEGFR2 activation adds a much broader control of this angiogenic pathway via CD47 and is consistent with the elevated Akt phosphorylation reported in retinas of TSP1-null mice (23). Akt phosphorylation is inhibited by engagement of CD47, which, in addition to controlling activation of eNOS, controls a broad range of prosurvival signaling in endothelial cells (2, 51). This suggests that inhibitors of CD47/VEGFR2 interactions would be potent angiogenesis inhibitors.
Regulation of VEGFR2 activation appears to involve a proximal interaction between CD47 and VEGFR2. This is supported by co-immunoprecipitation and immunostaining but more strongly by FRET between CD47-GFP and VEGFR2-mCherry. Interestingly, this association is maintained when VEGF binds to VEGFR2, but ligation of CD47 by TSP1 or a small peptide derived from TSP1 leads to a significant decrease in this association.
The detailed mechanism by which TSP1/CD47 signaling controls the association between CD47 and VEGFR2 and VEGFR2 phosphorylation remains to be defined. Future studies will examine whether TSP1 or CD47 alters the trafficking or stability of VEGFR2. Recently, VEGF stimulation was reported to induce proteolytic processing of VEGFR2 (52). Future studies will also focus on whether CD47 associates differentially with these processed forms of VEGFR2. Finally, the effects of CD47 ligation on the known associations of VEGFR2 with VE-cadherin, integrins, IQGAP1, and CD36 should be further examined (53–55).
Supplementary Material
Acknowledgments
We thank Drs. Deane Mosher, Frank Cuttitta, Dennis Discher, and Jack Lawler for providing reagents.
This work was supported, in whole or in part, by the Intramural Research Program of the National Institutes of Health/NCI (to D. D. R.) and by National Institutes of Health Grant CA128616 (to J. S. I.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- VEGFR2
- VEGF receptor-2
- TSP1
- thrombospondin-1
- eNOS
- endothelial NOS
- HUVEC
- human umbilical vein endothelial cell(s)
- BAEC
- bovine aortic endothelial cell(s)
- HDMVEC
- human dermal microvascular endothelial cell(s)
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tretrazolium (inner salt).
REFERENCES
- 1.Ku D. D., Zaleski J. K., Liu S., Brock T. A. (1993) Am. J. Physiol. 265, H586–H592 [DOI] [PubMed] [Google Scholar]
- 2.Shibuya M. (2006) J. Biochem. Mol. Biol. 39, 469–478 [DOI] [PubMed] [Google Scholar]
- 3.Lee S., Chen T. T., Barber C. L., Jordan M. C., Murdock J., Desai S., Ferrara N., Nagy A., Roos K. P., Iruela-Arispe M. L. (2007) Cell 130, 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curwen J. O., Musgrove H. L., Kendrew J., Richmond G. H., Ogilvie D. J., Wedge S. R. (2008) Clin. Cancer Res. 14, 3124–3131 [DOI] [PubMed] [Google Scholar]
- 5.Grothey A., Galanis E. (2009) Nat. Rev. Clin. Oncol. 6, 507–518 [DOI] [PubMed] [Google Scholar]
- 6.Crawford Y., Ferrara N. (2009) Cell Tissue Res. 335, 261–269 [DOI] [PubMed] [Google Scholar]
- 7.Folkman J. (2004) APMIS 112, 496–507 [DOI] [PubMed] [Google Scholar]
- 8.Bagavandoss P., Wilks J. W. (1990) Biochem. Biophys. Res. Commun. 170, 867–872 [DOI] [PubMed] [Google Scholar]
- 9.Good D. J., Polverini P. J., Rastinejad F., Le Beau M. M., Lemons R. S., Frazier W. A., Bouck N. P. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 6624–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taraboletti G., Roberts D., Liotta L. A., Giavazzi R. (1990) J. Cell Biol. 111, 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isenberg J. S., Martin-Manso G., Maxhimer J. B., Roberts D. D. (2009) Nat. Rev. Cancer 9, 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson D. W., Pearce S. F., Zhong R., Silverstein R. L., Frazier W. A., Bouck N. P. (1997) J. Cell Biol. 138, 707–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiménez B., Volpert O. V., Crawford S. E., Febbraio M., Silverstein R. L., Bouck N. (2000) Nat. Med. 6, 41–48 [DOI] [PubMed] [Google Scholar]
- 14.Isenberg J. S., Jia Y., Fukuyama J., Switzer C. H., Wink D. A., Roberts D. D. (2007) J. Biol. Chem. 282, 15404–15415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu W., Smart E. J. (2005) J. Biol. Chem. 280, 29543–29550 [DOI] [PubMed] [Google Scholar]
- 16.Zhang X., Kazerounian S., Duquette M., Perruzzi C., Nagy J. A., Dvorak H. F., Parangi S., Lawler J. (2009) FASEB J. 23, 3368–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta K., Gupta P., Wild R., Ramakrishnan S., Hebbel R. P. (1999) Angiogenesis 3, 147–158 [DOI] [PubMed] [Google Scholar]
- 18.Greenaway J., Lawler J., Moorehead R., Bornstein P., Lamarre J., Petrik J. (2007) J. Cell. Physiol. 210, 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isenberg J. S., Ridnour L. A., Perruccio E. M., Espey M. G., Wink D. A., Roberts D. D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13141–13146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isenberg J. S., Ridnour L. A., Dimitry J., Frazier W. A., Wink D. A., Roberts D. D. (2006) J. Biol. Chem. 281, 26069–26080 [DOI] [PubMed] [Google Scholar]
- 21.Isenberg J. S., Romeo M. J., Yu C., Yu C. K., Nghiem K., Monsale J., Rick M. E., Wink D. A., Frazier W. A., Roberts D. D. (2008) Blood 111, 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isenberg J. S., Pappan L. K., Romeo M. J., Abu-Asab M., Tsokos M., Wink D. A., Frazier W. A., Roberts D. D. (2008) Ann. Surg. 247, 180–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J., Hopkins B. D., Tsujikawa K., Perruzzi C., Adini I., Swerlick R., Bornstein P., Lawler J., Benjamin L. E. (2009) Am. J. Physiol. Heart Circ. Physiol. 296, H1344–H1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandrasekaran L., He C. Z., Al-Barazi H., Krutzsch H. C., Iruela-Arispe M. L., Roberts D. D. (2000) Mol. Biol. Cell 11, 2885–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barazi H. O., Li Z., Cashel J. A., Krutzsch H. C., Annis D. S., Mosher D. F., Roberts D. D. (2002) J. Biol. Chem. 277, 42859–42866 [DOI] [PubMed] [Google Scholar]
- 26.Roberts D. D., Cashel J., Guo N. (1994) J. Tissue Cult. Methods 16, 217–222 [Google Scholar]
- 27.Misenheimer T. M., Huwiler K. G., Annis D. S., Mosher D. F. (2000) J. Biol. Chem. 275, 40938–40945 [DOI] [PubMed] [Google Scholar]
- 28.Tan K., Duquette M., Liu J. H., Dong Y., Zhang R., Joachimiak A., Lawler J., Wang J. H. (2002) J. Cell Biol. 159, 373–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian S., Boder E. T., Discher D. E. (2007) J. Biol. Chem. 282, 1805–1818 [DOI] [PubMed] [Google Scholar]
- 30.Laklai H., Laval S., Dumartin L., Rochaix P., Hagedorn M., Bikfalvi A., Le Guellec S., Delisle M. B., Schally A. V., Susini C., Pyronnet S., Bousquet C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17769–17774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isenberg J. S., Frazier W. A., Roberts D. D. (2008) Cell. Mol. Life Sci. 65, 728–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isenberg J. S., Annis D. S., Pendrak M. L., Ptaszynska M., Frazier W. A., Mosher D. F., Roberts D. D. (2009) J. Biol. Chem. 284, 1116–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isenberg J. S., Romeo M. J., Abu-Asab M., Tsokos M., Oldenborg A., Pappan L., Wink D. A., Frazier W. A., Roberts D. D. (2007) Circ. Res. 100, 712–720 [DOI] [PubMed] [Google Scholar]
- 34.Papapetropoulos A., García-Cardeña G., Madri J. A., Sessa W. C. (1997) J. Clin. Invest. 100, 3131–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimmeler S., Fleming I., Fisslthaler B., Hermann C., Busse R., Zeiher A. M. (1999) Nature 399, 601–605 [DOI] [PubMed] [Google Scholar]
- 36.Dimmeler S., Dernbach E., Zeiher A. M. (2000) FEBS Lett. 477, 258–262 [DOI] [PubMed] [Google Scholar]
- 37.Hutchings H., Ortega N., Plouët J. (2003) FASEB J. 17, 1520–1522 [DOI] [PubMed] [Google Scholar]
- 38.Houck K. A., Leung D. W., Rowland A. M., Winer J., Ferrara N. (1992) J. Biol. Chem. 267, 26031–26037 [PubMed] [Google Scholar]
- 39.Krutzsch H. C., Choe B. J., Sipes J. M., Guo N., Roberts D. D. (1999) J. Biol. Chem. 274, 24080–24086 [DOI] [PubMed] [Google Scholar]
- 40.Brown E. J., Frazier W. A. (2001) Trends Cell Biol. 11, 130–135 [DOI] [PubMed] [Google Scholar]
- 41.Youvan D. C., Silva C. M., Bylina E. J., Coleman W. J., Dilworth M. R., Yang M. M. (1997) Biotechnology 3, 1–18 [Google Scholar]
- 42.Gordon G. W., Berry G., Liang X. H., Levine B., Herman B. (1998) Biophys. J. 74, 2702–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bornstein P. (2009) J. Cell Commun. Signal. 3, 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi T., Yamaguchi S., Chida K., Shibuya M. (2001) EMBO J. 20, 2768–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maile L. A., Clemmons D. R. (2003) Circ. Res. 93, 925–931 [DOI] [PubMed] [Google Scholar]
- 46.Lawler J. (2002) J. Cell. Mol. Med. 6, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Short S. M., Derrien A., Narsimhan R. P., Lawler J., Ingber D. E., Zetter B. R. (2005) J. Cell Biol. 168, 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calzada M. J., Zhou L., Sipes J. M., Zhang J., Krutzsch H. C., Iruela-Arispe M. L., Annis D. S., Mosher D. F., Roberts D. D. (2004) Circ. Res. 94, 462–470 [DOI] [PubMed] [Google Scholar]
- 49.Isenberg J. S., Romeo M. J., Maxhimer J. B., Smedley J., Frazier W. A., Roberts D. D. (2008) Ann. Surg. 247, 860–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bauer E. M., Qin Y., Miller T. W., Bandle R. W., Csanyi G., Pagano P. J., Bauer P. M., Schnermann J., Roberts D. D., Isenberg J. S. (2010) Cardiovasc Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiojima I., Walsh K. (2002) Circ. Res. 90, 1243–1250 [DOI] [PubMed] [Google Scholar]
- 52.Bruns A. F., Herbert S. P., Odell A. F., Jopling H. M., Hooper N. M., Zachary I. C., Walker J. H., Ponnambalam S. (2010) Traffic 11, 161–174 [DOI] [PubMed] [Google Scholar]
- 53.Somanath P. R., Malinin N. L., Byzova T. V. (2009) Angiogenesis 12, 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamaoka-Tojo M., Tojo T., Kim H. W., Hilenski L., Patrushev N. A., Zhang L., Fukai T., Ushio-Fukai M. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 1991–1997 [DOI] [PubMed] [Google Scholar]
- 55.Lampugnani M. G., Orsenigo F., Gagliani M. C., Tacchetti C., Dejana E. (2006) J. Cell Biol. 174, 593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.