Abstract
Dna2 endonuclease/helicase participates in eukaryotic DNA transactions including cleavage of long flaps generated during Okazaki fragment processing. Its unusual substrate interaction consists of recognition and binding of the flap base, then threading over the 5′-end of the flap, and cleaving periodically to produce a terminal product ∼5 nt in length. Blocking the 5′-end prevents cleavage. The Dna2 ATP-driven 5′ to 3′ DNA helicase function promotes motion of Dna2 on the flap, presumably aiding its nuclease function. Here we demonstrate using two different nuclease-dead Dna2 mutants that on substrates simulating Okazaki fragments, Dna2 must thread onto an unblocked 5′ flap to display helicase activity. This requirement is maintained on substrates with single-stranded regions thousands of nucleotides in length. To our knowledge this is the first description of a eukaryotic helicase that cannot load onto its tracking strand internally but instead must enter from the end. Biologically, the loading requirement likely helps the helicase to coordinate with the Dna2 nuclease function to prevent creation of undesirably long flaps during DNA transactions.
Keywords: DNA Enzymes, DNA Helicase, DNA Repair, DNA Replication, Enzyme Mechanisms, Dna2, Okazaki Fragment Maturation
Introduction
DNA and RNA perform their biological roles in either double-stranded, double helical or single-stranded configurations. Functions requiring access to the nucleotide bases involve conversion of double to single strands, a process that requires energy and is carried out by the helicases. These motor proteins derive their energy from nucleoside triphosphate, usually adenosine triphosphate (ATP), hydrolysis (1). Helicases have evolved to bind and act in a sequence-independent manner on DNA, RNA, or DNA-RNA hybrid duplexes. Each helicase exhibits a specific directionality of unwinding, defined by the tracking orientation, either 5′ to 3′ or 3′ to 5′ on one strand of the substrate. The basic mechanism involves binding to the strand on which tracking occurs and then translocation that pushes away the complementary strand.
Helicases display highly multimeric, hexameric, dimeric, or monomeric structures (2). Typically, multimeric helicases are loaded on the DNA with the help of helicase loaders which orient the DNA in the helicase active domain (3). Helicases participate in DNA replication and repair, RNA transcription, and maturation (4). In eukaryotes, the mini-chromosome maintenance (MCM) proteins 2–7, form the putative replicative helicase (5). Other helicases participate in DNA replication, removing double-stranded barriers to enzyme movement or preparing strands for the action of single-stranded nucleases.
Because DNA forms antiparallel double-stranded helices, on unwinding the duplex DNA for replication, nascent DNA is synthesized continuously on the leading strand and discontinuously on the lagging strand. On the lagging strand, DNA polymerase (pol)2 α synthesizes a short stretch of RNA/DNA primer, totaling 20–24 nt, which is subsequently extended by DNA polymerase δ (pol δ) to form Okazaki fragments of ∼150 nt (6). Pol δ, on encountering a downstream Okazaki fragment, performs strand displacement synthesis to generate a short single-stranded flap. The displaced flap is recognized and cleaved by flap endonuclease 1 (FEN1) creating a nick, which is joined by DNA ligase I (Lig I) completing Okazaki fragment maturation (7, 8). This mechanism is thought to remove the RNA/DNA primer synthesized by the error-prone pol α. Flaps escaping FEN1 cleavage may be further lengthened, requiring the “two-nuclease” pathway (8).
This latter pathway involves Dna2 helicase/endonuclease, a multifunctional protein, exhibiting 5′ to 3′ endonuclease, 3′ to 5′ exonuclease and 5′ to 3′ ATP-dependent helicase activities (9). It was first identified in a screen for DNA replication mutants (10). Pif1, another 5′ to 3′ helicase, has also been implicated in “two nuclease” Okazaki fragment maturation and telomere maintenance (11, 12). Flaps lengthened by these helicases may stably bind the single strand binding protein replication protein A (RPA), inhibiting FEN1 cleavage (13). The endonuclease activity of Dna2 is stimulated by RPA, enabling it to cleave faster on long flap substrates (14). Dna2 cleavage shortens the flap to 5–6 nt, precluding RPA, and thereby allowing for FEN1 nuclease activity and subsequent ligation (14).
Dna2 helicase is active as a monomer both in vitro and in vivo (15). The C-terminal domain of Saccharomyces cerevisiae Dna2 contains both its nuclease and helicase active sites suggesting a coupling of these functions (16). Whereas the helicase activity of yeast Dna2 is dispensable under certain growth conditions, its endonuclease activity is essential. However, growth defects in the helicase mutants demonstrate that the helicase is important for Dna2 physiological function in vivo (17). The helicase and nuclease activities can be differentially regulated for studies in vitro, by altering the ratios of [ATP]/[Mg2+] (16, 18, 19). Higher ATP concentrations over Mg2+ favor helicase function whereas higher Mg2+ over ATP supports nuclease activity.
The endonuclease activity of Dna2 has been shown to play a role in Okazaki maturation (20), telomere maintenance (21), double strand break repair (22–25), long patch base excision repair (26), and aging (27). However, with the exception of showing unwinding of G4 quartets by Dna2 helicase (28) and showing that mutation of the helicase relatively few studies have been focused on understanding its roles. In the current investigation, using nuclease-dead Dna2 mutants, we questioned whether the helicase function of Dna2 shared the strict free ssDNA 5′-end requirement described for Dna2 endonuclease activity on substrates simulating Okazaki fragment intermediates.
EXPERIMENTAL PROCEDURES
Materials
Synthetic oligonucleotides including those having a 5′ or internal biotin conjugation were synthesized by Integrated DNA Technologies (IDT, Coralville, IA) or Midland Certified Reagents Company (Midland, TX). Radioactive nucleotides [α-32P]dCTP and [γ-32P]ATP were purchased from Perkin Elmer Life Sciences. T4 DNA ligase used in the creation of the 154-nt strand was purchased from New England Biolabs (NEB). Phage plasmids ΦX174 and M13mp18 were obtained from NEB. Klenow fragment of Escherichia coli DNA polymerase I (for 3′ labeling) and T4 polynucleotide kinase (for 5′ labeling) were purchased from Roche Applied Science. Streptavidin and ATP were also purchased from Roche Applied Science. Other reagents were the best commercially available.
Purified Proteins
S. cerevisiae Dna2E675A was overexpressed and purified from baculovirus High Five cells as previously described (24, 29). Dna2K677R was expressed in a yeast expression vector and purified as described previously (29, 30). S. cerevisiae Pif1 was cloned into the pET-28b bacterial expression vector (Novagen/EMD Biosciences), expressed in the E. coli Rosetta strain (Novagen/EMD Biosciences), and purified as previously described (11).
Oligonucleotides
Experimental substrates were designed to represent intermediates made during the Okazaki maturation. The sequences of the oligonucleotides are listed in Table 1. The primer sequences are listed 5′ to 3′, and the template sequences are listed 3′ to 5′ to facilitate visual alignment. A “B” in the nucleotide sequence represents a biotin-conjugated strand. The 154-nt strand was created by ligating two strands; 80 nt (D2) and 74 nt (U2) containing a 40-nt (S1) splint as previously described (31). Following ligation the 154-nt strand was isolated on a 6% denaturing polyacrylamide gel containing 8 m urea, the correct product was identified by ethidium bromide staining, and the DNA was purified from the gel. Labels were added at the 5′-end of strands by incubating them with T4 polynucleotide kinase and [γ-32P]ATP. Labels at the 3′-end were added by first annealing 20 pmol of primer to 50 pmol of template having a 5′ G overhang. The annealed complex was then incubated with the Klenow fragment of E. coli DNA polymerase I and [α-32P]dCTP. The labeled primers were purified by electrophoresis and isolated from a 15% polyacrylamide, 7 m urea denaturing gel. Experimental substrates were created by combining primers in annealing buffer containing 10 mm Tris-HCl (pH 8.0), 50 mm NaCl, and 1 mm DTT, heated to 95 °C for 5 min, transferred to 70 °C, and then slowly cooled to room temperature. Substrates were annealed in a 1:2:4 ratio (labeled primer: template: second primer). Substrates containing a primer complementary to the 5′ flap end were annealed in a 1:2:4:6 ratio (labeled primer: template: second primer: 5′ flap complementary primer).
TABLE 1.
Oligonucleotide sequences
| Primer | Length | Sequencea,b |
|---|---|---|
| nt | ||
| Upstream | Listed 5′ to 3′ | |
| U1 | 25 | GTCCACCCGACGCCACCTCCTGCCT |
| U2 | 74 | ACAAGGCTGACTTTTCCTCCCCTTGTGCTGCCTTCTGGGGGGGGCCCAGCCGGATCCCGGGGCGAGCTCGAATT |
| U3 | 30 | AATTCGAGCTCGCCCCGGGATCCGGCTGGG |
| U3 | 26 | CGCCAGGGTTTTCCCAGTCACGACCA |
| Downstream | Listed 5′ to 3′ | |
| D1 | 60 | AGACGAATTCCGGATACGACGGCCAGTGCCGACCGTGCCAGCCTAAATTTCAATCCACCC |
| D2 | 80 | /p/ACT AAC CAG GCC CGA CCC TGC TTG GCT TCC GAG ATC AGA CGA TAT CGG GCA CTT TCA GGG TGG TAT GGC CGT AGG CGA GC |
| D3 | 30 | GGAAGCCAAGCAGGGTCGGGCCTGGTTAGT |
| D4 | 24 | ACGTTGTAAAACGACGGCCAGTG |
| D5 | 42 | AGCTAGCTCTTGATCGTAGACGTTGTAAAACGACGGCCAGTG |
| D6 | 76 | /B/GTACCGAGCTCGAATTCGCCCGTTTCACGCCTGTTAGTTAATTCACTGGCCGTCGTTTTACAACGACGTGACTGGG |
| D7 | 76 | GTACCGAGCTCGAATTCGCCCGTTTCACGCCTG/B/TTAGTTAATTCACTGGCCGTCGTTTTACAACGACGTGACTGGG |
| Template | Listed 3′ to 5′ | |
| T1 | 110 | /B/CAGGTGGGCTGCGGTGGAGGACGGAAGTTACACGACCCTAGGATGTTGGTTCTGCTTAAGGCCTATGCTGCCGGTCACGGCTGGCACGGTCGGATTTAAAGTTAGGTGGGTGGG/B/ |
| T2 | 154 | TTAAGCTCGAGCGGGGCCCTAGGCCGACCCGGGGGGGGTCTTCCGTCGTGTTCCCCTCCTTTTCAGTCGGAACACGAGCGGATGCCGGTATGGTGGGACTTTCACGGGCTATAGCAGACTAGAGCCTTCGGTTCGTCCCAGCCCGGACCAATCA |
| T3 | 49 | GCGGTCCCAAAAGGGTCAGTGCTGGGCAAAATGTTGCTGCACTGACCCG |
| 5′ Complement | Listed 3′ to 5′ | |
| C1 | 33 | CATGGCTCGAGCTTAAGCGGGCAAAGTGCGGAC |
| C2 | 28 | CTCGAGCTTAAGCGGGCAAAGTGCGGAC |
| C3 | 23 | GCTTAAGCGGGCAAAGTGCGGAC |
| Splint | Listed 5′ to 3′ | |
| S1 | 40 | GGA GGA AAA GTC AGC CTT GTG CTC GCC TAC GGC CAT ACC A |
a /p/ denoted 5′ phosphate.
b /B/ denotes biotin conjugation.
Helicase Assay
Substrates requiring streptavidin blockage were exposed to a 50 fold excess of streptavidin and held on ice for 20 min prior to start of the experiment. Helicase assays were performed using the nuclease-deficient mutants Dna2E675A and Dna2K677R, and Pif1. 5 fmol of experimental substrate was incubated with various amounts of proteins, as indicated in a reaction volume of 20 μl, at 37 °C for 15 min. The reaction buffer contained 50 mm Tris-HCl (pH 7.5), 25 mm NaCl, 2 mm dithiothreitol, 0.25 mg/ml bovine serum albumin, 2 mm MgCl2, and 4 mm ATP. The reactions were terminated using 6× helicase dye (50 mm EDTA, 0.9% SDS, 0.125% bromphenol blue, 0.125% xylene cyanole, 30% glycerol). After termination, samples were loaded on a pre-run 5% native polyacrylamide gel and resolved by electrophoresis for 1.5 h at 150 V. Each experiment was performed at least in triplicate, and representative gels are shown in the figures. Gels were analyzed as previously described (32).
RESULTS
Dna2 Cannot Load Internally on an Okazaki Fragment Substrate
Initial analysis of Dna2 helicase activity presented it as having 3′ to 5′ directionality (10). Subsequently it was confirmed that Dna2 translocated in the 5′ to 3′ direction (18). Confirmation of the directionality of the Dna2 translocation was carried out using a partial duplex DNA substrate that consisted of a linear ΦX174 ssDNA annealed with a 29-nt fragment at the 3′-end and a 23-nt fragment at the 5′-end (18). In the presence of ATP, Dna2 displaced the 29-nt fragment from the 3′-end presumably by translocating on the template in the 5′ to 3′ direction (18).
We revisited this experiment, using an experimental substrate similar in design to the substrate described above, with the exception of using a shorter template. Our experimental substrate, simulating an Okazaki fragment intermediate, had a 110-nt template to which we annealed a 25-nt primer at the 3′-end and a 60-nt primer at the 5′-end of the template. This substrate provides a gap of 25 nt on the template to which Dna2 could potentially bind and translocate. Dna2 contains a robust 5′ to 3′ endonuclease activity that interferes with analysis of its helicase functions. To eliminate this concern, we used the nuclease-deficient mutant Dna2E675A. The [ATP]/[Mg2+] ratio in the reactions was maintained at 2:1, because this was previously shown to be optimal for Dna2 helicase activity (33). Because Pif1 helicase accesses DNA internally and unwinds in the same direction as Dna2 (i.e. 5′ to 3′), we used it as a positive helicase control.
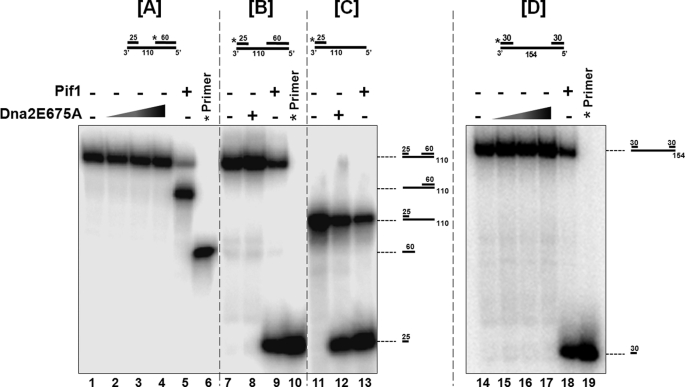
In the first set of experiments the substrate contained a 5′ label on the 60-nt primer bound at the 5′ template end. Titrating increasing amounts of Dna2E675A did not yield a helicase product (Fig. 1A, lanes 2–4). Pif1 was able to readily acquire the template internally, translocate on the template in the 5′ to 3′ direction and unwind the 25-nt primer bound at the 3′ template end. Because the 60-nt primer was labeled in this experimental substrate, we observed the 110-nt template bound to the 60-nt primer after Pif1 unwinding (Fig. 1A, lane 5). On the substrate containing a 5′ label on the 25-nt primer at the highest concentration, Dna2E675A did not yield a helicase product (Fig. 1B, lane 8). Unwinding of the 25-nt upstream primer by Pif1 was observed as noted previously (Fig. 1B, lane 9). Observation of a helicase product by Pif1 and not Dna2E675A suggested that Dna2 was unable to bind to the 25-nt gap on the template and consequently could not track and unwind in the 5′ to 3′ direction. To allow for Dna2E675A binding onto the 5′-end of the template, we used a substrate that did not contain the 60-nt primer. Dna2E675A was able to efficiently unwind the 25-nt primer on this substrate similar to Pif1 (Fig. 1C, compare lane 12 to lane 13). This suggested that in order to observe helicase activity, either Dna2 required binding to a longer sequence on the template strand, or it had a free 5′-end requirement, similar to that needed for endonuclease activity. To ensure that 25-nt gap was not below the minimal loading limit of Dna2, we used an additional substrate that contained a template 154 nt in length with a 5′-labeled 30-nt primer annealed to the 3′ of the template and 30-nt primer annealed to the 5′-end of the template, providing a 94-nt gapped substrate. Dna2E675A was unable to unwind even on this substrate suggesting that Dna2 could not internally load even on a longer gapped substrate (Fig. 1D, lanes 15–17).
FIGURE 1.
Dna2 cannot bind to a gap on the template and translocate in the 5′ to 3′ direction. Helicase activity was assayed as described under “Experimental Procedures.” Experimental substrates used were: (A) a substrate consisting of a 110-nt template with a 5′-labeled 60-nt primer annealed at the template 5′-end and a 25-nt primer annealed at the template 3′-end (U1:T1:D1). B, substrate had the same structure as in A, but the 5′ label was on the 25-nt primer. C, substrate was the same as in B but with no 60-nt primer (U1:T1). When two primers were present, the gap between them was 25 nt. D, substrate consisting of a 154-nt template with a 5′-labeled 30-nt primer annealed at the template 3′-end and a 30-nt primer annealed at the template 5′-end (U3:T2:D3). Reactions containing 5 fmol of each substrate were incubated with either Dna2E675A or Pif1 helicase for 15 min at 37 °C. Lanes 1, 7, 11, 14 are the substrate-alone controls, and lanes 6, 10, 19 are the labeled primer alone controls. Dna2E675A was titrated into the reactions at a concentration of 100 fmol (lanes 2, 15), 250 fmol (lanes 3, 16) and 500 fmol (lanes 4, 8, 12, 17). Lanes 5, 9, 13, and 18 contained 500 fmol of Pif 1. The asterisk on the substrates depicted above the figures denotes the position of the radiolabel. Positions of the substrate and helicase products are indicated in the figure.
Additionally, we used a different nuclease-dead mutant Dna2K677R in our assays to exclude the possibility that the results obtained in Fig. 1 derived from a property of the mutant Dna2E675A. The mutant Dna2K677R is completely nuclease dead similar to mutant Dna2E675A (supplemental Fig. S1). Consistent with the results obtained in Fig. 1, A and B, we did not observe any helicase products in the presence of Dna2K677R when the 5′-ends of the substrate were blocked, suggesting the helicase has a true free 5′-end requirement for helicase function (supplemental Fig. S2).
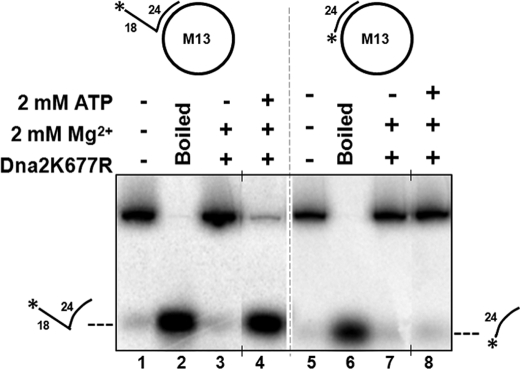
To understand the discrepancy between our helicase results and those previously reported (18), we examined the ΦX174 substrate obtained from New England Biolabs (NEB) that was used in the previous report and found that it contained ∼15% linear molecules (as noted in NEB catalogue). To overcome this issue, we used another phage template, M13mp18 (NEB) which contained a relatively lower fraction (∼2%) of linear molecules to assay for helicase function of Dna2 (data not shown). Using the circular M13mp18 template containing a complementary primer, which was either completely annealed or which contained an unannealed flap we studied the helicase activity of Dna2K677R. In both substrates, with or without the flap, in the absence of ATP we did not observe any helicase product (Fig. 2, lanes 3 and 7). However, at a concentration of 2:1 of [ATP]/[Mg2+], such that the helicase function of Dna2 is more active, we observed a helicase product only on the substrate that contained a flap (Fig. 2, compare lane 4 to lane 8). Presumably the Dna2 can load on the flap and displace the primer. However, without the flap the helicase would have to load on the long template. Lack of helicase activity in that case indicates that a single strand template cannot load Dna2 for helicase function even if that template is very long.
FIGURE 2.
Dna2 cannot track on a primed circular long single strand without a free 5′-end. Helicase activity was assayed as described under “Experimental Procedures.” Reactions containing 15 fmol of either (A) M13mp18 containing a completely annealed primer (M13mp18: D4) or (B) M13mp18 containing a primer with an unannealed 5′-flap (M13mp18:D5) were incubated with 300 fmol of Dna2K677R for 30 min at 37 °C. The reaction buffer contained 2 mm ATP and 2 mm Mg Cl2. Lanes 1 and 5 are substrate-alone controls, and lanes 2 and 6 are boiled substrate controls. Lanes 3 and 7 contained only 2 mm MgCl2 in the reaction buffer. Lanes 4 and 8 contained 2 mm ATP and 2 mm MgCl2 in the reaction buffer. The asterisk on the substrates depicted above the figures denotes the position of the radiolabel. Positions of the helicase products are indicated in the figure.
Dna2 Requires a Free 5′-End for Helicase Activity on Flap Substrates
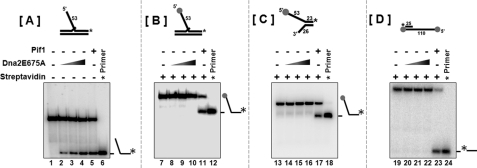
To clarify the free 5′-end requirement for helicase function, we used a 53-nt flap substrate that contained a conjugated biotin at the 5′-end of the flap. This substrate resembles the substrate used in Fig. 1A, because the biotin-conjugated streptavidin at the 5′-end of the flap acts similar to the complementary primer annealed to the 5′-end of the template in Fig. 1A to create a blocked 5′-end substrate. Increasing concentrations of Dna2E675A were titrated in the presence of either an unblocked 53-nt flap substrate (Fig. 3A) or a streptavidin blocked 53-nt flap substrate (Fig. 3B). Dna2E675A was able to efficiently unwind the 5′-unblocked flap similar to Pif1 helicase (Fig. 3B, lanes 2–5). However, when the flap was blocked at the 5′-end using streptavidin, we did not observe any helicase product with increasing Dna2E675 concentrations (Fig. 3B, lanes 8–10). However, Pif1 was still able to efficiently unwind the 5′-blocked flap (Fig. 3B, lane 11). Additionally, we also performed helicase assays on a forked substrate that was bound by streptavidin at the unannealed 5′-end and an overhang substrate containing streptavidin at the 5′-end of the overhang. In the case of both these substrates, since the streptavidin blocked the 5′-end, Dna2 could not load and unwind and hence we did not observe any helicase products (Fig. 3C, lanes 14–16 and Fig. 3D, lanes 20–22). A helicase product in the Pif1 control lanes suggested that unlike Dna2, Pif1 was capable of internally loading and unwinding the substrate (Fig. 3C, lane 17; Fig. 3D, lane 23).
FIGURE 3.
Dna2 requires a free 5′-end for helicase activity. Helicase activity was assayed as described under “Experimental Procedures.” Reactions containing 5 fmol of either (A) unblocked 53-nt flap substrate (U4:T3:D6); (B) streptavidin blocked 53-nt flap substrate; (C) forked substrate (T3:D6); (D) 5′ overhang substrate (T1:U1) were incubated with either Dna2E675A or Pif1 helicase for 15 min at 37 °C. Lanes 1, 7, 13, 19 are the substrate-alone controls, and lanes 6, 12, 18, 24 are the labeled primer alone controls. Dna2E675A was titrated into the reactions at a concentration of 100 fmol (lanes 2, 8, 14, 20), 250 fmol (lanes 3, 9, 15, 21), and 500 fmol (lanes 4, 10, 16, 22). Lanes 5, 11, 17, 23 contained 500 fmol of Pif 1. The asterisk on the substrates depicted above the figures, denote the position of the radiolabel. Positions of the substrate and helicase products are indicated in the figure.
These results confirmed that Dna2 required a free 5′-end for unwinding in the 5′ to 3′ direction. To eliminate any possible effect of the streptavidin moiety on the helicase activity of Dna2 we used an 18-nt fold-back flap substrate expected to block 5′-end loading in a similar manner as streptavidin. As with streptavidin, we did not observe unwinding of the hairpin flap substrate by Dna2E675A, however, Pif1 was able to unwind the entire downstream primer containing the 18-nt fold-back flap (data not shown). Data from the helicase assays using a variety of streptavidin-blocked 5′ substrates suggested that Dna2 requires an unobstructed 5′ flap end for helicase function.
Dna2 Threads the Flap for Unwinding Activity
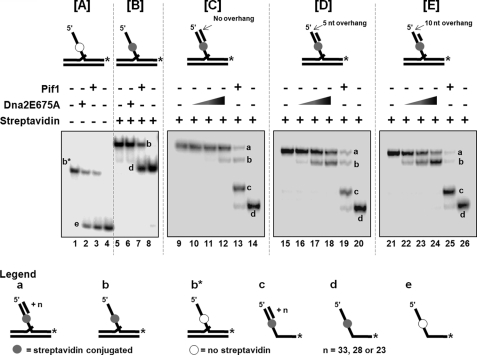
Because this is the first report of a eukaryotic helicase with an absolute free 5′-end requirement we designed additional substrates with blocked and unblocked 5′-ends to absolutely ascertain the free end requirement of Dna2 for helicase activity. Four different substrates were designed for these experiments. As the basis for these substrates, a presubstrate similar to the 53-nt flap substrate was constructed with the exception that instead of containing a biotin at the 5′-end of the flap, the biotin was conjugated internally at the 33rd nt from the 5′-end in the 53-nt flap sequence. The first experimental substrate was the presubstrate unaltered. The second substrate had a 33-nt primer annealed to the 5′-end region of the flap. This substrate therefore, would be similar to the streptavidin-blocked substrate, in that it would not contain a free 5′-end flap. The third substrate had a 28-nt annealed primer that provided a 5 nt single stranded overhang at the 5′-end of the flap. The fourth substrate had a 23-nt annealed primer providing a 10-nt overhang at the 5′-end of the flap. All the substrates were labeled on the 3′-end of the 53-nt flap primer.
The effect of Dna2E675A helicase activity was then observed for each of these substrates. To confirm that the internally labeled biotin did not interfere with the helicase activity of Dna2, we observed the unwinding function of Dna2 on the 53-nt internal biotin flap substrate. Both Dna2E675A and Pif1 efficiently unwound the 53-nt flap-containing primer, confirming the non-interference of the internal biotin (Fig. 4A, lanes 2 and 3). When streptavidin was conjugated with the internal biotin on the 53-nt flap substrate we observed a helicase product only with Pif1 (Fig. 4B, lane 7). Despite having a free 5′-end, this substrate was not unwound by Dna2, presumably because the streptavidin acted as an impediment to threading the flap through Dna2 before the helicase could act on the annealed region of the flap-containing primer (Fig. 4B, lane 6).
FIGURE 4.
A block in the 5′ flap can prevent threading and thereby inactivate Dna2 helicase activity. Helicase activity was assayed as described under “Experimental Procedures.” Reactions contained 5 fmol of either (A) unblocked 53-nt flap substrate (U4:T3:D7); (B) 53-nt flap substrate with streptavidin conjugated to the internal biotin (U4:T3:D7); (C) 53-nt flap substrate with streptavidin conjugated to the internal biotin containing a 33-nt primer complementary to the 5′ flap (U4:T3:D7:C1); (D) 53-nt flap substrate with streptavidin conjugated to the internal biotin containing a 28-nt primer complementary to the 5′ flap (U4:T3:D7:C2); (E) 53-nt flap substrate with streptavidin conjugated to the internal biotin containing a 23-nt primer complementary to the 5′ flap (U4:T3:D7:C3). They were incubated with either Dna2E675A or Pif1 helicase for 15 min at 37 °C. Lanes 1, 5, 9, 15, and 21 are the substrate-alone controls. Lanes 4, 8, 14, 20, and 26 are the labeled primer alone controls. Dna2E675A was titrated into the reactions at a concentration of 100 fmol (lanes 10, 16, 22), 250 fmol (lanes 11, 17, 23) and 500 fmol (lanes 2, 6, 12, 18, 24). Lanes 3, 7, 13, 19, and 25 contained 500 fmol of Pif 1. The asterisk on the substrates depicted above the figures, denote the position of the radiolabel. Positions of the substrate and helicase products are indicated in the figure. Gels shown in Fig. 3, A and B have been electrophoresed for a shorter period of time compared with Fig. 3, C–E.
We next measured helicase function of Dna2 on 53-nt flap substrates having both streptavidin conjugated to the internal biotin and the 5′-end region blocked using varying length complementary primers. Because all the substrates used in this series of experiments were labeled only on the 3′-end of the flap primer, unwinding of the 5′ complementary primer will be revealed on the gel as a band migrating faster than the original substrate, corresponding to the presubstrate (substrate 1) with streptavidin conjugated to the internal biotin, equivalent to the substrate in Fig. 3B.
Increasing concentrations of Dna2E675A were unable to unwind the 53-nt flap substrate with a 33-nt 5′ complementary primer (Fig. 4C, lanes 10 and 11). At the highest concentration of Dna2, we observed weak helicase activity, presumably from breathing of the 33nt primer (Fig. 4C, lane 12). On the substrate with the 5-nt 5′ overhang, Dna2 displayed good unwinding of the 28-nt 5′ complementary primer (Fig. 4D, lanes 16–18). Dna2 showed robust helicase activity on the substrate with the 10-nt 5′ overhang by displacing the 23-nt 5′ complementary primer (Fig. 4E, lanes 22–24). Similar to results in Fig. 4B (lane 6), the internal streptavidin conjugated biotin blocked the unwinding of the downstream primer by Dna2.
Interestingly the positive Pif 1 helicase control produced various helicase products in the case of all the substrates containing a complementary 5′ primer (Fig. 4, C–E, lanes 13, 19, 25, respectively). The first helicase product was the internal streptavidin-conjugated 53-nt flap substrate alone (a template with two primers), after removal of the 5′ complementary primers (denoted by b in the figure). Because Pif1 was able to unwind even in the presence of a streptavidin block, the second helicase product observed was the displaced 53-nt downstream flap with the 5′ complementary primer still annealed (denoted by c in the figure). The third product was the internal streptavidin conjugated 53-nt downstream flap primer alone (denoted by d in the figure). Presumably Pif1 sometimes initially unwound the entire 53-nt flap-primer with the 5′ complementary primer, then unwound the 5′ complementary primer from the flap primer. Other times it would unwind the 5′ complementary primer first. This set of experiments showed that the presence of the a streptavidin block in the middle of the 5′ flap prevented Dna2 helicase activity past the block, presumably by preventing threading of the blocked section of the flap through the active site of the Dna2 protein. Moreover, these results conclusively proved that Dna2 has a strict requirement for a free 5′-end for entry into the substrate, and can load on a short 5′ single strand. It must then thread from the entry point to the region of double strands, so that it can exhibit helicase activity. This was evident from displacement of the shorter 5′ complementary primers up to, but not beyond, the streptavidin.
DISCUSSION
While the endonuclease property of Dna2 is conserved among various species, the reported levels of helicase activity have greatly varied (34). Helicase activities of Xenopus laevis and human Dna2 were considered to be either non-existent or extremely weak (22, 35). The robust nuclease activity of Dna2 tends to obscure detection of its unwinding property. The development of nuclease-dead Dna2 mutants have aided in discovering the properties of Dna2 helicase activity in both Xenopus and humans (25, 36).
Proposed functional roles of the helicase activity of Dna2 include moving the active site of the nuclease to within several nt of the flap base for cleavage and disrupting the formation of secondary structures on the 5′-displaced flap (18). The main role of Dna2 during Okazaki fragment processing is to displace RPA from flap structures and cleave the flap to a length at which RPA cannot rebind, allowing for FEN1 cleavage (14, 37). However, the helicase function of Dna2 is dispensable for displacing RPA from the flap substrates (14). Significantly, Pif1 helicase has been reported to work along with Dna2 during lagging strand synthesis (8). The coordinate effort of helicase function of both these proteins may help in resolution of secondary structures, which in turn can help to reinitiate stalled replication forks (33).
While Dna2 has also been implicated in mitochondrial LP-BER, the exact mechanism of action of Dna2 in this pathway is currently unclear (26, 38). Additionally, Dna2 works in double-strand break repair by acting to resect the 5′-end on broken template strands. It was recently shown that the helicase function of Dna2 in this process is not necessary (24, 39). Both Dna2 and Pif1 have also been shown to play a significant role in telomere maintenance. Considering the higher occurrence of G-rich sequences at telomeres, Dna2 helicase activity has been suggested to resolve unusual structures such as G4 quartets with the help of RPA (28).
Dna2 functions to either cleave or unwind in the 5′ to 3′ direction, and there is evidence that it is a tracking enzyme, very similar to FEN1 (40), having to enter the substrate from the 5′-end of the flap. In the case of both FEN1 and Dna2, the tracking requirement would prevent unintended cleavage of the single-stranded region between Okazaki fragments. Recent Dna2 binding analysis data from our laboratory have shown that Dna2 initially recognizes the base of the flap, interacting with the double-stranded region on the downstream flap primer and the ssDNA flap itself (32). On stable binding, Dna2 likely recognizes the 5′-end of the flap and threads it through its active site for endonuclease activity. This variation on the original proposed mechanism still protects the genome from unintended cleavage.
Our results show that Dna2E675A and Dna2K677R were able to stably bind to all our experimental substrates (supplemental Fig. S3). However, when the 5′-ends of the substrates were blocked Dna2 could not perform its helicase activity (Figs. 1–4). Similar to the endonuclease activity, a threading requirement may allow the Dna2 helicase to carry out its function with minimal disruptive effects. We envision that the helicase has evolved to promote the proper function of the nuclease, which must thread. Helicase activity, functioning during threading in vivo can overcome natural blocks such as small hairpins, bubbles, in which the flap end anneals to nearby template, and recombination intermediates, in which the flap anneals to other regions of the genome, so that the nuclease can move near to the base of the flap for its final cleavage.
In considering the biological significance of the end-loading requirement, we have considered (a) the cellular pathways that utilize Dna2, (b) the catalytic properties of the protein, or (c) the potential advantages of an end-loading helicase. For (a), Dna2 is involved in Okazaki fragment maturation, telomere maintenance, double strand break repair and base excision repair. All of these processes involve DNA intermediates with single strands, some with ends and some not. A reasonable expectation is that the end-loading specificity directs the Dna2 to the portion of the substrates with single-stranded ends, the presumed sites where the actions of Dna2 are required. For (b), Dna2 has evolved an end-loading requirement for its endonuclease activity. Possibly structural constraints in the evolved design of the protein force the helicase to load at ends if the nuclease must. Alternatively, the need for end-loading may not be obligatory, but helps to coordinate the nuclease and helicase functions. For (c), the end-loading requirement may protect substrates from disruption by undesirable strand melting. For example, on an Okazaki fragment substrate, tracking of the helicase on the template toward the polymerase might cause premature polymerase dissociation. Binding of the helicase at the base of the flap might promote rapid flap displacement that could create a gap between the upstream primer being extended by the polymerase and the flap base. This structure would be a suboptimal substrate for FEN1 action (41), and any FEN1 cleavage that did occur would not create a nick for ligation. The flap could then become long enough to participate in undesirable recombination. In the case of double strand break repair, the end-loading requirement may prevent disruption of gapped intermediates of the joining process. Any or all of these factors might have supported the evolution of the end-loading requirement.
Previous analysis demonstrating the 5′ to 3′ directionality of the Dna2 helicase was performed on a phage-length template with short primers at either end (18). The ability of Dna2 helicase to use this substrate suggested that it displays an internal strand loading mechanism only evident on substrates with extremely long single stranded regions. However, using an M13 based primer-template, we could not detect internal loading onto a very long single strand. Commonly encountered issues with substrate construction may explain this discrepancy in the previous work (18). These include incomplete primer hybridization, a moderate level of template breakage or the presence of linear molecules in the circular plasmid preparation.
The nuclease active site of Dna2 shares sequence homology with the prokaryotic RecB helicase/nuclease and Dna2, in conjuction with Sgs1 helicase has recently been shown to be a component of the functional counterpart of the RecBCD double-strand break resection complex (24). RecBCD loads on blunt ends of double-stranded DNA and unwinds them for homologous recombination (32), with a mechanism that does not share many similarities to that of Dna2.
To date there has not been an extensive characterization of the helicase substrate requirements of Dna2. Our current report shows that on substrates with 5′ flaps that simulate Okazaki fragment processing intermediates, Dna2 has an absolute free 5′-end requirement, similar to its substrate entry requirement for endonuclease activity. This requirement is consistent with the proposed role of the helicase as a partner to the nuclease in the cell. To our knowledge, Dna2 is the only eukaryotic helicase that has an end-loading requirement.
Supplementary Material
Acknowledgments
We thank members of the Bambara and Campbell laboratories for helpful suggestions and discussions. Special thanks to Dr. Steven Matson for critical reading of our manuscript. We would also like to thank Dr. Robert Brosh for critical discussions about the helicase activity.
This work was supported, in whole or in part, by National Institutes of Health Grants GM024441 (to R. A. B.) and GM087666 (to J. L. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- pol
- DNA polymerase
- nt
- nucleotide(s)
- RPA
- replication protein A.
REFERENCES
- 1.West S. C. (1996) Cell 86, 177–180 [DOI] [PubMed] [Google Scholar]
- 2.Patel S. S., Picha K. M. (2000) Annu. Rev. Biochem. 69, 651–697 [DOI] [PubMed] [Google Scholar]
- 3.Davey M. J., O'Donnell M. (2003) Curr. Biol. 13, R594–596 [DOI] [PubMed] [Google Scholar]
- 4.Lohman T. M. (1992) Mol. Microbiol. 6, 5–14 [DOI] [PubMed] [Google Scholar]
- 5.Forsburg S. L. (2004) Microbiol Mol. Biol. Rev. 68, 109–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arezi B., Kuchta R. D. (2000) Trends Biochem. Sci. 25, 572–576 [DOI] [PubMed] [Google Scholar]
- 7.Ayyagari R., Gomes X. V., Gordenin D. A., Burgers P. M. (2003) J. Biol. Chem. 278, 1618–1625 [DOI] [PubMed] [Google Scholar]
- 8.Rossi M. L., Pike J. E., Wang W., Burgers P. M., Campbell J. L., Bambara R. A. (2008) J. Biol. Chem. 283, 27483–27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budd M. E., Campbell J. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7642–7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budd M. E., Choe W. C., Campbell J. L. (1995) J. Biol. Chem. 270, 26766–26769 [DOI] [PubMed] [Google Scholar]
- 11.Boulé J. B., Vega L. R., Zakian V. A. (2005) Nature 438, 57–61 [DOI] [PubMed] [Google Scholar]
- 12.Budd M. E., Reis C. C., Smith S., Myung K., Campbell J. L. (2006) Mol. Cell. Biol. 26, 2490–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bae S. H., Bae K. H., Kim J. A., Seo Y. S. (2001) Nature 412, 456–461 [DOI] [PubMed] [Google Scholar]
- 14.Stewart J. A., Miller A. S., Campbell J. L., Bambara R. A. (2008) J. Biol. Chem. 283, 31356–31365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bae S. H., Kim J. A., Choi E., Lee K. H., Kang H. Y., Kim H. D., Kim J. H., Bae K. H., Cho Y., Park C., Seo Y. S. (2001) Nucleic Acids Res. 29, 3069–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bae S. H., Kim D. W., Kim J., Kim J. H., Kim D. H., Kim H. D., Kang H. Y., Seo Y. S. (2002) J. Biol. Chem. 277, 26632–26641 [DOI] [PubMed] [Google Scholar]
- 17.Formosa T., Nittis T. (1999) Genetics 151, 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bae S. H., Seo Y. S. (2000) J. Biol. Chem. 275, 38022–38031 [DOI] [PubMed] [Google Scholar]
- 19.Kim D. H., Lee K. H., Kim J. H., Ryu G. H., Bae S. H., Lee B. C., Moon K. Y., Byun S. M., Koo H. S., Seo Y. S. (2005) Nucleic Acids Res. 33, 1372–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgers P. M. (2009) J. Biol. Chem. 284, 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choe W., Budd M., Imamura O., Hoopes L., Campbell J. L. (2002) Mol. Cell. Biol. 22, 4202–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao S., Toczylowski T., Yan H. (2008) Nucleic Acids Res. 36, 6091–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G. (2008) Cell 134, 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J. L., Kowalczykowski S. C. (2010) Nature 467, 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wawrousek K. E., Fortini B. K., Polaczek P., Chen L., Liu Q., Dunphy W. G., Campbell J. L. (2010) Cell. Cycle 9, 1156–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng L., Zhou M., Guo Z., Lu H., Qian L., Dai H., Qiu J., Yakubovskaya E., Bogenhagen D. F., Demple B., Shen B. (2008) Mol. Cell. 32, 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lesur I., Campbell J. L. (2004) Mol. Biol. Cell. 15, 1297–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masuda-Sasa T., Polaczek P., Peng X. P., Chen L., Campbell J. L. (2008) J. Biol. Chem. 283, 24359–24373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Budd M. E., Choe W., Campbell J. L. (2000) J. Biol. Chem. 275, 16518–16529 [DOI] [PubMed] [Google Scholar]
- 30.Pokharel S., Jayalath P., Maydanovych O., Goodman R. A., Wang S. C., Tantillo D. J., Beal P. A. (2009) J. Am. Chem Soc. 131, 11882–11891 [DOI] [PubMed] [Google Scholar]
- 31.Cole H. A., Tabor-Godwin J. M., Hayes J. J. (2010) J. Biol. Chem. 285, 2876–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart J. A., Campbell J. L., Bambara R. A. (2010) Nucleic Acids Res. 38, 920–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kao H. I., Campbell J. L., Bambara R. A. (2004) J. Biol. Chem. 279, 50840–50849 [DOI] [PubMed] [Google Scholar]
- 34.Kang Y. H., Lee C. H., Seo Y. S. (2010) Crit. Rev. Biochem. Mol. Biol. 45, 71–96 [DOI] [PubMed] [Google Scholar]
- 35.Kim J. H., Kim H. D., Ryu G. H., Kim D. H., Hurwitz J., Seo Y. S. (2006) Nucleic Acids Res. 34, 1854–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuda-Sasa T., Imamura O., Campbell J. L. (2006) Nucleic Acids Res. 34, 1865–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart J. A., Campbell J. L., Bambara R. A. (2009) J. Biol. Chem. 284, 8283–8291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Copeland W. C., Longley M. J. (2008) Mol. Cell. 32, 457–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niu H., Chung W. H., Zhu Z., Kwon Y., Zhao W., Chi P., Prakash R., Seong C., Liu D., Lu L., Ira G., Sung P. (2010) Nature 467, 108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murante R. S., Rust L., Bambara R. A. (1995) J. Biol. Chem. 270, 30377–30383 [DOI] [PubMed] [Google Scholar]
- 41.Murante R. S., Huang L., Turchi J. J., Bambara R. A. (1994) J. Biol. Chem. 269, 1191–1196 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.