Abstract
Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1), a GPI-anchored endothelial cell protein, binds lipoprotein lipase (LPL) and transports it into the lumen of capillaries where it hydrolyzes triglycerides in lipoproteins. GPIHBP1 is assumed to be expressed mainly within the heart, skeletal muscle, and adipose tissue, the sites where most lipolysis occurs, but the tissue pattern of GPIHBP1 expression has never been evaluated systematically. Because GPIHBP1 is found on the luminal face of capillaries, we predicted that it would be possible to define GPIHBP1 expression patterns with radiolabeled GPIHBP1-specific antibodies and positron emission tomography (PET) scanning. In Gpihbp1−/− mice, GPIHBP1-specific antibodies were cleared slowly from the blood, and PET imaging showed retention of the antibodies in the blood pools (heart and great vessels). In Gpihbp1+/+ mice, the antibodies were cleared extremely rapidly from the blood and, to our surprise, were taken up mainly by lung and liver. Immunofluorescence microscopy confirmed the presence of GPIHBP1 in the capillary endothelium of both lung and liver. In most tissues with high levels of Gpihbp1 expression, Lpl expression was also high, but the lung was an exception (very high Gpihbp1 expression and extremely low Lpl expression). Despite low Lpl transcript levels, however, LPL protein was readily detectable in the lung, suggesting that some of that LPL originates elsewhere and then is captured by GPIHBP1 in the lung. In support of this concept, lung LPL levels were significantly lower in Gpihbp1−/− mice than in Gpihbp1+/+ mice. In addition, Lpl−/− mice expressing human LPL exclusively in muscle contained high levels of human LPL in the lung.
Keywords: Cell Surface Receptor, Lipase, Lipolysis, Lipoprotein, Lipoprotein Receptor
Introduction
The triglyceride-rich lipoproteins (chylomicrons and very low density lipoproteins) undergo lipolytic processing in the capillaries of peripheral tissues, mainly in heart and skeletal muscle, where the lipids are used as fuel, and in adipose tissue, where the lipids are stored (1). Lipolysis depends on lipoprotein lipase (LPL),4 an enzyme that is synthesized and secreted at high levels by myocytes and adipocytes (1, 2). A deficiency of LPL results in extremely high levels of triglycerides in the blood, both in humans (2) and in animal models (3, 4).
For many decades, the mechanism by which LPL reached the lumen of capillaries was mysterious, but this puzzle was solved recently. GPIHBP1 (glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1), a GPI-anchored protein of capillary endothelial cells (5, 6), transports LPL from the interstitial spaces surrounding myocytes and adipocytes into the capillary lumen (7). In Gpihbp1 knock-out mice (Gpihbp1−/−), LPL cannot reach the capillary lumen and therefore remains mislocalized within the interstitial spaces surrounding myocytes and adipocytes (7). Because LPL is absent from capillaries in Gpihbp1−/− mice, the plasma triglyceride levels in those mice are extremely high (8), similar to those in mice with a complete deficiency of LPL (9, 10).
Only limited information has been published on the tissue sites of GPIHBP1 expression (5, 6), but the available data appears consistent with its function in lipoprotein processing. Thus, GPIHBP1 is expressed highly in heart and brown adipose tissue, tissues with prominent and well established roles in lipolysis (6, 7). In contrast, GPIHBP1 is not found in the capillaries of the brain, a tissue that relies primarily on glucose for fuel (6).
A recent study showed that a fluorescently labeled rat monoclonal antibody against GPIHBP1, when injected intravenously into wild-type mice, binds avidly to the luminal surface of capillaries in the heart and white adipose tissue (7). We reasoned that the existence of a GPIHBP1-specific antibody might make it possible to explore GPIHBP1 expression patterns in vivo. In the current study, we used 124I-labeled GPIHBP1-specific monoclonal antibodies, in combination with positron emission tomography (PET) imaging, to define sites of GPIHBP1 expression in mice. These imaging studies were complemented by analyses of GPIHBP1 and LPL transcript and protein levels in multiple tissues.
EXPERIMENTAL PROCEDURES
Mice
Gpihbp1−/− mice have been described previously (6). In some experiments, we used transgenic mice expressing a human LPL transgene exclusively in skeletal muscle and heart. In some cases, we examined human LPL transgenic mice that were homozygous for the knock-out mutation in the mouse Lpl gene. The latter mice express substantial amounts of human LPL activity in skeletal muscle, but the levels are undetectable in adipose tissue (11). Mice were fed a chow diet and housed in a barrier facility with a 12-h light-dark cycle. All studies were approved by the UCLA Animal Research Committee.
Antibodies
For the in vivo GPIHBP1 biodistribution studies, we used a pair of rat monoclonal antibodies (mAbs) against mouse GPIHBP1, 11A12 and 2A8 (12). Control antibodies included a rat monoclonal antibody of the same isotype, 16B10; a hamster monoclonal antibody against CD31, 2H8 (Millipore, Billerica, MA); and a hamster monoclonal antibody against EMR, 3D7 (Abcam, Cambridge, MA).
Immunohistochemistry
To detect GPIHBP1 in mouse tissues, 8–10-μm thick frozen sections were prepared and processed for immunohistochemistry as described (7). Alexa Fluor 555-labeled mAb11A12 (3 μg/ml) was used to detect GPIHBP1. Endothelial cells were identified with the hamster anti-CD31 monoclonal antibody (1:200) and Alexa Fluor 488-labeled goat anti-hamster IgG (1:200). Extracellular matrix was identified with a rabbit anti-collagen IV antibody (1:1000) and an Alexa Fluor 488-labeled donkey anti-rabbit IgG (1:200). Images were obtained with an Axiovert 200 MOT microscope equipped with an Apotome (Zeiss, Germany) or by confocal fluorescence microscopy with a Leica SP2 1P-FCS microscope (Heidelberg, Germany).
Western Blots
Tissue extracts were prepared in radioimmunoprecipitation assay buffer (RIPA: 1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) containing complete mini EDTA-free protease inhibitors (Roche Applied Science). Extracts were size-fractionated on 12% polyacrylamide BisTris gels (Invitrogen), and the separated proteins were transferred to nitrocellulose for Western blotting. Antibody dilutions were 1:200 for a goat antibody against lamin A/C (sc-6215, Santa Cruz Biotechnology); 1:1000 for mAb11A12 (12); and 1:1000 for a goat antibody against a recombinant mouse LPL fragment (13). The binding of IR-coupled secondary antibodies was detected and quantified with an Odyssey infrared imaging scanner (Li-Cor, Lincoln, NE).
Quantitative RT-PCR
Total RNA was prepared from mouse tissues with TriReagent (Sigma), treated with DNase I (Ambion, Austin, TX), and reversed transcribed into cDNA with a mixture of random primers and oligo(dT) and Superscript III (Invitrogen). Primers 5′-AGCAGGGACAGAGCACCTCT-3′ and 5′-AGACGAGCGTGATGCAGAAG-3′ were used to amplify the mouse Gpihbp1 cDNA; primers 5′-AGGTGGACATCGGAGAACTG-3′ and 5′-TCCCTAGCACAGAAGATGACC-3′ were used to amplify mouse Lpl cDNA; primers 5′-TAGCTGGTCAGACTGGTGGA-3′ and 5′-TTCACAAATACCGCAGGTG-3′ were used to amplify human LPL cDNA; and primers 5′-TGGTGCTTGTCTCACTGACC-3′ and 5′-TATGTTCGGCTTCCCATTCT-3′ were used to amplify mouse β2-microglobulin cDNA. Quantitative PCR was performed on 50 ng of cDNA, 200 nm of each primer, and 10 μl of SYBR Green PCR master mix (Qiagen, Valencia, CA). PCR were performed in triplicate on a 7900HT Fast Real Time PCR system (Applied Biosystems, Foster City, CA). Gene expression levels normalized to β2-microglobulin were calculated by the comparative CT method.
Measurement of LPL in Tissues
Mice were fasted overnight. After adding food to the cages for 1 h, the mice were fasted for 4 h before being euthanized. Tissue (100 mg) was homogenized with a Fisher Scientific PowerGen 125 in 1.0 ml of lysis solution (13). Samples were centrifuged at 20,000 × g for 30 min at 4 °C. The supernatant fractions were collected and stored at −80 °C. LPL levels in these samples were determined by ELISA (13).
Radioiodinations
Antibodies (25–100 μg) were radioiodinated with either Na131I (PerkinElmer Life Sciences) or Na124I (IBA Molecular, Sterling, VA) with IODO-GEN-coated tubes (Thermo Scientific, Rockford, IL) (14–18). Radiolabeling efficiencies were measured by instant thin-layer chromatography with Tec-Control chromatography strips (Biodex Medical Systems, Shirley, NY) and a running buffer of 0.9% NaCl. The mean labeling efficiency of mAb11A12 with 124I was 65% (n = 4; range, 35–79), whereas it was 93% with 131I. For mAb2A8, the labeling efficiencies with 124I and 131I were 69 and 96%, respectively. The labeling efficiencies for rat mAb16B10, hamster anti-CD31 mAb2H8, and hamster anti-EMR mAb3D7 with 131I were 90.3, 86.7, and 97.7%, respectively. Free iodine was removed with Micro Bio-Spin 6 columns, and the removal was documented by instant TLC. The functional integrity of the GPIHBP1-specific antibodies after radiolabeling was assessed by dot blotting using extracts of mouse GPIHBP1-transfected cells.
PET Imaging and Image Analyses
Imaging studies were performed with an Inveon scanner, a dedicated PET system (Siemens Preclinical Solutions, Knoxville, TN), and anatomic CT imaging was provided with 10-min scans using a microCATII scanner (Concorde Microsystems, Knoxville, TN). An anatomic CT image was also used for attenuation correction in generating the PET images. For dynamic PET scans, wild-type and Gpihbp1−/− mice (n = 4/group) were induced and maintained under anesthesia with 1–2% isoflurane, positioned in the scanner, and injected intravenously via the tail vein with 124I-mAb11A12 (151–217 μCi; specific activity, 9.6 μCi/μg). Each mouse was imaged continuously for 2 h, followed by a CT scan. An additional 10-min static PET scan was obtained 24 h after injection. After the final scan, each mouse was euthanized and perfused extensively with PBS (5 ml through the right ventricle and 15 ml through the left ventricle), and the liver removed. The hepatectomized mice and livers were then imaged separately (acquisition time, 20 min).
To assess tissue uptake at 30 min, wild-type and Gpihbp1−/− mice (n = 4/group) were injected intravenously with 124I-mAb11A12 (50–106 μCi; specific activity, 1.7–9.6 μCi/μg). After 30 min, the mice were euthanized and perfused with PBS. The livers were removed, and the hepatectomized mice were imaged for 30 min. The liver, kidneys, spleen, lungs, heart, brown adipose tissue, and quadriceps muscle were then imaged separately for 30 min. Wild-type and Gpihbp1−/− mice (n = 2/group) were also injected intravenously with 124I-mAb2A8 (80 μCi/injection; specific activity, 5.4 μCi/μg). After 30 min, the mice were euthanized, perfused, hepatectomized, and imaged for 30 min.
The PET and CT scans were co-registered to yield a single image with AMIDE software (19). The color-coded PET images were scaled to the level of accumulated radioactivity, expressed as %ID/cc (percentage of injected dose per cubic centimeter), determined by plotting the injected dose (μCi) and the empirically determined cylinder factor value 2 μCi/cc/image units into the modification dialog window of AMIDE.
Biodistribution and Blood Activity Studies
To determine the biodistribution of 124I-labeled mAb11A12 or mAb2A8, the blood and organs/tissues-of-interest were harvested at either 0.5 or 24 h post-injection, weighed, and counted in a WIZARD automatic γ-counter (PerkinElmer Life Sciences). Similarly, blood and tissue levels of 131I-labeled antibodies were determined in perfused mice 30 min after antibody injections (11–52 μCi; specific activity, 1.0–6.6 μCi/μg). Uptake of radioactivity into tissues was corrected for decay and expressed as a percentage of injected dose/g of tissue (% ID/g) (mean ± S.E. in cases where groups of four mice were evaluated).
After an injection of 131I-mAb11A12 (11 μCi; specific activity, 0.9 μCi/μg) into wild-type and Gpihbp1−/− mice (n = 4/group), blood samples (10 μl each) were collected at 1, 2.5, 5, 10, 15, 30, and 60 min, and at 4, 8, 12, 24, 48, and 72 h. At 72 h, mice were euthanized; tissues were harvested without perfusion; and % ID/g of tissue was determined. Blood radioactivity curves were generated, and two rate constants (k1 and k2) were calculated with GraphPad Prism 5 software (SHI, El Segundo, CA). The α and β half-lives of 131I-mAb11A12 were derived from the formula: t1/2α = ln2/k1 and t1/2β = ln2/k2.
RESULTS
Imaging and Biodistribution Studies with Gpihbp1-specific Monoclonal Antibodies
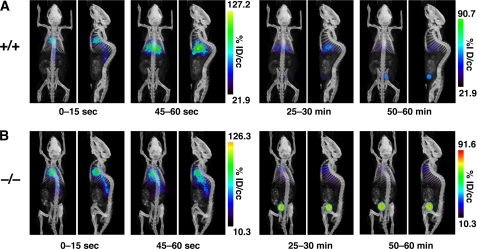
The ability to detect GPIHBP1 at the luminal surface of capillaries with a fluorescently labeled antibody against GPIHBP1 (7) raised the possibility that one could define the distribution of GPIHBP1 in vivo with PET imaging. To test this idea, we imaged mice after injecting a 124I-labeled rat monoclonal antibody against GPIHBP1 (124I-mAb11A12). Gpihbp1−/− and wild-type mice (n = 4/group) were injected intravenously with a small amount of mAb11A12 (14 μg; specific activity 9.6 μCi/μg), and then subjected to dynamic imaging by PET (Fig. 1). In the first few seconds after the injection, most of the radioactivity in both groups of mice was found in the blood pool (heart and great vessels). At 1 min, most of the radioactivity in wild-type mice was located in the thoracic cavity (heart/lung) and liver (Fig. 1A and supplemental Movie A), whereas it remained in the blood pool in Gpihbp1−/− mice (Fig. 1B and supplemental Movie B). After 1 h, radioactivity remained in the liver and heart/lung of wild-type mice and in the blood pool of Gpihbp1−/− mice (in addition, some radioactivity appeared in the bladder).
FIGURE 1.
Dynamic PET/CT images of wild-type and Gpihbp1−/− mice after an intravenous injection of a 124I-labeled GPIHBP1-specific monoclonal antibody (mAb11A12). A, images of the wild-type (+/+) mouse. B, images of the Gpihbp1−/− (−/−) mouse. The PET images are color-scaled to the level of radioactivity, expressed as percentage of injected dose per cubic centimeter (% ID/cc). Coronal and sagittal images are shown for specific time periods after the injection: 0–15 s, 45–60 s, 25–30 min, and 50–60 min. In the first 15 s after the injection, the radioactivity is located predominantly in blood pools (i.e. heart and large vessels). By 45–60 s (and also at the later time points), most of the radioactivity in the wild-type mouse was located in the liver, whereas the radioactivity in the Gpihbp1−/− mouse remained in the blood pool. Radioactivity was observed in the bladder at later time points.
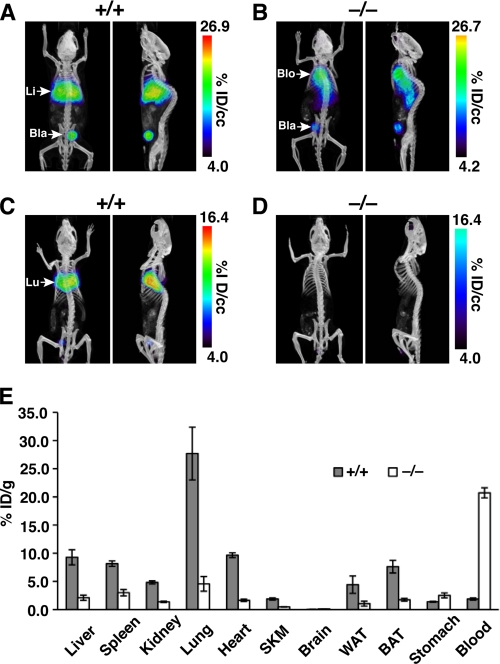
After 24 h, a strong signal persisted in the liver/lung/heart of the wild-type mouse (Fig. 2A), whereas most of the radioactivity in the Gpihbp1−/− mouse remained in the blood (Fig. 2B). To resolve the heart/lung signal from the liver signal with confidence, we euthanized the mice, perfused them with PBS, removed the liver, and then re-imaged the mice (Fig. 2, C and D). After the hepatectomy, a strong signal was observed in the thoracic cavity of wild-type mice (Fig. 2C), but not in Gpihbp1−/− mice (Fig. 2D). To quantify the specific uptake of the antibody in different tissues, radioactivity in different tissues was counted and plotted as the percentage of injected dose/g (% ID/g) of tissue (n = 4/group) (Fig. 2E). Large differences between wild-type and Gpihbp1−/− mice were observed in lungs (p = 0.0103) and blood (p = 0.00003); significant differences were also observed in liver, spleen, kidneys, heart, skeletal muscle, white adipose tissue, and brown adipose tissue (p ≤ 0.012 for all) (Fig. 2E). The levels of uptake of antibody 11A12 in the brain was extremely low in wild-type mice (and no different from the levels of uptake in Gpihbp1−/− mice), consistent with the absence of Gpihbp1 expression in the capillaries of the brain (6, 7).
FIGURE 2.
Biodistribution of 124I-mAb11A12 in wild-type and Gpihbp1−/− mice 24 h after the antibody injection. A and B, coronal and sagittal views of PET/CT images from living mice 24 h after the injection of 124I-mAb11A12. A strong signal was observed in the liver (Li) in the wild-type (+/+) mouse (A), whereas the radioactivity was located mainly in the blood pool (Blo) in the Gpihbp1−/− (−/−) mouse (B). Radioactivity was also observed in the bladder (Bla). C and D, coronal and sagittal views of the same mice after perfusion with PBS and hepatectomy. A strong signal was observed in the lungs (Lu) of the wild-type mouse (C), whereas no lung signal was observed in the Gpihbp1−/− mouse (D). E, biodistribution of 124I-mAb11A12 in tissues from PBS-perfused wild-type and Gpihbp1−/− mice (n = 4/group) 24 h post-injection. Error bars represent S.E.
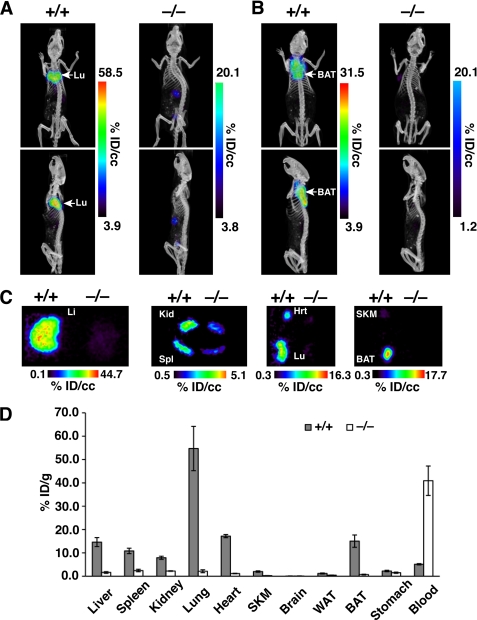
In a separate experiment, wild-type and Gpihbp1−/− mice (n = 4/group) were injected with 124I-mAb11A12 (10–20 μg; specific activity 1.7–3.1 μCi/μg). After 30 min, the mice were euthanized and perfused with PBS, hepatectomized, and imaged. A strong signal was observed in the thoracic cavity of wild-type mice, but not in Gpihbp1−/− mice (Fig. 3A). Next, the heart and lungs were excised and the mice were re-imaged. With the liver, heart, and lungs removed, a strong signal was observed in brown adipose tissue of wild-type mice (Fig. 3B), but not in Gpihbp1−/− mice (Fig. 3B). The excised lungs, heart, liver, and other tissues of interest were then imaged separately, revealing substantially higher amounts of radioactivity in wild-type tissues (Fig. 3C). Antibody uptake was also assessed by measuring radioactivity in tissue samples, adjusted for weight. Striking differences between wild-type and Gpihbp1−/− mice were observed in the lung (p = 0.013) and the blood (p = 0.0031); significant differences were also observed in the liver, spleen, kidneys, heart, skeletal muscle, and brown adipose tissue (p ≤ 0.013) (Fig. 3D).
FIGURE 3.
Biodistribution of 124I-mAb11A12 30 min after the antibody injection. A and B, ex vivo PET/CT images of a wild-type mouse and a Gpihbp1−/− mouse. Each mouse was given an intravenous injection of 124I-mAb11A12. After 30 min, the mice were euthanized and perfused with PBS. After removing the liver, the mouse carcasses were imaged. The top panels show coronal images; the lower panels show sagittal images. Intense radioactivity was observed in the lung (Lu) of the wild-type mouse, but not the Gpihbp1−/− mouse. B, images of the same mice after removing the spleen, kidney, lungs, and heart. An intense signal was observed in the brown adipose tissue (BAT) of the wild-type mouse, but not the Gpihbp1−/− mouse. C, PET images of the excised liver (Li), kidney (Kid), spleen (Spl), heart (Hrt), lung (Lu), skeletal muscle (SKM), and brown adipose tissue (BAT) from a wild-type mouse (left side of each image) and Gpihbp1−/− mouse (right side of each image). D, biodistribution at 30 min of 124I-mAb11A12 in different tissues of wild-type and Gpihbp1−/− mice (n = 4/group) adjusted for tissue weight. 30 min after the antibody injection, each mouse was euthanized and perfused with PBS. Error bars represent S.E.
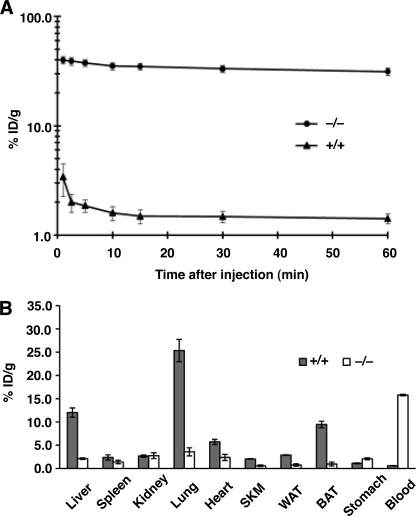
The rapid disappearance of 124I-mAb11A12 from the blood pool of wild-type mice (Fig. 1A) and the lower levels of 124I-mAb11A12 radioactivity in the blood of wild-type mice at 30 min (Fig. 3D) led us to suspect that mAb11A12 was cleared very rapidly from the blood of wild-type mice. Indeed, when mAb11A12 was labeled with 131I and injected intravenously into wild-type and Gpihbp1−/− mice (11 μg; specific activity 1.06 μCi/μg), the radioactivity was cleared rapidly in wild-type mice, with less than 2% remaining in the blood after 10 min (Fig. 4A). In contrast, >30% of the radioactivity remained in the plasma of Gpihbp1−/− mice after 60 min (Fig. 4A). Additional blood samples were taken at 4, 8, 12, 24, 48, and 72 h after injection. The terminal half-life of the radiolabeled antibody was 2.9 h in wild-type mice versus 56.8 h in Gpihbp1−/− mice. After the 72-h time point, the mice were euthanized, and the radioactivity in different tissues was quantified (Fig. 4B). Significant differences in 131I-mAb11A12 uptakes between wild-type and Gpihbp1−/− mice were noted in the liver (p < 0.001), lung (p < 0.001), brown adipose tissue (p < 0.0001), heart (p = 0.004), white adipose tissue (p = 0.0011), and blood (p < 0.00001) (Fig. 4B).
FIGURE 4.
Clearance and biodistribution of 131I-mAb11A12 in wild-type and Gpihbp1−/− mice. A, clearance of 131I-mAb11A12 from the bloodstream of wild-type (+/+) and Gpihbp1−/− (−/−) mice during the first 60 min after an intravenous injection of the antibody. The graph shows the percentage of injected dose/g of plasma at each time point. Data shows mean ± S.E. for each group of mice (n = 4/group). B, biodistribution of 131I-mAb11A12 in tissues of separate groups of wild-type and Gpihbp1−/− mice (n = 4/group) 72 h post-injection. In this experiment, the mice were not perfused with PBS. Error bars represent S.E.
Because we observed far slower clearance of mAb11A12 in Gpihbp1−/− mice than in wild-type mice, we suspected that the differences were specific (i.e. due to the absence of GPIHBP1 in Gpihbp1−/− mice). To confirm this suspicion, we labeled a different GPIHBP1-specific rat monoclonal antibody, mAb2A8, with 124I (specific activity 5.4 μCi/μg), and then injected a small amount (11 μg) intravenously into wild-type and Gpihbp1−/− mice. After 30 min, the mice were perfused with PBS; the liver was removed; and the mice were imaged (supplemental Fig. S1A). Consistent with the findings with mAb11A12, large amounts of 124I-mAb2A8 uptake were observed in the heart/lung of the wild-type mouse, but not in the Gpihbp1−/− mouse (supplemental Fig. S1A). When the amount of radioactivity in individual tissues was counted, large differences were observed in the liver, spleen, lung, heart, brown adipose tissue, skeletal muscle, and blood (supplemental Fig. S1B).
We also compared the specific tissue uptake of mAb2A8 and an irrelevant rat monoclonal antibody of the same isotype (mAb16B10) in wild-type and Gpihbp1−/− mice. After 30 min, the mice were perfused, and antibody uptake in different tissues was assessed (supplemental Fig. S2). We observed substantial differences in the tissue uptake of mAb2A8 in wild-type and Gpihbp1−/− mice, but no differences were observed in the uptake of the irrelevant antibody mAb16B10 (supplemental Fig. S2).
Both mAb11A12 and mAb2A8 were cleared quickly from the bloodstream in wild-type mice but not Gpihbp1−/− mice (Figs. 1–4 and supplemental Fig. S1). To be sure that this was simply due to the absence of GPIHBP1, and not to a global endothelial cell abnormality elicited by GPIHBP1 deficiency, we examined the biodistribution of an antibody against CD31 (another endothelial cell plasma membrane protein) in wild-type and Gpihbp1−/− mice. We injected a 131I-labeled hamster monoclonal antibody against CD31 (specific activity, 5.0 μCi/μg) and a 131I-labeled isotype-control hamster monoclonal antibody, mAb3D7 (specific activity, 6.6 μCi/μg), into wild-type and Gpihbp1−/− mice. After 30 min, the mice were perfused, and the organs harvested and counted. Of note, the biodistributions of both antibodies were similar in wild-type and Gpihbp1−/− mice (supplemental Fig. S3). When compared with the irrelevant isotype control antibody, there was greater uptake of the CD31-specific antibody in the lung (more than 10-fold) (supplemental Fig. S3), consistent with the large numbers of endothelial cells in the lung.
Molecular Analysis of GPIHBP1 Expression
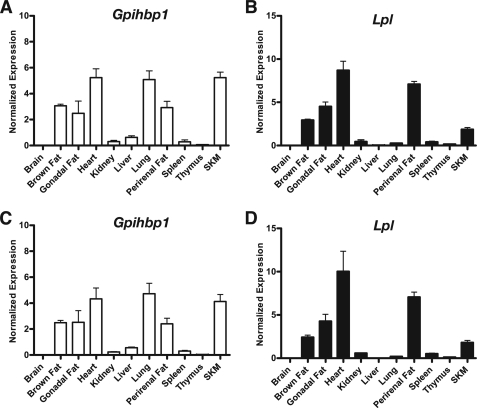
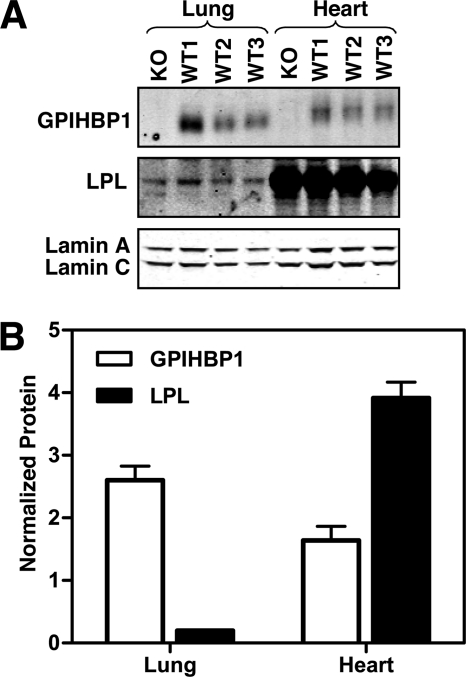
Finding high levels of antibody uptake in the liver and lung was surprising because earlier studies had pointed out that GPIHBP1 is expressed primarily in heart, skeletal muscle, and adipose tissue (tissues with high levels of LPL expression) (6). To investigate this finding, we examined GPIHBP1 and LPL expression by quantitative RT-PCR and Western blots. Gpihbp1 transcript levels, at both 4 and 10 weeks of age, were high in heart, skeletal muscle, and adipose tissue, but were also very high in the lung (Fig. 5). In tissues where Gpihbp1 transcript levels were high, Lpl expression was also high, except for the lung. In the lung, Lpl expression was extremely low. Gpihbp1 transcript levels were low to moderate in the liver (Fig. 5). Western blots showed high levels of LPL expression in the heart (Fig. 6), consistent with the mRNA results. Although Lpl transcripts were extremely low in the lung (Fig. 5), LPL protein was nevertheless, detectable by Western blot (a single band at 52 kDa) (Fig. 6).
FIGURE 5.
Gpihbp1 and Lpl transcript levels in tissues of wild-type mice. Gpihbp1 and Lpl expression levels were assessed by quantitative RT-PCR and normalized to the expression of β2-microglobulin. Bar graphs show the expression of Gpihbp1 (A and C) and Lpl (B and D) in 4-week-old (A and B) and 10-week-old (C and D) mice (mean ± S.E.). Error bars represent S.E.
FIGURE 6.
GPIHBP1 and LPL protein levels in mouse tissues, as judged by Western blots. A, Western blot detection of GPIHBP1 and LPL in lung and heart of a Gpihbp1−/− (KO) mouse and three wild-type (WT) mice. An antibody against lamin A/C was used as a loading control. B, bar graph showing quantitative analysis of protein levels in wild-type mice (from the Western blot shown in panel A). GPIHBP1, white bars; LPL, black bars. Signals on Western blots were detected with an Odyssey Li-Cor scanner and normalized to lamin C. Error bars represent S.E.
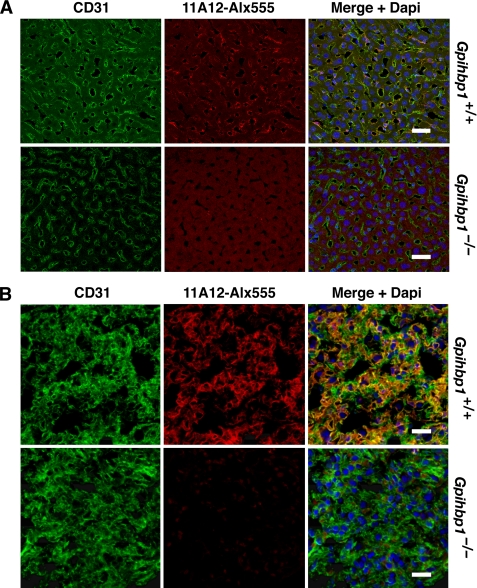
In heart, skeletal muscle, and adipose tissue, GPIHBP1 is expressed exclusively in capillary endothelial cells (6, 7). To define the site of GPIHBP1 expression in the liver and lung, we performed immunohistochemistry with the GPIHBP1-specific mAb11A12. In both liver and lung, GPIHBP1 was found in capillary endothelial cells, colocalizing with the endothelial cell marker CD31 (Fig. 7, A and B). GPIHBP1-specific staining was also present in the kidney and spleen of wild-type mice (supplemental Fig. S4).
FIGURE 7.
Confocal immunofluorescence microscopy of GPIHBP1 in the liver and lung of wild-type and Gpihbp1−/− mice. A, immunohistochemistry of liver sections with an Alexa Fluor 555-labeled rat monoclonal antibody against GPIHBP1 (mAb11A12) (red) and a hamster monoclonal antibody against CD31. Binding of hamster antibodies was detected with Alexa Fluor 488-labeled anti-hamster IgG (green). Nuclei were stained with DAPI (blue). B, immunolocalization of GPIHBP1 in the lung. GPIHBP1 and CD31 were detected as described for the liver in panel A. Images were taken with a ×63 objective. Scale bar represents distance of 30 μm.
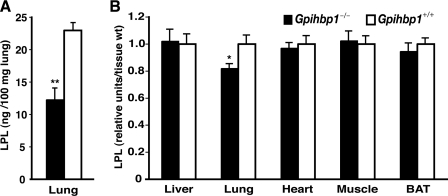
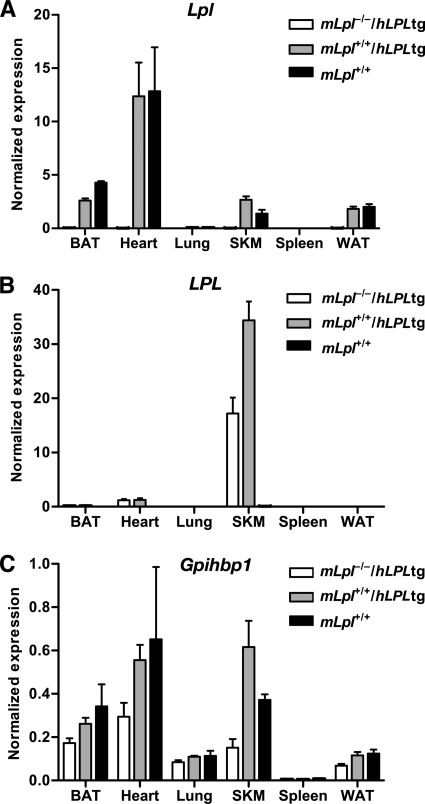
The lung had negligible levels of Lpl transcripts (Fig. 5), yet still had detectable levels of LPL protein (Fig. 6). These findings led us to hypothesize that some of the LPL in the lung might have originated in other tissues and subsequently captured by unoccupied GPIHBP1. If this hypothesis were correct, one would expect to find lower levels of LPL in the lungs of Gpihbp1−/− mice. Indeed, in two independent experiments, LPL mass levels in the lung were lower in Gpihbp1−/− mice than in wild-type mice (Fig. 8). In other tissues, LPL mass levels were identical in wild-type and Gpihbp1−/− mice (Fig. 8). Also, if this hypothesis were correct, we reasoned that it should be possible to identify LPL in the lung in cases where the LPL had been synthesized elsewhere. To examine this idea, we investigated whether human LPL was present in the lungs of mice that overexpressed a human LPL transgene exclusively in skeletal muscle (the transgene was driven by a skeletal muscle creatine kinase promoter) (20). As expected, the human LPL transgenic mice expressed high levels of human LPL in skeletal muscle, but expression was absent in the lung (Fig. 9). Despite an absence of human LPL expression in the lung, however, human LPL protein could be detected in the lungs of PBS-perfused transgenic mice (experiment 1, 714 ± 258 ng/ml in lung extracts versus <15 ng/ml in control mice; experiment 2, 331 ± 45 ng/ml of lung extract versus undetectable in control mice). (For each experiment, the tissue extracts contained 100 mg of tissue/ml.) In parallel, we found that the amount of mouse LPL in the lung of wild-type mice (n = 5) was 12 ± 8 ng/ml. In a third experiment, we measured human and mouse LPL in the lungs and white adipose tissue of MCK-LPL mice (n = 4/group). The amount of human LPL in the lung was 1364 ± 584 versus 106 ± 63 in white adipose tissue. In contrast, the amount of mouse LPL was higher in white adipose tissue than in lung (58 ± 12 versus 13 ± 7 ng/ml). These findings, along with the finding of significantly lower levels of mouse LPL in the lungs of Gpihbp1−/− mice, support the idea that some of the human LPL synthesized by the skeletal muscle and heart of MCK-LPL transgenic mice reaches the circulation and can be captured by GPIHBP1 in the lung.
FIGURE 8.
LPL protein levels in the tissues of wild-type and Gpihbp1−/− mice. Mice were fasted overnight; the next morning, after refeeding for 1 h, the mice were fasted for an additional 4 h and then euthanized and perfused with PBS. LPL mass levels in tissues were measured by ELISA (13). A, levels of LPL in multiple tissues of wild-type (n = 8) and Gpihbp1−/− (n = 8) mice. B, LPL levels in lungs of wild-type (n = 4) and Gpihbp1−/− (n = 6) mice. Error bars represent S.E.
FIGURE 9.
Expression of human LPL in tissues of Lpl knock-out mice that express a human LPL transgene driven by a muscle creatine kinase promoter (mLpl−/−/hLPL tg). A–C, expression of mouse Lpl, human LPL, and mouse Gpihbp1 transcripts in tissues of mLpl−/−/hLPL tg, mLpl+/+/hLPL tg, and Lpl+/+ mice were measured by RT-PCR and normalized to the expression of β2-microglobulin. Error bars represent S.E.
DISCUSSION
The milky plasma and high plasma triglyceride levels in Gpihbp1−/− mice (6, 8) demonstrated that GPIHBP1 was somehow important for lipolysis, but the mechanism was not initially clear. Within the past year, Davies and co-workers (7) made sense of the hyperlipidemia: they showed that GPIHBP1 shuttles LPL from the interstitial spaces of tissues, where it is secreted by parenchymal cells, into the capillary lumen, the site where it needs to be to hydrolyze triglycerides in lipoproteins. Although the function of GPIHBP1 at the molecular level has come into focus, little is known about GPIHBP1 at the whole animal level, including some of the fundamentals, such as the expression pattern of GPIHBP1 in different tissues. In the current study, we addressed that issue. Using PET imaging (16–18), we examined the uptake and clearance of GPIHBP1-specific monoclonal antibodies in both wild-type and Gpihbp1−/− mice. Because GPIHBP1 is an endothelial cell protein, we were not surprised to find that GPIHBP1-specific antibodies were cleared rapidly from the bloodstream of wild-type mice. Furthermore, because GPIHBP1 is entirely absent from Gpihbp1−/− mice, we were also not surprised by the slow clearance of antibodies in those animals. However, we were definitely surprised by the tissue sites of antibody uptake. We anticipated that GPIHBP1-specific antibodies would be cleared primarily in the heart, skeletal muscle, and adipose tissue (i.e. tissues that are most active in the lipolysis), but this was not the case. In wild-type mice, GPIHBP1-specific antibodies were taken up mainly by the liver, and after that, by the lungs (on a per organ basis). Remarkably, the uptake of GPIHBP1-specific antibodies, per gram of tissue, was greater in the lung than in any other tissue, and the uptake of antibodies in the liver, per gram of tissue, was equal or greater than in heart or adipose tissue. By PET imaging, antibody uptake in the liver uptake was dominant, a reflection of the large size of the liver (e.g. ∼9-fold greater weight than the heart). After removing the liver, the lungs were the predominant site of antibody uptake.
There is little doubt that the binding of GPIHBP1 antibodies to lung and liver was specific. First, uptake of the antibodies by the liver and heart of Gpihbp1−/− mice was negligible. Second, uptake of an irrelevant antibody by liver and heart was equally low in wild-type and Gpihbp1−/− mice. Third, both lung and liver contained GPIHBP1, as judged by immunohistochemistry. The GPIHBP1 was found in capillary endothelial cells, colocalizing with CD31 in the liver and the lung.
Different biodistributions of GPIHBP1-specific antibodies in wild-type and Gpihbp1−/− mice were observed very quickly after the antibody injection. Within 45–60 s of injecting the radiolabeled antibody, uptake of radioactivity was prominent in the liver of wild-type mice, whereas the radioactivity remained in the blood pools of Gpihbp1−/− mice. Finding large differences at the early time points makes sense, given that GPIHBP1 is expressed exclusively on capillary endothelial cells, meaning that the antibody does not need to percolate into the interstitial spaces to reach its target. Interestingly, the radiolabeled GPIHBP1 antibodies persisted in the tissues of wild-type mice for many hours, suggesting that antibody-GPIHBP1 complexes in endothelial cells are long-lived and not quickly degraded. This was quite evident by the biodistribution at 72 h, where the radioactivity uptake in the liver and lung were 5–6-fold higher in wild-type mice (∼12 and ∼25% ID/g, respectively) than in Gpihbp1−/− mice (∼2 and ∼5% ID/g, respectively).
The finding that the lungs and liver of wild-type mice were the main sites of antibody uptake did not mean that differences between wild-type and Gpihbp1−/− mice were absent in other tissues. Highly significant differences between Gpihbp1−/− and wild-type mice were observed in nearly every tissue, including the heart, adipose tissue, kidney, skeletal muscle, and spleen. No differences were observed for antibody uptake in the brain. These findings make sense because some GPIHBP1 can be found in virtually every mouse tissue by immunohistochemistry, aside from the brain, where its expression is undetectable (7).
Very high levels of Gpihbp1 transcripts were generally accompanied by high levels of Lpl transcripts, but the lung was an exception. Gpihbp1 transcript levels were as high as in the lung as in any other tissue, including the heart, but Lpl transcript levels in the lung were extremely low. Despite low levels of Lpl transcripts in the lung, we had no difficulty in detecting LPL protein in lungs by Western blotting. The fact that the LPL protein was detectable despite low levels of Lpl transcripts raised the possibility that some of the LPL in the lung might be produced elsewhere and then transported to the lung and captured by GPIHBP1. In support of this scenario, we found substantial amounts of human LPL in the lungs of muscle creatine kinase-human LPL transgenic mice, even though the transgene was not expressed in the lung. Also, the concept that the LPL in the lung might be captured by GPIHBP1 was supported by the fact that the levels of LPL in the lung were significantly lower in Gpihbp1−/− mice than in wild-type control mice. In contrast, there were no significant differences in LPL protein levels in other tissues. The proposition that LPL might be synthesized and secreted by one tissue and then be transported to another tissue is a new concept, as far as we are aware. However, this concept has been proposed previously for hepatic lipase, a structurally related lipase family member. Doolittle and co-workers (21) found that hepatic lipase was present in the adrenal gland, even though hepatic lipase transcripts were absent, and proposed that the hepatic lipase in the adrenal gland was synthesized elsewhere and then captured within the adrenal gland.
The function of LPL in the lungs is not clear, particularly because neither Gpihbp1−/− mice nor adult Lpl knock-out mice have noticeable pulmonary abnormalities. One possibility is that the high level of GPIHBP1 expression in the capillaries of the lungs functions to remove enzymatically active LPL from the bloodstream. Active LPL homodimers are secreted by adipocytes and myocytes into the interstitial fluid, and it seems possible that at least a small fraction of that LPL drains into the lymph and ultimately enters the circulation through the subclavian vein. The first capillary bed encountered by that LPL would be in the lung. Thus, it seems possible that GPIHBP1 expression in the lung could function to reduce levels of active LPL dimers that “leak” into the circulation. In humans and mice, some LPL protein can be detected in the plasma, but most of that LPL is in the form of monomers, which are enzymatically inactive and cannot bind GPIHBP1. Of course, it is also possible that the capture of LPL by GPIHBP1 in the lung could facilitate lipid metabolism in the lung. The pneumocytes of the lung synthesize large amounts of phospholipids for the surfactant lining of alveoli, and it is possible that lipolysis provides lipid substrates for this task.
We must acknowledge the possibility that GPIHBP1 serves some other role in the lung, perhaps one having nothing to do with LPL or lipid metabolism. The strongly negatively charged amino terminus of GPIHBP1 could allow it to bind some other, as-yet-unidentified protein with strong positively charged domains. Of course, we cannot exclude the possibility of another substrate, and another function, for GPIHBP1 in the lung, but it is noteworthy that Gpihbp1 knock-out mice appear quite healthy, aside from their hyperlipidemia.
Why the liver expresses GPIHBP1 is unclear. LPL expression in the liver is normally quite low, although its expression can be activated by high-cholesterol diets. The capillaries of the liver are fenestrated, so it is not clear that the capillaries of the liver would need a transporter to move LPL from one side of endothelial cells to the other. It seems possible that GPIHBP1 in the liver also functions to bind enzymatically active LPL homodimers and remove them from the bloodstream. Indeed, there is some evidence that the liver carries out this function (22, 23).
In summary, the use of 124I-labeled GPIHBP1-specific monoclonal antibodies, along with PET imaging, yielded new and unexpected insights into GPIHBP1 expression and function. First, we found substantial amounts of GPIHBP1 in the lungs and the liver, tissues that are not thought to have important roles in LPL-mediated processing of triglyceride-rich lipoproteins. Second, we present evidence that LPL in the lung may originate in other tissues and is captured, at least in part, by the GPIHBP1 in the lung. Third, our studies confirm that Gpihbp1 is expressed highly in all tissues with well established roles in the lipolytic processing of lipoproteins (e.g. heart, muscle, and adipose tissue), paralleling high levels of Lpl expression. Fourth, we show that GPIHBP1 expression is absent in the brain, a tissue that relies on glucose for fuel. Fifth, our studies show that specific antibody reagents, combined with PET scanning, are useful for understanding in vivo roles of key proteins in plasma lipid metabolism.
Supplementary Material
Acknowledgments
We thank Drs. David Stout and Waldemar Ladno (UCLA Crump Institute for Molecular Imaging) for the use of the microPET/CT equipment and for assisting with the imaging studies.
This work was supported, in whole or in part, by National Institutes of Health Grants 1RC1 HL100008, R01 HL094732, P01 HL090553, RO1 HL086683, R01 HL087228, R01 EB001943, P30 CA016042, U24 CA092865, and R25 CA098010 and a grant from the American Heart Association, Western States Affiliate.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Movies A and B.
- LPL
- lipoprotein lipase
- PET
- positron emission tomography
- GPIHBP1
- glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Havel R. J., Kane J. P. (2001) in The Metabolic and Molecular Bases of Inherited Disease (Scriver C. R., Beaudet A. L., Sly W. S., Valle D., Childs B., Kinzler K. W., Vogelstein B. eds) 8th Ed., McGraw-Hill, New York [Google Scholar]
- 2.Brunzell J. D., Deeb S. S. (2001) in The Metabolic and Molecular Bases of Inherited Disease (Scriver C. R., Beaudet A. L., Sly W. S., Valle D., Childs B., Kinzler K. W., Vogelstein B. eds) 8th Ed., McGraw-Hill, New York [Google Scholar]
- 3.Weinstock P. H., Bisgaier C. L., Aalto-Setälä K., Radner H., Ramakrishnan R., Levak-Frank S., Essenburg A. D., Zechner R., Breslow J. L. (1995) J. Clin. Invest. 96, 2555–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginzinger D. G., Clee S. M., Dallongeville J., Lewis M. E., Henderson H. E., Bauje E., Rogers Q. R., Jensen D. R., Eckel R. H., Dyer R., Innis S., Jones B., Fruchart J. C., Hayden M. R. (1999) Eur. J. Clin. Invest. 29, 17–26 [DOI] [PubMed] [Google Scholar]
- 5.Ioka R. X., Kang M. J., Kamiyama S., Kim D. H., Magoori K., Kamataki A., Ito Y., Takei Y. A., Sasaki M., Suzuki T., Sasano H., Takahashi S., Sakai J., Fujino T., Yamamoto T. T. (2003) J. Biol. Chem. 278, 7344–7349 [DOI] [PubMed] [Google Scholar]
- 6.Beigneux A. P., Davies B. S., Gin P., Weinstein M. M., Farber E., Qiao X., Peale F., Bunting S., Walzem R. L., Wong J. S., Blaner W. S., Ding Z. M., Melford K., Wongsiriroj N., Shu X., de Sauvage F., Ryan R. O., Fong L. G., Bensadoun A., Young S. G. (2007) Cell Metab. 5, 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies B. S., Beigneux A. P., Barnes R. H., 2nd, Tu Y., Gin P., Weinstein M. M., Nobumori C., Nyrén R., Goldberg I., Olivecrona G., Bensadoun A., Young S. G., Fong L. G. (2010) Cell Metab. 12, 42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinstein M. M., Yin L., Tu Y., Wang X., Wu X., Castellani L. W., Walzem R. L., Lusis A. J., Fong L. G., Beigneux A. P., Young S. G. (2010) Arterioscler. Thromb. Vasc. Biol. 30, 20–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strauss J. G., Frank S., Kratky D., Hämmerle G., Hrzenjak A., Knipping G., von Eckardstein A., Kostner G. M., Zechner R. (2001) J. Biol. Chem. 276, 36083–36090 [DOI] [PubMed] [Google Scholar]
- 10.Zhang X., Qi R., Xian X., Yang F., Blackstein M., Deng X., Fan J., Ross C., Karasinska J., Hayden M. R., Liu G. (2008) Circ. Res. 102, 250–256 [DOI] [PubMed] [Google Scholar]
- 11.Levak-Frank S., Weinstock P. H., Hayek T., Verdery R., Hofmann W., Ramakrishnan R., Sattler W., Breslow J. L., Zechner R. (1997) J. Biol. Chem. 272, 17182–17190 [DOI] [PubMed] [Google Scholar]
- 12.Beigneux A. P., Gin P., Davies B. S., Weinstein M. M., Bensadoun A., Fong L. G., Young S. G. (2009) J. Biol. Chem. 284, 30240–30247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinstein M. M., Yin L., Beigneux A. P., Davies B. S., Gin P., Estrada K., Melford K., Bishop J. R., Esko J. D., Dallinga-Thie G. M., Fong L. G., Bensadoun A., Young S. G. (2008) J. Biol. Chem. 283, 34511–34518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olafsen T., Kenanova V. E., Wu A. M. (2006) Nat. Protoc. 1, 2048–2060 [DOI] [PubMed] [Google Scholar]
- 15.Olafsen T., Kenanova V. E., Sundaresan G., Anderson A. L., Crow D., Yazaki P. J., Li L., Press M. F., Gambhir S. S., Williams L. E., Wong J. Y., Raubitschek A. A., Shively J. E., Wu A. M. (2005) Cancer Res. 65, 5907–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kenanova V., Olafsen T., Crow D. M., Sundaresan G., Subbarayan M., Carter N. H., Ikle D. N., Yazaki P. J., Chatziioannou A. F., Gambhir S. S., Williams L. E., Shively J. E., Colcher D., Raubitschek A. A., Wu A. M. (2005) Cancer Res. 65, 622–631 [PMC free article] [PubMed] [Google Scholar]
- 17.Cai W., Olafsen T., Zhang X., Cao Q., Gambhir S. S., Williams L. E., Wu A. M., Chen X. (2007) J. Nucl. Med. 48, 304–310 [PubMed] [Google Scholar]
- 18.Kenanova V., Olafsen T., Williams L. E., Ruel N. H., Longmate J., Yazaki P. J., Shively J. E., Colcher D., Raubitschek A. A., Wu A. M. (2007) Cancer Res. 67, 718–726 [DOI] [PubMed] [Google Scholar]
- 19.Loening A. M., Gambhir S. S. (2003) Mol. Imaging 2, 131–137 [DOI] [PubMed] [Google Scholar]
- 20.Levak-Frank S., Radner H., Walsh A., Stollberger R., Knipping G., Hoefler G., Sattler W., Weinstock P. H., Breslow J. L., Zechner R. (1995) J. Clin. Invest. 96, 976–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doolittle M. H., Wong H., Davis R. C., Schotz M. C. (1987) J. Lipid Res. 28, 1326–1334 [PubMed] [Google Scholar]
- 22.Vilaró S., Ramírez I., Bengtsson-Olivecrona G., Olivecrona T., Llobera M. (1988) Biochim. Biophys. Acta 959, 106–117 [DOI] [PubMed] [Google Scholar]
- 23.Wallinder L., Peterson J., Olivecrona T., Bengtsson-Olivecrona G. (1984) Biochim. Biophys. Acta 795, 513–524 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.