Abstract
Peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) is a transcriptional coactivator that regulates diverse aspects of energy metabolism in peripheral tissues. Mice deficient in PGC-1α have elevated metabolic rate and are resistant to diet-induced obesity. However, it remains unknown whether this alteration in energy balance is due to the action of PGC-1α in peripheral tissues or the central nervous system. In this study, we generated neuronal PGC-1α knock-out mice (BαKO) using calcium/calmodulin-dependent protein kinase IIα (CaMKIIα)-Cre to address its role in the regulation of energy balance and neuronal function. Unlike whole body PGC-1α null mice, BαKO mice have normal adaptive metabolic response to starvation and cold exposure in peripheral tissues. In contrast, BαKO mice are hypermetabolic, and similar to whole body PGC-1α null mice, are also resistant to diet-induced obesity, resulting in significantly improved metabolic profiles. Neuronal inactivation of PGC-1α leads to striatal lesions that are reminiscent of neurodegeneration in whole body PGC-1α null brain and impairs nutritional regulation of hypothalamic expression of genes that regulate systemic energy balance. Together, these studies have demonstrated a physiological role for neuronal PGC-1α in the control of energy balance. Our results also implicate CaMKIIα-positive neurons as an important part of the neural circuitry that governs energy expenditure in vivo.
Keywords: Energy Metabolism, Neurodegeneration, Nuclear Receptors, Obesity, Transcription Coactivators, PGC-1α
Introduction
Metabolic syndrome is emerging as a global epidemic in both industrialized and developing countries. Obesity is a central component of this syndrome and arises from a chronic imbalance of energy intake and energy expenditure (1, 2). Excess fat storage in the adipose tissue, and more importantly, ectopic lipid accumulation in skeletal muscle and the liver lead to development of insulin resistance and type 2 diabetes (3). Although there is no doubt that environmental factors, particularly high-calorie diets and sedentary life style, contribute to the pathogenesis of metabolic syndrome, various genetic and epigenetic factors underlie the predisposition of individuals to metabolic disorders. The homeostatic control of energy intake and expenditure is achieved through a complex network of nutritional, hormonal, and neural cues, which coordinates nutrient storage and fuel oxidation in peripheral tissues. In mammals, nuclear hormone receptors as well as their coactivators and corepressors play an important role in the regulation of diverse aspects of tissue metabolism (4–7).
The PPARγ coactivator 1 (PGC-1)4 family of transcriptional coactivators regulates glucose, lipid, and mitochondrial oxidative metabolism through physical interaction with a selective subset of nuclear hormone receptors and other transcription factors (8–11). PGC-1α and its close homolog PGC-1β are abundantly expressed in tissues with high oxidative capacity, including brown fat, brain, liver, as well as cardiac and skeletal muscle (12, 13). Their expression is highly regulated in response to various physiological and environmental signals. For example, PGC-1α is rapidly induced in skeletal muscle following physical exercise (14, 15), whereas its expression is cold-inducible in brown fat (13). In the skeletal muscle, PGC-1α induces mitochondrial biogenesis and activates a metabolic and contractile program characteristic of slow twitch myofibers (16, 17). In the brown fat, it promotes adaptive thermogenesis through stimulating UCP1 expression and mitochondrial respiration (13). In the liver, PGC-1α regulates hepatic gluconeogenesis and a broader program of metabolic response to starvation (18–20). The physiological role of this coactivator in adaptive energy metabolism has been demonstrated in multiple tissues in PGC-1α-deficient mice (21–25). Recent studies demonstrate that reduced PGC-1α expression in skeletal muscle is associated with insulin resistance in humans (26, 27).
Because PGC-1α stimulates mitochondrial fuel oxidation, a likely outcome of PGC-1α deficiency is increased susceptibility to the development of obesity. Paradoxically, whole body PGC-1α-null mice are resistant to high-fat diet-induced obesity (21). This alteration in systemic energy balance is associated with elevated metabolic rate and physical activity levels as well as disrupted circadian rhythm (28). However, it remains unknown whether PGC-1α influences energy balance and clock function directly through its action in peripheral tissues or via its function in the central nervous system. PGC-1α mRNA and protein expression has been observed in several brain areas and neuronal cell types (29, 30). Whole body PGC-1α null mice develop spongioform neurodegeneration, most notably in the striatum and the deep layers of the cerebral cortex. In addition, dysregulation of the PGC-1α pathway was recently implicated in Huntington and Parkinson disease (21, 31, 32). These studies strongly suggest PGC-1α plays a critical role in maintaining neuronal health. Whether neuronal PGC-1α is required for systemic metabolic homeostasis has not yet been explored.
To address the role of neuronal PGC-1α in the regulation of energy balance, we generated forebrain-specific PGC-1α null mice by crossing PGC-1αflox/flox mice with calcium/calmodulin-dependent protein kinase IIα (CaMKIIα)-Cre transgenic mice. We found that inactivation of PGC-1α in CaMKIIα-positive neurons leads to resistance to diet-induced obesity and results in neurodegenerative lesions in the striatum. Our studies reveal an essential role of PGC-1α in CaMKIIα-positive neurons in the regulation of energy balance and neuronal function.
EXPERIMENTAL PROCEDURES
Mice
All animal experiments were performed according to procedures approved by the University Committee on Use and Care of Animals. Mice carrying PGC-1αflox alleles were generated as described previously (21). These mice were mated with CaMKIIα-Cre transgenic mice to generate flox/flox control and BαKO mice. PCR analysis was performed on genomic DNA isolated from different tissues to assess Cre-mediated deletion. Wild type allele and Cre-mediated deletion allele was detected using primers 1 and 2 and primers 3 and 4, respectively: forward primer 1, GTCTAAGATGTCTGCTCTTGAGG; reverse primer 2, CCAGTTTCTTCATTGGTGTG; forward primer 3, TCCAGTAGGCAGAGATTTATGAC; reverse primer 4, CCAACTGTCTATAATTCCAGTTC. Mice were maintained on a standard rodent chow or a high-fat diet containing 60% fat-derived calories (D12492, Research Diets) with 12-h light and dark cycles. For cold exposure, 11-week-old female mice were individually housed in cages prechilled at 4 °C with free access to food and water. Core body temperature was monitored using a rectal thermometer 3 h after the initiation of cold exposure. Brown fat was dissected following cold exposure for gene expression and histological analyses. For hypothalamus analysis, 3–4-months-old control, whole body PGC-1α null, and BαKO mice were either fed or fasted for 48 h before harvest.
Metabolic Analysis
Metabolic rate and activity were measured using Comprehensive Lab Animal Monitoring System (CLAMS), which simultaneously measures whole-body O2 consumption and physical movements (33, 34). Mice were acclimated in the monitoring chambers for 3 days before the experiment to minimize the effects of housing environment changes on animal behaviors. Data were collected every 10 min for each mouse over a period of three light/dark cycles. CLAMS study was conducted by University of Michigan Animal Phenotyping Core. Plasma concentrations of triglyceride, insulin, and leptin were measured using commercially available assay kits. Liver triglyceride content was extracted and measured using previously described procedures (35).
Histological Analysis
Brains were fixed in situ by intracardiac perfusion with 15 ml PBS followed by 15 ml 4% PFA in PBS, post-fixed in 4% PFA in PBS overnight at 4 °C after dissection, dehydrated in 70% ethanol, and embedded in paraffin. Coronal sections (8 μm) were stained using an H&E staining method. Immunohistochemistry using antibody against neurofilament light chain was performed as described previously (21). Other tissues were fixed directly by 4% PFA in PBS overnight at 4 °C after dissection and underwent the same procedure as the brain for the H&E staining. Frozen livers were embedded in O.C.T. compound, Tissue-Tek (Sakura Finetek) sectioned into 12-μm sections, and stained using Oil Red O method.
RNA and Protein Analysis
Total RNA was isolated from tissues using TRIzol reagents (Invitrogen). For quantitative real-time PCR (qPCR) analysis, RNA samples were reverse transcribed and used in quantitative PCR reactions in the presence of SYBR Green (Applied Biosystems). Relative abundance of mRNA was normalized to ribosomal protein 36B4 or β-actin. Sequences for most qPCR primers used in this study were described previously (21, 36). The rest of primer sequences are available upon request. Immunoblotting studies were performed using specific antibodies for LC3 (LC3–5F10, Nanotools), p62 (PW9860, Enzo Life Sciences), and ubiquitin (catalog no. sc-8017, Santa Cruz Biotechnology).
RESULTS
Generation of Brain-specific PGC-1α-deficient Mice
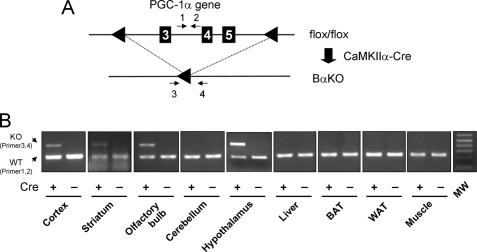
PGC-1α deficiency impairs adaptive metabolic responses in multiple tissues, including the liver, brown adipose tissue as well as skeletal and cardiac muscle (21, 22, 24). Paradoxically, whole body PGC-1α null mice are resistant to diet-induced obesity and have elevated metabolic rate. These findings raise the possibility that PGC-1α may play an important role in the regulation of energy balance through its action in the central nervous system. To test this possibility, we generated brain-specific PGC-1α null mice using the Cre-loxP system. We chose to use CaMKIIα-Cre transgenic mice because PGC-1α is abundantly expressed in forebrain, including cerebral cortex, hippocampus, basal ganglia, and hypothalamus (37). Transgenic expression of Cre recombinase under the control of CaMKIIα promoter has been widely used to inactivate genes in the forebrain (38, 39). We generated PGC-1αflox/flox and PGC-1αflox/flox; CaMKIIα-Cre (BαKO) mice for our studies (Fig. 1A). As shown in Fig. 1B, BαKO mice have selective deletion of exons 3–5 within the PGC-1α locus in several brain areas, including cerebral cortex, striatum, olfactory bulb, and hypothalamus, but not in any of the peripheral tissues examined.
FIGURE 1.
Generation of BαKO mice. A, strategy for Cre recombinase-mediated deletion of PGC-1α exons 3–5 in the brain. B, PCR analysis of genomic DNA isolated from tissues of flox/flox (−) and BαKO (+) mice. Note the deletion of PGC-1α exons is only detected in cortex, striatum, olfactory bulb, and hypothalamus. PCR primers are indicated in A. eWAT, epididymal white adipose tissue; BAT, brown adipose tissue.
BαKO Mice Have Normal Adaptive Metabolic Response
We next examined whether adaptive metabolic responses in peripheral tissues are affected in BαKO mice fed chow diet. In contrast to whole body PGC-1α null mice, which are cold-sensitive due to defects in adaptive thermogenesis, BαKO mice maintain their core body temperature following 3-h cold exposure at 4 °C. We observed a modest but similar decrease in body temperature in both control and BαKO groups (Fig. 2A). Histological analysis of brown adipose tissues reveals that brown adipocytes appear normal in size with similar accumulation of multilocular lipid droplets (Fig. 2B). Consistently, cold-inducible expression of genes involved in adaptive thermogenesis, including PGC-1α, deiodinase 2, Ucp1 (uncoupling protein 1), and mitochondrial genes, are similar in control and BαKO brown fat (Fig. 2C and data not shown). PGC-1α has been demonstrated to regulate multiple aspects of hepatic starvation response. PGC-1α deficient hepatocytes have defective gluconeogenic response and heme biosynthesis (18, 21). Compared with the control group, BαKO mice have normal induction of gluconeogenic genes following overnight fasting (data not shown). Plasma glucose levels are also similar in chow-fed control and BαKO mice under both fed and fasted conditions. These results indicate that adaptive energy metabolism in peripheral tissues is largely unperturbed in brain-specific PGC-1α-deficient mice.
FIGURE 2.
Adaptive thermogenesis in response to cold exposure. A, Rectal temperature of flox/flox (filled box) and BαKO (open box) mice kept at room temperature (RT) or exposed to 4 °C for 3 h. *, p < 0.05. B, H&E staining of paraffin-embedded brown fat sections. C, qPCR analysis of gene expression in brown fat. Data represent mean ± S.E. (n = 3 per group). *, p < 0.004; NS, not significant.
BαKO Mice Are Resistant to Diet-induced Obesity
PGC-1α-deficient mice have elevated metabolic rate and are resistant to diet-induced obesity. However, whether this can be attributed to neuronal PGC-1α function remains unknown. To determine the significance of central PGC-1α in energy balance, we subjected control and BαKO mice to high-fat diet feeding. Although body weight of these two groups of mice remains similar under chow-fed condition, BαKO mice are significantly resistant to weight gain when fed high-fat diet (Fig. 3A). The body weight of BαKO mice is ∼20% lower than control mice following 10 weeks of high-fat feeding. Transgenic expression of Cre recombinase alone does not affect body weight upon high-fat feeding (data not shown). Resistance to weight gain following high-fat feeding appears to be more pronounced in whole body PGC-1α null group, which weighs ∼40% less than the wild type group. Consistently, epididymal white adipose tissue weight and epididymal white adipose tissue/body weight ratio are significantly lower in the BαKO and whole body PGC-1α null mice compared with their respective control (Fig. 3C). In contrast to whole body PGC-1α null mice, which have lower plasma glucose concentrations, plasma glucose remains similar in control and BαKO mice following high-fat feeding (Fig. 3D). Resting core body temperature is also slightly but significantly higher in the BαKO mice but not whole body PGC-1α null mice (Fig. 3E). These results suggest that neuronal PGC-1α participates in the regulation of systemic energy balance and contributes to weight gain resistance in whole body PGC-1α null mice.
FIGURE 3.
High-fat diet-induced obesity. A, body weight of flox/flox, BαKO, WT, and whole body PGC-1α null (KO) mice fed high-fat diet for 10 weeks. *, p < 0.001; **, p < 0.01 BαKO versus KO group. B, appearance of control and BαKO mice following high-fat diet feeding. C, epididymal fat (eWAT) weight and epididymal white adipose tissue to body weight ratio in flox/flox, BαKO, WT, and whole body PGC-1α null (KO) mice. *, p < 0.02. D and E, plasma glucose (D) concentration and rectal temperature (E) in high-fat fed flox/flox, BαKO, WT, and whole body PGC-1α null mice (KO). Data represent mean ± S.E. (n = 7–8 per group). *, p < 0.02; NS, not significant.
A plausible explanation for the resistance to diet-induced obesity in BαKO mice is that they have elevated metabolic rate. We measured metabolic rate and physical movements in control and BαKO mice using the CLAMS. BαKO mice have increased food intake when normalized to their body weight (Fig. 4A). On a per-mouse basis, food intake is similar between these two groups. Oxygen consumption rate (VO2) is ∼30% higher in BαKO group than control (Fig. 4B). When normalized to total lean mass, VO2 remains significantly higher in BαKO mice. Respiratory exchange ratio, a measurement of in vivo fuel preference, is slightly lower in BαKO mice (data not shown), suggesting that BαKO mice may prefer fatty acid oxidation for energy production. Surprisingly, total physical activity level and diurnal locomotor profiles appear unaffected in these mice (Fig. 4C). Analyses of plasma hormones indicate that both insulin and leptin concentrations are significantly lower in BαKO mice, suggesting that they may have improved insulin sensitivity. However, we were unable to observe significant improvement in glucose tolerance in insulin and glucose tolerance tests (data not shown).
FIGURE 4.
CLAMS studies in high-fat diet fed flox/flox and BαKO mice. A, food intake in flox/flox and BαKO mice. Data represent food consumption per day following normalization to body weight (left) or on a per-mouse basis (right). *, p < 0.01. B, metabolic rate in flox/flox (filled box) and BαKO mice (open box). The oxygen consumption rate as normalized to body weight (left) or lean mass (right) is shown. *, p < 0.05; # p < 0.09. C, total activity level during night (N) and day (D) phases. Shown are average movement counts for flox/flox (filled box) and BαKO (open box) mice. D and E, plasma insulin (D) and leptin (E) concentrations in high-fat diet-fed flox/flox and BαKO mice. Data represent mean ± S.E. (n = 7–8 per group). *, p < 0.02.
To explore whether neuronal deficiency of PGC-1α alters the expression of key regulators of energy balance, we analyzed hypothalamic gene expression using qPCR. The expression of thyrotropin-releasing hormone, proopiomelanocortin, orexin, melanin-concentrating hormone, and prohormone convertase 2 remains similar in control and BαKO hypothalamus under both fed and fasted conditions (Fig. 5 and data not shown). In contrast, fasting induction of AgRP (Agouti-related protein) and neuropeptide Y (NPY), two factors known to regulate energy balance, is significantly diminished in hypothalamus from BαKO mice. Similar defects in AgRP and NPY expression were also observed in whole body PGC-1α null hypothalamus. These findings strongly suggest that neuronal PGC-1α may affect diet-induced obesity through its regulation of hypothalamic neuropeptides that control energy balance.
FIGURE 5.
Hypothalamic gene expression. A, qPCR analysis of gene expression in hypothalamus of WT and whole body PGC-1α null (KO) mice under fed and fasted conditions. Pooled RNA from 3–5 mice in each group was used in the analyses. Data represent mean ± S.D. *, p < 0.005. B, qPCR analysis of hypothalamic gene expression in flox/flox and BαKO mice under fed and fasted conditions. Pooled RNA from 6–8 mice in each group was used. Data represent mean ± S.D. *, p < 0.005. TRH, thyrotropin-releasing hormone; POMC, proopiomelanocortin; MCH, melanin concentrating hormone.
High-fat Diet-fed BαKO Mice Have Reduced Hepatic Steatosis
We further examined the impact of neuronal PGC-1α inactivation on hepatic metabolism following high-fat feeding. Analysis of hepatic triglyceride content indicates that BαKO mice have significantly less lipid accumulation in the liver (Fig. 6A). In fact, liver triglyceride is reduced by ∼57% in BαKO mice. Hepatocyte swelling and lipid accumulation is evident in high-fat-fed control mice (Fig. 6B). In contrast, the overall liver appearance and histology are significantly improved in the BαKO group. Analysis of hepatic gene expression indicates that the expression of several lipogenic genes, including fatty acid synthase and stearoyl-CoA desaturase as well as Fsp27, a gene involved in lipid droplet formation, are significantly decreased in BαKO mouse livers (Fig. 6C). The expression of gluconeogenic genes such as PEPCK and glucose-6-phosphatase and fatty acid β-oxidation genes remain largely unaltered. These results suggest that neuronal deletion of PGC-1α significantly improves the metabolic profile in the liver following high-fat diet feeding.
FIGURE 6.
Hepatic triglyceride content and gene expression. A, liver triglyceride content in control (filled box) and BαKO (open box) mice following 10 weeks of high-fat feeding. *, p < 0.01. B, H&E (top panel) and Oil Red O staining (lower panel) of liver sections. C, qPCR analysis of gene expression. Data represent mean ± S.E. (n = 7–8 per group). *, p < 0.05. FAS, fatty acid synthase.
Region-specific Degenerative Lesions in BαKO Mouse Brains
To determine the effects of PGC-1α inactivation on neuronal health, we performed histological analyses in several brain regions. Consistent with previous findings (21), whole body PGC-1α deficiency leads to spongiform neurodegeneration, most readily observed in striatum and deep layers of cerebral cortex (Fig. 7). Remarkably, striatum from BαKO mouse brain also contains numerous degenerative lesions that are similar but slightly smaller than those seen in whole body PGC-1α null mice. BαKO mice tend to have fewer lesions in both striatum and deep cortical layers. Immunohistochemical staining using an antibody against neurofilament light chain, a marker for nerve fibers, indicates that the lesions correlate with apparent degeneration of fibers in the striatum from BαKO mice (Fig. 8). Our results strongly suggest that PGC-1α is essential for maintaining axon integrity in CaMKIIα-positive neurons in the brain.
FIGURE 7.
H&E staining of brain sections from flox/flox, BαKO, whole body PGC-1α null (KO) mice. Shown are cerebral cortex (cx) and striatum (str) regions. Lower panel, high magnification of striatum sections. Scale bar, 500 μm. Note the absence of clear degenerative lesions in flox/flox mouse brain (arrow).
FIGURE 8.
Immunohistochemical staining using antibody against neurofilament light chain. Shown are cerebral cortex (cx), corpus callosum (cc), and striatum (str) regions in forebrain sections at low (top) and high (bottom) magnification in flox/flox and BαKO mice. Note the presence of small (arrowhead) and large (arrow) lesions in the striatum.
PGC-1α Deficiency Does Not Perturb Autophagy in Central Nervous System
Autophagy is responsible for bulk degradation of cytoplasmic components in the cell and plays an important role in nutrient homeostasis during starvation (40–42). Autophagy is also required for the clearance of organelles, such as mitochondria (43). Inhibition of autophagy leads to accumulation of ubiquitinated protein and results in neuronal death and neurodegeneration (44–46). To determine whether PGC-1α inactivation perturbs autophagy, we performed immunoblotting analyses using antibodies against LC3 and p62, two molecular markers of autophagy activity (47–49). LC3 is a cytosolic protein (LC3-I) that, upon autophagy induction, undergoes covalent lipid modification and translocates to the autophagosome membrane (LC3-II) (50). Relative abundance of these two LC3 isoforms is indicative of autophagy activity (45). As shown in Fig. 9, LC3-I and LC3-II levels are similar in the posterior cortex and striatum of wild type and whole body PGC-1α null mouse brains. Similarly, we did not observe differences in LC3-I and LC3-II levels in control and BαKO mouse brains. In addition, the protein levels of p62, an LC-3 binding protein that is involved in the formation of ubiquitin-containing inclusions, also remain largely unchanged (Fig. 9) (45). We further examined the expression of genes in the autophagy pathway and found that mRNA levels of autophagy genes are similar between control and PGC-1α null brains (data not shown). Together, these results suggest that neurodegeneration in PGC-1α deficient neurons is unlikely to be due to impaired neuronal autophagy.
FIGURE 9.
Immunoblotting analysis of proteins in the autophagy pathway. Total tissue lysates were prepared from posterior cortex and striatum dissected from WT, whole body PGC-1α null (KO), flox/flox, and BαKO mice. Immunoblotting was performed using indicated antibodies. Two different exposure times were included for LC3. Ponceau S stain was used as loading control.
DISCUSSION
The brain is a highly metabolically active tissue and relies on mitochondrial oxidative metabolism for ATP production under normal conditions. Not surprisingly, neurons are exquisitely sensitive to perturbations of mitochondrial function. Impaired mitochondrial energy metabolism has been implicated in numerous heritable neurological disorders as well as neurodegenerative diseases, including Huntington, Parkinson, and Alzheimer disease (51–53). PGC-1α is abundantly expressed in the brain and its deficiency leads to degenerative lesions in several brain regions. However, whether these lesions arise from cell-autonomous functions of PGC-1α in neurons remains unknown. In addition, the neuronal cell types that require PGC-1α for their normal function have not been identified. In this study, we demonstrate that PGC-1α in CaMKIIα-positive neurons plays an important role in maintaining systemic energy balance and neuronal health.
We have previously reported that mice lacking PGC-1α are resistant to high-fat diet-induced obesity and have significantly improved insulin sensitivity (21). This lean phenotype is associated with elevated metabolic rate and activity levels. Because PGC-1α is expressed in several key metabolic tissues, including skeletal muscle, adipose tissues, and liver (13), it was not immediately clear which tissue(s) underlies altered systemic energy balance in whole body PGC-1α null mice. To complicate the matter further, PGC-1α regulates distinct metabolic programs in different tissues, which influence systemic metabolism through crosstalk and secondary effects. To resolve these issues, we generated brain-specific PGC-1α null mice using CaMKIIα-Cre transgenic line. PGC-1α is abundantly expressed in pyramidal neurons, many of which also express CaMKIIα (54), as well as GABAergic neurons (29, 30). BαKO mice are capable of mounting normal metabolic responses in peripheral tissues. For example, the induction of genes involved in hepatic gluconeogenesis and fatty acid β-oxidation following starvation is similar in control and BαKO mice. In addition, the activation of adaptive thermogenesis in brown fat in response to cold exposure is normal in BαKO mice. Unlike whole body PGC-1α null mice, which are extremely cold-sensitive, BαKO mice are indistinguishable from control mice in maintaining their core body temperature. These observations allow us to assess the biological function of neuronal PGC-1α in the absence of the confounding metabolic perturbations in peripheral tissues caused by global PGC-1α deficiency.
Perhaps the most remarkable outcome of neuronal PGC-1α deficiency is its influence on systemic energy balance when the mice were fed a high-fat diet. Compared with control mice, BαKO mice gain significantly less body weight and have lower adiposity following 10 weeks of high-fat feeding. The resistance to weight gain in BαKO mice appears to be quantitatively less pronounced than whole body PGC-1α null mice, suggesting that PGC-1α in CaMKIIα-negative cells in nervous system and/or peripheral tissues may also contribute to its effects on systemic energy balance. Additionally, in both BαKO and whole body PGC-1α null mice, fasting-induced expression of AgRP and NPY in hypothalamus is impaired in the absence of PGC-1α. These observations are consistent with previous studies that implicate FoxO1, a transcriptional partner for PGC-1α, in the regulation of AgRP gene expression in hypothalamus (55, 56). A recent report showed that PGC-1α expression is present in NPY expressing neurons of the dorsalmedial hypothalamic nucleus (57). Together, these findings illustrate a potential role for the FoxO1/PGC-1α pathway in the regulation of hypothalamic transcription and function. The significance of AgRP and NPY in mediating the effects of PGC-1α on energy balance remains unknown at present.
Besides less weight gain under high-fat diet, BαKO mice also have reduced hepatic lipid content and lower plasma insulin levels, suggesting that these mice are more insulin-sensitive. However, BαKO mice are similar to control group in insulin tolerance and glucose tolerance tests. In contrast, whole body PGC-1α null mice have significantly lower blood glucose levels and improved glucose tolerance. A plausible explanation for these differences is that hepatic glucose output is impaired in whole body PGC-1α null mice, whereas it remains normal in BαKO mice. In fact, fasting glucose levels are similar between control and BαKO mice when kept on chow diet. As such, impaired hepatic gluconeogenesis may significantly contribute to improved glucose homeostasis in whole body PGC-1α null mice.
Surprisingly, although BαKO mice have increased oxygen consumption rate in CLAMS studies, these mice appear to have normal activity levels. In addition, diurnal regulation of locomotor activity is apparently unaffected in these mice. These results suggest that distinct neuronal populations might mediate the function of PGC-1α in the regulation of energy balance and circadian pacemaker. In this case, PGC-1α activity in CaMKIIα-positive neurons is essential for the control of metabolic rate and body weight homeostasis, whereas its regulation of biological clock is mediated by a distinct population of PGC-1α expressing neurons. Previous studies have shown that PGC-1α is also expressed in GABAergic neurons (29). Whether PGC-1α in GABAergic neurons is required for maintaining normal circadian metabolic rhythms remains to be addressed. It is also possible that certain CaMKIIα neurons may lack Cre expression. We cannot rule out the possibility that intact PGC-1α expression in a subset of CaMKIIα-positive neurons could mediate its role in circadian regulation.
We observed neurodenegerative lesions in the striatum of BαKO mouse brain. The striking similarity in the appearance of vacuoles in BαKO and whole body PGC-1α null mice strongly suggests that the deficiency of PGC-1α in CaMKIIα-positive neurons is the major neuronal population affected in whole body PGC-1α null mouse brain. Interestingly, we observed fewer lesions in the striatum and deep layers of cerebral cortex in BαKO mice. It remains to be determined whether this is due to partial deletion of PGC-1α in CaMKIIα-positive neurons or other cell types also contribute to neurodegeneration in whole body PGC-1α null mice. Because CaMKIIα is only modestly expressed in the striatum, our data suggest that the degenerative lesions are most likely due to the defects of neurons that reside in other brain areas, such as the cortex. Although autophagy is emerging as an important mechanism in neuronal homeostasis, we did not observe significant changes in autophagy activity in whole body PGC-1α null and BαKO mice compared with their respective control. It is likely that disruption of mitochondrial function and reactive oxygen species metabolism may be responsible for the development of neuronal lesions in the absence of PGC-1α.
In summary, we have demonstrated that PGC-1α activity in CaMKIIα neurons plays a key role in the regulation of energy balance and neuronal health. The resistance to diet-induced obesity and brain lesions in BαKO mice are strikingly similar to whole body PGC-1α null mice. These results strongly suggest that neuronal PGC-1α exerts profound effects on the neural circuitry that governs systemic energy balance.
Acknowledgments
We thank Drs. Jennifer Estall and Bruce Spiegelman for providing PGC-1α flox mice, Joseph Takahashi for CaMKIIα-Cre transgenic mice, and Geoffrey Murphy for discussions. We are grateful to Shengjuan Gu, Layla Yu, Fernanda Jimenez, and Matthew Molusky for technical assistance and other members of the laboratory for discussions. We thank Dr. Nathan Qi and the University of Michigan Animal Metabolic Phenotyping Core for performing CLAMS study.
This work was supported in part by National Institutes of Health Grant HL097738 (to J. D. L.).
- PGC-1α
- peroxisome proliferator-activated receptor γ, coactivator 1α
- qPCR
- quantitative real-time PCR
- CaMKIIα
- calcium/calmodulin-dependent protein kinase IIα
- BαKO
- brain PGC-1α knock-out
- NPY
- neuropeptide Y
- AgRP
- Agouti-related protein
- PEPCK
- phosphoenolpyruvate carboxykinase
- CLAMS
- Comprehensive Lab Animal Monitoring System.
REFERENCES
- 1.Flier J. S. (2004) Cell 116, 337–350 [DOI] [PubMed] [Google Scholar]
- 2.Spiegelman B. M., Flier J. S. (2001) Cell 104, 531–543 [DOI] [PubMed] [Google Scholar]
- 3.Erion D. M., Shulman G. I. (2010) Nat. Med. 16, 400–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaven S. W., Tontonoz P. (2006) Annu. Rev. Med. 57, 313–329 [DOI] [PubMed] [Google Scholar]
- 5.Chawla A., Repa J. J., Evans R. M., Mangelsdorf D. J. (2001) Science 294, 1866–1870 [DOI] [PubMed] [Google Scholar]
- 6.Feige J. N., Auwerx J. (2007) Trends Cell. Biol. 17, 292–301 [DOI] [PubMed] [Google Scholar]
- 7.Lin J. D. (2009) Mol. Endocrinol. 23, 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finck B. N., Kelly D. P. (2006) J. Clin. Invest. 116, 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handschin C. (2009) Trends Pharmacol Sci. 30, 322–329 [DOI] [PubMed] [Google Scholar]
- 10.Kelly D. P., Scarpulla R. C. (2004) Genes Dev. 18, 357–368 [DOI] [PubMed] [Google Scholar]
- 11.Lin J., Handschin C., Spiegelman B. M. (2005) Cell. Metab. 1, 361–370 [DOI] [PubMed] [Google Scholar]
- 12.Lin J., Puigserver P., Donovan J., Tarr P., Spiegelman B. M. (2002) J. Biol. Chem. 277, 1645–1648 [DOI] [PubMed] [Google Scholar]
- 13.Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. (1998) Cell 92, 829–839 [DOI] [PubMed] [Google Scholar]
- 14.Baar K., Wende A. R., Jones T. E., Marison M., Nolte L. A., Chen M., Kelly D. P., Holloszy J. O. (2002) Faseb J. 16, 1879–1886 [DOI] [PubMed] [Google Scholar]
- 15.Goto M., Terada S., Kato M., Katoh M., Yokozeki T., Tabata I., Shimokawa T. (2000) Biochem. Biophys. Res. Commun. 274, 350–354 [DOI] [PubMed] [Google Scholar]
- 16.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. (1999) Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- 17.Lin J., Wu H., Tarr P. T., Zhang C. Y., Wu Z., Boss O., Michael L. F., Puigserver P., Isotani E., Olson E. N., Lowell B. B., Bassel-Duby R., Spiegelman B. M. (2002) Nature 418, 797–801 [DOI] [PubMed] [Google Scholar]
- 18.Handschin C., Lin J., Rhee J., Peyer A. K., Chin S., Wu P. H., Meyer U. A., Spiegelman B. M. (2005) Cell 122, 505–515 [DOI] [PubMed] [Google Scholar]
- 19.Koo S. H., Satoh H., Herzig S., Lee C. H., Hedrick S., Kulkarni R., Evans R. M., Olefsky J., Montminy M. (2004) Nat. Med. 10, 530–534 [DOI] [PubMed] [Google Scholar]
- 20.Yoon J. C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C. R., Granner D. K., Newgard C. B., Spiegelman B. M. (2001) Nature 413, 131–138 [DOI] [PubMed] [Google Scholar]
- 21.Lin J., Wu P. H., Tarr P. T., Lindenberg K. S., St-Pierre J., Zhang C. Y., Mootha V. K., Jäger S., Vianna C. R., Reznick R. M., Cui L., Manieri M., Donovan M. X., Wu Z., Cooper M. P., Fan M. C., Rohas L. M., Zavacki A. M., Cinti S., Shulman G. I., Lowell B. B., Krainc D., Spiegelman B. M. (2004) Cell 119, 121–135 [DOI] [PubMed] [Google Scholar]
- 22.Leone T. C., Lehman J. J., Finck B. N., Schaeffer P. J., Wende A. R., Boudina S., Courtois M., Wozniak D. F., Sambandam N., Bernal-Mizrachi C., Chen Z., Holloszy J. O., Medeiros D. M., Schmidt R. E., Saffitz J. E., Abel E. D., Semenkovich C. F., Kelly D. P. (2005) PLoS Biol 3, e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huss J. M., Imahashi K., Dufour C. R., Weinheimer C. J., Courtois M., Kovacs A., Giguère V., Murphy E., Kelly D. P. (2007) Cell. Metab. 6, 25–37 [DOI] [PubMed] [Google Scholar]
- 24.Arany Z., He H., Lin J., Hoyer K., Handschin C., Toka O., Ahmad F., Matsui T., Chin S., Wu P. H., Rybkin II, Shelton J. M., Manieri M., Cinti S., Schoen F. J., Bassel-Duby R., Rosenzweig A., Ingwall J. S., Spiegelman B. M. (2005) Cell. Metab. 1, 259–271 [DOI] [PubMed] [Google Scholar]
- 25.Arany Z., Foo S. Y., Ma Y., Ruas J. L., Bommi-Reddy A., Girnun G., Cooper M., Laznik D., Chinsomboon J., Rangwala S. M., Baek K. H., Rosenzweig A., Spiegelman B. M. (2008) Nature 451, 1008–1012 [DOI] [PubMed] [Google Scholar]
- 26.Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. (2003) Nat. Genet. 34, 267–273 [DOI] [PubMed] [Google Scholar]
- 27.Patti M. E., Butte A. J., Crunkhorn S., Cusi K., Berria R., Kashyap S., Miyazaki Y., Kohane I., Costello M., Saccone R., Landaker E. J., Goldfine A. B., Mun E., DeFronzo R., Finlayson J., Kahn C. R., Mandarino L. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8466–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C., Li S., Liu T., Borjigin J., Lin J. D. (2007) Nature 447, 477–481 [DOI] [PubMed] [Google Scholar]
- 29.Cowell R. M., Blake K. R., Russell J. W. (2007) J. Comp. Neurol. 502, 1–18 [DOI] [PubMed] [Google Scholar]
- 30.Tritos N. A., Mastaitis J. W., Kokkotou E. G., Puigserver P., Spiegelman B. M., Maratos-Flier E. (2003) Brain Res. 961, 255–260 [DOI] [PubMed] [Google Scholar]
- 31.Cui L., Jeong H., Borovecki F., Parkhurst C. N., Tanese N., Krainc D. (2006) Cell 127, 59–69 [DOI] [PubMed] [Google Scholar]
- 32.St-Pierre J., Drori S., Uldry M., Silvaggi J. M., Rhee J., Jäger S., Handschin C., Zheng K., Lin J., Yang W., Simon D. K., Bachoo R., Spiegelman B. M. (2006) Cell 127, 397–408 [DOI] [PubMed] [Google Scholar]
- 33.Frayn K. N. (1983) J. Appl. Physiol. 55, 628–634 [DOI] [PubMed] [Google Scholar]
- 34.Simonson D. C., DeFronzo R. A. (1990) Am. J. Physiol. 258, E399–412 [DOI] [PubMed] [Google Scholar]
- 35.Lin J., Yang R., Tarr P. T., Wu P. H., Handschin C., Li S., Yang W., Pei L., Uldry M., Tontonoz P., Newgard C. B., Spiegelman B. M. (2005) Cell 120, 261–273 [DOI] [PubMed] [Google Scholar]
- 36.Li S., Liu C., Li N., Hao T., Han T., Hill D. E., Vidal M., Lin J. D. (2008) Cell Metab 8, 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X. B., Jones E. G. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7332–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsien J. Z., Chen D. F., Gerber D., Tom C., Mercer E. H., Anderson D. J., Mayford M., Kandel E. R., Tonegawa S. (1996) Cell 87, 1317–1326 [DOI] [PubMed] [Google Scholar]
- 39.Casanova E., Fehsenfeld S., Mantamadiotis T., Lemberger T., Greiner E., Stewart A. F., Schütz G. (2001) Genesis 31, 37–42 [DOI] [PubMed] [Google Scholar]
- 40.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 41.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. (2004) Mol. Biol. Cell. 15, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mortimore G. E., Schworer C. M. (1977) Nature 270, 174–176 [DOI] [PubMed] [Google Scholar]
- 43.Sandoval H., Thiagarajan P., Dasgupta S. K., Schumacher A., Prchal J. T., Chen M., Wang J. (2008) Nature 454, 232–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. (2006) Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 45.Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 46.Komatsu M., Wang Q. J., Holstein G. R., Friedrich V. L., Jr., Iwata J., Kominami E., Chait B. T., Tanaka K., Yue Z. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14489–14494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klionsky D. J., Cuervo A. M., Seglen P. O. (2007) Autophagy 3, 181–206 [DOI] [PubMed] [Google Scholar]
- 48.Mizushima N. (2004) Int. J. Biochem. Cell. Biol. 36, 2491–2502 [DOI] [PubMed] [Google Scholar]
- 49.Mizushima N., Yoshimori T., Levine B. (2010) Cell 140, 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DiMauro S., Schon E. A. (2008) Annu. Rev. Neurosci. 31, 91–123 [DOI] [PubMed] [Google Scholar]
- 52.Lin M. T., Beal M. F. (2006) Nature 443, 787–795 [DOI] [PubMed] [Google Scholar]
- 53.Schon E. A., Manfredi G. (2003) J. Clin. Invest. 111, 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ouimet C. C., McGuinness T. L., Greengard P. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 5604–5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kitamura T., Feng Y., Kitamura Y. I., Chua S. C., Jr., Xu A. W., Barsh G. S., Rossetti L., Accili D. (2006) Nat Med 12, 534–540 [DOI] [PubMed] [Google Scholar]
- 56.Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., Spiegelman B. M. (2003) Nature 423, 550–555 [DOI] [PubMed] [Google Scholar]
- 57.Draper S., Kirigiti M., Glavas M., Grayson B., Chong C. N., Jiang B., Smith M. S., Zeltser L. M., Grove K. L. (2010) Brain Res. 1350, 139–150 [DOI] [PMC free article] [PubMed] [Google Scholar]