Abstract
The formation of the functional mammalian cerebral cortex requires a concerted control of neurogenesis, neuronal migration, and neuronal morphogenesis. However, molecular mechanisms that control these processes are not well understood. We have found that the BMP signaling downstream transcription factor SMAD1 and CRMP2 (collapsin response mediator protein-2) are inversely and complementarily expressed in the developing neocortex. BMPs can suppress CRMP2 expression in cortical cells. Our ChIP assay demonstrates that both SMAD1 and -4 bind to CRMP2 promoter in the neocortex, and overexpression of SMAD1 and 4 in vivo suppresses CRMP2 expression. RNA interference of CRMP2 and overexpression of dominant negative forms of CRMP2 in utero cause accumulation of multipolar cells in the ventricular, subventricular, and intermediate zones and suppresses neurite outgrowth, suggesting that CRMP2 is required for multipolar to bipolar transition for directional neuronal migration and neurite outgrowth. Thus, our study reveals a novel mechanism that the BMP-SMAD signaling pathway controls neuronal migration and neurite outgrowth by suppressing the transcription of CRMP2.
Keywords: Bone Morphogenetic Protein (BMP), Cell Migration, Gene Transcription, Neurodevelopment, SMAD Transcription Factor, CRMP2
Introduction
During cortical development, neural progenitor cells undergo proliferation, differentiation, migration, and maturation in a distinct and sequential manner. Neurons migrate to different layers of the cortex via special routes to form a functional neural circuitry. The newborn neurons transiently become multipolar with multiple processes within the subventricular zone (SVZ)3 and lower intermediate zone (IZ), which is also known as the premigratory zone, and then change to bipolar to migrating out of the premigratory zone along radial fibers to the cortical plate (CP), where they further become mature neurons (1–5).
The TGF-β superfamily members, BMPs, are crucial regulators for the differentiation of ES cells as well as neural progenitor cells during development (6–9). In the canonical BMP-SMAD signaling pathway, BMP transduces its signal via the intracellular downstream mediators R-SMAD proteins (SMAD1, SMAD5, or SMAD8). The activated R-SMADs can form a complex with the Co-SMAD, SMAD4, to regulate target gene expression through cooperation with other DNA binding factors or transcription factors (6, 8). It has been shown that BMP2 and -4 induce neural stem cells to differentiate into a variety of cellular fates (10–12). However, the exact role of BMP2 and -4 in brain development and the underlying mechanism are poorly understood.
CRMP2 (collapsin response mediator protein 2) is one of five CRMP gene family members (CRMP1–5). CRMP2 is essential for axon-dendrite specification, axon outgrowth, and elongation (13, 14). CRMP2 has been shown to be able to bind to tubulin heterodimers to promote microtubule assembly (15). It has been proposed that CRMP2 promotes neurite elongation and axon specification by regulating microtubule assembly, endocytosis of adhesion molecules, reorganization of actin filaments, and axonal protein trafficking (14, 16–18). GSK3β (glycogen syntheses kinase 3β) plays an important role in the regulation of CRMP2 activity and neuron development. Phosphorylation of CRMP2 by GSK-3β can suppress the function of CRMP2 (14). NT-3 and BDNF can inhibit GSK-3β activity via the PI3K/Akt pathway and thereby reduce the phosphorylation levels of CRMP2, leading to axon elongation and branching (14, 19). Most of those studies were performed in cultured neurons. Thus, the physiological function of CRMP2 in mammalian brain remains elusive.
We have reported recently through chromatin immunoprecipitation-chip technology that the BMP-SMAD signaling pathway can regulate the transcription of CRMP2 in ES cells (20). In the present study, we investigated the role of the BMP-SMAD signaling pathway in brain development and found that this pathway can suppress the transcription of CRMP2. We then explored the in vivo function of CRMP2 by means of in utero electroporation in the developing murine brain. Utilizing RNAi and overexpression of dominant negative forms of CRMP2, we have discovered that interference with CRMP2 expression or function dramatically disturbs the redistribution and the morphology of newborn neurons, indicating that CRMP2 plays essential roles at multiple stages of neuronal development.
EXPERIMENTAL PROCEDURES
Animals
Pregnant Sprague-Dawley rats were provided by the Animal Center of the Institute of Genetics and Developmental Biology (Chinese Academy of Sciences). All experimental procedures involved were performed according to protocols approved by the Institutional Animal Care and Use Committee at the Institute of Genetics and Developmental Biology.
In Utero Electroporation
In utero electroporation was performed as described (21, 22). Briefly, pregnant Sprague-Dawley rats (E16) were anesthetized with sodium pentobarbital at 30 mg/kg of body weight. Plasmid DNA solution (2∼3 μg/μl) containing 0.01% Fast Green was injected (1∼2 μl) into the right side of the lateral ventricle with glass micropipettes. The heads of embryos in the uterus were placed between the Tweezertrode Electrode (7 mm in diameter, BTX Harvard Apparatus), and five electrical pulses (50 V for 50 ms in duration at intervals of 950 ms) were delivered using an electroporator (ECM830, Harvard Apparatus).
Chromatin Immunoprecipitation Assay (ChIP)
E16 rat or E14 mouse cerebral cortex are dissected. The cortex tissues were first flash-frozen by liquid nitrogen and then homogenized. Cell lysates collected in PBS were chemically cross-linked by addition of one-tenth volume of freshly prepared 11% formaldehyde solution for 15 min at room temperature. The following procedures were carried out as described previously (20). The ChIP antibody for Smad1 was purchased from Santa Cruz Biotechnology (catalog no. sc-7965), and Smad4 antibody was described previously (20). The primers used to amplify rat and mouse CRMP2 promoter were the same as follows: 5′-GCTACCCAAGGCTACCTCCAT-3′ and 5′-TCCACGCATCACGGTAAGTTTG-3′.
RNA Isolation, Quantitative Real-time PCR, and Analysis of Transcript Levels
Cells were treated with different concentrations of BMP4 or BMP2 for 4 h (R&D Systems, catalog no. 314-BP). Total RNA was isolated using TRIzol reagent (Invitrogen). Reverse transcriptase was employed for oligo(dT) primed first strand cDNA synthesis. Quantitative RT-PCR was carried out on a Mx3000P quantitative PCR system. The primers used were as follows: 5′-CATTGCCAATCAGACCAACT-3′ and 5′-CACCACAGTTCCCTTCTTCC-3′.
shRNA and Dominant Negative Constructs of CRMP2
The published target sequences against rat CRMP2 (5′-GTAAACTCCTTCCTCGTGT-3′ and 5′-GCCTATTGGCAGCCTTTGA-3′) were cloned into the EcoRI/BamHI site of pSIREN-RetroQ-DsRed (Clontech) to create shCRMP2a and shCRMP2b. Pll3.7 RNAi or Pll3.7 RNAi D2 (for knockdown of CRMP2), and full-length and dominant negative constructs of CRMP2 were described previously (15, 18, 20).
Immunocytochemistry
Rat embryos were perfused transcardially with ice-chilled saline followed by 4% paraformaldehyde in 0.1 m PBS, pH 7.4. Brains were postfixed in paraformaldehyde overnight and sectioned on a Cryostat (Leica, CM1900). Slices were blocked at room temperature for 1 h with 5% house serum, 0.1% Triton X-100, and 0.5% BSA in PBS. Primary antibodies were applied overnight at the following concentrations: anti-CRMP2 (1:100, Abcam); anti-SMAD1 (1:40, Santa Cruz Biotechnology); anti-nestin (1:1000, Chemicon); anti-MAP2 (1:500, Chemicon). Sections were then washed with PBS and incubated in Cy5-conjugated secondary antibodies (1:2000; Invitrogen).
Confocal Microscopy
Sections were imaged on an inverted laser-scanning confocal microscope (SP5; Leica) as described in Ref. 23.
Live Cell Imaging
Coronal slices were prepared 48 h after electroporation. Slices were placed on Millicell-CM inserts (Millipore) in culture medium as described (23). Multiple DsRed-positive cells were imaged on an inverted microscope (model ASMDW; Leica) with a 40× numerical aperture 0.55 objective. The ASMDW work station consisted of an inverted microscope (DMIRE2, Leica) and a multipoint time-lapse stage controlled by ASMDW software (Leica). Time-lapse images were captured at intervals of 6 or 10 min for 10 h.
Cell Culture
Neural progenitor cells were cultured as described previously with slightly modifications (9). Briefly, telencephalons from E16 Sprague-Dawley rats were cut into 1–3-mm pieces, mechanically dissected, and cultured in N2-supplement (GIBCO) DMEM/F12 with basic fibroblast growth factor at 10 ng/ml basic fibroblast growth factor (R&D Systems). Cells were cultured at 37 °C with 5% CO2.
Statistical Analysis
Data represented the mean ± S.D. Statistical differences between groups were performed with analysis of variance. Student's t test was used to value the significance of differences between means. An asterisk represented p value < 0.05.
RESULTS
BMP2 and BMP4 Regulate Expression of CRMP2 in Cortical Cells
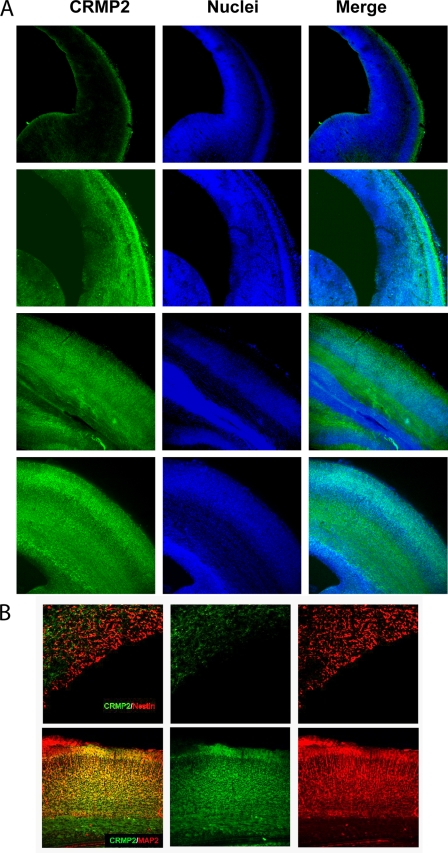
CRMP2 is expressed at high levels in the brain and has been shown to be essential for axon outgrowth and axon-dendrite specification in vitro (19). We have found that BMP2 and -4 regulate the transcription of CRMP2 in ES cells. To investigate the relationship between BMPs and CRMP2 during brain development, we first inspected the expression pattern of CRMP2 during cortical embryogenesis. As shown in Fig. 1A, low expression levels of CRMP2 in cerebral cortex began to appear at E14. From E16 on, high levels of CRMP2 began to be present in the cortical plate of the cerebral cortex (Fig. 1A). In addition, CRMP2 started to be expressed in the IZ at E16 and was intensively expressed in the IZ at E18 where many migrating neurons were present. The expression of CRMP2 continued to increase in the cortical plate, whereas it was still weak in VZ during the late stages of development. We also did double staining of both CRMP2 and nestin or MAP2 to determine the identity of CRMP2-positive cells. Few nestin-positive cells in the VZ and SVZ were positive for CRMP2 (Fig. 1B), whereas almost all of the MAP2-positive cells (mature neurons) were positive for CRMP2 (Fig. 1F). This is in agreement with previous reports that CRMP2 is mainly expressed in postmitotic neurons but not in progenitor cells. However, there were a large number of CRMP2-positive cells below the cortical plate in the IZ and SVZ, indicating that CRMP2 may have functions in immature neurons that are located in the IZ and SVZ.
FIGURE 1.
Expression pattern of CRMP2 during cortical development. A, brain slices from E14, E16, E18, and E16 rats were subjected to immunofluorescent staining for CRMP2 and propidium iodide staining for the nuclei. B, double immunofluorescent staining of both CRMP2 and the progenitor cell marker nestin in E20 rat brain (upper panels) or of CRMP2 and mature neuron marker MAP2 in E18 mouse brain to determine the identity of CRMP2-positive cells.
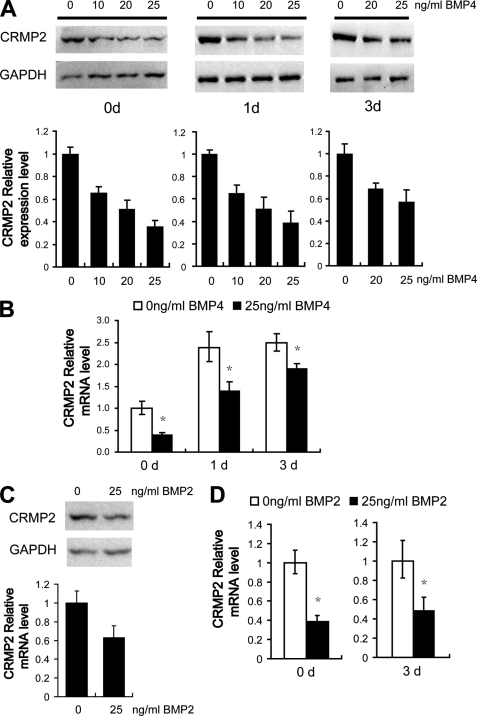
We dissected primary cortical cells from E16 rat brain cerebral cortex and cultured them for 0–3 days. Cells were then treated with BMP4 or BMP2 at different concentrations for 4 h and CRMP2 expression was examined thereafter. BMP signal activation in the primary cultured cortical cells was confirmed first by detecting the phosphorylation of SMAD1 in response to BMP stimulation (supplemental Fig. S1). As shown in Fig. 2A, the expression of CRMP2 declined significantly in all the BMP4-treated samples in a dose-dependent manner. It appeared that cells cultured for 3 days were not as sensitive to BMP4 as those cells that were just plated. Similar to BMP4, BMP2 could down-regulate the expression of CRMP2 (Fig. 2C).
FIGURE 2.
BMPs regulate CRMP2 transcription and expression in cortical cells. E16 primary cortical cells were cultured for 0–3 days and treated with different concentrations of BMP4 (A and B) or BMP2 (C and D) as described. A, Western blotting analysis of CRMP2 protein levels. GAPDH was used as loading control. The quantitative summarization of Western blots demonstrates that BMP4 can suppress CRMP2 protein expression in primary cortical cells. B, quantification of CRMP2 mRNA levels by quantitative RT-PCR shows that the transcription of CRMP2 is suppressed by BMP4 when primary cortical cells were treated with BMP4. C, Western blotting analysis of CRMP2 protein levels is shown. GAPDH was used as loading control. D, quantitative RT-PCR analysis shows that BMP2 can down-regulate the expression and transcription of CRMP2. Error bars represent S.E. t test, *, p < 0.05.
To test whether BMP2 and BMP4 regulate CRMP2 expression at the transcriptional level, real-time quantitative RT-PCR analysis was performed. As shown in Fig. 2B, the levels of CRMP2 mRNA appeared to increase after plating. However, the relative levels of CRMP2 mRNA decreased substantially in those primary cortical cells treated with either BMP2 or BMP4 (Fig. 2, B and D). This indicates that both BMP2 and BMP4 can suppress the expression of CRMP2 in primary cortical cells at the transcriptional level.
SMAD1 and SMAD4 Regulate CRMP2 Transcription in Developing Neocortex
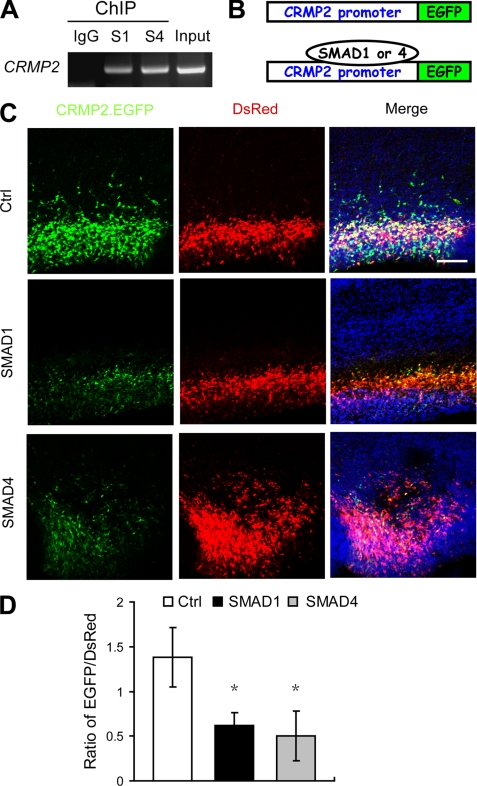
Because SMAD1 and SMAD4 are the transcription factors downstream of BMP signaling, we have shown recently that BMPs can induce the activation of SMAD1 and -4 to regulate target gene expression in ES cells. To determine whether SMAD1 and -4 play a role in regulating the expression of CRMP2 in the cerebral cortex, we performed ChIP assays in both E16 rat cerebral cortex and E14 mouse cerebral cortex and found that both SMAD1 and -4 bind to the CRMP2 promoter region in both rat and mouse cortical cells (Fig. 3A and supplemental Fig. S2). Together with the fact that rat and mouse share the same predicted SMAD-binding elements in the CRMP2 promoter region (data not shown), we may conclude that the underlying regulatory mechanism is conserved in both rat and mouse.
FIGURE 3.
Both SMAD1 and SMAD4 can regulate CRMP2 transcription directly in the developing neocortex. A, both SMAD1 and SMAD4 bind to the CRMP2 promoter region as evaluated by ChIP analysis as described under “Experimental Procedures.” B, a diagram illustrates the CRMP2 promoter-driven EGFP expression vector. There are SMAD1 and SMAD4 binding sites in the CRMP2 promoter region. C, ectopic expression of either SMAD1 or SMAD4 can suppress the expression of CRMP2.EGFP. Images of cortices co-transfected with CRMP2.EGFP (green) and DsRed (as a transfection efficiency control, red) together with control vector (upper panel), SMAD1 (middle panel), or SMAD4 (lower panel) expression vector and counterstained with Hoechst dye (for nuclei, blue) 2 days after electroporation. D, quantification of the ratio of EGFP/DsRed fluorescence intensities as analyzed by ImageJ. Error bars represent S.E. t test, *, p < 0.01. Ctrl, control.
To demonstrate further that the transcription of CRMP2 is regulated by SMADs directly in the developing neocortex, we made a construct in which the CRMP2 promoter (−968 to +619 of the CRMP2 promoter region covering a predicted SMAD-binding element) was put in front of the enhanced green fluorescent protein (EGFP) coding sequence (Fig. 3B). The resulting construct, named CRMP2.EGFP, was co-transfected together with pSIREN-RetroQ-DsRed into neuronal progenitor cells of the cortical VZ of brains at E16 via in utero electroporation with or without SMAD1- or SMAD4-expressing vectors. The CRMP2 transcription level was monitored and quantified based on EGFP expression under different conditions. If SMADs regulate CRMP2 directly by binding to the CRMP2 promoter, ectopic expression of SMADs should change CRMP2 transcriptional activity and, in turn, alter the expression of EGFP in the developing neocortex. As expected, expression of either SMAD1 or SMAD4 led to a significant suppression of EGFP expression 2 days (Fig. 3, C and D) and 4 days (supplemental Fig. S2) after transfection. These results suggest that SMAD1 and SMAD4 regulate CRMP2 transcription effectively in vivo by binding to the SMAD binding element in the CRMP2 promoter region.
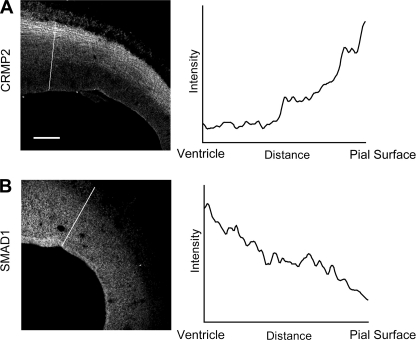
In support of our conclusion, it was of interest to notice that the elevated expression of CRMP2 was correlated with the decrease of SMADs during cortex development (Fig. 4). As shown Fig. 4A, the expression level of CRMP2 increased gradually from SVZ to CP in the E16 cortex. In contrast, the expression level of SMAD1 decreased gradually from SVZ to CP (Fig. 4B). At E18, the highest expression level of CRMP2 was in the IZ, whereas the expression level of SMAD1 was lowest in that area (supplemental Fig. S4). This indicates that SMAD1 and CRMP2 are inversely and complementarily expressed in general in the developing cerebral cortex.
FIGURE 4.
Negative correlation between the gradient SMAD1 and CRMP2 expression during cortical development. A and B, left panels, the expression patterns of SMAD1 and CRMP2. Brain slices at E16 were subjected to immunofluorescent staining for SMAD1 and CRMP2. A and B, right panels, graphs corresponding to intensities in arbitrary units for each pixel of the line drawn through the axis in A and B, respectively. X axes show the distance from ventricle to pial surface. Y axes show arbitrary units representing relative expression levels of CRMP2 and SMAD1, respectively.
CRMP2 Plays Important Roles in Neuronal Radial Migration and Neuronal Neurite Outgrowth
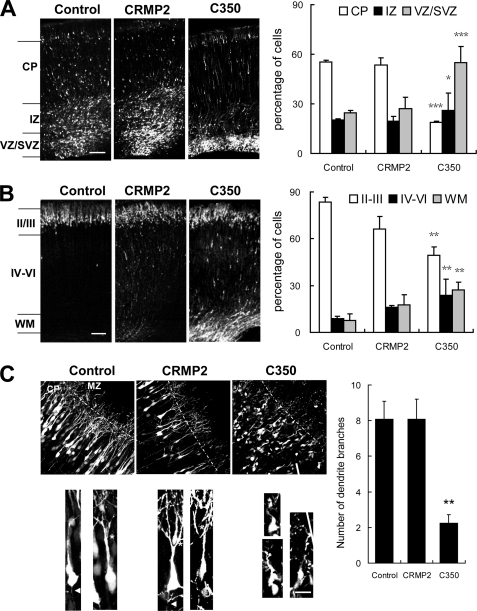
Although CRMP2 has been well studied for its role on axon outgrowth and axon-dendrite specification in vitro, its role in vivo has not been reported. To investigate the role of CRMP2 during brain development in vivo, vectors expressing full-length CRMP2 and truncated forms of CRMP2 (CRMP2 C350 and C381) had been reported to block neuronal axon outgrowth in a dominant negative manner, (13, 15) were introduced into neuronal progenitor cells at the ventricular zone of the rat brain by in utero electroporation at E16. Animals were sacrificed at subsequent developmental stages to observe the transfected cells and their progeny by means of the EGFP. Both the morphology of the transfected cells and their locations in the developing cortex were assessed. It was of interest to observe first that the distribution of neuronal cells was apparently affected by manipulating CRMP2 during development (Fig. 5, A and B). The distribution was statistically estimated by the fluorescence intensities in different layers of the cortex. At E20, the cerebral cortex can be regionally divided into CP, IZ, and VZ/SVZ. More than half of the cells in the control had migrated to the CP, whereas ∼21% of them were in the IZ and 25% were in the SVZ/VZ. The expression of full-length CRMP2 did not have apparent effects on the distribution of the transfected cells. However, <25% of the CRMP2 C350-expressing cells had migrated to the CP, whereas about 20% of them were in the IZ and ∼50% were in the SVZ/VZ (Fig. 5A). The effect of dominant negative (DN)-CRMP2 was still apparent even at postnatal day 2 (P2). Although >80% of control transfected cells migrated normally to superficial layers II/III, only ∼25% of DN-CRMP2 expressing cells had reached there (Fig. 5B). We observed similar phenotypes for another DN form of CRMP2, CRMP2 C381 (supplemental Fig. S5). These results suggest that DN-CRMP2 can inhibit the radial migration of neuronal cells.
FIGURE 5.
CRMP2 plays important roles in neural progenitor cell distribution and neuronal polarity in the neocortex. pCAGGS-EGFP, pCAGGS-CRMP2-EGFP, pCAGGS-C350-CRMP2-EGFP was introduced into neuronal progenitor cells at the ventricular zone of the rat brain by in utero electroporation at E16 (A–C). A, coronal sections of rat brain 4 days after electroporation (left panels). Cells expressing CAGGS-C350-CRMP2-EGFP were largely restricted to the VZ/SVZ, although some appeared within the IZ and CP at E20. Percentages of cells transfected with the above expression vectors in different regions of the neocortex (right panels). B, coronal sections of P2 rat brain (left panels). Compared with control transfected cells, only a fraction of the cells expressing CAGGS-C350-CRMP2-EGFP reached superficial layers II/III. Percentages of transfected cells in different regions of the neocortex (right panels). Scale bars: 250 μm. C, CRMP2 plays a role in neurite outgrowth. Coronal sections of P2 rat brain at superficial layers II/III. Some of the WT CRMP2-expressing cells had more branching processes than the control cells. In contrast, the DN-CRMP2-expressing cells did not show typical axons or dendrites (left lower panels). Percentages of transfected cells with multiple branching processes in the CP (right panels). Error bars represent S.E. t test, *, p < 0.0565307; **, p < 0.0165307; ***, p < 0.001. Scale bars: 5 μm.
To investigate the role of CRMP2 in neurite outgrowth in vivo, we analyzed the morphology of transfected cells that reached the superficial layers of the cortex at P2. As shown in Fig. 5C, some of the WT CRMP2-expressing cells seemed to have more branching neurites than the control cells, although it is not statistically significant. In contrast, the DN-CRMP2-expressing cells did not show typical axons or dendrites. This is in agreement with previous in vitro studies that CRMP2 plays a role in neurite outgrowth.
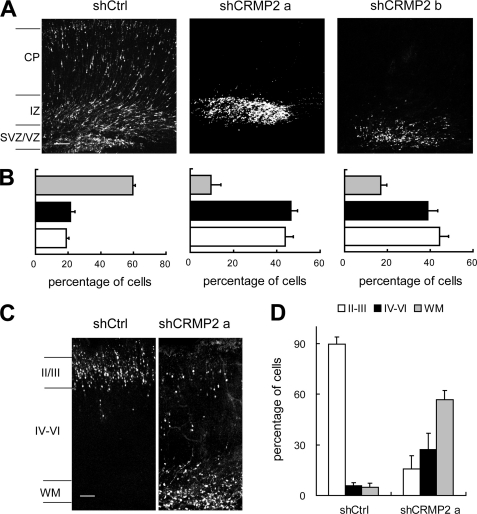
Expression of DN forms of certain molecules may interfere with the function of other endogenous proteins, leading to phenotypes different from loss-of-function experiments. To avoid potential artifactual effects of overexpression, we used RNA interference by adopting a shRNA vector, pSIREN-RetroQ-DsRed, a bicistronic construct encoding a shRNA and DsRed. Different shRNA sequences corresponding to different regions within CRMP2 were cloned into pSIREN-RetroQ-DsRed. Different constructs were transfected into neural progenitor cells of rat brain at E16. The knockdown of endogenous CRMP2 was confirmed in vivo (supplemental Fig. S8), and the specificity of the shRNAs was confirmed by rescue experiment (supplemental Fig. 9). The extent of cell migration was statistically estimated by the fluorescence intensities at E20. In brains transfected with control shRNA, ∼60% of transfected cells were detected in the CP (Fig. 6, A and B), whereas ∼20% were in either the IZ or VZ. In contrast, <10 and 20% of DsRed fluorescent cells were detected, respectively, in the CP for brains transfected with two CRMP2 shRNAs, whereas the intensities had more than doubled in both IZ and SVZ/VZ (>40%)(Fig. 6, A and B, and supplemental Fig. S6). At P2, compared with cells transfected with control shRNA, most cells transfected with shCRMP2 failed to enter cortical layers II/III but were stalled in layers IV–VI and the WM zone instead (Fig. 6, C and D). We got similar results using shRNAs different from those used in Fig. 6 (data not shown). These findings indicate that CRMP2 is essential for normal cortical neuron radial migration and correct lamination.
FIGURE 6.
Disruption of cell redistribution in the neocortex by CRMP2 RNAi. Rat embryos electroporated in utero with pSIREN-RetroQ-DsRed (shCtrl), pSIREN-RetroQ-dsRed-shCRMP2a (shCRMP2a), pSIREN-RetroQ-DsRed-shCRMP2b (shCRMP2b) at E16. A, coronal sections of rat brain 4 days after electroporation. Down-regulation of CRMP2 caused an evident accumulation of cells in the VZ/SVZ and IZ at E20. B, percentages of transfected cells in different regions of the neocortex in A. C, coronal sections of P2 rat brain. The majority of the shCRMP2-transfected cells were arrested in the WM, whereas a much fewer number of the shCRMP2 cells have reached layers II-III. D, percentages of transfected cells in different regions of the neocortex in C.
Although most CRMP2 knockdown cells accumulated in the VZ/SVZ, some cells with a bipolar morphology reached the IZ. To test whether these cells represented a motile subpopulation of CRMP2 shRNA transfectants, slices from brain tissue that had been transfected with CRMP2 or control shRNA constructs were placed in culture for live cell imaging. In control brains, migrating bipolar cells in the IZ extended a leading process toward the CP (supplemental Fig. S7A). Movement of the cell bodies was discontinuous as the cells underwent locomotion toward the pial surface as reported previously (23). In CRMP2 shRNA-transfected brains, we examine the cells with an overall bipolar morphology. The somas of all monitored cells were mostly immobile (supplemental Fig. S7B).
CRMP2 Is Essential for Progression from Multipolar to Migratory Stage and for Proper Orientation of Newly Emergent Bipolar Cells
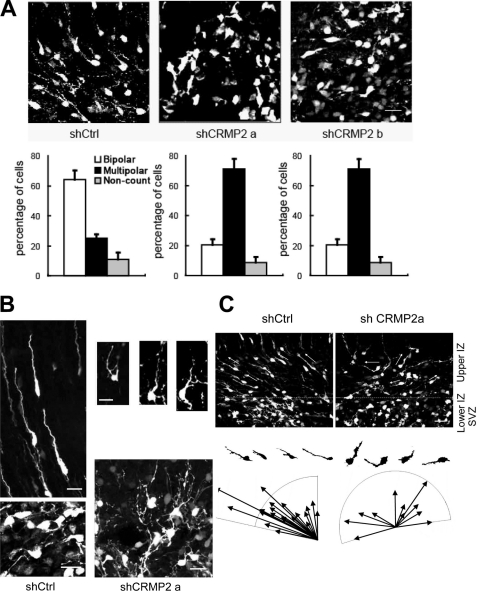
During development, immature neurons transiently become multipolar after the final division, with multiple processes being present within the SVZ and IZ, which is also known as the premigratory zone. Later, these cells change their shape from multipolar to bipolar just before migrating from the premigratory zone to the CP (Fig. 7A) (23).
FIGURE 7.
CRMP2 RNAi blocks the progression from the multipolar to the migratory stage and the proper orientation of newly emergent bipolar cells. Control (Ctrl) and CRMP2 shRNA vectors were electroporated as in Fig. 6 and analyzed 4 days later. A, morphology of cells in the upper SVZ and lower IZ transfected with control shRNA (shCtrl) and CRMP2 shRNAs (shCRMP2) (upper panels). The majority of control cells became bipolar, whereas most shCRMP2 cells remained in a multipolar morphology. Quantitative effects of shCRMP2 on cell morphology showed that the percentage of multipolar cells in shCRMP2-transfected brains is much higher than control shRNA-transfected brains (lower panels). B, typical morphology of cells transfected with control shRNA (left) and the abnormal morphology of CRMP2 shRNA transfected cells in the SVZ (lower panels) and IZ (right upper panels). Note that some multipolar cells transfected with shCRMP2 had extra long and thin processes compared with the control cells. Many of the bipolar cells transfected with shCRMP2 had curved and much thinner leading processes. In addition, there were several short and thin branches on these leading processes. C, misorientation of newly emergent bipolar cells by CRMP2 RNAi. Some of the cell bodies and leading processes of bipolar cells in the lower IZ are marked by arrows.
Because overexpression of DN forms of CRMP2 and knockdown of CRMP2 expression with different CRMP2 shRNAs led to the accumulation of cells in the VZ/SVZ/IZ, we next looked at their morphology in detail in the SVZ and lower IZ to understand the role of CRMP2 in radial migration. The ratio of cells exhibiting multipolar to bipolar morphologies were analyzed. When control shRNA was transfected, nearly 65% of the cells were bipolar and ∼25% were multipolar. In contrast, the percentages of bipolar and multipolar cells became ∼20 and 70%, respectively, when different CRMP2 shRNAs were transfected (Fig. 7A). We noticed that some of the multipolar cells in CRMP2 knockdown cells had unusual long and thin processes compared with those of control multipolar cells that had only short processes (Fig. 7B). Even for those cells transfected with shCRMP2 that did become bipolar, many of these were apparently abnormal when compared with bipolar cells in the controls (Fig. 7B). Some of the shCRMP2-transfected cells had curved, much thinner or sometimes longer leading processes. In addition, there were several short and thin branches on some of the leading processes.
To evaluate the differentiation state of SVZ cells, sections were stained with the neuronal marker TuJ1 and the progenitor cell marker nestin. Multipolar and bipolar cells that were transfected with CRMP2 shRNA all expressed the neuronal marker TuJ1, but not nestin, as the transfected control cells (data not shown). This suggests that the differentiation of cells in this stage is not affected by reduced CRMP2 expression.
More notably, we found that almost all of the control shRNA transfected cells were oriented toward the pial surface shortly after their switch from the multipolar to the bipolar migratory stage in the lower IZ. In contrast, many shCRMP2-transfected cells orientated in an apparently random manner even after they had switched to the bipolar state (Fig. 7C).
DISCUSSION
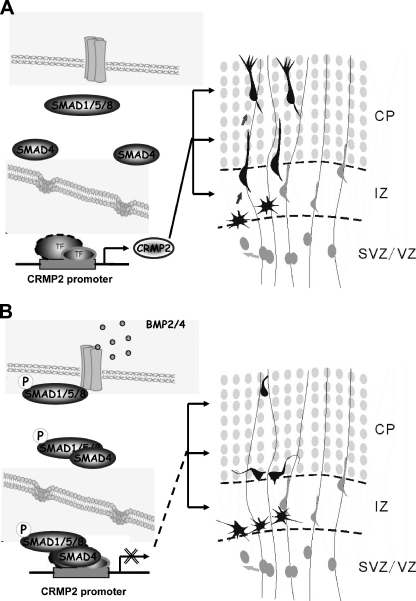
In this study, we have demonstrated that the BMP-SMAD signaling pathway regulates the transcription and expression of CRMP2 (collapsin response mediator protein-2) during brain development both in vitro and in vivo. We went on to study the biological function of CRMP2 during brain development in vivo by utilizing an in utero electroporation technique and using RNAi and overexpression of DN forms of CRMP2. We have discovered that acute interference with CRMP2 expression or function in wild-type background rat brain dramatically inhibits the distribution of newborn neurons during neocortical development. Our static and live imaging analysis of CRMP2 knockdown cells provided evidence for roles of CRMP2 at multiple steps of neuron development: 1) conversion of multipolar SVZ cells to the bipolar migratory state; 2) orientation and radial migration of bipolar cells through the IZ; and 3) axon/dendrite outgrowth in the cerebral cortex (Fig. 8A). We noticed that there were significant differences on neuron distribution between brains transfected with DN forms of CRMP2 and shCRMP2s. It is possible that the DN forms of CRMP2 took effect much faster than the shRNA. It is, therefore, intriguing to investigate in the future whether CRMP2 plays a role in neural progenitor cell differentiation by performing in utero electroporation earlier than E16 or by knocking out CRMP2 gene in mouse. The apparent multiple effects on different stages of newborn neurons by RNAi can be explained as the result of distinct effects at of CRMP2 knockdown at different stages during cerebral cortex development. We speculate that this is likely due to the uptake of various amounts of shRNA-expressing vector and transfection at different cell cycle stages of neural progenitor cells at the ventricular surface. This technique provides a unique way to gain direct insight into the complete range of functions of the BMP-SMAD signaling pathway and of CRMP2 during brain development.
FIGURE 8.
Schematic diagram showing that CRMP2 expression is regulated by BMP-SMAD signaling during brain development and CRMP2 is essential for multiple stages of neuron development. A, In the absence of BMP, SMADs are in the inactivated form so that CRMP2 is expressed and plays roles in multiple stages of neuron development, including the following: 1) conversion of multipolar SVZ cells to the bipolar migratory state; 2) orientation and radial migration of bipolar cells through the IZ; and 3) neurite outgrowth in the cerebral cortex. B, it can be speculated that SMADs can be activated by BMP to suppress the expression of CRMP2, which, in turn, prevents premature neuron development during brain development. TF, transcription factor.
BMP-SMAD Signaling Pathway Regulates CRMP2 Expression in Developing Cerebral Cortex
Although previous studies have indicated that BMP2/4 may play a role in brain development by regulating the differentiation of neural stem cells, the role of BMP2/4 during brain development and the underlying mechanism have not been studied in detail in vivo. We provided evidence here that BMP2/4 could regulate the expression of CRMP2 by suppressing the transcription of CRMP2 during neocortical development (Fig. 2). Through ChIP and a reporter assay, we found that the downstream mediators of BMP signaling SMAD1 and SMAD4 could bind to the promoter of CRMP2 directly and suppress the expression of EGFP driven by the CRMP2 promoter in the cerebral cortex in vivo (Fig. 3).
BMP4 expression has been shown to decline over time from E11.5 to E13.5 during brain development (24), and we have found that the expression of CRMP2 started at ∼E14 in rat brain and continued to increase during the late stages of development (Fig. 1). Thus, there is a negative correlation between BMP2/4 and CRMP2 expression during brain development. It was intriguing to notice the expression of both CRMP2 and SMAD1 is in gradient manner. In addition, there is also a negative correlation between expression of CRMP2 and SMAD1 in the developing cerebral cortex (Fig. 4). Together with the evidence that primary cortical cells that were cultured for 3 days were not as sensitive to BMP4 as those cells just after plating (Fig. 2), it can be speculated that BMPs may provide a checkpoint during brain development by suppressing the expression of CRMP2 to prevent the premature development of cortical neurons (Fig. 8B).
CRMP2 Plays Essential Roles at Different Stages of Neuron Development
The expression of CRMP2 in the IZ, SVZ, and CP at E16 (Fig. 1) implies that CRMP2 is likely to play a role in neuron development. By using RNAi and overexpression of DN constructs, we had confirmed in vivo that CRMP2 plays a role in neuronal polarity in cerebral cortex. In addition, we found that acute interference with CRMP2 expression or function substantially disturbed the redistribution of neural progenitor cells during neocortical development, leading to the accumulation of most cells within the SVZ and IZ. The phenotype is similar to what happens after interference with the expression of the dynein-interacting protein, LIS1 (23).
To understand the underlying mechanisms, we conducted both static and in situ live cell imaging analysis of brain slices. Small RNA interference with CRMP2 expression caused an apparent block of the progression from the multipolar to the migratory stage in the SVZ and lower IZ. Those multipolar cells had thin and highly dynamic processes as the controls, but a lot of these thin processes were much longer than those of the controls (Fig. 7 and data not shown).
The first detectable event in the conversion of multipolar to bipolar cell processes is a thickening of the presumptive migratory process, which the nucleus subsequently enters (25). Although the specific role of CRMP2 in the conversion of multipolar to bipolar cells is still uncertain, our observation of unusual long and thin processes in some of the multipolar cells and the newly emergent bipolar cells suggests that CRPM2 is involved in the shift of cytoplasmic contents into the differentiating migratory process. In support of this assumption, CRMP2 has been shown to link tubulin heterodimers or Sra-1/WAVE1 to kinesin-1 and regulates the transport of proteins to the distal part of the growing axon (16).
shCRMP2 transfected bipolar cells that reached the intermediate zone exhibited an apparent block in somal translocation (supplemental Fig. S7). When inspected carefully, such bipolar cells usually had obvious abnormalities including curved, much thinner, and at times much longer leading processes (Fig. 7 and data not shown). In addition, there were multiple short, thin, and highly dynamic branches on these processes. More notably, the shCRMP2 transfected bipolar cells orientated in an apparently random manner, in contrast to the control cells that were oriented toward the pial surface (Fig. 7). Therefore, one may speculate that the CRMP2 knockdown cells in the IZ would not be guided and supported by radial glial fibers, leading them to the stall in somal translocation and in migration of the whole cell.
Microtubule assembly and reorganization of actin filaments has been shown recently to be essential for neuronal radial migration (25–29). Based on evidence from primary culture systems, CRMP2 can regulate microtubule assembly, reorganization of actin filaments, and protein trafficking during neurite elongation and axon specification (15, 16, 18). It is therefore very likely that CRMP2 plays an essential role in neuronal radial migration through the regulation of microtubule assembly and reorganization of actin filaments.
Together with our previous studies, we can conclude that the BMP-SMAD signaling pathway is likely to play crucial roles through the regulation of CRMP2 expression during both early embryogenesis and mammalian brain development. CRMP2 has been reported to be associated with different brain disorders including Alzheimer disease, schizophrenia, and ischemic stroke, which involve either neuron (neurite) degeneration or neuron development (30–34). Therefore, our study would provide a clue in understanding the underlying pathogenesis and in the development of potential therapeutic treatment of these diseases.
Supplementary Material
Acknowledgments
We thank Dr. L Greene for constructive comments and editing during the preparation of this manuscript and Dr. Tsai and Dr. Yuan for help in establishing the in utero electroporation system.
This work was supported in part by grants from MOST of China, “973” program (2007CB947202 and 2006CB500701), “863” program (2006AA02Z173, 2009DFA32450, and 2009R0002); NSF (China) (30725007 and 30670663); and the Chinese Academy of Sciences (Bairen plan and KSCX1-YW-R-62/84). This work also was supported in part by grants from MOST (Ministry of Science and Technology) of China, “973” program (2007CB947202 and 2006CBS00701), “863” program (2006AA02Z173, 2009DFA32450, and 2009R0002); National Natural Science Foundation of China (30725007, 30670663 and 30921004); and the Chinese Academy of Sciences (Bairen plan and KSCX1-YW-R-62/84).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–9.
- SVZ
- subventricular zone
- BMP
- bone morphogenetic protein
- IZ
- intermediate zone
- CP
- cortical plate
- EGFP
- enhanced green fluorescent protein
- DN
- dominant negative
- P2
- postnatal day 2
- E16
- embryonic day 16
- WM
- white matter.
REFERENCES
- 1.Marín O., Rubenstein J. L. (2003) Ann. Rev. Neurosci. 26, 441–483 [DOI] [PubMed] [Google Scholar]
- 2.Hatten M. E. (1999) Ann. Rev. Neurosci. 22, 511–539 [DOI] [PubMed] [Google Scholar]
- 3.LoTurco J. J., Bai J. (2006) Trends Neurosci. 29, 407–413 [DOI] [PubMed] [Google Scholar]
- 4.Tabata H., Nakajima K. (2003) J. Neurosci. 23, 9996–10001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo X., Wang X. F. (2009) Cell Res. 19, 71–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Massagué J., Blain S. W., Lo R. S. (2000) Cell 103, 295–309 [DOI] [PubMed] [Google Scholar]
- 7.Watabe T., Miyazono K. (2009) Cell Res. 19, 103–115 [DOI] [PubMed] [Google Scholar]
- 8.Massagué J., Chen Y. G. (2000) Genes Dev. 14, 627–644 [PubMed] [Google Scholar]
- 9.Nakashima K., Takizawa T., Ochiai W., Yanagisawa M., Hisatsune T., Nakafuku M., Miyazono K., Kishimoto T., Kageyama R., Taga T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5868–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimogori T., Banuchi V., Ng H. Y., Strauss J. B., Grove E. A. (2004) Development 131, 5639–5647 [DOI] [PubMed] [Google Scholar]
- 11.Panchision D. M., Pickel J. M., Studer L., Lee S. H., Turner P. A., Hazel T. G., McKay R. D. (2001) Genes Dev. 15, 2094–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajan P., Panchision D. M., Newell L. F., McKay R. D. (2003) J. Cell Biol. 161, 911–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inagaki N., Chihara K., Arimura N., Ménager C., Kawano Y., Matsuo N., Nishimura T., Amano M., Kaibuchi K. (2001) Nat. Neurosci. 4, 781–782 [DOI] [PubMed] [Google Scholar]
- 14.Yoshimura T., Kawano Y., Arimura N., Kawabata S., Kikuchi A., Kaibuchi K. (2005) Cell 120, 137–149 [DOI] [PubMed] [Google Scholar]
- 15.Fukata Y., Itoh T. J., Kimura T., Ménager C., Nishimura T., Shiromizu T., Watanabe H., Inagaki N., Iwamatsu A., Hotani H., Kaibuchi K. (2002) Nat. Cell Biol. 4, 583–591 [DOI] [PubMed] [Google Scholar]
- 16.Kawano Y., Yoshimura T., Tsuboi D., Kawabata S., Kaneko-Kawano T., Shirataki H., Takenawa T., Kaibuchi K. (2005) Mol. Cell. Biol. 25, 9920–9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arimura N., Ménager C., Kawano Y., Yoshimura T., Kawabata S., Hattori A., Fukata Y., Amano M., Goshima Y., Inagaki M., Morone N., Usukura J., Kaibuchi K. (2005) Mol. Cell. Biol. 25, 9973–9984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimura T., Fukata Y., Kato K., Yamaguchi T., Matsuura Y., Kamiguchi H., Kaibuchi K. (2003) Nat. Cell Biol. 5, 819–826 [DOI] [PubMed] [Google Scholar]
- 19.Yoshimura T., Arimura N., Kaibuchi K. (2006) Ann. N.Y. Acad. Sci. 1086, 116–125 [DOI] [PubMed] [Google Scholar]
- 20.Fei T., Xia K., Li Z., Zhou B., Zhu S., Chen H., Zhang J., Chen Z., Xiao H., Han J. D., Chen Y. G. (2010) Genome Res. 20, 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito T., Nakatsuji N. (2001) Dev. Biol. 240, 237–246 [DOI] [PubMed] [Google Scholar]
- 22.Tabata H., Nakajima K. (2001) Neuroscience 103, 865–872 [DOI] [PubMed] [Google Scholar]
- 23.Tsai J. W., Chen Y., Kriegstein A. R., Vallee R. B. (2005) J. Cell Biol. 170, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furuta Y., Piston D. W., Hogan B. L. (1997) Development 124, 2203–2212 [DOI] [PubMed] [Google Scholar]
- 25.Ayala R., Shu T., Tsai L. H. (2007) Cell 128, 29–43 [DOI] [PubMed] [Google Scholar]
- 26.Conde C., Caceres A. (2009) Nat. Rev. 10, 319–332 [DOI] [PubMed] [Google Scholar]
- 27.Rakic P., Knyihar-Csillik E., Csillik B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9218–9222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tischfield M. A., Baris H. N., Wu C., Rudolph G., Van Maldergem L., He W., Chan W. M., Andrews C., Demer J. L., Robertson R. L., Mackey D. A., Ruddle J. B., Bird T. D., Gottlob I., Pieh C., Traboulsi E. I., Pomeroy S. L., Hunter D. G., Soul J. S., Newlin A., Sabol L. J., Doherty E. J., de, Uzcátegui C. E., de Uzcátegui N., Collins M. L., Sener E. C., Wabbels B., Hellebrand H., Meitinger T., de Berardinis T., Magli A., Schiavi C., Pastore-Trossello M., Koc F., Wong A. M., Levin A. V., Geraghty M. T., Descartes M., Flaherty M., Jamieson R. V., Møller H. U., Meuthen I., Callen D. F., Kerwin J., Lindsay S., Meindl A., Gupta M. L., Jr., Pellman D., Engle E. C. (2010) Cell 140, 74–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawauchi T., Hoshino M. (2008) Dev. Neurosci. 30, 36–46 [DOI] [PubMed] [Google Scholar]
- 30.Koide T., Aleksic B., Ito Y., Usui H., Yoshimi A., Inada T., Suzuki M., Hashimoto R., Takeda M., Iwata N., Ozaki N. (2010) J. Human Genet. 55, 469–472 [DOI] [PubMed] [Google Scholar]
- 31.Indraswari F., Wong P. T., Yap E., Ng Y. K., Dheen S. T. (2009) Neurochem. Int. 55, 235–242 [DOI] [PubMed] [Google Scholar]
- 32.Zhao X., Tang R., Xiao Z., Shi Y., Feng G., Gu N., Shi J., Xing Y., Yan L., Sang H., Zhu S., Liu H., Chen W., Liu J., Tang W., Zhang J., He L. (2006) Int. J. Neuropsychopharmacol. 9, 705–712 [DOI] [PubMed] [Google Scholar]
- 33.Soutar M. P., Thornhill P., Cole A. R., Sutherland C. (2009) Curr. Alzheimer Res. 6, 269–278 [DOI] [PubMed] [Google Scholar]
- 34.Takata K., Kitamura Y., Nakata Y., Matsuoka Y., Tomimoto H., Taniguchi T., Shimohama S. (2009) Am. J. Pathol. 175, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.