Abstract
Aminoacylhistidine dipeptidases (PepD, EC 3.4.13.3) belong to the family of M20 metallopeptidases from the metallopeptidase H clan that catalyze a broad range of dipeptide and tripeptide substrates, including l-carnosine and l-homocarnosine. Homocarnosine has been suggested as a precursor for the neurotransmitter γ-aminobutyric acid (GABA) and may mediate the antiseizure effects of GABAergic therapies. Here, we report the crystal structure of PepD from Vibrio alginolyticus and the results of mutational analysis of substrate-binding residues in the C-terminal as well as substrate specificity of the PepD catalytic domain-alone truncated protein PepDCAT. The structure of PepD was found to exist as a homodimer, in which each monomer comprises a catalytic domain containing two zinc ions at the active site center for its hydrolytic function and a lid domain utilizing hydrogen bonds between helices to form the dimer interface. Although the PepD is structurally similar to PepV, which exists as a monomer, putative substrate-binding residues reside in different topological regions of the polypeptide chain. In addition, the lid domain of the PepD contains an “extra” domain not observed in related M20 family metallopeptidases with a dimeric structure. Mutational assays confirmed both the putative di-zinc allocations and the architecture of substrate recognition. In addition, the catalytic domain-alone truncated PepDCAT exhibited substrate specificity to l-homocarnosine compared with that of the wild-type PepD, indicating a potential value in applications of PepDCAT for GABAergic therapies or neuroprotection.
Keywords: Amino Acid, Carboxypeptidase, Crystal Structure, Enzyme Structure, Metalloprotease, Peptidases
Introduction
Metallopeptidases of the M20 family from the metallopeptidase H clan play diverse functions throughout all kingdoms of life, ranging from a general role in the hydrolysis of late products of protein degradation to specific biochemical functions in protein maturation, tissue repair, and cell-cycle control (1). These enzymes hold great potential for biotechnological applications and therapeutic significance (2–11). For example, Lactobacillus sp. aminopeptidase V (PepV)3 and Salmonella typhimurium peptidase T (PepT) function in amino acid utilization, whereas Escherichia coli allantoate amidohydrolase and yeast β-alanine synthase (βAS) are enzymes of the nucleotides catabolic pathway, respectively. Furthermore, Escherichia coli K12 peptidase D (PepD) (12), human brain-specific carnosinase (CN1) and nonspecific carnosinase (CN2) (13), and mouse CN2 (14) exhibit dipeptidase activity on unusual dipeptides such as l-carnosine (β-Ala-His) and l-homocarnosine (γ-amino-butyl-His), as well as on a few other distinct tripeptides. Deficiency of serum carnosinase has been described in several sib-ships in conjunction with tremor, myoclonic seizures, hypotonia, and profound psychomotor retardation (15–19). Helicobacter pylori succinyldiaminopimelate desuccinylase is considered a potential target of antimicrobial agents (20), and Pseudomonas sp. strain RS-16 carboxypeptidase G2 (CPG2) has been proposed for use in antibody-directed enzyme prodrug therapy for the development of a rescue agent in cases of methotrexate overdoses (21–25).
The substrates of PepD and carnosinases, l-carnosine and related Xaa-His dipeptides, are naturally occurring dipeptides involved in many biological pathways. Although the physiological role of l-carnosine is still uncertain, studies have indicated that l-carnosine exhibits a range of cytoprotective properties (26) that support its role as a cytosolic buffer (27), an antioxidant and free radical scavenger (28), and an antiglycation agent (29). Particularly, it is a potent and selective scavenger of α,β-unsaturated aldehydes and is known to inhibit aldehyde-induced protein-protein and DNA-protein cross-linking in neurodegenerative disorders such as Alzheimer disease, in cardiovascular ischemic damage, and in inflammatory diseases (30). Other roles ascribed to the small molecule include neuroprotective functions or action as neurotransmitters, modulation of enzymatic activities, formation of complexes with transition metals, regulation of macrophage function, and retardation of senescence in cultured fibroblasts (31, 32). In particular, homocarnosine has been suggested as a precursor for the neurotransmitter GABA and acts as a GABA reservoir and may mediate the antiseizure effects of GABAergic therapies (33, 34). Recently, Sauerhöfer et al. (35) also reported that glucose metabolism could be influenced by l-carnosine.
We have previously cloned and sequenced a biofilm-related and carnosine-hydrolyzing pepD gene from Vibrio alginolyticus (36, 37). The V. alginolyticus PepD has a broader specificity for Xaa-His dipeptides and is sensitive to inhibition by bestatine and l-3,4-dihydroxyphenylalanine (36, 37). Bioinformatic analysis of the V. alginolyticus PepD protein revealed high sequence homology to that from other Vibrio species (94–76% identity) and bacteria (75–63%). In contrast, sequence-based alignment of PepD with proteins from the metallopeptidase H Clan showed low sequence identities and similarities in the range of 7–20 and 13–34%, respectively (6–8, 23, 36, 38–40). Nevertheless, sequence analysis revealed that putative active site residues for catalysis are conserved in PepD and related di-zinc enzymes in the M20 family (6, 23). His80, Asp119, Glu150, Asp173, and His461 were predicted to be involved in metal binding, whereas Asp82 and Glu149 were predicted to be necessary for catalysis. Each of these residues were completely conserved, with the exception of Asp173. Asp173 was present in homologs with aminopeptidase/dipeptidase specificity, although members of aminoacylase/carboxypeptidase contained a glutamic acid in the same position (36).
Several available crystal structures for the M20 family of enzymes, including PepV from Lactobacillus delbrueckii (6), CPG2 from Pseudomonas sp. strain RS-16 (23), and CN2 from mice (14) have been reported. These enzymes have all exhibited an overall two-domain organization, a di-zinc binding catalytic domain, and a typical smaller domain. Nevertheless, CPG2 and mouse CN2 are able to form homodimers, whereas PepV appears to exist only as a monomer (6, 14, 23). Although the biochemical activity of V. alginolyticus PepD has been well characterized, the structure-reaction mechanisms have not been studied in detail. Furthermore, little is known concerning the substrate specificity between l-carnosine and l-homocarnosine in PepD-catalyzed reactions.
We present herein the three-dimensional structure and putative substrate-binding residues of the V. alginolyticus PepD. We performed site-directed mutagenesis to determine which residues are involved in di-zinc metal ion binding and contribute to the architecture necessary for substrate C terminus binding and recognition. Furthermore, we found that the catalytic domain alone was sufficient for altering substrate specificity toward l-homocarnosine. Characterization of the PepD enzyme active-site architecture will aid in future studies to identify residues that may be modified to yield alternative substrate recognition properties and improve the potential therapeutic value of this protein and its closely related family members.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis of V. alginolyticus pepD Gene
Site-directed mutagenesis of the V. alginolyticus pepD gene was carried out using the Stratagene QuikChange site-directed mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA). Mutagenic primers were designed for use with the pET-28a(+)-pepD plasmid (wild-type) template (as described below). Mutations were confirmed by DNA sequencing using the dideoxy chain termination method and the ABI PRISM 3100 autosequencer (Applied Biosystems, Foster City, CA). The recombinant mutant plasmids were transformed into E. coli BL21(DE3) pLysS competent cells for expression of the mutated PepD proteins.
Construction of the Truncated V. alginolyticus PepD Catalytic Domain Gene
The truncated V. alginolyticus PepD catalytic domain gene (pepDCAT) was composed of the pepD gene sequence of nucleotides 1–558 and 1203–1470. An 826-bp fragment, which included 558 bp of the 5′-end and 268 bp of the 3′-end of the pepD gene, was amplified by PCR using the following primer pairs: CYC-PepD-BamHI-1 (sense primer, 5′-CGGGATCCGTGTCTGAGTTCCATTCTG-3′) and CYC-PepDcat-4 (antisense primer, 5′-TCCAGCCTGGTCCTGCACAACCCATGTACAC-3′), and by CYC-PepDcat-3 (sense primer, 5′-TGGGTTGTGCAGGACCAGGCTGGAAACCAGATG-3′) and CYC-PepD-XhoI-2 (antisense primer, 5′-CGCTCGAGTTACGCCTTTTCAGGAATG-3′). The PCR product was subcloned into pET-28a(+) to generate a recombinant plasmid, pET-28a(+)-pepDCAT, which was then transformed into E. coli BL21(DE3) pLysS competent cells for the production of the PepD catalytic domain protein (PepDCAT).
Protein Expression and Purification
Both the PepD and PepDCAT proteins were produced and purified in the same manner. Colonies grown on an LB plate were inoculated into LB broth supplemented with 50 μg/ml of kanamycin and grown at 37 °C until A600 of 0.5–0.6 was reached. At this point, protein production was induced by the addition of isopropyl thio-β-d-galactoside to a final concentration of 0.5 mm, and the culture was incubated at 37 °C for an additional 4 h before harvest. The cells were collected by centrifugation and then resuspended in 15 ml of 20 mm Tris-HCl (pH 7.0) buffer containing 0.5 m NaCl. The mixture was sonicated, and the cell debris was removed by centrifugation at 11,000 × g for 30 min at 4 °C.
The supernatant containing recombinant proteins was loaded onto a Ni-SepharoseTM 6 Fast Flow column (GH Healthcare) previously prepared by washing with 10 column volumes of buffer A (20 mm Tris-HCl, 0.5 m NaCl, pH 7.0) containing 20 mm imidazole. The protein-loaded column was first washed with 5 column volumes of buffer A + 20 mm imidazole, then with 5 column volumes of buffer A containing 70, 200, or 500 mm imidazole. Fractions of 1 ml each were collected, and the protein concentration in each fraction was determined using the Pierce BCA Protein Assay Reagent (Thermo Fisher Scientific) with BSA as the standard. Fractions containing PepD enzymatic activity were pooled and dialyzed twice against 2 liters of 50 mm Tris-HCl (pH 7.0). The purified recombinant proteins were stored at −80 °C for up to 6 months without loss of activity.
Enzymatic Activity Assay
The enzymatic activity was assayed according to the method described by Teufel et al. (13), which is based on the measurement of a histidine product by O-phthaldialdehyde (OPA) modification. Substrate hydrolysis was carried out in a final volume of 200 μl containing 80 μl of 50 mm Tris-HCl buffer (pH 7.0), 100 μl of 2 mm l-carnosine (dissolved in 50 mm Tris-HCl, pH 7.0), and 20 μl of purified enzyme (20 μm). The reaction was initiated by the addition of substrate and terminated by the addition of 50 μl of 1% trichloroacetic acid after a 30-min incubation at room temperature. Next, 50 μl of 5 mg/ml of OPA dissolved in 2 m NaOH was added to derivatize the liberated histidine, and the reaction was incubated for 15 min at 37 °C in darkness. The fluorescence of the OPA-derivatized l-histidine was measured using Fluoroskan Ascent FL (Thermo Scientific, Waltham, MA) (λExc, 355 nm and λEm, 460 nm). Reactions with only l-histidine or only l-carnosine were treated in parallel to serve as positive and negative controls, respectively. All reactions were carried out in triplicate.
Enzyme Kinetics of PepD
For determination of Vmax, Km, and kcat of V. alginolyticus PepD wild-type and mutant proteins, the method described by Csámpai et al. (41) was slightly modified for use with HPLC and fluorescence detection. Different concentrations of substrate (0.25, 0.5, 1, 1.5, 2, 5, and 10 mm) were added to a nanomolar concentration of enzyme solution in 200 μl at pH 7.4 for 20 min at 37 °C. The liberated histidine was derivatized with 100 μl of OPA reagent for 5 min at 37 °C, and the fluorescence was detected as described previously. The substrate conversion did not exceed 20%. A total of nine substrate concentration points were used for each determination. The data collected were applied to the Lineweaver-Burk equation. The kcat/Km values reflect values assuming 100% activity of the enzyme preparation. All reactions were carried out in triplicate.
Substrate Specificity of PepDCAT
Various Xaa-His dipeptides (100 μl, 2 mm) including β-Ala-l-His (l-carnosine), α-Ala-His, Gly-His, Val-His, Leu-His, Ile-His, Tyr-His, Ser-His, β-Asp-His, and γ-aminobutyryl-His (GABA-His, l-homocarnosine), two His-Xaa dipeptides (His-Asp and His-Arg), and two histidine-containing tripeptides (Gly-Gly-His and Gly-His-Gly) were used to investigate the substrate specificity of PepDCAT. The reaction solutions were incubated for 25 min at 37 °C and terminated by the addition of 50 μl of OPA solution. Catalytic activity was analyzed spectrofluorimetrically, as described above. The activity measured for l-carnosine was defined as 100%.
Crystallization and Data Collection
Crystallization of PepD was performed at 291 K by the hanging-drop vapor-diffusion method against a reservoir solution containing PEG 400 (28%, v/v), 0.2 m CaCl2 and 0.1 m Na-HEPES buffer (pH 7.5), as described previously (37). Crystals of a diamond shape appeared within six months and grew to maximum dimensions of 0.3 × 0.2 × 0.1 mm3. The protein crystals were transferred to the cryoprotectant solution containing glycerol (15%, v/v) prior to the x-ray diffraction experiment. Diffraction data were collected to 3.0-Å resolution on SPXF beamline BL13B1 at the National Synchrotron Radiation Research Center (NSRRC) in Taiwan and beamline BL12B2 at SPring-8 in Japan. The data were processed using the HKL2000 suite (42). The redundancy independent merging R factor (Rr.i.m.) and the precision indicating merging R factor (Rp.i.m.) were calculated using the program RMERGE (43, 44). The crystals belong to space group P65 with unit cell parameters a = 80.42 Å and c = 303.11 Å. The asymmetric unit contained two protein molecules, corresponding to a solvent content of 53.4%.
Structure Resolution and Refinement
The structure was solved by molecular replacement with MOLREP (CCP4) using the structure of Xaa-His dipeptidase from Haemophilus somnus 129PT (PDB code 2QYV) as the search model. The 2QYV was solved and deposited with the PDB by the Joint Center of Structure Genomics (JCSG), but was never published. The orientation of the lid domain was first located and fixed, subsequently leading to the determination of the relative position of the single catalytic domain. For structural refinement, the model was built using WinCOOT and refined using REFMAC5 (CCP4) to give the final Rwork = 0.231 and Rfree = 0.274, respectively (45). The Ramachandran results were determined using MOLPROBITY, and the percentage of residues in favored, allowed, and disallowed were 94.5, 98.6, and 1.4%, respectively (46). The structure found to have good stereochemistry was fully defined from Glu3 to Glu488, with all main chain angles in the most favorable or generally allowed regions (47). All figures were produced using PyMOL. Data collection and refinement statistics are shown in Table 1.
TABLE 1.
Data collection and refinement statistics for the PepD structure
| Data collection | |
| Space group | P65 |
| Cell dimensions | |
| a, b, c (Å) | 80.4, 80.4, 303.1 |
| α, β, γ (°) | 90, 90, 120 |
| Wavelength (Å) | 1.00 |
| Resolutiona (Å) | 30-3.0 (3.14-3.00) |
| 〈I/σI〉a | 17.6 (4.8) |
| Completenessa (%) | 99.8 (99.9) |
| Redundancya | 4.2 (4.4) |
| Rsyma,b | 4.6 (32.0) |
| Rr.i.m.a,c | 8.8 (45.0) |
| Rp.i.m.a,c | 2.7 (13.1) |
| Refinement | |
| Resolution (Å) | 30-3.0 |
| No. of reflections | 43,703 |
| Rworkd/Rfreee (%) | 23.1/27.4 |
| No. of atoms | 7,528 |
| Protein | 7,524 |
| Ion | 4 (Zn) |
| B-factors (Å2) | 86.27 |
| Protein | 86.28 |
| Ion | 90.25 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.9 |
a Values in parentheses correspond to the highest-resolution shell.
b Rsym = ∑h ∑i [|Ii(h) − 〈I(h)〉|/∑h ∑iIi(h)], where Ii is the ith measurement and 〈I(h)〉 is the weighted mean of all measurements of I(h).
c Rr.i.m. = ∑h [N/(N − 1)]1/2∑i|Ihi − 〈Ih〉|/∑h∑iIh,i. Rp.i.m. = ∑h [1/(N − 1)]1/2∑i|Ihi − 〈Ih〉|/∑h∑iIh,i. Rr.i.m. and Rp.i.m. are as defined by Weiss (43).
d Rwork = ∑h|Fo − Fc|/∑h Fo, where Fo and Fc are the observed and calculated structure factor amplitudes of reflection h.
e Rfree is as Rwork, but calculated with 10% of randomly chosen reflection omitted from the refinement.
RESULTS AND DISCUSSION
Overall Structure
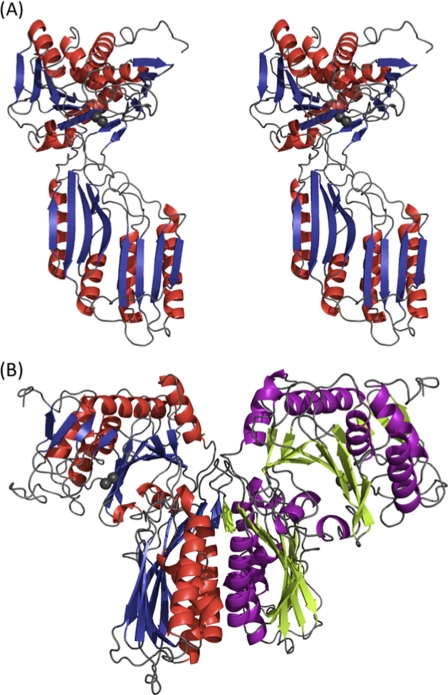
The crystal structure of V. alginolyticus PepD was solved by the molecular replacement method and refined to a resolution of 3.0 Å with an R factor of 23.1% and an Rfree factor of 27.4% (Table 1). The overall structure of the PepD monomer is comprised of a total of 486 residues in two domains: an N-terminal catalytic domain harboring two zinc ions for catalysis and a C-terminal lid domain for substrate binding and protein dimerization (Fig. 1A). The x-ray absorption measurement and electron density map confirmed the presence and locations of Zn2+ ions (supplemental Fig. S1). The high B-factors could be due to the flexible opened conformation between the catalytic and lid domains. PepD was also found to share similar structural folds with the PepV and related di-zinc-dependent M20/M28 family of enzymes, despite low sequence similarities among each. PepD and PepV showed root mean square deviations of 4.0 and 4.3 Å for Cα atoms of the catalytic and lid domains, respectively. Two PepD molecules were found to be packed as a dimer in the asymmetric unit with dimensions of ∼90 × 90 × 95 Å (Fig. 1B). The dimeric and monomeric characteristics of native and denatured PepD were also supported by evidence from analytical ultracentrifugation, which revealed molecular masses of 96.8 and 51.1 kDa under physiological and denatured conditions, respectively. The lid domain was found to utilize a hydrogen-bonding network between helices from each monomer to form the dimer interface. PepD and related di-zinc-dependent enzymes of the M20/M28 family were determined to be dimers, whereas PepV was determined to exist as a monomer.
FIGURE 1.
Overall structure of V. alginolyticus PepD. A, stereo view of the subunit of V. alginolyticus PepD. Secondary structure elements are shown in red for α-helices and blue for β-strands. Gray spheres represent the zinc ions. B, ribbon diagram of the PepD dimer. The same color scheme in A is used for the left subunit, whereas the helices and strands for the right subunit are purple and green, respectively.
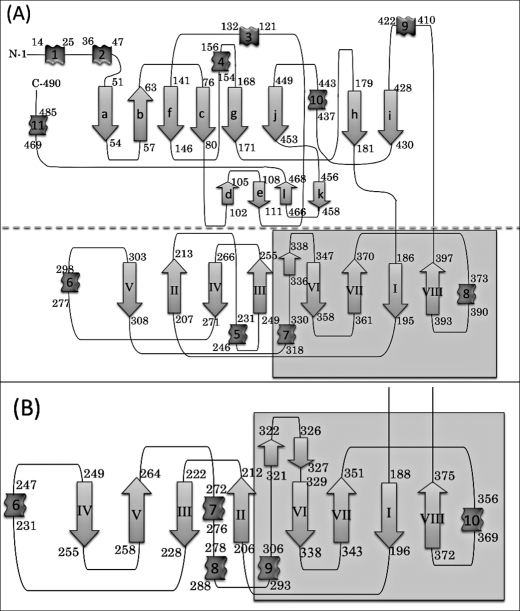
The Catalytic Domain
The topology of PepD and PepV is illustrated in Fig. 2. The PepD catalytic domain has a fold similar to that of PepV and the related di-zinc-dependent M20/M28 family of enzymes, including CPG2, βAS, mouse CN2, PepT, APAP, and SGAP (6, 7, 14, 23, 38, 39). The catalytic domain consists of residues 3–186 and 401–488 and has mixed three-layer α/β/α-sandwich architecture with two β-sheet groups and seven α-helices (Fig. 3). The large sheet group contains eight strands arranged in the order a-b-f-c-g-j-h-i, in which b is the only antiparallel strand. The small sheet group is composed of four shorter antiparallel strands arranged in the order of d-e-l-k and located on the surface of the catalytic domain. The zinc ions are located at the C-terminal end of the four central parallel strands. The active site is located within a deep cleft formed between the lid and the catalytic domain (Fig. 1). Two active sites in the dimer are ∼57 Å apart, suggesting that each protomer can function independently. No distinct zinc-bound water molecule was found; however, a higher electron density peak was observed with the closest zinc-water contact of 2.5 Å. The absence of the zinc-bound water could be due to the limited resolution of the data. The N- and C-terminal ends are located on the top of the catalytic domain, opposite to the lid domain and the active site.
FIGURE 2.
Topological diagrams of (A) V. alginolyticus PepD and (B) the lid domain of L. delbrueckii PepV. The secondary structural elements of α-helices and β-strands are represented by ribbons and arrows, respectively. The diagram (A) has been divided into two parts, separated by the dotted line; the region with gray in the lid domain represents the extra domain.
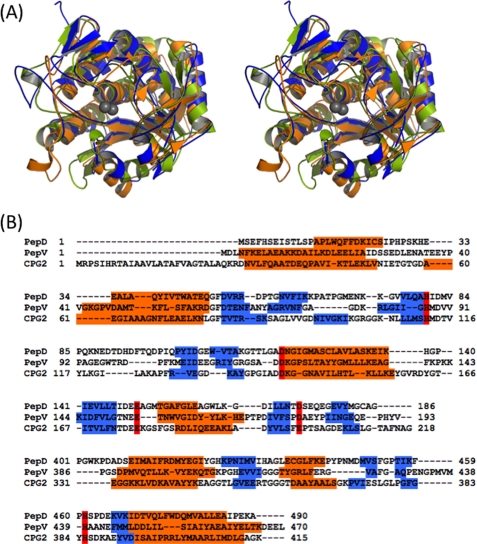
FIGURE 3.
A, superposition of the stereo view of catalytic domains of PepD (blue), PepV (orange), and CPG2 (yellow green) after optimal fit. The zinc ions of PepD are depicted by two gray spheres. B, structure-based sequence alignment of the PepD catalytic domain with PepV and CPG2. The secondary structures, α-helices and β-strands, are depicted in orange and blue, respectively. The conserved metal ion binding residues are shown in red. Each of the three catalytic domains has the same di-zinc binding residues, with the exception of Glu200 in CPG2.
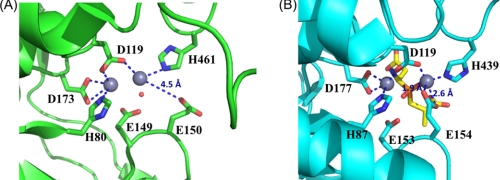
Biochemical studies of V. alginolyticus PepD revealed its metal-dependent characteristics (36) and optimal activation of apo-PepD has been observed with various divalent metal ions, including Mn2+, Co2+, Ni2+, Cu2+, and Cd2+ (36). Previous studies have shown that the addition of Co2+ ions to apo-PepD increases the enzymatic activity by a factor of ∼1.4, compared with that of the wild-type PepD containing Zn2+. Moreover, Zn2+ did not inhibit Co2+-loaded PepD activity. Substitution of Zn2+ with Mg2+ resulted in an approximate 80% restoration of the optimal enzymatic activity. In our study, the presence and locations of zinc atoms were confirmed by x-ray absorption and electron density map performed at beamline 13B1 (supplemental Fig. S1). The di-zinc center was found to be situated on the surface of the cleft between the catalytic and lid domains and, thus, is solvent-accessible. The crystal structure of PepD also revealed that several functional residues interact and fix two zinc ions (Zn1 and Zn2), which are separated by a distance of 2.8 Å (Fig. 4). Zn1 is coordinated by one of the carboxylate oxygens of Asp119, Nϵ2 from His461, and a single putative water molecule bound by hydrogen bonding with the carboxylate group of Glu149. Zn2 is coordinated by Nϵ2 of His80, the other carboxylate oxygen of Asp119, and two carboxylate oxygens of Asp173. Asp119 is positioned as a bridging ligand between the two zinc ions. Notably, this residue is followed by an asparagine residue through a cis peptide bond as observed in many di-zinc-dependent enzymes of the M20/M28 family (23). This peptide bond is able to break the α-helix at the N-terminal end to position the Asp-Asn dipeptide closer in space than they would otherwise have been. An apparent difference in the orientation of the side chain carboxylate group of Glu150 was noticed between the active-site center of PepD and that of the related M20/M28 family metallopeptidases. In PepV, the carboxylate oxygens of Glu154 point inward to Zn1 with a distance of 1.9 and 2.6 Å, respectively; whereas in PepD the carboxylate oxygens of Glu150 point away from the Zn1, and the distance between the carboxylate oxygen and Zn1 is 4.5 Å. Thus, the role of metal ion binding for Glu150 in PepD remains ambiguous.
FIGURE 4.
Comparison of the active sites in V. alginolyticus PepD and structural homologs. A, local view of the di-zinc center of PepD. The residues involved in coordination of Zn1 and Zn2 (gray spheres) are shown as green sticks. B, local view of the di-zinc center of PepV. The residues involved in metal coordination are shown as cyan sticks, and the phosphinic inhibitor (AspΨ[PO2CH2]AlaOH) is represented by yellow sticks. In the PepD active site, Zn1 is coordinated by Asp119, His461, and a single putative water molecule hydrogen-bonded to the Glu149, whereas Zn2 is coordinated by His80, Asp119, and Asp173. The putative water molecule of PepD is depicted by a red dot. Asp119 of PepD serves as a bridging ligand for metal coordination.
The Lid Domain
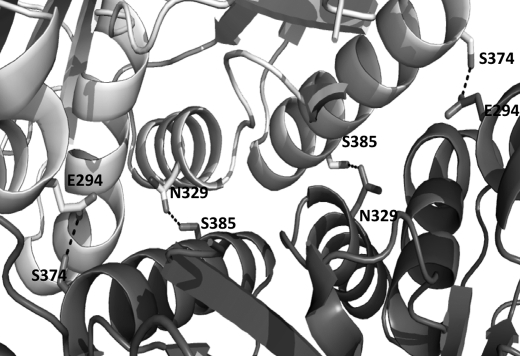
The lid domain of PepD consists of 214 residues (residues 187–400) between strands h and i of the β-sheet in the catalytic domain. It folds into a central eight-stranded antiparallel β-sheet flanked on one side by four α-helices packed in alternating orientations (Fig. 5). The antiparallel β-sheets are arranged in the order of V-II-IV-III and VI-VII-I-VIII, respectively. Interestingly, the structure of the lid domain of PepD resembles that of PepV, but shares only a portion of the structure of related dimeric M20/M28 family enzymes, including CPG2, βAS, mouse CN2, and PepT. The CPG2 dimer exhibits continuous β-sheets across the two monomers to form the dimer interface, whereas the lid domain of PepD formed the dimeric interface through hydrogen bonding between helices. Moreover, PepD formed a unique crisscross configuration via the interface interaction of the respective lid domains. Helices 6, 7, and 8 were found to participate in monomer-monomer contacts. Specifically, the carboxylate oxygens of Glu294 and the hydroxyl group of Ser374, as well as the C = O from the amide side chain of Asn329 and the hydroxyl group of Ser385, are hydrogen-bonded to each other and form the dimeric interface (Fig. 6).
FIGURE 5.
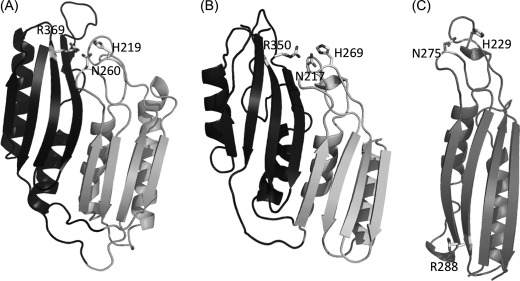
Comparison of the lid domain structures of PepV, PepD, and CPG2. A, the lid domain of PepD. Residues putatively involved in substrate C-terminal and/or transition state binding, including His219, Asn260, and Arg369 of PepD, are shown as sticks and annotated by labels. The “extra” domain region of PepD, which is absent in CPG2, is shown in black. B, the lid domain of PepV. The analogous residues present in PepV, including His269, Asn217, and Arg350, are shown as sticks and annotated by labels. The extra domain of PepV is shown in black. C, the lid domain of CPG2. Residues putatively involved in substrate C-terminal and/or transition state binding, including His229, Asn275, and Arg288, are shown as sticks and annotated. Notably, the Arg288 of CPG2 is located on the opposite side of the monomer lid domain, which is spatially different from that of Arg369 of PepD and Arg350 of PepV.
FIGURE 6.
PepD dimeric interface. Residues involved in the dimeric interface, including Glu294, Asn329, Ser374, and Ser385 of PepD, are shown as sticks and annotated by labels.
Structure Comparison of V. alginolyticus PepD and Related Di-zinc-dependent M20/M28 Family Enzymes
To further characterize the structural features of PepD, superimposition was carried out with the related M20/M28 metallopeptidases family members. The structure of PepD shows a close overall similarity to the uncharacterized PDB code 2QYV protein solved by the Joint Center for Structural Genomics (JCSG). The sequence alignment of these two proteins showed 50.9% sequence identity. The root mean square deviation of structure similarity between PepD and 2QYV for Cα atoms was 0.63 and 0.73 Å among the catalytic and lid domains, respectively. Although both proteins share a structurally conserved active site, two notable regions in PepD connecting catalytic and lid domains (PepD residues 183–187 and 400–403 versus code 2QYV residues 179–183 and 397–400, respectively) showed minor differences in loop conformations between the proteins. In addition, the PepD protein also exhibited limited amino acid yet overall folding similarity to the M20/M28 metallopeptidases, except in the region of the dimer topology. The catalytic domain of PepD superpositioned well with the single domain structures of the Streptomyces griseus SGAP (39) and Aeromonas proteolytica APAP (38), in addition to the two domain structures of the Pseudomonas sp. CPG2 (23), S. typhimurium PepT (7), Saccharomyces kluyveri βAS (8), human Acy1 (40), and mouse CN2 (14), as well as the counterpart of L. delbrueckii PepV (6). A major structural difference between PepD and the related di-zinc-dependent M20/M28 metallopeptidases is that the lid domain of PepD consists of an eight-stranded β-sheet and four α-helices similar to that of PepV, whereas the enzymes from the di-zinc-dependent M20/M28 family are composed of only one four-stranded antiparallel β-sheet flanked by two α-helices. Furthermore, part of the lid domain of the PepD structure is superimposed to that of the two domain structures of Pseudomonas sp. CPG2 (23), S. typhimurium PepT (7), S. kluyveri βAS (8), human Acy1 (40), and mouse CN2 (14). These proteins are known to form a dimer interface through hydrophobic interactions between helices, as well as through hydrogen bonds between the two β-strands within the lid domain. Nevertheless, PepD exhibited a different dimeric architecture from that of the compared dimeric proteins in that the two lid domains of the dimeric proteins mediate enzyme dimerization through side by side packing of their four-stranded β-sheets to form contiguous extended eight-stranded sheets. In contrast, a crisscross configuration was observed in PepD, wherein the lid domain formed the dimeric interface through hydrogen bonds between two sets of four α-helices (Fig. 1). Although the abovementioned M20/M28 family metallopeptidases all consist of homodimer structures similar to CPG2, no report in literature to date has discussed the PepD-like crisscross dimeric architecture. On the other hand, the structure of PepD aligns well with the counterpart of PepV (6), which is a monomer and does not have a known function in subunit dimerization. The lid domain of the PepV partially resembles the lid domain of CPG2 but is about twice as large as that of CPG2. Moreover, the PepV lid domain extends itself away from the active site of the catalytic domain, folding over the active site and assisting the catalytic domain to form a cavity that is uniquely involved in substrate specificity (6). Surprisingly, the lid domain of PepD, which is also about twice as large as the lid domain of the CPG2 and related dimeric proteins, is able to form a dimer instead of a monomer. Lindner et al. (40) reported that the eight-stranded β-sheets lid domain of PepV can be divided into two subdomains, both of which exhibit the same topology as the lid domain of CPG2 and together can mimic the arrangement of the two lid domains within the CPG2 and PepT dimers. However, dimerization of the subunits in PepD was mediated through hydrogen bonding of the α-helices instead of the side by side packing of the β-sheets. Moreover, one additional region (residues 186–203 and 311–400) within the lid domain of PepD was similar to the lid domain of related dimeric proteins, and was renamed as the “extra domain.” Although the function of the extra domain has been hypothesized, the true physiological function of the extra domain of PepD remains unclear.
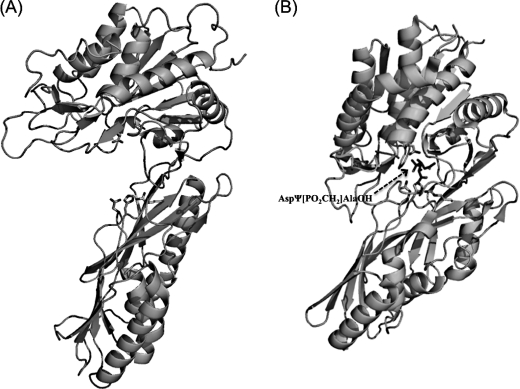
Currently, the crystal structures of the M20 family of proteins have been reported for two different (open and closed) conformations. When the protein is crystallized in a free form, the catalytic and lid domains are in an orientation that exposes the active site to bulky water; whereas when the protein is complexed with an inhibitor, a closed conformation has been observed. In the PepV-inhibitor complex, a fixed “bridging” water molecule was found to be located between both zinc ions and close to the carboxylate group of the catalytic Glu153, which corresponds to Glu149 of PepD and has been proposed as necessary for substrate hydrolysis (Fig. 7). Upon binding of the substrate, the water molecule will be positioned between the zinc ions and the carbonyl carbon of the scissile peptide bond. Then, an attacking hydroxyl ion nucleophile is able to be subsequently generated through activation of the water molecule by both the zinc ions and transfer of the resultant proton to the Glu153. Proximal to the Glu153 of PepV is the conserved metal-binding residue, Glu154, which utilizes its carboxylate oxygen to bind to the zinc ion with a distance of less than 3.0 Å. The carboxylate oxygen of Glu154 of PepV is directed toward the Zn1. Nevertheless, our structural analysis of PepD in an open conformation revealed that the carboxylate oxygen of the corresponding Glu150 residue of PepD is directed away from the Zn1 at a distance of 4.5 Å. It has been suggested that dipeptidases of M20 families can change their conformation from opened to closed during enzymatic catalysis (40). The conformational change could be achieved by a movement of the catalytic and lid domains. Consistent with this is the presence of a large clearance between the two domains that would allow a peptide chain to move to the opened active site, as was observed for the PepD structure. We, therefore, speculated that upon substrate binding, the PepD protein may change the metal ions' coordination and/or its protein conformation; the carboxylate oxygen of Glu150 would be subsequently swung toward Zn1 and would push the Glu149-bound water molecule toward Zn2, effectively bridging the water between the two zinc ions. However, the precise molecular interactions between the enzyme active site and the substrate or inhibitor still await final x-ray structure determination.
FIGURE 7.
Opened and closed conformations of PepD and PepV. A, the overall structure of PepD in the opened conformation. B, the overall structure of PepV in the closed conformation. The substrate C-terminal binding and/or transition state binding residues, including Arg, His, and Asn, are shown as sticks from left to right in the lid domain; in the catalytic domains, Glu149 (PepD) and Glu153 (PepV) are shown, respectively. The phosphinic AspΨ[PO2CH2]AlaOH inhibitor in PepV is represented by sticks.
The di-zinc binding ligands, His80, Asp119, Glu150, and His461, are conserved among all of the proteins compared in this study, but the Asp173 was found to be replaced by Glu in CPG2 and hACy1. This finding is consistent with the observation reported by Lindner et al. (40) that all homologs with proven aminopeptidase or dipeptidase specificity contain an aspartic acid, whereas a glutamic acid residue has been identified in the same position in Acyl1/M20 family members that exhibit either aminoacylase or carboxypeptidase specificity. In addition, four additional residues were found to be conserved among all of the compared proteins: 1) Asp82, which is two residues downstream from the His80 in the vicinity of the zinc center and is assumed to clamp the imidazolium ring of His80; 2) Glu149, a putative general base for enzyme catalysis; and 3) His219 and Arg369, putative substrate C-terminal and/or transition state binding residues. On the other hand, within the PepV, CPG2, and related M20/M28 family metallopeptidases, a cis-peptide bond exists between the bridging Asp and the proximal residue. In CPG2, APAP, PepT, and PepV, this residue is an Asp, which is replaced by Asn in PepD (Asn120) and SGAP (Asn98). The cis-peptide has been proposed to have a function in forcing the bridging carboxylate to conform to the correct geometry for metal binding (23).
Mutational Analysis on Metal-binding and Catalytic Residues of V. alginolyticus PepD
Previously, His80, Asp119, Glu150, Asp173, and His461 were described as being putatively involved in metal binding in PepD. We individually mutated each of these residues using alanine-scanning mutagenesis and characterized the expressed proteins with CD spectrometry (supplemental Fig. S2). Each of the mutations produced similar quantities of the protein and exhibited homologous CD spectra. Nevertheless, no activity could be detected, indicating the essential role of these residues in the enzymatic activity of PepD (Tables 2 and 3). In a parallel experimental procedure, Asp119 was substituted with Glu, Met, Leu, Ile, Arg, Phe, Ala, Ser, Thr, Cys, Pro or Asn, Glu150 was replaced with Arg or His, and Asp173 was mutated to a Glu residue. As expected, substitution of Asp119 with other proteinogenic amino acid residues completely abolished enzymatic activity. On the other hand, substitution of Glu150 with Asp led to the retention of ∼60% of the maximal hydrolytic activity of the wild-type enzyme, whereas substitution of Glu150 with Arg or His completely abolished enzymatic activity. Substitution of Asp173 with Glu also completely abolished the enzymatic activity.
TABLE 2.
Enzymatic studies of V. alginolyticus PepD wild-type and mutant proteins
(A) Kinetic parameters for the hydrolysis of l-carnosine at 37 °C and pH 7.4 by the V. alginolyticus wild-type and mutants.
| PepD | kcat | Km | kcat/Km |
|---|---|---|---|
| min−1 | mm | mm−1s−1 | |
| WT | 10.84 ± 0.22 | 0.244 ± 0.004 | 0.737 ± 0.007 |
| H80A | NDa | ND | ND |
| H82X (X = G, V,F,Y,H,E) | ND | ND | ND |
| D119X (X = A,E,LR,F,P,M,I,F,S,C,N) | ND | ND | ND |
| N120A | ND | ND | ND |
| E149D | 7.07 ± 0.07 | 0.408 ± 0.005 | 0.288 ± 0.001 |
| E149X (X = A,G,I,S,H,W) | ND | ND | ND |
| E150D | 7.3 ± 0.4 | 0.905 ± 0.195 | 0.139 ± 0.02 |
| E150X (X = A,R,H) | ND | ND | ND |
| D173X (X = A,E) | ND | ND | ND |
| S374A/S385A | 11.54 ± 0.34 | 0.221 ± 0.012 | 0.87 ± 0.02 |
| H461A | ND | ND | ND |
a ND, not determined.
TABLE 3.
Enzymatic studies of V. alginolyticus PepD wild-type and mutant proteins
Residual activity determination for the hydrolysis of l-carnosine at 37 °C and pH 7.4 by the V. alginolyticus wild-type and mutants.
| PepD | Related activity |
|---|---|
| % | |
| WT | 100 ± 0.5 |
| H80A | NDa |
| H82X (X = G,V,F,Y,H,E) | ND |
| D119X (X = A,E,L,R,F,P,M,I,S,C,N) | ND |
| N120A | ND |
| E149D | 53.1 ± 3.6 |
| E149X (X = A,G,I,S,H,W) | ND |
| E150D | 60.4 ± 1.3 |
| E150X (X = A,R,H) | ND |
| D173X (X = A,E) | ND |
| H219A | 119.1 ± 1.5 |
| N260A | 60.6 ± 0.3 |
| R369A | ND |
| S374A,S385A | 113 ± 0.8 |
| H461A | ND |
a ND, not determined.
Next the Asp82 and Glu149 residues, which are both putatively involved in catalysis, were subjected to site-directed mutagenesis. Asp82 was substituted for Gly, Val, Phe, Tyr, His, or Glu, whereas the Glu149 was replaced with Gly, Ala, Ile, Ser, His, Trp, or Asp. No activity was detected for any of the Asp82 mutants. Substituting Glu149 of PepD with Gly, Ala, Ile, Ser, His, Trp, or Asp also resulted in the abolishment of enzymatic activity, except for the Asp mutant, which retained ∼55% of the wild-type activity (Tables 2 and 3). It is interesting to note that replacement of Glu149 or Glu150 with aspartic acid led to partial retention of enzymatic activity despite being only one carbon shorter and having the same negative charge. We speculate that shortening the amino acid side chain in this position may allow for its acidic group to move away from an optimum position to promote activation of the catalytic water molecule, or perhaps the replacement of Glu with Asp at this position may partially affect the metal ligand-binding affinity and subsequent activation of the catalytic water for substrate-enzyme tetrahedral intermediate formation. This may have, in turn, resulted in partial loss of the enzymatic activity.
To investigate whether Asn120 of the cis-peptide is involved in catalysis or protein folding/stabilization, Asn120 was substituted with Ala and its enzymatic activity was examined. As expected, the activity was not detected in the PepDN120A mutant. In addition, the CD spectra of the PepD wild-type and PepDN120A mutant proteins presented almost the same shape in the range of 198–250 nm (supplemental Fig. S2), implying that the PepDN120A mutant protein was not perturbed in either its stability or folding properties. These results indicate that Asn120 plays an essential role in the enzyme reaction.
Mutational Analysis on Probable Substrate C-terminal Binding Residues within the Lid Domain of V. alginolyticus PepD
Jozic et al. (6) have previously identified three residues, Asn217, His269, and Arg350, within the lid domain of PepV that are putatively involved in the substrate C-terminal and/or transition state binding through hydrogen bonding. Due to the different topology of the β-sheet order, a simple primary sequence alignment was not able to identify the corresponding residues in the lid domain of PepD, except for Arg369, which aligned with Arg350 of PepV. This residue also superimposed with Arg324, Arg280, and Arg276 in the small domains of the dimeric CPG2, PepT, and hACy1, respectively. We then used structure-based sequence alignment to identify the other equivalent residues in PepD. A structure superimposition but inversed sequence order, in which the Asn217 and His269 residues of PepV superimposed with the Asn260 and His219 residues of PepD, was noticed. Asn260 is conserved among PepV, CPG2, βAS, and PepT, but is substituted by Thr in human CN1 and mouse CN2 as well as by Tyr in PepT. Remarkably, the His219, Asn260, and Arg369 residues are located on the same side of the lid domain for both PepD dimers, but the corresponding residues are located on the opposite side of the lid domain of the same monomer for CPG2 and related dimeric proteins (Fig. 5). Therefore, in CPG2, the Arg288 from the lid domain of one monomer interacts with the His229 and Asn275 from the lid domain of the other monomer; in contrast, the Arg369 from the lid domain of the PepD monomer interacts with the Asn260 and His219 from the lid domain of the same monomer.
We also performed site-directed mutagenesis experiments to test the roles of these equivalent lid domain residues. The mutated PepD proteins were produced in a procedure similar to that of the wild-type PepD. All mutants exhibited similar purification characteristics and the same electrophoretic mobility as the wild-type enzyme in SDS-PAGE. Although each of the mutations produced similar quantities of the protein, the Arg369 to Ala mutation resulted in complete loss of the enzymatic activity for hydrolyzing l-carnosine, whereas the Asn260 to Ala mutation decreased the catalytic activity to almost half. Interestingly, the His219 to Ala mutation did not affect the enzymatic activity significantly, yielding only a slight increase in activity of ∼10% as compared with the wild-type PepD. In PepV, the Arg350 was located near the C terminus of the bound inhibitor (2.7 Å) but appeared to be too far away from the zinc ions (∼8 Å), indicating a role in substrate binding but not in catalysis. The replacement of Arg with Ala might disrupt the hydrogen bond network between the Arg369 side chain and Asn260 Nδ with the carboxylate group of the substrate. In the case of PepV, Jozic et al. (6) have argued that domain flexibility is required to allow substrate access. Moreover, the bad diffraction and high mosaicity observed in the inhibitor-free PepV crystal have been attributed to conformational variability between open and closed states. A significant opening of the protein conformation would clearly benefit access of the peptides to the active site cavity. It is conceivable that even the whole lid domain might move away from its site to allow for easier substrate access and product egress. Therefore, although the Arg369 guanidinium side chain and the Asn260 Nδ within the active site of PepD are both ∼16 Å away from the zinc ion, a conformational change between the open and closed states might have contributed to the movement of both Arg369 and Asn260 upon substrate binding and subsequent transition state stabilization. Furthermore, binding of the His219 in PepD to the substrate likely persists during the conformational change between the open and closed states and contributes to transition-state stabilization through an electrostatic interaction between His219 and the free carboxyl group of the ligand, as shown in the PepV-inhibitor complex (Fig. 7).
Mutational Analysis on Dimeric Interface of V. alginolyticus PepD
Based on the crystal structure, PepD and PepV revealed similar architecture, except that PepD is a dimer and PepV is a monomer. From the structural illustration of PepD, the dimeric interface appears to be formed by hydrogen bonds through Ser374 and Ser385 of one subunit to Glu294 and Asn329 of the other subunit, respectively (Fig. 6). We then performed site-directed mutagenesis experiments on these residues to investigate the putative dimeric interface interactions. According to the results obtained from analytical sedimentation velocity ultracentrifugation, the molecular mass of the PepDS374A/S385A double mutant was determined to be 54.4 ± 0.02 kDa, whereas the PepD wild-type was 96.8 ± 0.11 kDa. Size exclusion chromatography of PepDWT and the PepDS374A/S385A double mutant also revealed the corresponding sizes of ∼103.7 and 50.6 kDa, respectively (supplemental Fig. S3). These results suggested that the PepDS374A/S385A mutant existed as a monomer in solution. Interestingly, the PepDS374A/S385A mutant exhibited ∼130% activity of the wild-type, indicating independent function for the monomer. However, the exact reason for forming the dimeric structure remains unclear, but a physiochemical or regulatory function of PepD may be involved.
Substrate Specificity Alteration of the Truncated PepD Catalytic Domain
We have previously observed that the catalytic domain of PepD contains metal binding sites and is responsible for substrate hydrolysis, whereas the lid domain plays a role in substrate recognition. To further substantiate the functional role of the catalytic domain, we constructed a truncated enzyme of the PepD catalytic domain alone (PepDCAT) and investigated the preference for substrate specificity. The catalytic domain regions, comprised of residues 1–186 and 401–490, were PCR amplified, ligated, and subcloned into the pET28a(+) vector to construct the PepDCAT recombinant plasmid. The truncated protein was produced similar to the wild-type PepD and exhibited the expected size of ∼31 kDa. The substrate specificity of PepDCAT was determined at pH 7.4 and 37 °C with 14 peptides, including 10 Xaa-His dipeptides, two His-Xaa dipeptides, and two His-containing tripeptides (Table 4). Compared with the enzymatic activity of wild-type PepD, the activity of PepDCAT was significantly reduced or not measurable, except for l-carnosine or l-homocarnosine. The PepDCAT exhibited ∼20% of the wild-type activity toward the l-carnosine substrate. Unexpectedly, the PepDCAT protein exhibited altered substrate specificity to l-homocarnosineas compared with that of the full-length PepD protein, and with ∼6% of the activity. The results suggested that the substrate selectivity of the PepDCAT protein hold the potential for application in GABAergic therapies or as a neuroprotector, because l-homocarnosine is a precursor for GABA and acts as a GABA reservoir.
TABLE 4.
Substrate specificity of PepDWT and PepDCAT for 10 Xaa-His dipeptides, 2 His-Xaa dipeptides, and 2 His-containing tripepides
Values are expressed as relative activity setting the degradation of PepDWT for carnosine to 100%.
| PepDWT | PepDCAT | |
|---|---|---|
| % | % | |
| l-Carnosine | 100 | 24.5 |
| α-Ala-His | 155.7 | 4.7 |
| Gly-His | 118.1 | 4.2 |
| Val-His | 117.2 | 5.5 |
| Leu-His | 157.5 | 3.7 |
| Ile-His | 146.1 | 5.8 |
| Tyr-His | 125 | 5.3 |
| Ser-His | 126.8 | 5.5 |
| His-Arg | 112.3 | 4.7 |
| His-Asp | 131.7 | 5.9 |
| β-Asp-His | 3.9 | 3.8 |
| l-Homocarnosine | 1.6 | 6.8 |
| Gly-His-Gly | 4.8 | 4.8 |
| Gly-Gly-His | 3.7 | 4.2 |
Proposed Catalytic Mechanism
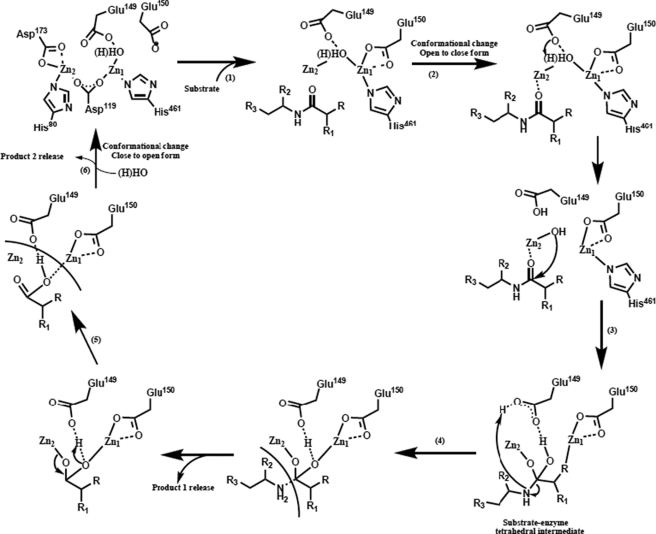
The structural similarity between PepD and related M20/M28 family metallopeptidases led to the hypothesis that these enzymes share a common catalytic mechanism. Based on superimposition of putative metal- and substrate-binding residues among these enzymes, a general mechanism may be described that: (a) before substrate binding, a bridging water molecule is positioned between two Zn2+ ions and spatially close to the carboxylate group of the catalytic Glu149; (b) the PepD undergoes a conformational change upon substrate binding and hydrolysis; (c) during catalysis, the catalytic Glu149 acts as a general base by promoting the nucleophilic attack of the metal-bound water on the substrate carbonyl carbon and transfers the proton to the Glu149; (d) the carbonyl oxygen then binds in an “oxyanion binding hole” formed by Zn1 and the imidazole group of His219 in PepD, resulting in polarization of the carbonyl group and facilitating the nucleophilic attack of the scissile bond by the zinc-oriented hydroxyl group; (e) this leads to a tetrahedral intermediate, which subsequently decays to the product after one additional proton transfer from the catalytic Glu149 carboxylate to the amide nitrogen in His219. In addition to the catalytic Glu149, mutational analyses indicated that putative substrate binding residues Asp82, Glu149, and Arg369 play essential roles in the hydrolysis reaction (Fig. 8).
FIGURE 8.
Proposed reaction mechanism of V. alginolyticus PepD. (1) Binding of the substrate to the substrate C-terminal anchoring residues; (2) protein conformational change from the open to closed form upon substrate binding, displacing the zinc-bound water by the substrate and activating the bridging water molecule by Glu149; (3) nucleophilic attack of the carbonyl carbon of the substrate by the activated water molecule and formation of the tetrahedral intermediate; (4) cleavage of the carbon-nitrogen bond and formation of product; (5) opening of the active site and release of the product; (6) addition of a new bridging water molecule.
In summary, despite the lack of detectable sequence homology, the PepD has clear structural homology to other di-zinc-dependent M20 and M28 family of enzymes. The structure of V. alginolyticus PepD reveals it to be a dimer with two domains in each subunit. The catalytic domain of PepD contains two zinc ions and is structurally homologous to other proteolytic enzymes with dinuclear zinc catalytic sites. The lid domain of PepD is structurally homologous to that of PepV, but with different topology of β-sheet order. Interestingly, part of the lid domain of the PepD structure is also homologous to the lid domain of the dimeric proteins. Nevertheless, PepV exists as a monomer, whereas the PepD and related di-zinc-dependent M20/M28 family of enzymes were determined to be dimers. Structural comparisons between PepD and related di-zinc-dependent metallopeptidases suggest that formation of the catalytically competent active site in the PepD family of enzymes may be associated with transition from an open to a closed enzyme conformation. In parallel, the site-directed mutation of the putative substrate C-terminal binding residues, D260A and R369A, resulted in complete loss or partial decrease of the enzymatic activity. Furthermore, the enzymatic assay of the truncated PepD catalytic domain, PepDCAT, further demonstrated the functional role of the lid domain in substrate binding and selectivity. Finally, the structural data on PepD reported here may inspire strategies for the improvement of the PepD family of enzymes toward applications in biotechnology and allow the design of targeted disease peptidases or prodrugs with altered specificity.
Supplementary Material
Acknowledgments
We are grateful to our colleagues, Yuch-Cheng Jean, and Chun-Shiun Chao and supporting staff members for technical assistance at the Synchrotron-Radiation X-ray Facility during data collection at BL13B1 of NSRRC in Taiwan, and Kuan-Li Yu at BL12B2 of SPring-8 in Japan.
This work was supported by the National Chiao Tung University, the Ministry of Education, Aiming for Top University Plan program, and National Science Council Grants NSC-97-2627-M-009-002 and NSC-97-2627-M-009-003, National Synchrotron Radiation Research Center Grant 963RSB02, and National Science Council Grant NSC 93-2321-B-213-001) (to C.-J. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
The atomic coordinates and structure factors (code 3MRU) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PepV
- aminopeptidase
- PepD
- aminoacylhistidine dipeptidases
- GABA
- γ-aminobutyric acid
- PepT
- peptidase T
- βAS
- β-alanine synthase
- CN
- carnosinase
- OPA
- O-phthaldialdehyde.
REFERENCES
- 1.Chen S. L., Marino T., Fang W. H., Russo N., Himo F. (2008) J. Phys. Chem. B 112, 2494–2500 [DOI] [PubMed] [Google Scholar]
- 2.Vongerichten K. F., Klein J. R., Matern H., Plapp R. (1994) Microbiology 140, 2591–2600 [DOI] [PubMed] [Google Scholar]
- 3.Hellendoorn M. A., Franke-Fayard B. M., Mierau I., Venema G., Kok J. (1997) J. Bacteriol. 179, 3410–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biagini A., Puigserver A. (2001) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 128, 469–481 [DOI] [PubMed] [Google Scholar]
- 5.Bommarius A. S. (2002) in Enzyme Catalysis in Organic Synthesis (Drauz K., Waldmann H. eds) pp. 741–749, Wiley-VCH, Weinheim [Google Scholar]
- 6.Jozic D., Bourenkow G., Bartunik H., Scholze H., Dive V., Henrich B., Huber R., Bode W., Maskos K. (2002) Structure 10, 1097–1106 [DOI] [PubMed] [Google Scholar]
- 7.Håkansson K., Miller C. G. (2002) Eur. J. Biochem. 269, 443–450 [DOI] [PubMed] [Google Scholar]
- 8.Lundgren S., Gojković Z., Piskur J., Dobritzsch D. (2003) J. Biol. Chem. 278, 51851–51862 [DOI] [PubMed] [Google Scholar]
- 9.Klein J., Henrich B. (2004) in Handbook of Proteolytic Enzymes (Barrett A. J., Rawlings N. D., Woessner J. F. eds) 2nd Ed., pp. 948–949, Elsevier, London [Google Scholar]
- 10.Miller C. G., Boder D. H. (2004) in Handbook of Proteolytic Enzymes (Barrett A. J., Rawlings N. D., Woessner J. F. eds) pp. 946–948, Elsevier, London [Google Scholar]
- 11.Agarwal R., Burley S. K., Swaminathan S. (2007) J. Mol. Biol. 368, 450–463 [DOI] [PubMed] [Google Scholar]
- 12.Schroeder U., Henrich B., Fink J., Plapp R. (1994) FEMS Microbiol. Lett. 123, 153–159 [DOI] [PubMed] [Google Scholar]
- 13.Teufel M., Saudek V., Ledig J. P., Bernhardt A., Boularand S., Carreau A., Cairns N. J., Carter C., Cowley D. J., Duverger D., Ganzhorn A. J., Guenet C., Heintzelmann B., Laucher V., Sauvage C., Smirnova T. (2003) J. Biol. Chem. 278, 6521–6531 [DOI] [PubMed] [Google Scholar]
- 14.Unno H., Yamashita T., Ujita S., Okumura N., Otani H., Okumura A., Nagai K., Kusunoki M. (2008) J. Biol. Chem. 283, 27289–27299 [DOI] [PubMed] [Google Scholar]
- 15.Perry T. L., Hansen S., Love D. L. (1968) Lancet 1, 1229–1230 [DOI] [PubMed] [Google Scholar]
- 16.Perry T. L., Hansen S., Tischler B., Bunting R., Berry K. (1967) N. Engl. J. Med. 277, 1219–1227 [DOI] [PubMed] [Google Scholar]
- 17.Terplan K. L., Cares H. L. (1972) Neurology 22, 644–655 [DOI] [PubMed] [Google Scholar]
- 18.Murphey W. H., Lindmark D. G., Patchen L. I., Housler M. E., Harrod E. K., Mosovich L. (1973) Pediatr. Res. 7, 601–606 [DOI] [PubMed] [Google Scholar]
- 19.Lunde H., Sjaastad O., Gjessing L. (1982) J. Neurochem. 38, 242–245 [DOI] [PubMed] [Google Scholar]
- 20.Karita M., Etterbeek M. L., Forsyth M. H., Tummuru M. K., Blaser M. J. (1997) Infect. Immun. 65, 4158–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minton N. P., Atkinson T., Bruton C. J., Sherwood R. F. (1984) Gene 31, 31–38 [DOI] [PubMed] [Google Scholar]
- 22.Sherwood R. F., Melton R. G., Alwan S. M., Hughes P. (1985) Eur. J. Biochem. 148, 447–453 [DOI] [PubMed] [Google Scholar]
- 23.Rowsell S., Pauptit R. A., Tucker A. D., Melton R. G., Blow D. M., Brick P. (1997) Structure 5, 337–347 [DOI] [PubMed] [Google Scholar]
- 24.Khan T. H., Eno-Amooquaye E. A., Searle F., Browne P. J., Osborn H. M., Burke P. J. (1999) J. Med. Chem. 42, 951–956 [DOI] [PubMed] [Google Scholar]
- 25.Krause A. S., Weihrauch M. R., Bode U., Fleischhack G., Elter T., Heuer T., Engert A., Diehl V., Josting A. (2002) Leuk. Lymphoma 43, 2139–2143 [DOI] [PubMed] [Google Scholar]
- 26.Boldyrev A. A. (2000) Biochemistry 65, 751–756 [PubMed] [Google Scholar]
- 27.Vaughan F. L., Hughes E. A., Jones R. S., Woods R. T., Tipper S. P. (2006) J. Int. Neuropsychol. Soc. 12, 416–423 [DOI] [PubMed] [Google Scholar]
- 28.Decker E. A., Livisay S. A., Zhou S. (2000) Biochemistry 65, 766–770 [PubMed] [Google Scholar]
- 29.Seidler N. W. (2000) J. Biochem. Mol. Toxicol. 14, 215–220 [DOI] [PubMed] [Google Scholar]
- 30.Guiotto A., Calderan A., Ruzza P., Borin G. (2005) Curr. Med. Chem. 12, 2293–2315 [DOI] [PubMed] [Google Scholar]
- 31.Hipkiss A. R. (1998) Int. J. Biochem. Cell Biol. 30, 863–868 [DOI] [PubMed] [Google Scholar]
- 32.Tabakman R., Lazarovici P., Kohen R. (2002) J. Neurosci. Res. 68, 463–469 [DOI] [PubMed] [Google Scholar]
- 33.Petroff O. A., Hyder F., Mattson R. H., Rothman D. L. (1999) Neurology 52, 473–478 [DOI] [PubMed] [Google Scholar]
- 34.Petroff O. A., Hyder F., Rothman D. L., Mattson R. H. (2001) Neurology 56, 709–715 [DOI] [PubMed] [Google Scholar]
- 35.Sauerhöfer S., Yuan G., Braun G. S., Deinzer M., Neumaier M., Gretz N., Floege J., Kriz W., van der Woude F., Moeller M. J. (2007) Diabetes 56, 2425–2432 [DOI] [PubMed] [Google Scholar]
- 36.Wang T. Y., Chen Y. C., Kao L. W., Chang C. Y., Wang Y. K., Liu Y. H., Feng J. M., Wu T. K. (2008) FEBS J. 275, 5007–5020 [DOI] [PubMed] [Google Scholar]
- 37.Chang C. Y., Hsieh Y. C., Wang T. Y., Chen C. J., Wu T. K. (2009) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 216–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chevrier B., Schalk C., D'Orchymont H., Rondeau J. M., Moras D., Tarnus C. (1994) Structure 2, 283–291 [DOI] [PubMed] [Google Scholar]
- 39.Greenblatt H. M., Almog O., Maras B., Spungin-Bialik A., Barra D., Blumberg S., Shoham G. (1997) J. Mol. Biol. 265, 620–636 [DOI] [PubMed] [Google Scholar]
- 40.Lindner H. A., Lunin V. V., Alary A., Hecker R., Cygler M., Ménard R. (2003) J. Biol. Chem. 278, 44496–44504 [DOI] [PubMed] [Google Scholar]
- 41.Csámpai A., Kutlán D., Tóth F., Molnár-Perl I. (2004) J. Chromatogr. A 1031, 67–78 [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 43.Weiss M. S. (2001) J. Appl. Cryst. 34, 130–135 [Google Scholar]
- 44.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 45.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 46.Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 47.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Cryst. 26, 283–291 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.