Abstract
The three members of the p160 family of steroid receptor coactivators (SRC-1, SRC-2, and SRC-3) steer the functional output of numerous genetic programs and serve as pleiotropic rheostats for diverse physiological processes. Since their discovery ∼15 years ago, the extraordinary sum of examination of SRC function has shaped the foundation of our knowledge for the now 350+ coregulators that have been identified to date. In this perspective, we retrace our steps into the field of coregulators and provide a summary of selected seminal work that helped define the SRCs as masters of systems biology.
Keywords: Coregulator Transcription, Drug Design, Metabolism, Nuclear Receptors, Transcription Coactivators, Biomarkers
A Stroll Down Memory Lane
More than half a century ago, Britten and Davidson first proposed their theory of master genes (1). Since then, countless attempts have been made to crown genes as master regulators. However, the criteria set forth by Britten and Davidson, that a true master gene integrates the transcription of many “producer” genes in response to a single molecular event, has been satisfied only by an incredibly finite set of genes. Now, more than a decade and a half after the discovery of the first steroid receptor coactivator (SRC),2 we believe that the genes that encode the coregulators have evolved as bona fide master genes of physiology in eukaryotes.
Our own journey into the field of coregulators began in the early 1970s, when we discovered that nuclear receptors (NRs) bound to nuclear DNA as a complex associated with proteins that we coined “acceptor proteins” (2). These acceptor proteins were first identified in non-histone nuclear fractions, and we originally viewed them as simple adapter molecules that did not bind ligand themselves but rather accepted the NR-ligand complex into chromatin and facilitated the downstream transcriptional actions of the receptor. At the time, we hypothesized there were a limited number of these adapter proteins that served to bridge the basal transcriptional machinery to the liganded receptor. Our extensive efforts to purify an acceptor protein via size and charge exclusion chromatography produced numerous perplexing peaks, and we eventually abandoned the project. Around this time, the laboratory of Murray and Towle made similar observations working with the thyroid receptor, which associated differentially with proteins from tissue nuclear extracts in response to ligand (3). Little did we know at the time that, taken together, these complex observations would come to represent the vast heterogeneity of NR coregulators that are now known to exist in mammalian cells.
Our contributions to the field of coregulator function and NR biology are undoubtedly built upon the efforts and findings of numerous laboratories. Here, we provide our account of our work on the SRC family of coregulators that shaped our thinking and honed our understanding of coregulator function in general. From the mid-80s to the early 90s, our laboratory continued to work on NR action in cell-free transcription systems. We were consistently forced to revisit these elusive acceptor proteins when we realized that they were not only sufficient to promote NR activity but were required for optimal receptor activation. A further motivation to identify these acceptor proteins came when Ma and Ptashne published that yeast Gal80, an inhibitor of Gal4, could be transformed from a transcriptional repressor to an activator by inserting an acidic activating sequence (4). In 1991, studies followed in Drosophila showing that TATA-binding protein-associated factors interact through TATA-binding protein to regulate basal promoter activity (5). A year later, the Roeder laboratory found that OCA-B (Oct coactivator from B cells) stimulated transcription from an IgH promoter with Oct-1/2 (6). Importantly, two additional studies from our laboratory in 1992 defined a ligand-controlled repressor domain in the C terminus of NRs that suppresses its transcriptional activity (7, 8). In line with these results, our laboratory described the concept of a “transcriptional switch” in yeast when we found that SSN6 binds to and suppresses the activation domain of estrogen (ER) and progesterone receptors (9). Subsequently, the Yamamoto laboratory reported that a derivative of the glucocorticoid receptor coprecipitated with the SWI3 complex, substantiating that NRs are part of a regulatory protein complex (10). Collectively, these studies helped to set the stage for the ensuing race to clone and characterize transcriptional coregulators.
Almost 2 years later in 1994, the Goodman laboratory identified the first general coregulator, cAMP-response element-binding protein-binding protein (CBP) (11) and went on to identify p300 as a functional homolog of CBP (12). Although the activities of CBP and p300 are consistent with the accepted functions of a coactivator, these proteins are now considered to be ubiquitous integrative components of virtually every eukaryotic transcription complex (13, 14). The Brown laboratory followed with the identification of ERAP160, which they identified as a ligand-dependent ER-associated protein in gel-fractionated cell extracts (15). At that time, our laboratory was working on a potential coactivator termed Spt6 in yeast. SPT6 bound to and activated the ER via its TAF2 domain, which helped us to define in part the criteria for a transcriptional coactivator (16). Conversely, that same month we presented biochemical evidence for the first NR corepressor, which allowed us to publish the now accepted mechanistic concept of coregulator function: that ligand-induced activation of NRs mediates the exchange of corepressor for coactivator to initiate transcription (17). Subsequent publications from the Glass/Rosenfeld and Evans laboratories confirmed the existence of corepressors with the cloning of SMRT (silencing mediator of retinoid and thyroid receptor) (18) and the NR corepressor, respectively (19).
Simultaneously, with our biochemical characterization of NR corepressors (17), the missing pieces of the inductive transcriptional machinery that exchanged and opposed their function were being identified. Also in 1995, our laboratory cloned an authentic NR coactivator with the discovery and characterization of SRC-1 (20). The findings from this seminal work established the criteria to which all future coactivators would be held. Published studies in 1996 from the Stallcup and Gronemeyer laboratories independently identified SRC-2 (TIF2, GRIP1) as the second member of the SRC family (21, 22). Two months later, Meltzer et al. published their findings on the amplification of three genes in human breast carcinomas, one of which, termed AIB1, turned out to be SRC-3 (23). Five laboratories subsequently characterized SRC-3 as a bona fide coactivator (ACTR, RAC-3, pCIP, TRAM-1), thus substantiating the ternion of the SRC family (24–28).
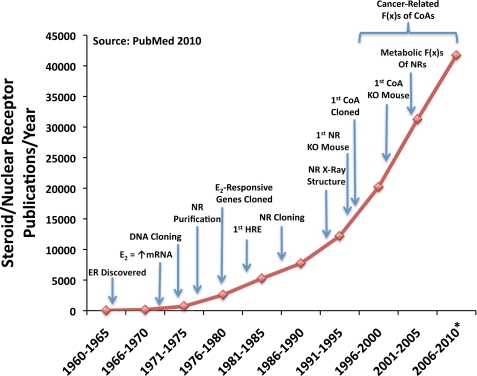
We now realize that our original prediction of the existence of only a handful of coregulators was a gross underestimate of the ∼350+ coregulators reported to date. Fig. 1 illustrates the explosion in NR publications in relation to important discoveries and key technologies, beginning with the uncovering of the ER and the discovery that steroid hormones (e.g. estrogen) acted at target genes to induce synthesis of specific mRNAs. Although the SRC family accounts for <1% of the total known coregulators, since their discovery in 1995, the collective body of work on the p160 family represents nearly 20% of the total publications on coregulators and serves as a bedrock for their enormous biological potential. Indeed, the spike in steroid receptor-related publications following the discovery of SRC-1 provides convincing evidence for the immense mechanistic and physiological importance of this evolutionarily essential class of molecules (Fig. 1). As such, this minireview focuses heavily on the SRC family of coactivators (in particular, SRC-3) to integrate the most recent and influential work on mechanism and regulation, molecular cross-talk, physiology and pathology, genetic functions, biomarkers for pathologies, and drug discovery.
FIGURE 1.
Steroid/nuclear receptor publications. A graphical analysis of publications on steroid/nuclear receptors as they relate to influential findings in the field is presented. Discoveries listed include identification of a receptor for estrogens (1962) (82), identification of estrogen stimulation of gene expression (1968–1972) (83, 84), molecular DNA cloning of the first gene (1972) (85), partial purification of NRs (1974) (86–88), identification of estrogen-responsive genes (1976) (89), identification of hormone-response elements (HRE) (1982) (90), cloning of the first full-length NRs (1985) (91), determination of the first x-ray crystal structure of NRs (1991) (92–94), development of the first nuclear receptor knock-out (KO) mouse (1993) (95), cloning of the first SRC (1995) (20), development of the first coactivator (CoA) knock-out mouse (1998) (60), and identification of metabolic functions (F(x)s) for nuclear receptors (1997) (96). These data were compiled from PubMed. The asterisk indicates a predicted estimate for the number of publications for 2010.
Mechanism and Regulation
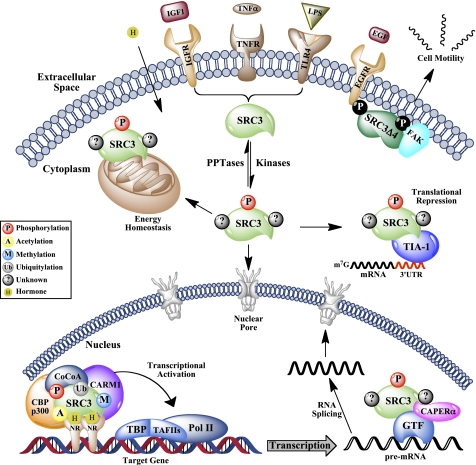
Although originally thought of as molecular “bridges” for NR/transcription factor assembly, we now appreciate that coactivator function goes well beyond the role of a simple adapter. In fact, not only do the coactivators serve as power boosters for transcription, but they integrate virtually all of the substeps of gene expression. To date, the SRCs have been shown to function in transcription initiation, elongation, RNA splicing, receptor and coregulator turnover, and even mRNA translation, suggesting that these molecules have evolved as the regulatory amalgam of higher ordered eukaryotes (summarized in Fig. 3) (20, 29–32). Recent evidence supporting this evolutionary selection emanates from the International HapMap Project, which calculates positive selection pressures for various alleles across independent ethnic populations (33). This study identified SRC-1 (NCoA1) as the gene with the strongest selective pressure among all populations analyzed, suggesting the importance of coactivators as key instruments for human evolutionary adaptation (34).
FIGURE 3.
Molecular functions of SRC-3. Presented is a schematic representation of how PTMs selectively code the numerous molecular functions of SRC-3, which include, but are not limited to, amplification of steroid- and mitogen-mediated gene transcription, regulation of RNA splicing, translational corepression, modulation of energy homeostasis, and control of cellular motility. TNFR, TNF receptor; EGFR, EGF receptor; FAK, focal adhesion kinase; CoCoA, co-coactivator; Ub, ubiquitin; TBP, TATA-binding protein; TAFIIs, TATA-binding protein-associated factors; Pol II, RNA polymerase II; PPTases, phosphatases; GTF, general transcription factor.
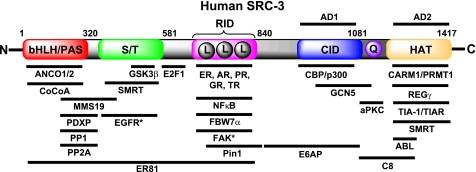
Much of our general understanding of coactivator function stems from various characterizations of the structural domains of the SRCs. The SRC family shares five fundamental and structurally conserved motifs (Fig. 2). Of these, the N-terminal bHLH-PAS (basic helix-loop-helix-Per/ARNT/Sim) domain is the most highly conserved and is necessary for several protein-protein interactions with other co-coregulators (35, 36). The bHLH-PAS domain also houses multiple nuclear localization signals, which are essential for subcellular localization and trafficking (37). The SRCs contain a serine/threonine-rich region, which is a hotspot for post-translational modification (PTM) of the coactivators (31, 38). The midregion of the SRC proteins contains three highly conserved LXXLL (where X is any amino acid) motifs, which form amphipathic α-helices and are essential for NR interaction and activation (reviewed in Ref. 39). In addition to these three LXXLL motifs, SRC family members contain three other highly conserved structural motifs. As examples, a region termed activation domain 1 binds CBP/p300 to effect histone acetylation (24). More C-terminal, activation domain 2 interacts with CARM1 (coactivator-associated arginine methyltransferase 1) (40) and PRMT1 (41) and promotes histone methylation and subsequent chromatin remodeling (Fig. 3). Interestingly, SRC-1 and SRC-3 contain an acetyltransferase domain, although its precise functional in vivo targets have not been clarified (26, 42).
FIGURE 2.
Molecular structure and interacting partners of human SRC-3. Conserved functional domains of human SRC-3 include the bHLH/PAS domain, a serine/threonine (S/T)-rich domain, a nuclear receptor-interacting domain (RID) (where L = LXXLL), the CBP/p300 interaction domain (CID), a polyglutamate region (Q), and a histone acetyltransferase (HAT) domain. This representation is not to scale and is an incomplete list of known interacting proteins. Asterisks indicate proteins that have been validated to interact specifically with SRC-3Δ4. AD, activation domain; AR, androgen receptor; PR, progesterone receptor; GR, glucocorticoid receptor; TR, thyroid receptor; CoCoA, co-coactivator; aPKC, atypical PKC; EGFR, EGF receptor; FAK, focal adhesion kinase.
The evolutionary selection of these structural domains provides the SRCs with great potential for coordinately regulating myriad cellular functions. Akin to the way an automobile engine is an assembly of hundreds of interworking parts, the coactivators function not independently but as an integral cog within a highly tuned multiprotein machine (43). We now appreciate that the coactivators function in large multiprotein complexes composed of other co-coregulators, chromatin modifiers, general transcription factors, splicing regulators, and even proteasomal components to successfully coordinate the panoply of reactions involved in gene transcription (summarized in Fig. 2) (44). Although initially characterized for their potent transcriptional activation of NR-dependent transcription, the coactivators also function to coordinately promote the activity of many other transcription factors (e.g. NFκB, HIF1α, Rb, STATs (signal transducers and activators of transcription), p53, AP-1, and E2F1) (reviewed in Ref. 39). Because the SRCs can interact with and coactivate a wide assortment of transcription factors, their perceived promiscuity actually represents the capacity of the coactivators to dynamically respond to numerous extracellular stimuli and stresses by regulating disparate genes that form functional groupings. These signaling inputs activate the cell's enzymatic transcriptional machinery by modulating the PTM code of target coactivators. Reprogramming the PTM code changes the strata of higher order coactivator complexes to provide gene-specific outputs that efficiently execute physiological programs such as reproduction, growth, motility, inflammation, and metabolism.
PTMs serve as the cell's directives for protein function and are sufficient even to dictate diametrically opposing functions (e.g. corepression) by the coactivator, creating a chemical fingerprint for localization, activity, and stability. Coactivators are targeted by diverse enzymatic machineries that alter the levels of phosphorylation, acetylation, methylation, ubiquitylation, and SUMOylation that define the PTM code. Although all three p160 family members are tightly regulated by PTMs, SRC-3 has served as an extreme model for how PTMs influence the dynamic functions of a coactivator. The early observation that SRC-3 localization and transcriptional activity could be regulated by IκB kinase β phosphorylation provided the initial impetus for identification and characterization of other PTMs (45). Since that time, work from our laboratory has helped to identify >50 unique PTM sites on SRC-3 that provide a plethora of combinations that coordinately regulate its many functions (46).
Reorganization of the coactivator PTM code changes the complement of proteins that associate with the coactivator, which in turn sets the parameters for localized concentrations and genetic activities of the coactivator complex. For example, phosphorylation of SRC-3 by GSK3β coordinately increases the transcriptionally active pool of SRC-3 by promoting its subsequent ubiquitylation (31). By comparison, atypical PKC phosphorylation shields SRC-3 from the proteasome, leading to cellular accumulation of the coactivator and enhancement of downstream gene expression and cell growth (47). In contrast, SRC-3 is targeted by phosphatases (PP1, PP2A, and pyridoxal phosphatase) that differentially regulate its ligand-dependent transcriptional activity, protein stability, and oncogenic potential (48). In addition to these functions, PTMs also regulate the intracellular localization of the coactivator. Identification of two key PTM sites within the N terminus of SRC-3 is sufficient to drive nuclear localization (37), suggesting the cooperative functions of PTMs for synchronizing location, activity, and stability of the coactivator. In contrast, phosphorylation of an SRC-3 isoform drives it to the cytoplasmic membrane for cross-talk functions (see below and Fig. 3). In the nucleus, SRC-3 is acetylated by CBP/p300, which disrupts the coactivator-NR complex and attenuates hormone-induced gene expression (49). Similarly, the hormone-regulated actions of CARM1 lead to methylation of SRC-3, acting as a molecular switch that triggers coactivator complex disassembly and decreased transcriptional activity (50). To say that the combinatorial potential of the PTM code is enormous is a gross understatement (51), and although intricate, it is precisely this complexity that governs every facet of coactivator function. The plethora of PTMs on SRC-3 are a vivid testimony to the diversity and regulatory power of the mammalian proteome.
Molecular Cross-talk
On the basis simply of their name, we are tempted to think of the SRCs as one-dimensional executers of steroid-induced transcriptional programs. However, we now recognize the value of these master gene products in functions that extend far beyond their roles in transcription. For example, SRC-3 was identified as a translational corepressor with TIA-1/TIAR (T cell-restricted intracellular antigen-1), which dampens the production of pro-inflammatory cytokines in response to an inflammatory insult (Fig. 3) (32). This elegant system underscores the ability of SRC-3 to dovetail its regulation of NFκB transcription with translational output to simultaneously provide a therapeutic inflammatory response and mitigate the deleterious potency of this response (52).
The SRCs are powerful responders to the extracellular milieu, and although many of these functions culminate to drive transcriptional programs, an SRC-3 gene product has been shown to localize to the membrane where signaling events are initiated. The relatively understudied splicing isoform of SRC-3, termed SRC-3Δ4, lacks the nuclear localization sequence-containing N-terminal bHLH domain (37), which permits its cellular membrane localization (53, 54). Stimulation with EGF enhances SRC-3Δ4 membrane localization through activation of PAK1 kinase, which phosphorylates it. At the membrane, phospho-SRC-3Δ4 directly interacts with both the EGF receptor and focal adhesion kinase to regulate cell migration and motility (Fig. 3), which consequently promotes breast tumor cell migration and metastasis to the lymph nodes and lungs (55). In this manner, the EGF-stimulated transcriptional output mediated through activation of full-length SRC-3 induces changes in genetic output while simultaneously cross-talking with SRC-3Δ4 to coordinate cell migration and invasion of cancer cells.
Physiology and Pathology
Reproductive Functions of the SRCs
As we advance our mechanistic understanding of coregulators, we also improve our knowledge of how their cellular functions marry in vitro observations with physiological outcomes. One might easily have predicted the coactivators to be intimately involved in endocrine-related processes simply due to their abundant expression in various reproductive tissues (i.e. uterus, ovary, breast, prostate) (reviewed in Ref. 56). Indeed, the coactivators are fundamental to the proper function of reproductive events such as fertility (57), uterine growth, blastocyst implantation (58), and mammary gland development (59, 60). Understandably, deficiencies in the amount or mutations that alter coactivator activity result in reproductive tissue dysfunctions. Because these tissues are tightly controlled by the mitogenic potency of steroid hormones, any abnormal change that increases coactivator expression or activity often results in the onset and progression of cancer. The breadth of studies examining the reproductive functions of the SRCs and their aberrant roles in human reproductive cancers extends well beyond the scope of this minireview, and we direct the reader to a recent review by Xu and O'Malley for a comprehensive evaluation of these findings (61).
Cancer Biology of the SRCs
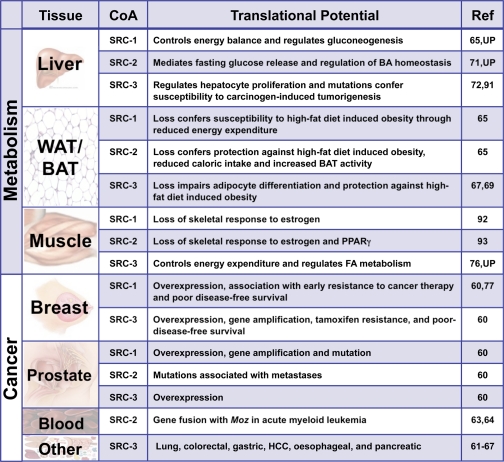
In addition to their importance in reproductive cancers, the SRCs are emerging as extremely prominent players in human cancers of non-endocrine tissues (Table 1). Although all SRCs have been thoroughly associated with breast, endometrial, ovarian, prostate, and meningioma tumors, SRC-3 figures heavily in a variety of distinct tumor types (reviewed in Ref. 61). Specifically, SRC-3 has been linked with lung, colorectal, esophageal, gastric, hepatocellular, oral squamous cell, and pancreatic cancers (reviewed in Ref. 61). Along with their roles in tumor initiation, the SRCs are known to be important regulators of cancer metastasis (see above). In a murine model for breast cancer, SRC-1 appears to be required for metastasis to the lung (62). In human prostate, mutations in SRC-2 have emerged as a predictor of prostate cancer metastasis (63). Additionally, an abnormal chromosomal rearrangement involving a fusion between the 5′-MOZ mRNA and the 3′-NCoA2 (SRC-2) mRNA positively correlates with acute myeloid leukemia (64, 65). Not only do these observations speak to the incredible mitogenic responsiveness of the coactivators, but they also dispute the misconception of explicit compensation of one SRC for another.
TABLE 1.
SRCs in Metabolism
As the coactivators continue to evolve as lynchpins of cancer biology, so too are they gaining attention as key modulators of the metabolic landscape (Table 1). The bulk of our current understanding of the metabolic functions of the SRCs emanates primarily from the characterization of knock-out mouse models. Ablation of SRC-2 protects mice against high-fat diet-induced obesity by increasing peroxisome proliferator-activated receptor γ (PPARγ) target gene expression in brown adipose tissue through the thermogenic activation of PGC-1α (66). In contrast, SRC-1−/− mice are susceptible to obesity due to decreased energy expenditure (66). This obese phenotype is partially mirrored by the combined ablation of SRC-1 and SRC-3, which arrests brown adipose activity and impairs adaptive thermogenesis. Mechanistically, these data are explained by failure to induce PPARγ target gene expression (67). Similarly, loss of SRC-3 alone impairs the white adipogenic program through decreased PPARγ2 activity (68, 69). Interestingly, the net effects of loss of SRC-1 differ starkly from ablation of SRC-3, which yields a lean body type that is highly resistant to high-fat diet-induced obesity (70). Collectively, these data suggest that the SRCs play critical yet distinct roles in controlling the energy equilibrium between brown and white adipose tissues.
Relevant to the alarming proliferation of obesity and related metabolic disorders in developed countries, the SRCs may have major implications in the pathophysiological aspects of these diseases. Of particular importance, type 2 diabetes, which is preceded by hyperglycemia, follows the inability of insulin to suppress gluconeogenesis. The ensuing compensatory elevation of insulin production is believed to promote dyslipidemia, which further compromises the insulin sensitivity of the patient. The molecular basis for this mode of insulin resistance remains ill defined but might involve the dysfunction of select key coregulators. In fact, mice deficient for SRC-1 are hypoglycemic and display markedly improved insulin sensitivity. Importantly, SRC-1 directly coordinates gene expression for the rate-limiting enzymes of the hepatic gluconeogenic program (e.g. pyruvate carboxylase) and is essential for the maintenance of glucose homeostasis (71). Distinct from this mechanism, SRC-2 has emerged as a critical regulator of hepatic glucose release by controlling the expression of the glucose-6-phosphatase gene. The absence of SRC-2 leads to a deficiency in glucose-6-phosphatase gene expression, resulting in a constellation of phenotypes that mimic glycogen storage disease type 1a (von Gierke disease) (72). In line with this function, recently published data3 suggest that SRC-2 also acts as a fulcrum to coordinately regulate the balance between dietary fuel absorption and energy utilization.
Unlike SRC-2, ablation of SRC-3 affords protection against obesity and improves insulin sensitivity, partly through regulating the acetylation of PGC-1α (70). In addition to the collective role of SRC-3 in the maintenance of glucose homeostasis, recent evidence suggests that even functional impairment of the PTM code for SRC-3 is sufficient to alter glucose homeostasis and peripheral insulin sensitivity (73). Characterization of a knock-in mouse model containing mutations at four functionally conserved phosphorylation sites in SRC-3 emphasizes the general importance of PTMs to whole animal systems biology, particularly as they relate to metabolism. The sum of these findings highlights the broad capacity of the SRCs to dynamically modulate the metabolic landscape and has identified new targets for potential therapeutic intervention in metabolic diseases.
Genetic Functions of the SRCs
The fact that coactivator functions are effected through multiprotein complexes argues that combinations of even weakly penetrant alleles within the complex may coalesce into deleterious effects that mimic polygenic diseases (74). This idea implies that improvements in our mechanistic and physiological knowledge of the SRC family might provide a path to better understand polygenic diseases secondary to a variety of coactivator complexes. The multifunctional capacity of the SRCs is often exploited to execute the directives of the cell. As such, the SRCs have been clearly implicated in a variety of human diseases, many of which are undoubtedly polygenic, likely arising from the combined misregulation of several genes. This concept is observed in the SRC-3 knock-in mouse, where even subtle genetic changes that alter coactivator function can manifest phenotypes resembling polygenic diseases like obesity and type 2 diabetes (73). In humans, we can predict that dysfunction of multiple downstream target genes would result from dysregulation of a single coactivator complex. Consequently, if a minimal defect exists in more than one coactivator in a given complex, synergism may lead to a more pronounced dysfunction of the intact complex.
Alternatively, the possibility exists that genetic changes in components of a coactivator complex could manifest phenotypes that resemble monogenic disease states. Evidence supporting this concept shows that ablation of SRC-2 phenotypically mirrors von Gierke disease, which is a genetically inherited glycogen storage disorder arising from inactivating monogenic mutations in the glucose-6-phosphatase gene. Mechanistically, SRC-2 cooperates with retinoid-related orphan receptor α to directly regulate glucose-6-phosphatase gene transcription (72). In line with these findings, genetic ablation of retinoid-related orphan receptor α confers a number of metabolic derangements that parallel loss of SRC-2 (75). Similarly, unpublished studies4 from our laboratory suggest that SRC-3 is indispensable for muscle-specific fatty acid metabolism, which we have traced to the dysregulation of an essential mitochondrial long-chain fatty acid transporter. Interestingly, genetic mutations of this transporter in humans leads to hypoglycemia, enhanced muscle-specific glucose uptake, and drastically improved insulin sensitivity, all of which we have characterized in our SRC-3−/− mice. Taken together, these findings situate the coactivators and the complexes they regulate at the nexus between monogenic and polygenic diseases.
Biomarkers for Pathologies and Drug Discovery
The known pleiotropic actions of the coactivators make them attractive candidates as biomarkers for a multitude of pathologies. In relation to cancer, SRC-3 overexpression results in aggressive breast and lung cancers (76, 77), SRC-2 mutations are associated with prostate cancer metastases (63), and SRC-1 overexpression leads to early resistance to cancer therapy (78). Also, the significance of SRC activity in shaping the metabolic landscape suggests that they may be etiological predictors for numerous metabolic diseases. Given these observations, we predict that technological advances in genome-wide association studies, protein antibody meta-arrays, and high-throughput proteomics should promote the use of SRCs as excellent biomarkers for human disease. This idea is supported by multiple findings that, unlike normal cells that tightly manage the pools of coactivator, cancer cells often become addicted to the mitogenic power afforded by coactivator overexpression or gene amplification (79–81). In addition to their utility as biomarkers in personalized medicine, we strongly view the SRCs as feasible targets for pharmacological intervention. As many of the detrimental effects of coactivator function arise from their aberrant expression or activity, we recently initiated the first small molecule chemical screens aimed at identifying pharmacological candidates that regulate the stability and/or transcriptional activity of the SRCs. Armed with the recent improvements in high-throughput screening tools and the relatively short explosion of knowledge on the SRCs, we are encouraged as we look ahead in anticipation of what the next 15 years will reveal about this amazing family of proteins.
Supplementary Material
This is the first article in the Thematic Minireview Series on Nuclear Receptors in Biology and Diseases. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
Chopra, A. R., Kommagani, R., Saha, P., Louet, J. F., Salazar, C., Song, J., Jeong, J., Finegold, M., Viollet, B., DeMayo, F., Chan, L., Moore, D. D., and O'Malley, B. W. (2011) Cell Metab., in press.
B. York and B. W. O'Malley, unpublished data.
- SRC
- steroid receptor coactivator
- NR
- nuclear receptor
- ER
- estrogen receptor
- CBP
- cAMP-response element-binding protein-binding protein
- PTM
- post-translational modification
- PPARγ
- peroxisome proliferator-activated receptor γ.
REFERENCES
- 1.Britten R. J., Davidson E. H. (1969) Science 165, 349–357 [DOI] [PubMed] [Google Scholar]
- 2.Spelsberg T. C., Steggles A. W., O'Malley B. W. (1971) J. Biol. Chem. 246, 4188–4197 [PubMed] [Google Scholar]
- 3.Murray M. B., Towle H. C. (1989) Mol. Endocrinol. 3, 1434–1442 [DOI] [PubMed] [Google Scholar]
- 4.Ma J., Ptashne M. (1988) Cell 55, 443–446 [DOI] [PubMed] [Google Scholar]
- 5.Dynlacht B. D., Hoey T., Tjian R. (1991) Cell 66, 563–576 [DOI] [PubMed] [Google Scholar]
- 6.Luo Y., Fujii H., Gerster T., Roeder R. G. (1992) Cell 71, 231–241 [DOI] [PubMed] [Google Scholar]
- 7.Vegeto E., Allan G. F., Schrader W. T., Tsai M. J., McDonnell D. P., O'Malley B. W. (1992) Cell 69, 703–713 [DOI] [PubMed] [Google Scholar]
- 8.Allan G. F., Leng X., Tsai S. Y., Weigel N. L., Edwards D. P., Tsai M. J., O'Malley B. W. (1992) J. Biol. Chem. 267, 19513–19520 [PubMed] [Google Scholar]
- 9.McDonnell D. P., Vegeto E., O'Malley B. W. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10563–10567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshinaga S. K., Peterson C. L., Herskowitz I., Yamamoto K. R. (1992) Science 258, 1598–1604 [DOI] [PubMed] [Google Scholar]
- 11.Kwok R. P., Lundblad J. R., Chrivia J. C., Richards J. P., Bächinger H. P., Brennan R. G., Roberts S. G., Green M. R., Goodman R. H. (1994) Nature 370, 223–226 [DOI] [PubMed] [Google Scholar]
- 12.Lundblad J. R., Kwok R. P., Laurance M. E., Harter M. L., Goodman R. H. (1995) Nature 374, 85–88 [DOI] [PubMed] [Google Scholar]
- 13.Pugh B. F. (2006) Mol. Cell 23, 776–777 [DOI] [PubMed] [Google Scholar]
- 14.Tyteca S., Legube G., Trouche D. (2006) Mol. Cell 24, 807–808 [DOI] [PubMed] [Google Scholar]
- 15.Halachmi S., Marden E., Martin G., MacKay H., Abbondanza C., Brown M. (1994) Science 264, 1455–1458 [DOI] [PubMed] [Google Scholar]
- 16.Baniahmad C., Nawaz Z., Baniahmad A., Gleeson M. A., Tsai M. J., O'Malley B. W. (1995) Mol. Endocrinol. 9, 34–43 [DOI] [PubMed] [Google Scholar]
- 17.Baniahmad A., Leng X., Burris T. P., Tsai S. Y., Tsai M. J., O'Malley B. W. (1995) Mol. Cell. Biol. 15, 76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurokawa R., Söderström M., Hörlein A., Halachmi S., Brown M., Rosenfeld M. G., Glass C. K. (1995) Nature 377, 451–454 [DOI] [PubMed] [Google Scholar]
- 19.Chen J. D., Evans R. M. (1995) Nature 377, 454–457 [DOI] [PubMed] [Google Scholar]
- 20.Oñate S. A., Tsai S. Y., Tsai M. J., O'Malley B. W. (1995) Science 270, 1354–1357 [DOI] [PubMed] [Google Scholar]
- 21.Hong H., Kohli K., Trivedi A., Johnson D. L., Stallcup M. R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4948–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voegel J. J., Heine M. J., Zechel C., Chambon P., Gronemeyer H. (1996) EMBO J. 15, 3667–3675 [PMC free article] [PubMed] [Google Scholar]
- 23.Guan X. Y., Xu J., Anzick S. L., Zhang H., Trent J. M., Meltzer P. S. (1996) Cancer Res. 56, 3446–3450 [PubMed] [Google Scholar]
- 24.Torchia J., Rose D. W., Inostroza J., Kamei Y., Westin S., Glass C. K., Rosenfeld M. G. (1997) Nature 387, 677–684 [DOI] [PubMed] [Google Scholar]
- 25.Li H., Gomes P. J., Chen J. D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8479–8484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H., Lin R. J., Schiltz R. L., Chakravarti D., Nash A., Nagy L., Privalsky M. L., Nakatani Y., Evans R. M. (1997) Cell 90, 569–580 [DOI] [PubMed] [Google Scholar]
- 27.Anzick S. L., Kononen J., Walker R. L., Azorsa D. O., Tanner M. M., Guan X. Y., Sauter G., Kallioniemi O. P., Trent J. M., Meltzer P. S. (1997) Science 277, 965–968 [DOI] [PubMed] [Google Scholar]
- 28.Takeshita A., Cardona G. R., Koibuchi N., Suen C. S., Chin W. W. (1997) J. Biol. Chem. 272, 27629–27634 [DOI] [PubMed] [Google Scholar]
- 29.Lefebvre B., Brand C., Lefebvre P., Ozato K. (2002) Mol. Cell. Biol. 22, 1446–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auboeuf D., Dowhan D. H., Kang Y. K., Larkin K., Lee J. W., Berget S. M., O'Malley B. W. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2270–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu R. C., Feng Q., Lonard D. M., O'Malley B. W. (2007) Cell 129, 1125–1140 [DOI] [PubMed] [Google Scholar]
- 32.Yu C., York B., Wang S., Feng Q., Xu J., O'Malley B. W. (2007) Mol. Cell 25, 765–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voight B. F., Kudaravalli S., Wen X., Pritchard J. K. (2006) PLoS Biol. 4, e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lonard D. M., Lanz R. B., O'Malley B. W. (2007) Endocr. Rev. 28, 575–587 [DOI] [PubMed] [Google Scholar]
- 35.Kim J. H., Li H., Stallcup M. R. (2003) Mol. Cell 12, 1537–1549 [DOI] [PubMed] [Google Scholar]
- 36.Chen Y. H., Kim J. H., Stallcup M. R. (2005) Mol. Cell. Biol. 25, 5965–5972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li C., Wu R. C., Amazit L., Tsai S. Y., Tsai M. J., O'Malley B. W. (2007) Mol. Cell. Biol. 27, 1296–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goel A., Janknecht R. (2004) J. Biol. Chem. 279, 14909–14916 [DOI] [PubMed] [Google Scholar]
- 39.Leo C., Chen J. D. (2000) Gene 245, 1–11 [DOI] [PubMed] [Google Scholar]
- 40.Koh S. S., Chen D., Lee Y. H., Stallcup M. R. (2001) J. Biol. Chem. 276, 1089–1098 [DOI] [PubMed] [Google Scholar]
- 41.Anafi M., Yang Y. F., Barlev N. A., Govindan M. V., Berger S. L., Butt T. R., Walfish P. G. (2000) Mol. Endocrinol. 14, 718–732 [DOI] [PubMed] [Google Scholar]
- 42.Spencer T. E., Jenster G., Burcin M. M., Allis C. D., Zhou J., Mizzen C. A., McKenna N. J., Onate S. A., Tsai S. Y., Tsai M. J., O'Malley B. W. (1997) Nature 389, 194–198 [DOI] [PubMed] [Google Scholar]
- 43.McKenna N. J., Nawaz Z., Tsai S. Y., Tsai M. J., O'Malley B. W. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 11697–11702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malovannaya A., Li Y., Bulynko Y., Jung S. Y., Wang Y., Lanz R. B., O'Malley B. W., Qin J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2431–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu R. C., Qin J., Hashimoto Y., Wong J., Xu J., Tsai S. Y., Tsai M. J., O'Malley B. W. (2002) Mol. Cell. Biol. 22, 3549–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han S. J., Lonard D. M., O'Malley B. W. (2009) Trends Endocrinol. Metab. 20, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi P., Feng Q., Amazit L., Lonard D. M., Tsai S. Y., Tsai M. J., O'Malley B. W. (2008) Mol. Cell 29, 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li C., Liang Y. Y., Feng X. H., Tsai S. Y., Tsai M. J., O'Malley B. W. (2008) Mol. Cell 31, 835–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen H., Lin R. J., Xie W., Wilpitz D., Evans R. M. (1999) Cell 98, 675–686 [DOI] [PubMed] [Google Scholar]
- 50.Naeem H., Cheng D., Zhao Q., Underhill C., Tini M., Bedford M. T., Torchia J. (2007) Mol. Cell. Biol. 27, 120–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lonard D. M., O'Malley B. W. (2007) Mol. Cell 27, 691–700 [DOI] [PubMed] [Google Scholar]
- 52.Gao Z., Chiao P., Zhang X., Zhang X., Lazar M. A., Seto E., Young H. A., Ye J. (2005) J. Biol. Chem. 280, 21091–21098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reiter R., Wellstein A., Riegel A. T. (2001) J. Biol. Chem. 276, 39736–39741 [DOI] [PubMed] [Google Scholar]
- 54.Reiter R., Oh A. S., Wellstein A., Riegel A. T. (2004) Oncogene 23, 403–409 [DOI] [PubMed] [Google Scholar]
- 55.Long W., Yi P., Amazit L., LaMarca H. L., Ashcroft F., Kumar R., Mancini M. A., Tsai S. Y., Tsai M. J., O'Malley B. W. (2010) Mol. Cell 37, 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Molenda H. A., Kilts C. P., Allen R. L., Tetel M. J. (2003) Biol. Reprod. 69, 1449–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukherjee A., Amato P., Allred D. C., DeMayo F. J., Lydon J. P. (2007) Nucl. Recept. Signal. 5, e011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mukherjee A., Soyal S. M., Fernandez-Valdivia R., Gehin M., Chambon P., Demayo F. J., Lydon J. P., O'Malley B. W. (2006) Mol. Cell. Biol. 26, 6571–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu J., Liao L., Ning G., Yoshida-Komiya H., Deng C., O'Malley B. W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6379–6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu J., Qiu Y., DeMayo F. J., Tsai S. Y., Tsai M. J., O'Malley B. W. (1998) Science 279, 1922–1925 [DOI] [PubMed] [Google Scholar]
- 61.Xu J., Wu R. C., O'Malley B. W. (2009) Nat. Rev. Cancer 9, 615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang S., Yuan Y., Liao L., Kuang S. Q., Tien J. C., O'Malley B. W., Xu J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor B. S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B. S., Arora V. K., Kaushik P., Cerami E., Reva B., Antipin Y., Mitsiades N., Landers T., Dolgalev I., Major J. E., Wilson M., Socci N. D., Lash A. E., Heguy A., Eastham J. A., Scher H. I., Reuter V. E., Scardino P. T., Sander C., Sawyers C. L., Gerald W. L. (2010) Cancer Cell 18, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liang J., Prouty L., Williams B. J., Dayton M. A., Blanchard K. L. (1998) Blood 92, 2118–2122 [PubMed] [Google Scholar]
- 65.Carapeti M., Aguiar R. C., Goldman J. M., Cross N. C. (1998) Blood 91, 3127–3133 [PubMed] [Google Scholar]
- 66.Picard F., Géhin M., Annicotte J., Rocchi S., Champy M. F., O'Malley B. W., Chambon P., Auwerx J. (2002) Cell 111, 931–941 [DOI] [PubMed] [Google Scholar]
- 67.Wang Z., Qi C., Krones A., Woodring P., Zhu X., Reddy J. K., Evans R. M., Rosenfeld M. G., Hunter T. (2006) Cell Metab. 3, 111–122 [DOI] [PubMed] [Google Scholar]
- 68.Louet J. F., Coste A., Amazit L., Tannour-Louet M., Wu R. C., Tsai S. Y., Tsai M. J., Auwerx J., O'Malley B. W. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17868–17873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Louet J. F., O'Malley B. W. (2007) Cell Cycle 6, 2448–2452 [DOI] [PubMed] [Google Scholar]
- 70.Coste A., Louet J. F., Lagouge M., Lerin C., Antal M. C., Meziane H., Schoonjans K., Puigserver P., O'Malley B. W., Auwerx J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17187–17192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Louet J. F., Chopra A. R., Sagen J. V., York B., Stevens R. D., Wenner B. R., Ilkayeva O. R., Bain J. R., DeMayo F., Xu J., Newgard C. B., O'Malley B. W. (2011) Cell Metab., in press [Google Scholar]
- 72.Chopra A. R., Louet J. F., Saha P., An J., Demayo F., Xu J., York B., Karpen S., Finegold M., Moore D., Chan L., Newgard C. B., O'Malley B. W. (2008) Science 322, 1395–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.York B., Yu C., Sagen J. V., Liu Z., Nikolai B. C., Wu R. C., Finegold M., Xu J., O'Malley B. W. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 11122–11127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lonard D. M., Kumar R., O'Malley B. W. (2010) Mol. Endocrinol. 24, 279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dussault I., Fawcett D., Matthyssen A., Bader J. A., Giguère V. (1998) Mech. Dev. 70, 147–153 [DOI] [PubMed] [Google Scholar]
- 76.Osborne C. K., Bardou V., Hopp T. A., Chamness G. C., Hilsenbeck S. G., Fuqua S. A., Wong J., Allred D. C., Clark G. M., Schiff R. (2003) J. Natl. Cancer Inst. 95, 353–361 [DOI] [PubMed] [Google Scholar]
- 77.Wang H., Zhang D., Wu W., Zhang J., Guo D., Wang Q., Jing T., Xu C., Bian X., Yang K. (2011) J. Histochem. Cytochem., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Redmond A. M., Bane F. T., Stafford A. T., McIlroy M., Dillon M. F., Crotty T. B., Hill A. D., Young L. S. (2009) Clin. Cancer Res. 15, 2098–2106 [DOI] [PubMed] [Google Scholar]
- 79.Cavarretta I. T., Mukopadhyay R., Lonard D. M., Cowsert L. M., Bennett C. F., O'Malley B. W., Smith C. L. (2002) Mol. Endocrinol. 16, 253–270 [DOI] [PubMed] [Google Scholar]
- 80.Tai H., Kubota N., Kato S. (2000) Biochem. Biophys. Res. Commun. 267, 311–316 [DOI] [PubMed] [Google Scholar]
- 81.Planas-Silva M. D., Shang Y., Donaher J. L., Brown M., Weinberg R. A. (2001) Cancer Res. 61, 3858–3862 [PubMed] [Google Scholar]
- 82.Jensen E., Jacobson H. I. (1962) Recent Prog. Horm. Res. 18, 387–414 [Google Scholar]
- 83.O'Malley B. W., McGuire W. L. (1968) Proc. Natl. Acad. Sci. U.S.A. 60, 1527–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Means A. R., Comstock J. P., Rosenfeld G. C., O'Malley B. W. (1972) Proc. Natl. Acad. Sci. U.S.A. 69, 1146–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jackson D. A., Symons R. H., Berg P. (1972) Proc. Natl. Acad. Sci. U.S.A. 69, 2904–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Puca G. A., Sica V., Nola E. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 979–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith R. G., Iramain C. A., Buttram V. C., Jr., O'Malley B. W. (1975) Nature 253, 271–272 [DOI] [PubMed] [Google Scholar]
- 88.Wrange O., Okret S., Radojćić M., Carlstedt-Duke J., Gustafsson J. A. (1984) J. Biol. Chem. 259, 4534–4541 [PubMed] [Google Scholar]
- 89.Woo S. L., Smith R. G., Means A. R., O'Malley B. W. (1976) J. Biol. Chem. 251, 3868–3874 [PubMed] [Google Scholar]
- 90.Payvar F., Firestone G. L., Ross S. R., Chandler V. L., Wrange O., Carlstedt-Duke J., Gustafsson J. A., Yamamoto K. R. (1982) J. Cell. Biochem. 19, 241–247 [DOI] [PubMed] [Google Scholar]
- 91.Hollenberg S. M., Weinberger C., Ong E. S., Cerelli G., Oro A., Lebo R., Thompson E. B., Rosenfeld M. G., Evans R. M. (1985) Nature 318, 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Luisi B. F., Xu W. X., Otwinowski Z., Freedman L. P., Yamamoto K. R., Sigler P. B. (1991) Nature 352, 497–505 [DOI] [PubMed] [Google Scholar]
- 93.McGrath M. E., Wagner R. L., Apriletti J. W., West B. L., Ramalingam V., Baxter J. D., Fletterick R. J. (1994) J. Mol. Biol. 237, 236–239 [DOI] [PubMed] [Google Scholar]
- 94.Bourguet W., Ruff M., Chambon P., Gronemeyer H., Moras D. (1995) Nature 375, 377–382 [DOI] [PubMed] [Google Scholar]
- 95.Lubahn D. B., Moyer J. S., Golding T. S., Couse J. F., Korach K. S., Smithies O. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 11162–11166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peters J. M., Hennuyer N., Staels B., Fruchart J. C., Fievet C., Gonzalez F. J., Auwerx J. (1997) J. Biol. Chem. 272, 27307–27312 [DOI] [PubMed] [Google Scholar]
- 97.Xu Y., Chen Q., Li W., Su X., Chen T., Liu Y., Zhao Y., Yu C. (2010) Oncogene 29, 3386–3397 [DOI] [PubMed] [Google Scholar]
- 98.Mödder U. I., Sanyal A., Kearns A. E., Sibonga J. D., Nishihara E., Xu J., O'Malley B. W., Ritman E. L., Riggs B. L., Spelsberg T. C., Khosla S. (2004) Endocrinology 145, 913–921 [DOI] [PubMed] [Google Scholar]
- 99.Mödder U. I., Monroe D. G., Fraser D. G., Spelsberg T. C., Rosen C. J., Géhin M., Chambon P., O'Malley B. W., Khosla S. (2009) J. Biol. Chem. 284, 18767–18777 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.