Abstract
In Saccharomyces cerevisiae, the mitochondrial inner membrane readily allows transport of cytosolic NAD+, but not NADPH, to the matrix. Pos5p is the only known NADH kinase in the mitochondrial matrix. The enzyme phosphorylates NADH to NADPH and is the major source of NADPH in the matrix. The importance of mitochondrial NADPH for cellular physiology is underscored by the phenotypes of the Δpos5 mutant, characterized by oxidative stress sensitivity and iron-sulfur (Fe-S) cluster deficiency. Fe-S clusters are essential cofactors of proteins such as aconitase [4Fe-4S] and ferredoxin [2Fe-2S] in mitochondria. Intact mitochondria isolated from wild-type yeast can synthesize these clusters and insert them into the corresponding apoproteins. Here, we show that this process of Fe-S cluster biogenesis in wild-type mitochondria is greatly stimulated and kinetically favored by the addition of NAD+ or NADH in a dose-dependent manner, probably via transport into mitochondria and subsequent conversion into NADPH. Unlike wild-type mitochondria, Δpos5 mitochondria cannot efficiently synthesize Fe-S clusters on endogenous aconitase or imported ferredoxin, although cluster biogenesis in isolated Δpos5 mitochondria is restored to a significant extent by a small amount of imported Pos5p. Interestingly, Fe-S cluster biogenesis in wild-type mitochondria is further enhanced by overexpression of Pos5p. The effects of Pos5p on Fe-S cluster generation in mitochondria indicate that one or more steps in the biosynthetic process require NADPH. The role of mitochondrial NADPH in Fe-S cluster biogenesis appears to be distinct from its function in anti-oxidant defense.
Keywords: Iron Metabolism, Iron-Sulfur Protein, Metals, Mitochondria, Mitochondrial Metabolism, Yeast
Introduction
The pyridine nucleotides play vital roles in numerous biological processes, including energy metabolism, mitochondrial functions, anti-oxidant defense, aging, and cell death (1). In eukaryotic cells, nicotinamide adenine dinucleotide (NAD) is primarily present in the oxidized state (NAD+), but its phosphorylated counterpart (NADP) is mostly found in the reduced state (NADPH). NAD+ is well known for its role as an electron carrier in oxidoreductase reactions, particularly as the mediator of hydrogen transfer from substrates to the respiratory chain of mitochondria. On the other hand, NADPH is required for oxidative stress defense, for reductive syntheses, and for maintaining the cellular redox state (2, 3). NAD+- and NADPH-dependent processes occur both inside and outside of the mitochondria. In Saccharomyces cerevisiae, NAD+ is synthesized outside, and not inside, of mitochondria. Cytosolic NAD(H) is then transported across the mitochondrial inner membrane into the matrix via carrier proteins in this membrane (4). By contrast, NADP(H) is generated separately in mitochondria and the cytosol by distinct NAD(H) kinases in these compartments (5–7), and no transporter for NADP(H) has been found in the mitochondrial inner membrane.
The NAD(H) kinases are the sole enzymes able to convert NAD(H) to NADP(H) using ATP as a phosphate donor (8). In yeast, Pos5p is the only known NADH kinase in the mitochondrial matrix, and therefore it is the major source of mitochondrial NADPH (5). The Δpos5 mutant is highly sensitive to H2O2, high O2 conditions, and paraquat, a superoxide-generating agent (5, 9–12). These phenotypes can be understood in terms of the mitochondrial source of reactive oxygen species leaking from the respiratory chain and the strong reliance of mitochondrial anti-oxidant defenses on enzymes that utilize NADPH. In particular, glutathione and thioredoxin are maintained in their reduced states by dedicated mitochondrial reductases that use NADPH. These reduced molecules in turn serve as cofactors for peroxidases, glutaredoxins, and peroxiredoxins involved in oxidant stress protection (1, 5, 13–16). A biosynthetic role of NADPH in mitochondria was uncovered following observation of the arginine auxotrophy of the Δpos5 mutant, and the defect was explained by the lack of NADPH required for an NADPH-dependent step in arginine biosynthesis catalyzed by the mitochondrial N-acetylglutamyl phosphate reductase (5). Interestingly, the presence or overexpression of cytosolic NAD(H) kinases (Utr1p or Yef1p) does not rescue the arginine auxotrophy of the Δpos5 mutant, indicating that NADPH generated in the cytoplasm is unable to cross into the mitochondrial matrix (17).
Another phenotype of the Δpos5 mutant is a marked dysregulation of iron homeostasis, with constitutive activation of the cellular iron uptake system and accumulation of iron in mitochondria (5). This phenotype has been associated with deficiency of iron-sulfur (Fe-S) clusters (18–26), and indeed, the Δpos5 mutant exhibits deficient activities of aconitase and succinate dehydrogenase, two Fe-S cluster proteins of mitochondria (5). Fe-S clusters are essential cofactors of proteins that participate in numerous important cellular processes, including catalysis, respiration, redox reactions, DNA repair, protein translation, and O2-sensing (27). Mitochondria contain a complete machinery for Fe-S cluster synthesis, and subsequent transfer of these cluster intermediates to apoproteins forming active holoproteins. A conundrum arises because Fe-S clusters may be sensitive to oxidants, and thus the deficient mitochondrial Fe-S cluster proteins in the Δpos5 mutant could result from excessive oxidative stress in the mutant leading to destruction of Fe-S clusters. Alternatively, the deficiency could result from failure to efficiently synthesize the Fe-S clusters. In the latter case, a direct role for NADPH in Fe-S cluster synthesis in mitochondria would be implied, perhaps implicating a specific NADPH-requiring reductase in the process.
To distinguish these possibilities, we used kinetic assays able to track formation of newly made Fe-S clusters in isolated and intact mitochondria. The assays were performed with wild-type or Δpos5 mutant mitochondria, and cells as well as isolated mitochondria were manipulated in terms of oxygen exposure to decrease the level of oxidant stress. NAD(H) availability was also manipulated, and iron was added or withheld during the assays. Aconitase is an abundant [4Fe-4S] cluster-containing protein of the tricarboxylic acid cycle. Upon incubation of wild-type mitochondria with [35S]cysteine, aconitase was rapidly radiolabeled indicating new Fe-S cluster synthesis. The rate and efficiency of the process in wild-type mitochondria were dependent on added NAD(H). By contrast, cluster biogenesis of aconitase was markedly impaired and retarded in the Δpos5 mutant, and low oxygen and/or NAD(H) could not rescue the defect. Similar defects were also observed for [2Fe-2S] cluster biogenesis of ferredoxin. Apoferredoxin precursor protein was imported and yet was very poorly loaded with its Fe-S cluster in Δpos5 mitochondria. These results indicate that Pos5p and by extension NADPH plays a direct role in Fe-S cluster synthesis in mitochondria.
EXPERIMENTAL PROCEDURES
Yeast Strains, Growth, and Mitochondria Isolation
Two S. cerevisiae strains, D273-10B (ATCC 24657) and BY4741 (Invitrogen), were used for the experiments described here. Mitochondria isolated from these wild-type (WT) strains can synthesize new Fe-S clusters and insert them into apoproteins with comparable efficiency, thereby ruling out the possibility of strain-specific results (28). Wild-type D273-10B cells were grown in rich media (0.3% yeast extract, 0.1% glucose, 2% lactate, 7.4 mm KH2PO4, 18.7 mm NH4Cl, 8.6 mm NaCl, 4 mm CaCl2, 4 mm MgSO4, and 19 μm FeCl3) at 30 °C under normal aerobic conditions with shaking at 200 rpm for 14–16 h to an A600 of 1.2–1.5, and mitochondria were isolated as described previously (29). D273-10B mitochondria were used for experiments in Figs. 1–3; these experiments required the use of only wild-type mitochondria.
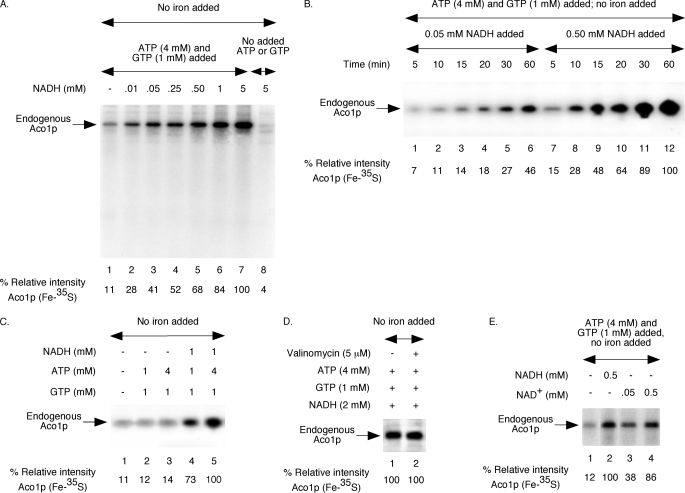
FIGURE 1.
NAD(H)-dependent Fe-35S labeling of endogenous aconitase in wild-type mitochondria. Each reaction mixture contained wild-type mitochondria (200 μg of proteins) and [35S]cysteine (10 μCi), and all assays were performed without added iron. A, assay mixtures were supplemented with ATP (4 mm) and GTP (1 mm) and/or different concentrations of NADH as indicated. Samples were incubated at 30 °C for 30 min and diluted with buffer A, and mitochondria were reisolated by centrifugation. Mitochondrial membranes were ruptured, and soluble proteins were analyzed by native PAGE followed by autoradiography (28, 29). The intensity of Aco1p (Fe-35S) in the presence of ATP (4 mm), GTP (1 mm), and NADH (5 mm) was arbitrarily considered 100% (lane 7). B, reaction mixtures were supplemented with ATP (4 mm), GTP (1 mm), and NADH (0.05 or 0.5 mm) as indicated. Samples were incubated at 30 °C for different time periods and analyzed as in A. C, NADH, ATP, and GTP were included as indicated. Samples were incubated at 30 °C for 30 min and analyzed as in A. D, mitochondria were pretreated with valinomycin for 5 min on ice as indicated. Following addition of ATP, GTP, and NADH, samples were incubated at 30 °C for 30 min and analyzed. E, reaction mixtures were supplemented with ATP (4 mm) and GTP (1 mm). NADH or NAD+ was included as indicated. Following incubation at 30 °C for 30 min, samples were analyzed.
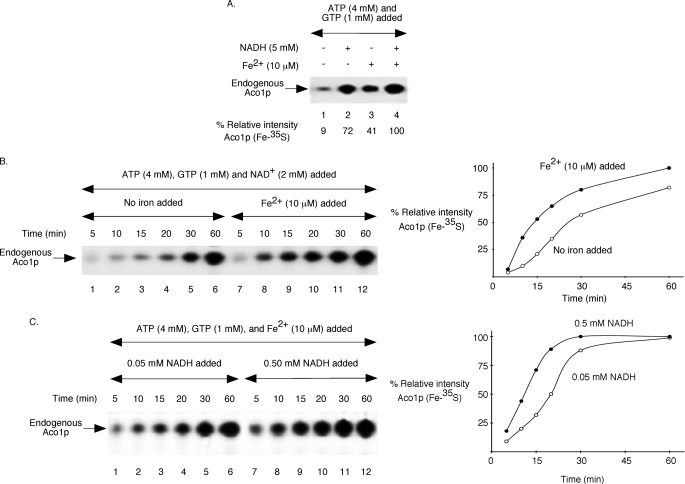
FIGURE 2.
Effects of NAD(H) addition on Fe-35S labeling of endogenous aconitase in wild-type mitochondria in the absence or presence of supplementary iron. Each reaction mixture contained wild-type mitochondria (200 μg of proteins), ATP (4 mm), GTP (1 mm), and [35S]cysteine (10 μCi). A, reaction mixtures were supplemented with NADH (5 mm) and/or ferrous ascorbate (10 μm) as indicated. Following incubation at 30 °C for 30 min, samples were analyzed by native PAGE followed by autoradiography. B, NAD+ (2 mm) was included, and assays were performed with or without added ferrous ascorbate (10 μm). Samples were incubated at 30 °C for different time periods and analyzed. The right panel shows quantitation of data presented on the left panel. The intensity of Aco1p (Fe-35S) in the presence of added iron at the 60-min time point (lane 12) was considered 100%. C, ferrous ascorbate (10 μm) and NADH (0.05 or 0.5 mm) were included as indicated. Samples were analyzed after incubation at 30 °C for different time periods. The right panel shows quantitation of the autoradiogram (left panel). The intensity of Aco1p (Fe-35S) in the presence of 0.5 mm NADH at the 60-min time point (lane 12) was considered 100%.
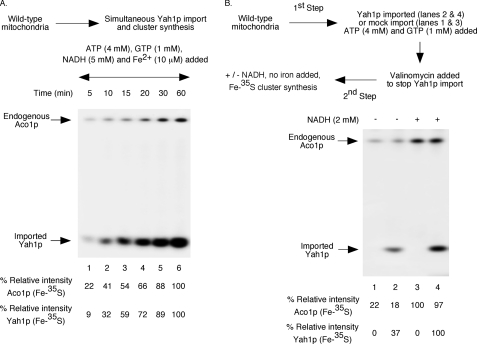
FIGURE 3.
Addition of NADH stimulates Fe-35S labeling of imported ferredoxin in wild-type mitochondria. A, precursor form of ferredoxin was expressed in bacteria, and the protein was solubilized from inclusion bodies with 8 m urea. Wild-type mitochondria (200 μg of proteins) were supplemented with ATP, GTP, NADH, and ferrous ascorbate as indicated. Following addition of [35S]cysteine (10 μCi) and unlabeled ferredoxin (Yah1p) precursor protein (200 ng), reaction mixtures were incubated at 30 °C for different time periods and diluted with buffer A, and mitochondria were reisolated by centrifugation. Samples were analyzed by native PAGE followed by autoradiography. B, wild-type mitochondria were supplemented with ATP (4 mm) and GTP (1 mm) and incubated at 30 °C for 15 min with or without added unlabeled Yah1p precursor protein (200 ng). Valinomycin (5 μm) was added to stop any further import, and reaction mixtures were supplemented with [35S]cysteine (10 μCi). Samples were incubated at 30 °C for 30 min with or without added NADH (2 mm) and analyzed as in A.
BY4741 was used as the congenic wild-type strain for experiments in Figs. 4–8; these experiments were carried out to compare Fe-S cluster biogenesis in mitochondria of various modified strains. The Δpos5::KanMX mutant in the BY4741 background was generated by tetrad dissection of the corresponding heterozygous diploid purchased from Open Biosystems and was reconfirmed by PCR analysis. Another strain was also purchased from Open Biosystems in which a tandem affinity purification (TAP)3 tag was inserted at the POS5 locus so as to generate a Pos5p variant with a C-terminal TAP tag in the genome, and this strain is called WT (Pos5p-TAP). Two plasmids for Pos5p expression were constructed, using URA3-marked pRS416 as a starting point. In these constructs, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH or TDH3) promoter is followed by the POS5 open reading frame with or without a C-terminal TAP tag. As controls, cells were transformed with the empty plasmid pRS416. These BY4741 transformants were grown in defined synthetic media lacking uracil and containing 2% raffinose and 0.5% glucose at 30 °C under normal aerobic conditions, and mitochondria were isolated as described above. For growth of these strains under reduced oxygen conditions, argon gas was bubbled through cultures (750 ml) in bottles (1 liter) for at least 5 min; the caps were tightly closed, and cells were grown at 30 °C for 40–48 h without shaking to A600 of 1.4–1.6. Mitochondria were isolated from these cells with no further manipulation of oxygen availability.
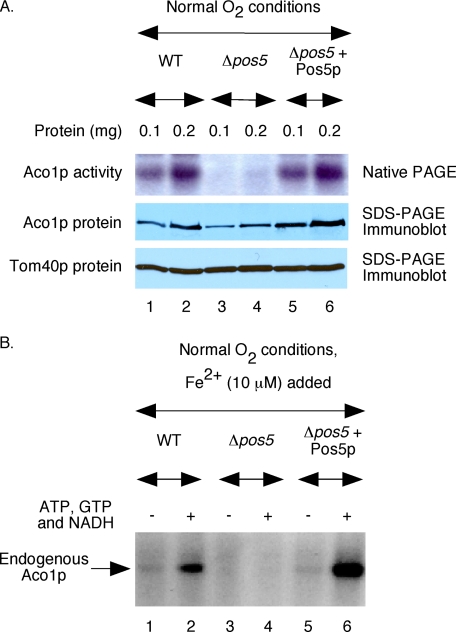
FIGURE 4.
In vivo expression of Pos5p restores Fe-S cluster biogenesis in Δpos5 mitochondria. A, WT, Δpos5, and Δpos5 with reintroduced Pos5p (Δpos5 + Pos5p) cells were grown under aerobic conditions, and mitochondria were isolated. Aconitase activity in these mitochondria (top panel) was determined by a native in-gel assay as described previously (28). Identical samples were also analyzed by SDS-PAGE followed by immunoblotting using anti-Aco1p (middle panel) and anti-Tom40p (bottom panel) antibodies. B, mitochondria were supplemented with ferrous ascorbate (10 μm) and [35S]cysteine (10 μCi). Reaction mixtures were incubated at 30 °C for 30 min, with or without added nucleotides (4 mm ATP, 1 mm GTP, and 5 mm NADH). Samples were analyzed by native PAGE followed by autoradiography.
FIGURE 5.
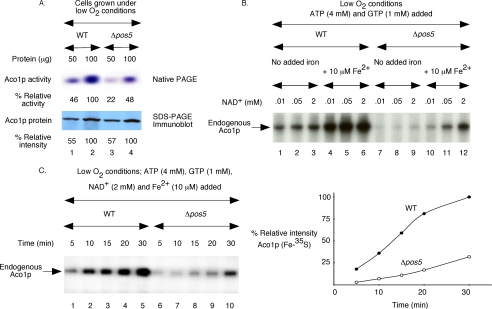
Deficient Fe-S cluster synthesis in Δpos5 mitochondria persists even under reduced oxidative stress conditions. Mitochondria were isolated from WT and Δpos5 cells grown under low oxygen conditions. A, aconitase activity (top panel) and protein (bottom panel) levels were determined as in Fig. 4A. B, mitochondria were supplemented with ATP (4 mm), GTP (1 mm), and [35S]cysteine (10 μCi). Following addition of NAD+ and/or ferrous ascorbate as indicated, reaction mixtures were incubated at 30 °C for 60 min under low O2 conditions. Samples were analyzed by native PAGE followed by autoradiography. C, mitochondria were supplemented with ATP, GTP, NAD+, and ferrous ascorbate as indicated. Following addition of [35S]cysteine (10 μCi), samples were incubated at 30 °C for different time periods under reduced O2 conditions and analyzed. The right panel shows quantitation of data presented on the left panel. The intensity of Aco1p (Fe-35S) in WT mitochondria at the 30-min time point (lane 5) was considered 100%.
FIGURE 6.
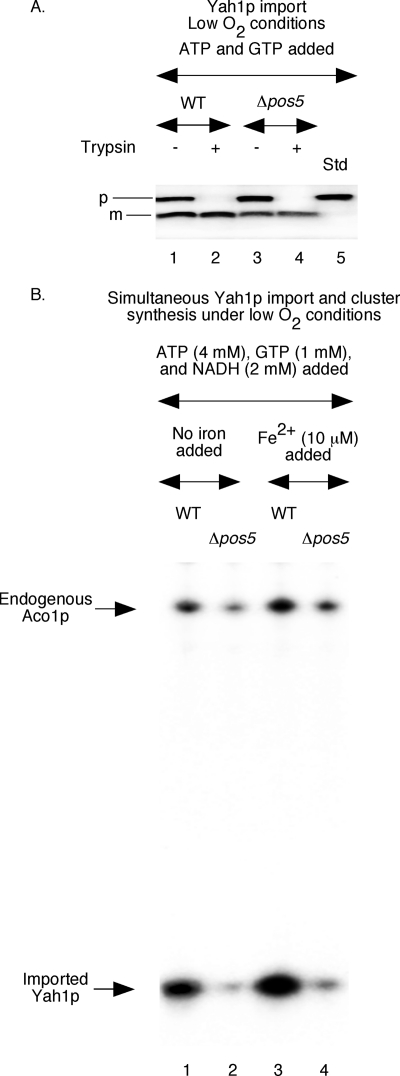
Impaired [2Fe-2S] cluster biogenesis in Δpos5 mitochondria. Mitochondria were isolated from WT and Δpos5 cells grown under reduced O2 conditions. A, precursor form of ferredoxin with a C-terminal His6 tag was expressed in bacteria in the presence of [35S]methionine, solubilized with 8 m urea, and purified by Ni-NTA chromatography. The radiolabeled protein (100 ng) was added to mitochondria supplemented with ATP (4 mm) and GTP (1 mm), and incubated at 30 °C for 15 min. Following import, samples were treated with trypsin (0.2 mg/ml) as indicated and analyzed by SDS-PAGE followed by autoradiography. The precursor and mature forms of Yah1p are indicated by p and m, respectively. Std indicates 50% of the precursor protein used for import experiments. B, mitochondria were supplemented with ATP, GTP, and NADH. Ferrous ascorbate was included as indicated. Following addition of [35S]cysteine (10 μCi) and unlabeled ferredoxin (Yah1p) precursor protein (200 ng), reaction mixtures were incubated at 30 °C for 30 min and diluted with buffer A, and mitochondria were reisolated by centrifugation. Samples were analyzed by native PAGE followed by autoradiography.
FIGURE 7.
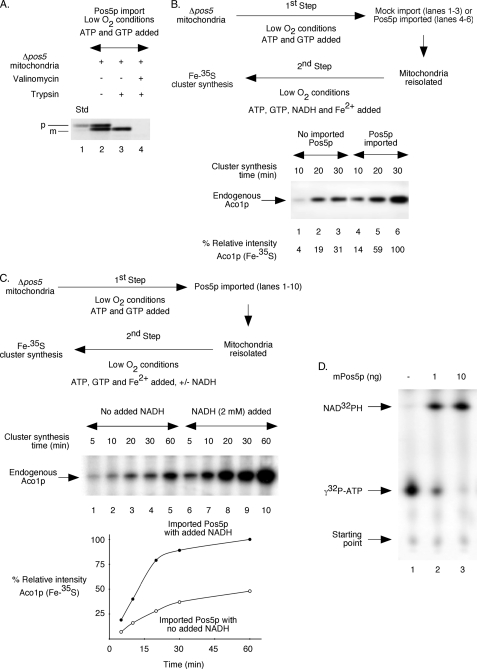
Pos5p imported into isolated Δpos5 mitochondria stimulates Fe-S cluster biogenesis. For experiments in A–C, mitochondria were isolated from Δpos5 cells grown under reduced O2 conditions. Likewise, import and Fe-S cluster insertion assays were also performed under low O2 conditions. A, [35S]methionine-labeled Pos5p precursor protein was synthesized in reticulocyte lysate. A post-ribosomal supernatant (1 μl) containing the radiolabeled protein was incubated with Δpos5 mitochondria in the presence of ATP (4 mm) and GTP (1 mm) at 30 °C for 30 min. Valinomycin (5 μm) was included as indicated. Following import, samples were treated with trypsin (0.2 mg/ml) where indicated and analyzed by SDS-PAGE followed by autoradiography. The precursor and mature forms of Pos5p are indicated by p and m, respectively. Std indicates 10% of the precursor protein used for import experiments. B, post-ribosomal reticulocyte lysate (10 μl) with or without newly synthesized Pos5p precursor protein was added to Δpos5 mitochondria and incubated in the presence of ATP (4 mm) and GTP (1 mm) at 30 °C for 30 min. Samples were diluted with buffer A, and mitochondria with or without imported Pos5p were isolated by centrifugation. Mitochondria were resuspended in HSB buffer and supplemented with ATP (4 mm), GTP (1 mm), NADH (2 mm), ferrous ascorbate (10 μm), and [35S]cysteine (10 μCi). After incubation at 30 °C for different time periods, samples were analyzed by native PAGE followed by autoradiography. Note that [35S]methionine-labeled and imported Pos5p was not detected by native gels under these conditions. C, Pos5p was imported into Δpos5 mitochondria as in B. Mitochondria with imported Pos5p were isolated and supplemented with ATP (4 mm), GTP (1 mm), ferrous ascorbate (10 μm), and [35S]cysteine (10 μCi). NADH (2 mm) was included as indicated. Samples were incubated at 30 °C for different time periods and analyzed. The intensity of Aco1p (Fe-35S) in the presence of NADH at the 60-min time point (lane 10) was considered 100%. D, bacterially expressed and purified Pos5p was incubated with [γ-32P]ATP (0.5 μCi) plus unlabeled ATP (0.5 μm) in the presence of NADH (2 mm) at 30 °C for 5 min. Samples were analyzed by TLC followed by autoradiography.
FIGURE 8.
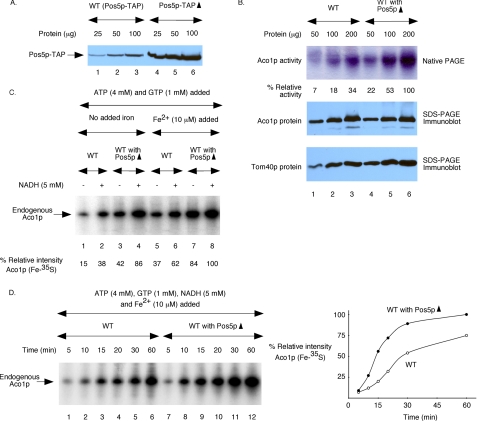
Pos5p overexpression enhances Fe-S cluster biogenesis in wild-type mitochondria. A, wild-type strain, called WT (Pos5p-TAP), expressed Pos5p-TAP from the POS5 promoter in the genome. This strain was transformed with a plasmid for overexpression of Pos5p-TAP from the GAPDH promoter (Pos5p-TAP▴). Mitochondria isolated from these strains were evaluated for Pos5p-TAP protein levels by immunoblotting using affinity-purified rabbit IgG as the first antibody. B, aconitase activity (top panel) was determined in WT and wild-type with overexpressed and untagged Pos5p (WT with Pos5p▴) mitochondria by a native in-gel assay (28). Identical samples were also analyzed by SDS-PAGE followed by immunoblotting using anti-Aco1p (middle panel) and anti-Tom40p (bottom panel) antibodies. C, mitochondria were supplemented with ATP (4 mm) and GTP (1 mm). As indicated, reaction mixtures also contained NADH (5 mm) and/or ferrous ascorbate (10 μm). Following addition of [35S]cysteine, samples were incubated at 30 °C for 60 min and analyzed by native PAGE and autoradiography. The intensity of Aco1p (Fe-35S) in WT with Pos5p▴ mitochondria with both NADH and iron added (lane 8) was considered 100%. D, mitochondria were supplemented with ATP, GTP, NADH, ferrous ascorbate, and [35S]cysteine as in C. Samples were analyzed after incubation at 30 °C for different time periods. The right panel shows quantitation of the autoradiogram presented on the left panel. The intensity of Aco1p (Fe-35S) in WT with Pos5p▴ mitochondria at the 60-min time point (lane 12) was considered 100%.
Insertion of Newly Formed Fe-35S Clusters into Apoproteins
To deplete endogenous nucleotides, mitochondria were preincubated at 30 °C for 10 min. Depletion of nucleotides in this manner does not cause any noticeable changes in mitochondrial integrity and function (29). Isolated mitochondria contain a stored pool of iron that can be used for Fe-S cluster synthesis (28), and experiments were performed with or without addition of chemically reduced iron as ferrous ascorbate. Insertion of newly formed Fe-35S clusters into a pool of endogenous apoaconitase (apoAco1p) in isolated and intact mitochondria was examined as described before (28, 29) with some modifications. A typical assay mixture (100 μl) contained mitochondria (200 μg of proteins) in HSB buffer (20 mm Hepes/KOH, pH 7.5, 0.6 m sorbitol, 0.1 mg/ml bovine serum albumin, 10 mm Mg(OAc)2, and 40 mm KOAc) containing 1–4 mm ATP, 1 mm GTP, and 1 mm DTT. NADH or NAD+ was included at various concentrations. As needed, the reaction mixtures were also supplemented with chemically reduced iron as ferrous ascorbate (10 μm) from a stock mixture of ferrous ammonium sulfate (0.5 mm) and sodium ascorbate (5 mm). Following addition of [35S]cysteine (10 μCi, 1075 Ci/mmol), samples were incubated at 30 °C under normal or low oxygen conditions for different time periods (5–60 min). To achieve reduced oxygen conditions, air from the top of the reaction mixtures in Eppendorf tubes was forced out by passing argon gas, and caps were tightly closed and further sealed with parafilm. After incubation, reaction mixtures were diluted 10-fold with ice-cold buffer A (20 mm Hepes/KOH, pH 7.5, 0.6 m sorbitol, 0.1 mg/ml bovine serum albumin). Mitochondria were reisolated by centrifugation and processed for 12% native PAGE followed by autoradiography as described previously (28, 29). Radiolabeled protein bands were quantitated using the software NIH Image. Experiments were repeated at least three times, and the variation between experiments was in the range of 5–10%.
The precursor form of ferredoxin with a C-terminal His6 tag (pYah1p-His6) was expressed in bacteria, and the protein was found in inclusion bodies. The protein was solubilized with 8 m urea in 50 mm Tris/HCl, pH 8.0, and centrifuged at 250,000 × g for 20 min at 20 °C to remove insoluble material. The supernatant fraction contained pYah1p-His6 with ∼95% purity and was stored at −80 °C until further use for import and Fe-S cluster assembly experiments (28–30). The requirement of nucleoside triphosphates (NTPs) for these processes is very similar (28, 29, 31), and this allowed us to examine insertion of radiolabeled Fe-S clusters into imported apoferredoxin (apoYah1p) in two different ways. One assay involved simultaneous import and cluster assembly of apoferredoxin. In this case, urea-denatured apoYah1p precursor protein (200 ng) was added to mitochondria (200 μg of proteins) in HSB buffer containing ATP (4 mm), GTP (1 mm), NADH (2–5 mm), DTT (1 mm), and [35S]cysteine (10 μCi). The total volume was 100 μl, and the final urea concentration was 160 mm. Samples were incubated at 30 °C for different time periods and then diluted with buffer A. Mitochondria were reisolated and analyzed by native PAGE followed by autoradiography. These assays were performed with or without added ferrous ascorbate (10 μm) and under normal or low O2 conditions as described above.
The one-step assay cannot distinguish the effects of NADH addition on import versus cluster biogenesis. To determine the effects of NADH specifically on [2Fe-2S] cluster assembly, assays were therefore performed in two steps. In the first step, apoferredoxin was imported into mitochondria. Import reactions were stopped, and then insertion of radiolabeled clusters into already imported apoferredoxin was tested. Briefly, urea-denatured apoferredoxin precursor protein (200 ng) was added to mitochondria (200 μg of proteins) in HSB buffer containing 4 mm ATP, 1 mm GTP, and 1 mm DTT and incubated at 30 °C for 15 min. Valinomycin (5 μm) was added to dissipate the membrane potential across the mitochondrial inner membrane, thereby blocking further import (31). Reaction mixtures were then supplemented with 10 μCi of [35S]cysteine and again incubated at 30 °C for 30 min with or without added NADH (2 mm). Samples were analyzed by native PAGE followed by autoradiography.
As described above, unlabeled ferredoxin precursor protein was used for import and subsequent Fe-35S cluster labeling. To directly demonstrate ferredoxin import, pYah1p-His6 was expressed in bacteria in the presence of [35S]methionine and purified by Ni-NTA chromatography as described for other proteins (31–33). Import reactions were performed under normal or low O2 conditions as indicated in the figures. Radiolabeled and urea-denatured ferredoxin precursor protein (100 ng) was added to mitochondria (200 μg of proteins) in HSB buffer (100 μl) containing 4 mm ATP, 1 mm GTP, and 1 mm DTT. Following incubation at 30 °C for 15 min, reaction mixtures were placed on ice and treated with trypsin (0.2 mg/ml) for 30 min. Samples were diluted with ice-cold buffer A containing a mixture of protease inhibitors. Mitochondria were reisolated and washed with 10% trichloroacetic acid. Samples were analyzed by SDS-PAGE followed by autoradiography (31, 32, 34).
Pos5p Import into Δpos5 Mitochondria and Fe-S Cluster Biogenesis
The POS5 open reading frame was amplified by PCR from the yeast genomic DNA. The product was digested with NdeI and XhoI, cloned into the same sites of pSP64T (32, 34), and sequenced. The resulting plasmid (pSP64T/pPos5p) was linearized with BamHI for transcription using the Ribomax-SP6 kit, and [35S]methionine-labeled protein was generated in reticulocyte lysate according to the manufacturer's protocol (Promega). A post-ribosomal supernatant containing the radiolabeled Pos5p precursor protein was incubated with Δpos5 mitochondria. Samples were treated with trypsin as necessary and were analyzed by SDS-PAGE followed by autoradiography as described above for import of [35S]methionine-labeled ferredoxin. To determine the effects of imported Pos5p on Fe-S cluster biogenesis in Δpos5 mitochondria, experiments were performed in two steps, separating import from cluster insertion. These assays are outlined in the corresponding figures.
Bacterial Expression and NADH Kinase Activity of Pos5p
The Pos5p precursor protein (414 amino acids) contains a mitochondrial targeting signal of 17 amino acids at the N terminus of the protein (11). Upon import, the targeting signal is removed generating the mature form of Pos5p (mPos5p). The DNA encoding mPos5p was amplified by PCR using the plasmid pSP64T/pPos5p as the template, digested with NdeI and XhoI, cloned into the same sites of pET21b, and sequenced. This introduces a His6 tag in-frame at the C terminus of the protein. BL21DE3 cells carrying the resulting plasmid pET21b/mPos5p were cultured at 37 °C in LB media supplemented with 0.1 mg/ml ampicillin to A600 of 0.6. The culture was switched to 16 °C, and sorbitol was added to 0.5 m, prior to induction of the protein by the addition of isopropyl 1-thio-β-d-galactopyranoside to 0.2 mm. Cells were harvested 48 h after induction, and mature Pos5p-His6 was found expressed mostly in a soluble form. The protein was purified to homogeneity by Ni-NTA chromatography. To determine the NADH kinase activity, purified mPos5p was added to 50 mm Hepes/KOH, pH 7.5, containing 4 mm MgCl2, 0.1 mg/ml bovine serum albumin, 2 mm NADH, 0.5 μm unlabeled ATP, and 0.5 μCi of [γ-32P]ATP (4500 Ci/mmol, PerkinElmer Life Sciences) in a total volume of 20 μl. Samples were incubated at 30 °C for 5 min, and reactions were stopped by the addition of EDTA to a final concentration of 25 mm. One-tenth of the reaction mixture was spotted onto a polyethyleneimine cellulose TLC plate (Sigma). The plate was developed using 0.75 m potassium phosphate as a solvent, air-dried, and exposed to a film for autoradiography (35).
RESULTS
Insertion of Newly Formed Fe-35S Clusters into Endogenous Apoaconitase in Isolated Wild-type Mitochondria, Stimulatory Effects of Added NAD(H) with No Supplementary Iron
In our assays of Fe-S cluster biogenesis, isolated and intact mitochondria are incubated with [35S]cysteine. Radioactive sulfur is generated within mitochondria as persulfide on the enzyme cysteine desulfurase and is subsequently utilized for Fe-35S cluster synthesis. The biosynthetic machinery for these clusters can use either a pool of stored iron or iron supplemented in a reduced form. Newly formed Fe-35S clusters are subsequently incorporated into apoproteins within mitochondria. Samples are analyzed by native PAGE followed by autoradiography, thereby allowing direct visualization and quantitation of radiolabeled proteins. The proteins become radiolabeled because they contain Fe-35S clusters; polypeptide backbones of the proteins are unlabeled. The clusters are destroyed by denaturants such as SDS, and therefore, Fe-35S-labeled proteins can only be analyzed by native gels and not by SDS gels. These assays allowed us to demonstrate that mitochondria isolated from wild-type yeast contain a pool of apoaconitase (apoAco1p) and that the mitochondrial Fe-S cluster machinery is capable of inserting newly formed and radiolabeled clusters into this endogenous apoAco1p pool in an NTP (ATP and GTP)-dependent manner (28, 29).
To determine the NAD(H) requirement for Fe-S cluster biogenesis utilizing endogenous iron, assays were performed with wild-type mitochondria with no added iron (Fig. 1). In one set of experiments (Fig. 1A), mitochondria were incubated with [35S]cysteine and increasing concentrations of NADH. In the presence of added NTPs (ATP and GTP), radiolabeling of endogenous aconitase (Aco1p) was greatly enhanced by the addition of NADH in a dose-dependent manner (Fig. 1A, lanes 1–7). In the absence of added NTPs, however, NADH failed to stimulate cluster biogenesis, and no significant radiolabeled Aco1p was detected even at the highest concentration of NADH (5 mm) tested (Fig. 1A, lane 8). These results suggest that neither NTPs (ATP and GTP) nor NADH alone is sufficient for [4Fe-4S] cluster biogenesis of aconitase. Addition of NTPs cannot replace NADH requirement and vice versa, and both are required for efficient cluster biogenesis in mitochondria.
The experiment in Fig. 1A was performed for a fixed time point (30 min). The kinetics of cluster biogenesis was therefore determined at different concentrations of NADH as judged by the formation of radiolabeled aconitase at various time points (Fig. 1B). At a relatively higher concentration of NADH (0.5 mm), cluster biogenesis occurred significantly faster and more efficiently compared with the time course observed at a lower concentration of NADH (0.05 mm), even though all reaction mixtures were otherwise identical, including added ATP and GTP. These results suggest that an adequate level of NADH is critical for efficient and rapid [4Fe-4S] cluster biogenesis of aconitase in mitochondria.
NADH can be used as a substrate for the respiratory chain, generating ATP by oxidative phosphorylation. We therefore considered the possibility that added NADH was acting by stimulating ATP production. However, this seemed very unlikely because the assay mixtures contained as much as 4 mm ATP, and yet addition of NADH at a low concentration (e.g. 0.5 mm) stimulated cluster biogenesis by ∼6-fold (Fig. 1A, compare lanes 1 and 5). Furthermore, Aco1p radiolabeling was ∼5-fold more efficient with 1 mm ATP plus 1 mm NADH compared with 4 mm ATP alone (Fig. 1C, lanes 3 and 4). Theoretically, one molecule of NADH can be used to generate a maximum of three molecules of ATP through the respiratory chain and oxidative phosphorylation under optimum conditions (36, 37). Thus, the stimulatory effects of NADH on Fe-S cluster biogenesis are not due to its effects on mitochondrial ATP synthesis. We also tested if a stronger membrane potential across the mitochondrial inner membrane can explain the NADH stimulatory effects on Fe-S cluster biogenesis. NADH is often included in several assays with isolated mitochondria, such as protein import, with the idea of generating a stronger membrane potential. When the membrane potential is dissipated by valinomycin, protein import into the matrix is completely blocked even in the presence of NADH, ATP, and GTP (34). By contrast, Fe-S cluster biogenesis was not affected by valinomycin under similar conditions (Fig. 1D). Thus, the stimulatory effects of NADH on Fe-S cluster biogenesis are not mediated through generation of a stronger membrane potential. Interestingly, the stimulatory effects of added NADH on Fe-S cluster biogenesis was not restricted to the reduced form; the oxidized form (NAD+) was also equally effective (Fig. 1E). Experiments in the following sections were therefore performed with added NADH or NAD+, and the results were very similar. As discussed below, added NAD(H) was likely imported into the mitochondrial matrix and was used by the NADH kinase Pos5p to generate NADPH, which in turn facilitated cluster synthesis. Direct addition of NADP+ or NADPH was less effective (data not shown) consistent with their poor transport across the mitochondrial inner membrane into the matrix (4).
Comparison of Stimulatory Effects of Added NAD(H) on Fe-35S Cluster Labeling of Endogenous Apoaconitase in Wild-type Mitochondria in the Absence or Presence of Supplementary Iron
All cluster insertion experiments in Fig. 1 were performed with no added iron. These assays most likely utilized an endogenous pool of stored iron for Fe-S cluster synthesis. The stimulatory effects of NADH on cluster biogenesis in these experiments could represent physiological reduction, mobilization, and/or utilization of stored iron, a critical process required for efficient Fe-S cluster synthesis. Interestingly, when wild-type mitochondria were supplemented with ferrous ascorbate, a reduced iron chelate, significant radiolabeling of endogenous aconitase was observed even in the absence of added NADH (Fig. 2A, compare lanes 1 and 3). However, the process was less efficient than that observed in the presence of NADH with no added iron (Fig. 2A, compare lanes 2 and 3). Thus, addition of reduced iron can promote cluster biogenesis to a reasonable extent, but it cannot completely bypass the NADH requirement for an efficient process. In agreement with this conclusion, the most effective radiolabeling of aconitase was observed when both NADH and reduced iron were added (Fig. 2A, lane 4).
We determined the kinetics of cluster biogenesis in wild-type mitochondria in the presence of NAD+, with and without added iron. Compared with the samples with no iron added, cluster biogenesis in iron-added samples occurred significantly faster at earlier time points (Fig. 2B, lanes 1–4 versus lanes 7–10, respectively), and the difference was less pronounced at later time points (lanes 5 and 6 versus lanes 11 and 12, respectively). An implication of these results is that isolated wild-type mitochondria contain sufficient but not optimum levels of iron for cluster synthesis. Added NAD+ (or NADH) may facilitate utilization of this stored pool of iron. It is, however, a relatively slow process compared with the use of newly imported and chemically reduced iron. We then determined aconitase radiolabeling in wild-type mitochondria in the presence of added iron at two different concentrations of NADH (0.05 and 0.5 mm). At the higher concentration of NADH, the process was kinetically faster and saturated earlier (Fig. 2C). These results suggest that an optimum level of NADH must be added for efficient and rapid Fe-S cluster biogenesis of aconitase even when mitochondria are supplemented with chemically reduced iron.
Stimulatory Effects of Added NADH on Fe-35S Cluster Labeling of Imported Apoferredoxin
Like aconitase, ferredoxin (Yah1p) is also localized to the mitochondrial matrix. Both proteins are encoded by the nuclear genome and synthesized on cytoplasmic ribosomes as precursor proteins with N-terminal targeting signals. Upon import into mitochondria, the targeting signal of each precursor protein is removed by matrix processing peptidases, generating the corresponding mature form. Fe-S clusters are then inserted into each mature apoprotein, forming the corresponding holoprotein. Whereas holoaconitase contains a [4Fe-4S] cluster, holoferredoxin contains a [2Fe-2S] cluster. Compared with aconitase, ferredoxin is much less abundant in mitochondria, and unlike in the case of aconitase, Fe-35S labeling of endogenous apoferredoxin was not detected in our assays of cluster biogenesis (e.g. Fig. 1A). However, when the purified apoferredoxin (apoYah1p) precursor protein was imported into isolated mitochondria, radiolabeled cluster was efficiently inserted into ferredoxin in an NTP (ATP and GTP)-dependent manner as in the case of aconitase (28, 29). These observations allowed us to investigate the requirement of NADH addition for [2Fe-2S] cluster biogenesis of ferredoxin in isolated wild-type mitochondria. For this purpose, we first compared the kinetics of [4Fe-4S] and [2Fe-2S] cluster biogenesis using endogenous apoaconitase and newly imported apoferredoxin as substrates. These assays were performed in one step, i.e. ferredoxin import and cluster labeling were coupled. Briefly, wild-type mitochondria were supplemented with ATP, GTP, NADH, reduced iron, and [35S]cysteine. Following addition of bacterially expressed and unlabeled apoYah1p (urea-denatured), samples were incubated at 30 °C for different time periods. As shown in Fig. 3A, the kinetics of radiolabeling of endogenous aconitase and imported ferredoxin were very similar. Thus, the cluster assembly machinery in isolated mitochondria can handle proteins with different clusters in a similar manner. Note that urea-denatured precursor proteins are mostly unfolded and therefore rapidly imported (31). Thus, the lag between import and cluster loading of ferredoxin is minimal.
To determine the effects of NADH addition on ferredoxin cluster biogenesis, assays were performed in two steps, separating import from cluster insertion. This was necessary to eliminate any effects of NADH on ferredoxin import. In the first step, unlabeled and urea-denatured ferredoxin was imported into wild-type mitochondria in the presence of ATP and GTP with no added NADH. Valinomycin was then added to stop any further import. In the second step, reaction mixtures were supplemented with [35S]cysteine and incubated with or without added NADH. Radiolabeling of imported Yah1p was found to be enhanced by the addition of NADH (Fig. 3B, compare lanes 2 and 4). Radiolabeling of endogenous aconitase served as internal control. These assays were performed with no added iron. Thus, as in the case of [4Fe-4S] cluster, the [2Fe-2S] cluster can also be synthesized from endogenous stored iron, and the process is stimulated by NADH addition. Under these experimental conditions, imported ferredoxin had no significant effect on radiolabeling of aconitase (Fig. 3B, compare lanes 1 and 2 and also lanes 3 and 4). As expected for cluster biogenesis in vivo, biogenesis of both [2Fe-2S] and [4Fe-4S] clusters can occur simultaneously in our assay system with isolated mitochondria.
Lack of Aconitase Activity and Fe-S Cluster Biogenesis in Δpos5 Mitochondria under Normal Oxygen Conditions
Pos5p is the only known NADH kinase in the mitochondrial matrix and serves as the local source of NADPH. The Δpos5 mutant is highly sensitive to oxidative stress and lacks aconitase activity (5). We asked if these phenotypic defects are specifically due to the absence of Pos5p. For this purpose, a plasmid carrying POS5 was introduced into the mutant, and the protein was expressed from a strong promoter to ∼15-fold wild-type level (data not shown, but see Fig. 8A). Cells were grown under aerobic conditions, and mitochondria were isolated. Virtually undetectable aconitase activity in the Δpos5 mutant was completely restored to wild-type level or even better (Fig. 4A, top panel). Likewise, decreased Aco1p protein level in Δpos5 also returned to wild-type level (Fig. 4A, middle panel). Tom40p is the major component of the mitochondrial protein import machinery at the outer membrane (33) and served as the loading control (Fig. 4A, bottom panel). These results suggest that the in vivo phenotypes of the Δpos5 mutant related to aconitase activity are directly linked to the absence of Pos5p; the defects are reversible and can be corrected by reintroduction of Pos5p. This conclusion is in agreement with the previous observation by others that Pos5p expression from a plasmid in the Δpos5 mutant complements both hyperoxia sensitivity and growth defects on glycerol (5).
We then compared Fe-S cluster biogenesis in mitochondria isolated from wild-type, Δpos5, and the mutant with reintroduced Pos5p strains grown under aerobic conditions. No radiolabeling of aconitase was observed in Δpos5 mitochondria (Fig. 4B, lane 4) even though Aco1p protein was present at a significant level (see Fig. 4A, middle panel). In any case, the process was restored in mutant mitochondria containing reintroduced Pos5p (Fig. 4B, lane 6). Thus, like in the case for in vivo phenotypes, defects in cluster biogenesis in isolated Δpos5 mitochondria are also corrected by reintroduced Pos5p. Interestingly, aconitase radiolabeling was significantly higher in Δpos5 + Pos5p mitochondria than in wild-type mitochondria (Fig. 4B, compare lanes 2 and 6), and this issue is further addressed later (see Fig. 8).
Aconitase Activity in Δpos5 Is Partially Restored under Reduced Oxygen Conditions
The hyperoxia-sensitive phenotype of the Δpos5 mutant might explain the decreased aconitase activity, particularly when the cells are grown under aerobic conditions. The cubane [4Fe-4S]2+ cluster in the active site of aconitase is essential for its enzymatic activity, but it also makes the enzyme highly susceptible to oxidative stress. Furthermore, the Δpos5 mutant (grown under aerobic conditions) exhibits high cellular iron uptake and mitochondrial iron overload (5). Accumulated iron in mitochondria may exacerbate oxidative stress through Fenton chemistry particularly under aerobic conditions. Under low oxygen growth conditions, the overall oxidative stress should be minimal. With this idea in mind, cells were grown under reduced O2 conditions, and mitochondria were isolated. Aconitase activity was partially rescued in Δpos5 mitochondria (Fig. 5A, top panel) even though aconitase protein level was fully restored (bottom panel). Thus, a reduction in oxidative stress in vivo did not abrogate the requirement of Pos5p for fully active aconitase.
Deficient Fe-35S Cluster Labeling of Endogenous Apoaconitase in Isolated Δpos5 Mitochondria Persists Even under Reduced Oxygen Conditions
Fe-S cluster biogenesis in wild-type versus Δpos5 mitochondria isolated from cells grown under reduced O2 conditions were compared. Assays were also performed in tubes saturated with argon to maintain low O2 conditions during synthesis of Fe-35S clusters. Reaction mixtures contained ATP, GTP, [35S]cysteine, and increasing concentrations of NAD+, with or without added ferrous ascorbate. As expected, radiolabeled Aco1p was detected in wild-type mitochondria even in the absence of added iron (Fig. 5B, lanes 1–3), and this radiolabeling was enhanced by the addition of reduced iron (lanes 4–6). By contrast, radiolabeled Aco1p was almost undetectable in Δpos5 mitochondria in the absence of added iron even with the highest concentration of NAD+ tested (Fig. 5B, lanes 7–9). Several possibilities exist. One is that even under reduced oxidative stress conditions, Δpos5 mitochondria do not have sufficient endogenous iron available for Fe-S cluster synthesis. Another possibility is that for efficient Fe-S cluster biogenesis, added NAD+ or NADH must be converted to NADPH by the NADH kinase activity of Pos5p, a process that does not occur in Δpos5 mitochondria. To distinguish between these two possibilities, assays were performed with added iron. Some radiolabeled Aco1p was detected in Δpos5 mitochondria at higher concentrations of NAD+ and 10 μm ferrous ascorbate (Fig. 5B, lanes 10–12). However, the process was still highly inefficient (Fig. 5B, compare lanes 4–6 with 10–12, respectively). Likewise, the kinetics of cluster biogenesis in Δpos5 mitochondria with added NAD+ and iron was much slower compared with that in wild-type mitochondria (Fig. 5C). These results suggest that low O2 (and hence reduced oxidative stress) conditions cannot bypass the Pos5p NADH kinase activity required for efficient Fe-S cluster biogenesis in isolated mitochondria.
Isolated Δpos5 Mitochondria Can Import Apoferredoxin but Cannot Efficiently Insert Fe-35S Clusters into the Imported Protein Even under Reduced Oxygen Conditions
Unlike the [4Fe-4S] cluster of aconitase, the [2Fe-2S] cluster of ferredoxin is less sensitive to oxidative damage (38). This prompted us to determine whether isolated Δpos5 mitochondria can insert Fe-S clusters into ferredoxin. For this purpose, we first tested if apoferredoxin can be imported into Δpos5 mitochondria. The precursor form of apoferredoxin was expressed in bacteria in the presence of [35S]methionine and purified. In this case, the protein backbone was radiolabeled. Mitochondria were isolated from cells grown under reduced O2 conditions and were incubated with urea-denatured and radiolabeled ferredoxin precursor protein in the presence of ATP and GTP and treated with trypsin as needed. These import reactions were performed under low O2 conditions. Samples were analyzed by SDS-PAGE followed by autoradiography. Like in the case of wild-type mitochondria (Fig. 6A, lanes 1 and 2), ferredoxin was imported into Δpos5 mitochondria (lane 3), and the mature form thus generated remained protected from external trypsin (lanes 4) with comparable efficiency. Thus, isolated Δpos5 mitochondria are functionally active, and the organellar integrity is maintained, at least in terms of protein import.
For testing Fe-S cluster biogenesis of imported ferredoxin, assays were performed in one step (Fig. 6B). Mitochondria (isolated from cells grown under low O2 conditions) were supplemented with ATP, GTP, NADH, and [35S]cysteine. Ferrous ascorbate was included as indicated. Following addition of bacterially expressed and unlabeled apoYah1p (urea-denatured), samples were incubated under reduced O2 conditions and analyzed by native PAGE followed by autoradiography. Radiolabeling of endogenous aconitase served as internal control. Compared with wild-type mitochondria, very little imported Yah1p was radiolabeled in Δpos5 mitochondria, and the pattern was very similar in the absence (Fig. 6B, lanes 1 and 2) or presence (lanes 3 and 4) of added iron. These results suggest that as for the [4Fe-4S] cluster of aconitase, efficient biogenesis of the [2Fe-2S] cluster of ferredoxin also requires the presence of Pos5p, and this requirement cannot be effectively bypassed by reduced oxidative stress conditions even with the addition of NADH and/or reduced iron.
Imported Pos5p Enhances the Kinetics and Efficiency of Fe-35S Cluster Labeling of Endogenous Apoaconitase in Isolated Δpos5 Mitochondria
In isolated wild-type mitochondria, biogenesis of both [4Fe-4S] and [2Fe-2S] clusters was stimulated by NAD(H) addition. On the other hand, biogenesis of these clusters in isolated Δpos5 mitochondria was impaired even with added NAD(H). Together, these results suggest that added NAD(H) must be converted to NADPH by the NADH kinase activity of Pos5p within mitochondria and that NADPH thus generated participates in a critical process required for efficient Fe-S cluster synthesis. Pos5p is the local source of NADPH in mitochondria because cytosolic NADPH cannot efficiently enter into mitochondria. In agreement with this notion, we found that direct addition of NADPH failed to restore cluster biogenesis in intact Δpos5 mitochondria (data not shown).
Pos5p is an enzyme, and a small amount of Pos5p may be sufficient to generate an adequate level of NADPH required for cluster synthesis. We therefore asked if Pos5p imported into isolated Δpos5 mitochondria was able to enhance Fe-S cluster biogenesis. For this purpose, we first tested Pos5p import (Fig. 7A). The precursor form of Pos5p was synthesized in a reticulocyte cell-free translation system in the presence of [35S]methionine. Mitochondria were isolated from Δpos5 cells grown under reduced O2 conditions, and import reactions were also performed under low O2 conditions. A post-ribosomal supernatant containing the radiolabeled Pos5p precursor protein was incubated with Δpos5 mitochondria in the presence of ATP and GTP, with no added NADH. The precursor protein (Fig. 7A, lane 1) was imported into the matrix, and the N-terminal targeting signal was cleaved by the matrix processing peptidase (lane 2). The mature protein thus generated remained completely protected from digestion by external trypsin. However, the unimported precursor protein was digested (Fig. 7A, lane 3). As an additional control, mitochondria were treated with valinomycin to dissipate the membrane potential across the inner membrane and then incubated with the precursor protein. As expected, import did not occur, and the radiolabeled protein was completely degraded by trypsin (Fig. 7A, lane 4). Thus, Pos5p can be imported into Δpos5 mitochondria (Fig. 7A) in agreement with ferredoxin import into these mutant mitochondria (Fig. 6A).
The effect of imported Pos5p on cluster biogenesis of endogenous Aco1p in Δpos5 mitochondria was then investigated in two steps under reduced O2 conditions as outlined (Fig. 7B, top panel). In the first step, mitochondria were supplemented with ATP and GTP but not NADH and incubated with a reticulocyte post-ribosomal supernatant with or without the Pos5p precursor protein. Samples were then diluted with isotonic buffer, and mitochondria with or without imported Pos5p were isolated. In the second step, mitochondria were supplemented with ATP, GTP, NADH, reduced iron, and [35S]cysteine and incubated to synthesize Fe-35S clusters. Compared with Δpos5 mitochondria containing no imported Pos5p (Fig. 7B, bottom panel, lanes 1–3), radiolabeling of aconitase in Δpos5 mitochondria with imported Pos5p (bottom panel, lanes 4–6) was at least three times more efficient at every time point that we tested. Thus, like in the case for Pos5p introduced into Δpos5 in vivo (Fig. 4), Pos5p imported into isolated Δpos5 mitochondria also restored cluster biogenesis to a significant extent (Fig. 7B), suggesting a direct role of Pos5p in the process.
We also tested if imported Pos5p-mediated stimulatory effects on cluster biogenesis are dependent on addition of its substrate NADH. Assays were performed under reduced O2 conditions in two steps as outlined (Fig. 7C, top panel). Briefly, Pos5p was imported into Δpos5 mitochondria in the presence of ATP and GTP but no added NADH (Fig. 7C, top panel, 1st step). Mitochondria were reisolated and then allowed to synthesize Fe-35S clusters in the absence or presence of added NADH (Fig. 7C, top panel, 2nd step). Radiolabeling of aconitase occurred much faster and more efficiently in NADH-supplemented mitochondria (lanes 6–10) than in mitochondria with no added NADH (lanes 1–5) even though all samples contained imported Pos5p (Fig. 7C, middle and bottom panels). These results suggest that added NADH is converted to NADPH by the NADH kinase Pos5p and that NADPH thus generated participates in cluster biogenesis. In the absence of Pos5p, NADH addition is ineffective. Likewise, Pos5p is of little use without added NADH.
The results described above imply that imported Pos5p was able to generate some NADPH for cluster biogenesis. Ideally, the NADH kinase activity of imported Pos5p should be determined to validate this notion. However, mitochondria contain numerous dehydrogenases, ATPases, and phosphatases, making it very difficult to determine Pos5p activity in mitochondrial extracts. We therefore tackled this issue in a different way. The mature form of Pos5p with a C-terminal His6 tag was expressed in bacteria in a soluble form, and the protein was purified to homogeneity by Ni-NTA chromatography (data not shown). To determine the NADH kinase activity, the purified enzyme was incubated with a mixture of [γ-32P]ATP (0.5 μCi), unlabeled ATP (0.5 μm), and NADH (2 mm) for 5 min at 30 °C. Samples were analyzed by thin layer chromatography looking for NAD32PH formation (Fig. 7D). As little as 1 ng of the purified enzyme was able to convert most of the ATP to NADPH during such a short period of incubation (Fig. 7D, compare lanes 1 and 2). Depending on the size and other factors, 1–10 ng of proteins can be synthesized per μl of the reticulocyte lysate translation system (Promega). We used 10 μl of Pos5p translation product for import followed by cluster loading assays (Fig. 7, B and C). Assuming 50% import efficiency, Δpos5 mitochondria in these experiments should contain 5–50 ng of imported Pos5p. Thus, imported Pos5p could generate 0.5–5 μm NADPH per min during cluster synthesis over a period of 30–60 min. Such a level of NADPH synthesis may not be optimum but appears to be sufficient for promoting Fe-S cluster biogenesis to a significant extent. These estimations are based on activity of the purified enzyme, which may or may not be fully active. The NADH kinase activity of imported Pos5p may be substantially more in its native environment within mitochondria. In any case, these calculations provide an explanation for imported Pos5p-mediated cluster synthesis in Δpos5 mitochondria when supplemented with NADH.
Pos5p Overexpression in Wild-type Mitochondria Enhances Fe-S Cluster Biogenesis Both in Vivo and in Vitro
Pos5p overexpression in Δpos5 cells not only restored Fe-S cluster biogenesis but appeared to confer augmented biosynthetic activity compared with the wild type (Fig. 4). To further investigate this finding, we determined the effects of Pos5p overexpression on cluster biogenesis in wild-type cells. Because we did not have antibodies against Pos5p, we used Pos5p with a C-terminal TAP tag to monitor the level of overexpression. Pos5p-TAP is completely functional, and a haploid yeast strain that expresses Pos5p-TAP from the POS5 promoter in the genome, called WT (Pos5p-TAP), behaves just like the wild-type strain without the tag (data not shown). A plasmid with a strong (GAPDH) promoter driving Pos5p-TAP was introduced into the parent strain. The resulting strain was called Pos5p-TAP▴. As judged by immunoblot analysis (Fig. 8A), the Pos5p-TAP protein was ∼15-fold overexpressed in Pos5p-TAP▴ mitochondria compared with WT (Pos5p-TAP) mitochondria. To determine the effects on Fe-S cluster biogenesis, experiments were performed with overexpressed Pos5p with or without the TAP tag. Results from experiments with untagged Pos5p are presented below. As expected, the TAP tag did not alter these results (data not shown).
Interestingly, Pos5p overexpression in wild-type cells (WT with Pos5p▴) led to ∼3-fold higher aconitase activity (Fig. 8B, top panel) even though aconitase (middle panel) and Tom40p control (bottom panel) protein levels remained the same. This effect could reflect enhanced biogenesis or decreased turnover of the aconitase Fe-S cluster. However, stabilization of the Fe-S cluster on aconitase due to decreased oxidative stress is unlikely here, because wild-type cells are already well protected from oxidative stress. Furthermore, no significant change in aconitase activity was observed in wild-type cells grown under aerobic versus low O2 conditions (data not shown). We then compared cluster biogenesis in isolated wild type versus wild type with Pos5p▴ mitochondria (Fig. 8C). Regardless of the assay conditions, radiolabeling of endogenous aconitase was more efficient in Pos5p-overexpressed mitochondria. For example, when NADH but no iron was added, more than 2-fold radiolabeled aconitase was detected in WT with Pos5p▴ mitochondria than in WT mitochondria (Fig. 8C, compare lanes 2 and 4). These results suggest a greater pool of usable stored iron and/or enhanced utilization of endogenous iron for cluster biogenesis in Pos5p▴ mitochondria. Likewise, when iron but no NADH was added, cluster biogenesis in Pos5p▴ mitochondria still occurred more effectively (Fig. 8C, compare lanes 5 and 7), perhaps because of an increased level of endogenous NADPH in these mitochondria. In the presence of both NADH and iron added, cluster biogenesis occurred faster in Pos5p▴ mitochondria than in wild-type mitochondria, and the differences were greatly pronounced particularly at earlier time points (Fig. 8D). We conclude that Pos5p/NADPH plays a direct and important role in Fe-S cluster biogenesis in mitochondria.
DISCUSSION
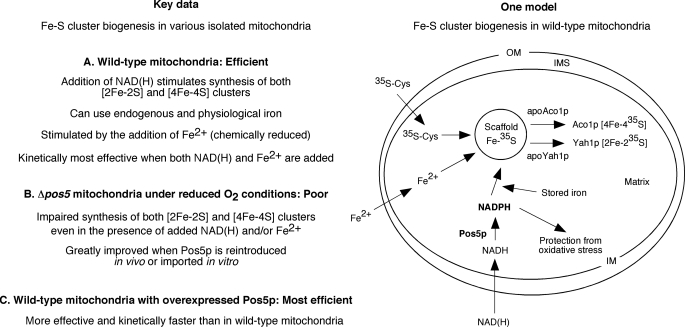
Several proteins in mitochondria contain Fe-S clusters as essential cofactors for their important functions in various processes such as enzyme catalysis and respiration. Synthesis and insertion of these clusters take place in the mitochondrial matrix. In S. cerevisiae, Pos5p is the only known mitochondrial NADH kinase. The enzyme is localized to the matrix and serves as the major source of NADPH in this compartment. Here, we provide strong evidence that NADPH, generated by Pos5p, plays an important role in Fe-S cluster biogenesis in mitochondria. The key results of this study are summarized in Fig. 9, left panel.
FIGURE 9.
Model for Pos5p/NADPH-dependent Fe-S cluster biogenesis in mitochondria. The left panel summarizes the key data reported here. The right panel shows a model depicting how Pos5p/NADPH might be involved in Fe-S cluster biogenesis in mitochondria.
In S. cerevisiae, the enzymes required for NAD+ synthesis are localized outside of mitochondria. Therefore, NAD+ must be transported from the cytosol into mitochondria. Transport of NAD+ across the mitochondrial inner membrane appears to be mediated by two carrier proteins (Ndt1p and Ndt2p) in this membrane (4). When bacterially expressed and purified Ndt1p was reconstituted into liposomes, it transported NAD+, to a lesser extent NADH, and virtually no NADP+ or NADPH. We observed that addition of NAD+ or NADH to isolated and intact mitochondria greatly stimulated Fe-S cluster biogenesis. In these experiments, NAD+ was likely imported into the matrix via the carrier proteins (Ndt1p/Ndt2p) and converted to NADH through the tricarboxylic acid cycle. Added NADH might also have been directly imported via Ndt1p/Ndt2p carrier proteins. Alternatively, NADH might have been first converted to NAD+ by dehydrogenases outside of the inner membrane (39), followed by NAD+ transport into the matrix. These possibilities remain to be distinguished. Once in the matrix, NADH served as the substrate for Pos5p, enabling local production of NADPH. NADPH in turn was able to promote Fe-S cluster synthesis.
Fe-S cluster biogenesis is a multistep process that requires coordinated action of multiple proteins (40, 41). Briefly, the enzyme responsible for the sulfur contribution is the cysteine desulfurase (Nfs1p), which forms a complex with another protein (Isd11p) of unknown function (20, 24, 42, 43). Initially, a persulfide is formed on a conserved cysteine residue of Nfs1p with the conversion of cysteine to alanine. Pyridine nucleotides are not required for Nfs1p/Isd11p desulfurase activity.4 The persulfide is then transferred to a scaffold protein Isu1p (or to the homologous protein Isu2p), where it is assembled together with iron to form an Fe-S cluster intermediate. Iron is presumed to be imported via specific carrier proteins (Mrs3p/Mrs4p) in the mitochondrial inner membrane. The conserved iron-binding protein, frataxin, may be involved here. Frataxin genetically interacts with Mrs3p/Mrs4p (30, 44) and physically interacts with Isu1p, Nfs1p, and Isd11p (45). The donor for iron has not been definitely established. Likewise, the physiological forms of imported and/or stored iron are unknown (46). However, reduced iron (ferrous) seems to be required, and thus an iron reductase could be involved at this step. After formation of the Fe-S cluster intermediate on scaffold proteins in mitochondria, it must be transferred to apoproteins, and this process involves another set of mediators, including the Hsp70 chaperone Ssq1p, a co-chaperone Jac1p, and a nucleotide exchange factor Mge1p. A monothiol glutaredoxin Grx5p has also been implicated at this stage, although the physiological role of Grx5p is unknown, and a corresponding reductase has not been identified (40, 41).
We have previously demonstrated that ATP and GTP in the mitochondrial matrix are required for cluster biogenesis (28, 29). Data presented here suggest that adequate levels of both NADPH and NTPs (ATP and GTP) are crucial for efficient Fe-S cluster assembly, and neither can replace the other (e.g. Figs. 1, A and C, 3B, and 7C). NADPH may participate in one or more reduction steps involved in Fe-S cluster synthesis. Adrenodoxin reductase homolog (Arh1p) is a mitochondrial inner membrane protein homologous to the mammalian adrenodoxin reductase and able to transfer electrons from NAD(P)H to ferredoxin (Yah1p) (47). Both Arh1p and Yah1p are essential for cell viability, and depletion of either of these proteins leads to deficiency of aconitase and other Fe-S cluster enzyme activities. Arh1p- or Yah1p-depleted cells also exhibit high cellular iron uptake and accumulate iron in mitochondria (21, 48). These iron-related defects mimic those of the Δpos5 mutant (5). Arh1p and Yah1p may therefore mediate NADPH-dependent processes required for efficient Fe-S cluster biogenesis in mitochondria. However, the recipient of reducing equivalents from these proteins has not been ascertained. The involvement of an NADH-dependent reduction step in release of the terminal sulfane (S0) of the Nfs1p persulfide and transfer to Isu1p/Isu2p as sulfide anion (S2−) has been suggested (41), although no data have been presented to support this idea. The data reported here neither support nor refute such a possibility, and much work is needed to determine whether NAD(P)H is required for the reduction of sulfane prior to use on the scaffold proteins. An additional reduction step, called reductive coupling, has been proposed specifically for formation of [4Fe-4S] clusters from [2Fe-2S] clusters on scaffold proteins (49). This is unlikely to be the exclusive function of NADPH in mitochondrial Fe-S cluster synthesis, because a block of this step should cause deficient [4Fe-4S] clusters while sparing [2Fe-2S] clusters. However, both types were defective in the Δpos5 mutant.
In many respects, both in vivo and in vitro phenotypes of the Δpos5 mutant resemble those of Δyfh1. Yfh1p is the yeast frataxin homolog and is thought to serve as iron chaperone/donor for Fe-S cluster synthesis on the Isu1p/Isu2p scaffold proteins in mitochondria. Like Δpos5, the Δyfh1 mutant was also sensitive to oxidative stress and was deficient in Fe-S cluster proteins (30, 50, 51). Both mutants activated the cellular iron uptake system and accumulated iron in mitochondria (5, 19), and both showed similar gene expression profiles (10, 52). Like in the case of Δpos5, Δyfh1 cells grown under aerobic conditions also exhibited practically no aconitase activity, and no significant Fe-S cluster biogenesis occurred in isolated Δyfh1 mitochondria. When grown under reduced O2 conditions, aconitase activity in Δyfh1 was partially restored. Mitochondria isolated from these cells continued to be deficient in Fe-S cluster biogenesis, and a small amount of imported Yfh1p was able to restore the process to a significant extent (30, 40). Pos5p imported into isolated Δpos5 mitochondria (or Yfh1p imported into isolated Δyfh1 mitochondria) could not reverse oxidative damage to other proteins that had already occurred but could restore Fe-S cluster biogenesis to a significant extent. The close resemblance of Δpos5 and Δyfh1 mitochondrial phenotypes (more so than for any of the other Fe-S cluster assembly mutants) could indicate that Pos5p functions together with Yfh1p or at a similar step in Fe-S cluster assembly.
The Δpos5 mutant exhibits hyperoxia sensitivity. In a previous study, reduction in mitochondrial iron did not restore growth of the Δpos5 mutant at high oxygen conditions. Furthermore, an Fe-S cluster assembly mutant (Δisa2) accumulated iron in mitochondria and yet grew just like the wild-type cell even at 100% O2. It was therefore proposed that the defects in mitochondrial iron homeostasis and hyperoxia sensitivity of the Δpos5 mutant are not linked and represent different effects of mitochondrial NADPH depletion (5). Data presented here are in agreement with this notion and show that reduced O2 conditions cannot efficiently restore cluster biogenesis in Δpos5 mitochondria.
A hypothesis (Fig. 9, right panel) that fits with the available data is as follows. The NADH kinase activity of Pos5p generates NADPH inside mitochondria. NADPH in mitochondria activates a reductase, perhaps Arh1p/Yah1p, facilitating the utilization of iron for Fe-S cluster synthesis. Although bacterially expressed Arh1p can use both NADPH and NADH as the substrate (53), NADPH may be the preferred supplier of reducing equivalents to Arh1p/Yah1p in mitochondria. This model is based on several observations reported here. First, isolated wild-type mitochondria were able to efficiently use a pool of stored iron for new Fe-S cluster synthesis only after NAD(H) addition (Fig. 1). NADPH was likely generated from added NAD(H) and was used for reduction, mobilization, and/or utilization of stored iron for cluster synthesis. In the absence of NADPH synthesis in Δpos5 mitochondria, stored iron could not be utilized for cluster synthesis (Fig. 5B). Second, addition of chemically reduced iron to wild-type mitochondria was able to bypass the NADPH requirement for cluster biogenesis to some extent (Fig. 2A). NADPH may therefore be involved in physiological reduction of stored iron for Fe-S cluster synthesis. Third, cluster synthesis in wild-type mitochondria in the presence of reduced iron was further stimulated by NADH addition (Fig. 2, A and C). At least two possibilities exist, and they may not be mutually exclusive. NADH may facilitate iron transport into mitochondria through generation of a stronger membrane potential (54), thereby enhancing cluster synthesis. The other possibility is that once iron is imported into mitochondria, it must be maintained in usable form for cluster synthesis, and NADPH generated from added NADH facilitates this process. In the absence of adequate levels of mitochondrial NADPH, imported iron may become unusable. This notion is consistent with poor cluster biogenesis in Δpos5 mitochondria even when supplemented with both NAD(H) and reduced iron (Figs. 4B, 5, B and C, and 6B). Finally, significant cluster biogenesis was observed in isolated wild-type mitochondria with overexpressed Pos5p even in the absence of added NADH and reduced iron (Fig. 8C). These mitochondria likely contained a higher level of endogenous NADPH, which facilitated utilization of endogenous iron for cluster synthesis.
The effects of Pos5p overexpression on Fe-S cluster assembly were striking and unanticipated. The final Fe-S cluster levels in vivo (e.g. aconitase activity) were increased (Fig. 8B). The rate of new Fe-S cluster synthesis in isolated mitochondria was also accelerated (Fig. 8D), indicating enhanced efficiency of the process. These results were unanticipated, because for a multistep biochemical process such as Fe-S cluster assembly one would have thought that overexpression of a single component of the pathway would not be sufficient to overdrive the entire pathway. For example, in the heme biosynthetic pathway, overexpression of individual enzymatic components generally does not result in increased heme synthesis and, in fact, may produce porphyria due to uncoupled formation of porphyrin intermediates (55). The effect of Pos5p overexpression on yielding more and faster Fe-S cluster synthesis is unlikely to be due to improved oxidative stress protection, because the wild-type cell is already well defended against oxidative stress. Possibly the entire Fe-S cluster biosynthetic apparatus is controlled and limited by pyridine nucleotide availability, such that more NADPH in mitochondria drives more Fe-S cluster synthesis and yields higher levels of active Fe-S cluster enzymes. If our observations on the effects of overexpression of Pos5p in yeast mitochondria also hold true for human mitochondria, more NADPH in mitochondria might be beneficial for human mitochondrial function and for counteracting diseases associated with mitochondrial decline.
Acknowledgments
We thank Donna M. Gordon and Boominathan Amutha for their valuable contributions in the early stage of this work.
This work was supported, in whole or in part, by National Institutes of Health Grant AG030504 from NIA (to D. P.) and Grant R37DK053953 (to A. D.). This work was also supported by American Heart Association Grant 09GRNT2260364 (to D. P.).
A. Dancis and D. Pain, unpublished observations.
- TAP
- tandem affinity purification
- NTP
- nucleoside triphosphate
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1.Ying W. (2008) Antioxid. Redox. Signal. 10, 179–206 [DOI] [PubMed] [Google Scholar]
- 2.Pollak N., Dölle C., Ziegler M. (2007) Biochem. J. 402, 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jamieson D. J. (1998) Yeast 14, 1511–1527 [DOI] [PubMed] [Google Scholar]
- 4.Todisco S., Agrimi G., Castegna A., Palmieri F. (2006) J. Biol. Chem. 281, 1524–1531 [DOI] [PubMed] [Google Scholar]
- 5.Outten C. E., Culotta V. C. (2003) EMBO J. 22, 2015–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawai S., Suzuki S., Mori S., Murata K. (2001) FEMS Microbiol. Lett. 200, 181–184 [DOI] [PubMed] [Google Scholar]
- 7.Shi F., Kawai S., Mori S., Kono E., Murata K. (2005) FEBS J. 272, 3337–3349 [DOI] [PubMed] [Google Scholar]
- 8.McGuinness E. T., Butler J. R. (1985) Int. J. Biochem. 17, 1–11 [DOI] [PubMed] [Google Scholar]
- 9.Krems B., Charizanis C., Entian K. D. (1995) Curr. Genet. 27, 427–434 [DOI] [PubMed] [Google Scholar]
- 10.Shianna K. V., Marchuk D. A., Strand M. K. (2006) Mitochondrion 6, 99–106 [DOI] [PubMed] [Google Scholar]
- 11.Strand M. K., Stuart G. R., Longley M. J., Graziewicz M. A., Dominick O. C., Copeland W. C. (2003) Eukaryot. Cell 2, 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuart G. R., Humble M. M., Strand M. K., Copeland W. C. (2009) Mitochondrion 9, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Outten C. E., Culotta V. C. (2004) J. Biol. Chem. 279, 7785–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedrajas J. R., Kosmidou E., Miranda-Vizuete A., Gustafsson J. A., Wright A. P., Spyrou G. (1999) J. Biol. Chem. 274, 6366–6373 [DOI] [PubMed] [Google Scholar]
- 15.Pedrajas J. R., Miranda-Vizuete A., Javanmardy N., Gustafsson J. A., Spyrou G. (2000) J. Biol. Chem. 275, 16296–16301 [DOI] [PubMed] [Google Scholar]
- 16.Pedrajas J. R., Porras P., Martínez-Galisteo E., Padilla C. A., Miranda-Vizuete A., Bárcena J. A. (2002) Biochem. J. 364, 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bieganowski P., Seidle H. F., Wojcik M., Brenner C. (2006) J. Biol. Chem. 281, 22439–22445 [DOI] [PubMed] [Google Scholar]
- 18.Chen O. S., Crisp R. J., Valachovic M., Bard M., Winge D. R., Kaplan J. (2004) J. Biol. Chem. 279, 29513–29518 [DOI] [PubMed] [Google Scholar]
- 19.Babcock M., de Silva D., Oaks R., Davis-Kaplan S., Jiralerspong S., Montermini L., Pandolfo M., Kaplan J. (1997) Science 276, 1709–1712 [DOI] [PubMed] [Google Scholar]
- 20.Li J., Kogan M., Knight S. A., Pain D., Dancis A. (1999) J. Biol. Chem. 274, 33025–33034 [DOI] [PubMed] [Google Scholar]
- 21.Li J., Saxena S., Pain D., Dancis A. (2001) J. Biol. Chem. 276, 1503–1509 [DOI] [PubMed] [Google Scholar]
- 22.Knight S. A., Sepuri N. B., Pain D., Dancis A. (1998) J. Biol. Chem. 273, 18389–18393 [DOI] [PubMed] [Google Scholar]
- 23.Kim R., Saxena S., Gordon D. M., Pain D., Dancis A. (2001) J. Biol. Chem. 276, 17524–17532 [DOI] [PubMed] [Google Scholar]
- 24.Kispal G., Csere P., Prohl C., Lill R. (1999) EMBO J. 18, 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garland S. A., Hoff K., Vickery L. E., Culotta V. C. (1999) J. Mol. Biol. 294, 897–907 [DOI] [PubMed] [Google Scholar]
- 26.Kaut A., Lange H., Diekert K., Kispal G., Lill R. (2000) J. Biol. Chem. 275, 15955–15961 [DOI] [PubMed] [Google Scholar]
- 27.Johnson D. C., Dean D. R., Smith A. D., Johnson M. K. (2005) Annu. Rev. Biochem. 74, 247–281 [DOI] [PubMed] [Google Scholar]
- 28.Amutha B., Gordon D. M., Gu Y., Lyver E. R., Dancis A., Pain D. (2008) J. Biol. Chem. 283, 1362–1371 [DOI] [PubMed] [Google Scholar]
- 29.Amutha B., Gordon D. M., Dancis A., Pain D. (2009) Methods Enzymol. 456, 247–266 [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Lyver E. R., Knight S. A., Pain D., Lesuisse E., Dancis A. (2006) J. Biol. Chem. 281, 22493–22502 [DOI] [PubMed] [Google Scholar]
- 31.Sepuri N. B., Gordon D. M., Pain D. (1998) J. Biol. Chem. 273, 20941–20950 [DOI] [PubMed] [Google Scholar]
- 32.Gu Y., Gordon D. M., Amutha B., Pain D. (2005) J. Biol. Chem. 280, 18604–18609 [DOI] [PubMed] [Google Scholar]
- 33.Gordon D. M., Wang J., Amutha B., Pain D. (2001) Biochem. J. 356, 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordon D. M., Kogan M., Knight S. A., Dancis A., Pain D. (2001) Hum. Mol. Genet. 10, 259–269 [DOI] [PubMed] [Google Scholar]
- 35.Gordon D. M., Lyver E. R., Lesuisse E., Dancis A., Pain D. (2006) Biochem. J. 400, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinkle P. C., Kumar M. A., Resetar A., Harris D. L. (1991) Biochemistry 30, 3576–3582 [DOI] [PubMed] [Google Scholar]
- 37.Tzagoloff A. (1982) Mitochondria (Siekevitz P. ed) Chapter 9, pp. 199–233, Plenum Publishing Corp., New York [Google Scholar]
- 38.Khoroshilova N., Popescu C., Münck E., Beinert H., Kiley P. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6087–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luttik M. A., Overkamp K. M., Kötter P., de Vries S., van Dijken J. P., Pronk J. T. (1998) J. Biol. Chem. 273, 24529–24534 [DOI] [PubMed] [Google Scholar]
- 40.Stemmler T. L., Lesuisse E., Pain D., Dancis A. (2010) J. Biol. Chem. 285, 26737–26743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lill R., Mühlenhoff U. (2008) Annu. Rev. Biochem. 77, 669–700 [DOI] [PubMed] [Google Scholar]
- 42.Wiedemann N., Urzica E., Guiard B., Müller H., Lohaus C., Meyer H. E., Ryan M. T., Meisinger C., Mühlenhoff U., Lill R., Pfanner N. (2006) EMBO J. 25, 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adam A. C., Bornhövd C., Prokisch H., Neupert W., Hell K. (2006) EMBO J. 25, 174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y., Lyver E. R., Knight S. A., Lesuisse E., Dancis A. (2005) J. Biol. Chem. 280, 19794–19807 [DOI] [PubMed] [Google Scholar]
- 45.Wang T., Craig E. A. (2008) J. Biol. Chem. 283, 12674–12679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atkinson A., Winge D. R. (2009) Chem. Rev. 109, 4708–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mühlenhoff U., Richhardt N., Gerber J., Lill R. (2002) J. Biol. Chem. 277, 29810–29816 [DOI] [PubMed] [Google Scholar]
- 48.Lange H., Kaut A., Kispal G., Lill R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1050–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chandramouli K., Unciuleac M. C., Naik S., Dean D. R., Huynh B. H., Johnson M. K. (2007) Biochemistry 46, 6804–6811 [DOI] [PubMed] [Google Scholar]
- 50.Karthikeyan G., Lewis L. K., Resnick M. A. (2002) Hum. Mol. Genet. 11, 1351–1362 [DOI] [PubMed] [Google Scholar]
- 51.Gakh O., Park S., Liu G., Macomber L., Imlay J. A., Ferreira G. C., Isaya G. (2006) Hum. Mol. Genet. 15, 467–479 [DOI] [PubMed] [Google Scholar]
- 52.Foury F., Talibi D. (2001) J. Biol. Chem. 276, 7762–7768 [DOI] [PubMed] [Google Scholar]
- 53.Lacour T., Achstetter T., Dumas B. (1998) J. Biol. Chem. 273, 23984–23992 [DOI] [PubMed] [Google Scholar]
- 54.Lange H., Kispal G., Lill R. (1999) J. Biol. Chem. 274, 18989–18996 [DOI] [PubMed] [Google Scholar]
- 55.Labbe-Bois R., Camadro J. M. (1994) in Metal Ions in Fungi (Winkelmann G., Winge D. R. eds) pp. 413–453, Marcel Dekker, Inc., New York [Google Scholar]