Abstract
Unlike the prototype transforming growth factor-β (TGF-β), bone morphogenetic protein-6 (BMP-6) activates macrophages. Here, we report that BMP-6 induces the expression of IL-6 in macrophages. Using overexpression and knockdown experiments, we demonstrate that BMP receptor type II and activin-like kinase-2 are necessary for IL-6 induction by BMP-6. At the intracellular level, both Smad and p38 signaling pathways are required for the induction of IL-6. The cross-talk between the two pathways occurs at the level of transcription factor GATA4 and Smad 1/4. These results, taken together, demonstrate a novel BMP-6 signaling mechanism in which both the Smad and non-Smad pathways directly interact to activate the transcription of a target gene.
Keywords: Bone Morphogenetic Protein (BMP), Interleukin, Macrophage, p38 MAPK, Signal Transduction
Introduction
Bone morphogenetic proteins (BMPs)3 have been initially identified as factors critical for bone and cartilage formation (1). Subsequent investigations have revealed that BMPs are members of the TGF-β superfamily that regulate embryonic development as well as normal tissue homeostasis (2). To date, more than 20 subtypes of BMPs have been identified. Results of knock-out studies in mice suggest that these ligands have different functions (2). BMPs signal through a heteromeric, most likely heterotetrameric, complex of type I and type II receptors (3, 4). There are three type I (ALK-2/Act-RIA, ALK-3/BMP-RIA, and ALK-6/BMP-RIB) and three type II receptors (Act-RIIA, Act-RIIB, and BMP-RII) (3). The ligand binds to type I and II receptors, which then phosphorylate receptor-activated Smads (R-Smads) (Smads 1, 5, and 8) (5, 6). Subsequently, the phosphorylated R-Smads interact with the common mediator Smad (Co-Smad) (Smad 4) and translocate into the nucleus and regulate specific gene expressions. Currently, the precise role of each BMP receptor and R-Smad remains unclear.
In addition to the canonical Smad signaling pathway, BMPs can signal independent of Smads (7). For example, BMP-4 has been reported to stimulate vascular endothelial growth factor synthesis in osteoblasts via p38 (8). Similarly, BMP-7 stimulates renal epithelial cell morphogenesis through the activation of p38 that is negatively regulated by Smad 1 (9), whereas BMP-2 activates both p38 and c-Jun-NH2-terminal kinase (JNK) (10). Thus, upon activation of the receptors by BMPs, both Smad and non-Smad signaling pathways can be activated simultaneously. At the present time, the precise mechanism as well as the biological consequences of the concurrent activation of Smad and non-Smad signal transduction cascades by BMPs remain largely unknown.
In the context of immune regulation, BMP-6 has been reported to suppress both B and T cells (11, 12). More recently, we have reported that BMP-6 activates macrophages (13, 14). Consistent with our previous publications, we demonstrate here that BMP-6 induces the expression of inteleukin-6 (IL-6). In investigating the mechanism of IL-6 induction by BMP-6 in macrophages, we have unexpectedly uncovered a novel mechanism in which a cross-talk between the canonical Smad pathway and the noncanonical p38 pathway is required for IL-6 expression.
EXPERIMENTAL PROCEDURES
Cell Culture
RAW 264.7 and THP-1, cells were purchased from American Type Tissue Collection. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 0.1 mg/ml streptomycin. BMPs were purchased from R&D Systems.
Peritoneal macrophages were obtained from C57BL/6 and IL-6 KO mice (Jackson Laboratory) and cultured in 10% FBS/20% L929 culture medium supplemented DMEM. Cycloheximide and actinomycin D were used at 50 μg/ml and 1 μg/ml, respectively. Where indicated, SB 203580 (Sigma), BAY11-7082 (Sigma), and αIL-6Ab (R&D Systems) were added to the co-culture system.
Plasmids
All BMP receptors and Smad constructs were obtained from Dr. Joan Massague (Memorial Sloan Kettering Cancer Center, New York, NY), Dr. Rik Derynck (UCSF, San Francisco, CA) and Dr. Kohei Miyazono (Tokyo University, Tokyo, Japan). Smad 4DN, p38DN, GATA4, and GATA4DN were purchased from Addgene (Cambridge, MA). IL-6-Luc plasmid was kindly provided by Dr. Yoshio Yamaoka (Baylor College of Medicine, Houston, TX).
RNA Isolation and RT-PCR
Cells were collected at the indicated time points, and total RNA was extracted using TRIzol reagent (Invitrogen). Unless indicated, the standard number of PCR cycles was 30. Primer sequences for RT-PCR are shown in supplemental Table 1.
Immunoblot Analysis
Primary antibodies were purchased from the following sources: IL-6 (R&D Systems); Smad 1, pSmad 1/5, p38, p-p38, GATA4, myc, and FLAG (Cell Signaling Technology, Danvers, MA). Cells were cultured under the designated conditions and harvested at the indicated times points. Twenty to fifty μg of protein was loaded onto SDS-polyacrylamide gel. Immunoreactive bands were visualized using enhanced chemiluminescence.
Immunoprecipitation
Cells were collected, centrifuged, and lysed with denaturing lysis buffer. Immunoprecipitation was carried out using the standard protocol using protein A-Sepharose bead slurry.
Transient Transfection and Luciferase Assay
Cells were transfected using 1 μg/ml Lipofectamine 2000 (Invitrogen) and 1 μg/ml plasmid. Then, cells were treated with BMP-6 (0–100 ng) for the indicated lengths of time. The Dual-Luciferase Reporter Assay System (Promega, Madison, WI), which controls the variations in transfection efficiency by co-transfecting Renilla luciferase plasmid, was used.
Knockdown of Receptors, R-Smads, and GATA4
Three shRNA oliognucleotides (supplemental Table 2) were designed and synthesized for each target (receptors and R-Smads) and cloned into pLKO.1-puro lentiviral vector. Subsequently, lentiviral particles were generated using the ViraPowerTM T-RexTM Lentivral Expression System (Invitrogen) and infected into RAW 264.7 cells. The level of expression of the target gene was measured using semiquantitative RT-PCR. To knock down GATA4, siRNA was used (Qiagen). After plating 5 × 105 cells/well onto 6-well plates, 10 μl of control, GATA3, and GATA4 siRNA solution were transfected using Oligofectamine (Invitrogen). After transfection, cells were treated with BMP-6 (100 ng/ml) and harvested. Semiquantitative RT-PCR was used to confirm the specificity of knockdowns.
Serially Deleted IL-6 Promoter Constructs
Serial deletions of IL-6 promoter were carried out using PCR (supplemental Table 3) and cloned into the luciferase reporter vector (pGL3 Basic, Promega). The reporter constructs were transiently transfected into RAW 264.7 cells, and luciferase activity was measured.
ChIP assay
ChIP assay was carried out using the EZ ChIP kit (Upstate, VA). RAW 264.7 cells were treated with formaldehyde, lysed, and sonicated to generate DNA fragments. After immunoprecipitation with appropriate antibodies, DNA was purified, and PCR was performed.
Statistical Analysis
For all analyses, Student's t tests were performed. A p value of <0.05 was considered to indicate significant differences between datasets.
RESULTS
BMP-6 Induces IL-6 Expression in Macrophages
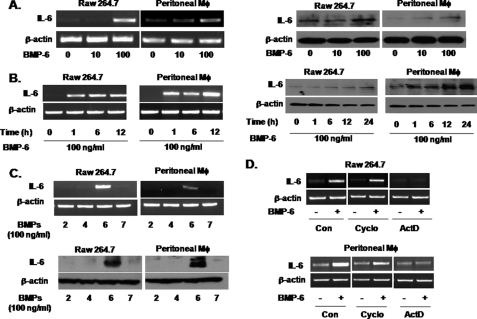
We initially treated the murine macrophage cell line RAW 264.7 with BMP-6 and performed a multiplex RT-PCR for cytokines. This revealed that BMP-6 altered the expression of multiple cytokines in RAW 264.7 cells (data not shown). Among the cytokines induced by BMP-6, we focused on IL-6. BMP-6 induced IL-6 expression in a concentration-dependent manner in both RAW 264.7 and murine peritoneal macrophages at the RNA and protein level (Fig. 1A). A similar degree of IL-6 mRNA induction by BMP-6 was observed in the human monocyte cell line THP-1 (supplemental Fig. 1). The consistent induction of IL-6 by BMP-6 in RAW 264.7, murine peritoneal macrophages, and THP-1 demonstrates that RAW 264.7 is a reasonable model for investigating the mechanism of BMP-6-induced IL-6 expression in macrophages. Kinetic studies using 100 ng/ml BMP-6 showed IL-6 induction at the mRNA level within 1 h, whereas protein induction was seen in 1–6 h (Fig. 1B). Among BMP-2, -4, -6, and -7, only BMP-6 induced the expression of IL-6 in RAW 264.7 and murine peritoneal macrophages (Fig. 1C). This induction of IL-6 was blocked by actinomycin D but not by cycloheximide, demonstrating that BMP-6 induces IL-6 expression directly at the transcription level (Fig. 1D).
FIGURE 1.
Induction of IL-6 expression by BMP-6 in macrophages. A, murine macrophage (Mφ) cell line RAW 264.7 and murine peritoneal macrophages were treated with increasing concentrations of BMP-6. RT-PCR (left panel) and immunoblot (right panel) analysis demonstrated that BMP-6 induced expression of IL-6 in macrophages in a concentration-dependent manner. B, RT-PCR (left panel) demonstrated that BMP-6 at 100 ng/ml induced expression of IL-6 mRNA within 1 h after treatment in RAW 264.7 and murine peritoneal macrophages. Immunoblot analysis (right panel) supported the results of RT-PCR. C, RAW 264.7 and murine peritoneal macrophages were treated with BMP-2, -4, -6, and -7 at 100 ng/ml, and IL-6 expression was measured by RT-PCR (top panel) and immunoblot analysis (bottom panel). Only BMP-6 induced IL-6 expression. D, RAW 264.7 and murine peritoneal macrophages were treated with cycloheximide (Cyclo) and actinomycin D (ActD) along with BMP-6 and IL-6 expression level was measured by RT-PCR. IL-6 mRNA induction by BMP-6 was blocked by actinomycin D but not by cycloheximide, suggesting that BMP-6 directly activates IL-6 promoter in macrophages. Con, control.
ALK-2 and BMP-RII Are the Functional BMP-6 Receptors in the Context of IL-6 Induction
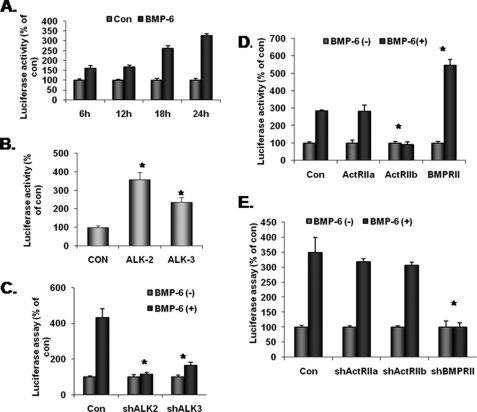
Because there are three each of type I and II BMP receptors, we next investigated the functional BMP-6 receptors in the context of IL-6 in macrophages. To this end, we obtained the previously reported luciferase reporter construct containing the 1.2-kb human IL-6 promoter (IL-6-Luc) (15). When this construct was transfected into RAW 264.7 cells, BMP-6 increased luciferase activity more than 3.5-fold in 24 h (Fig. 2A). Next, constitutively active type I BMP receptors were co-transfected with the IL-6-Luc plasmid. Among the three known type I BMP receptors, we have reported that macrophages express only ALK-2 (Act-RIA) and -3 (BMP-RIA) (13). Thus, only ALK-2 and -3 were studied. The results demonstrated that 18 h after the co-transfection of constitutively active ALK-2, luciferase activity increased more than 3.5-fold (Fig. 2B). Co-transfection of constitutively active ALK-3 also increased luciferase activity but to a more modest level. When the expression of ALK-2 and -3 was knocked down using the lentivirus-based shRNA approach (13), BMP-6-induced IL-6 expression was blocked only in cells infected with the ALK-2 shRNA lentivirus (Fig. 2C).
FIGURE 2.
BMP-6 receptors and IL-6 induction in macrophages. A, plasmid IL-6-Luc containing IL-6 promoter and luciferase reporter was transfected into RAW 264.7 cells and treated with 100 ng/ml BMP-6. Twenty-four hours after adding BMP-6, IL-6 promoter activity increased more than 3-fold. B, constitutively active ALK-2 and -3 were co-transfected with IL-6-Luc reporter. Although both ALK-2 and -3 induced IL-6 promoter activity to a statistically significant level, ALK-2 increased IL-6 promoter activity more than 3-fold. C, lentiviruses containing shRNA sequences targeting ALK-2 and -3 were infected into RAW 264.7 cells. Statistically significant knockdown of target gene expression was confirmed previously and published (13). When transfected with IL-6-Luc and treated with BMP-6, only the knockdown of ALK-2 blocked the induction of luciferase activity. D, macrophages express all three known BMP type II receptors (BMP-RII, Act-RIIA, and Act-RIIB). Thus, each of the three type II BMP receptors was co-transfected with IL-6-Luc into RAW 264.7 cells and treated with BMP-6. Only in cells transfected with BMP-RII was a significant increase in IL-6 promoter activity observed. Interestingly, overexpression of Act-RIIB suppressed IL-6 promoter activity consistently. E, lentiviruses containing shRNA sequences targeting each of the three type II BMP receptors were infected into RAW 264.7 cells. We have previously confirmed the knockdown of the target gene expression (13). Among the three type II BMP receptors, knockdown of BMP-RII reversed the induction of IL-6 promoter. *, statistically significant.
In contrast to type I receptors, type II BMP receptors are constitutively active serine/threonine kinases. Thus, RAW 264.7 cells were co-transfected with each of the three known BMP type II receptors along with IL-6-Luc plasmid and treated with BMP-6. As shown in Fig. 2D, cells transfected with BMP-RII exhibited the highest level of luciferase activity following treatment with BMP-6. Interestingly, co-transfection with ActRIIB repeatedly decreased the luciferase activity, suggesting that receptor stoichiometry may play a role in BMP-6 signaling. As a complementary approach, cells were infected with lentivirus containing shRNA sequence targeting each of the type II BMP receptors (13). Only BMP-RII shRNA significantly blocked the induction of luciferase activity in RAW 264.7 cells (Fig. 2E). These results suggest that ALK-2 and BMP-RII are the optimal and functional BMP-6 receptors in the context of IL-6 expression in macrophages.
Smad 1 Transduces BMP-6 Signaling for IL-6 Expression
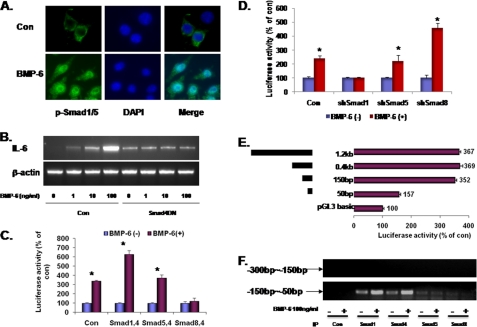
The canonical BMP signaling pathway requires R-Smads (Smad 1, 5, and 8) and Co-Smad (Smad4). When RAW 264.7 cells were treated with BMP-6, confocal immunofluorescence microscopy demonstrated a dramatic nuclear translocation of phospho-Smad 1/5 (pSmad 1/5) (Fig. 3A). In addition, transfection of dominant negative Smad 4 (Smad4DN) successfully blocked the induction of IL-6 mRNA (Fig. 3B). These results demonstrate that the classical Smad pathway is necessary for BMP-6-induced IL-6 expression in macrophages.
FIGURE 3.
Smads and IL-6 induction in macrophages. A, BMPs signal through the R-Smads 1, 5, and 8. Confocal microscopy in peritoneal macrophages demonstrated that BMP-6 induced increased levels as well as nuclear translocation of phospho-Smad 1/5. B, dominant negative Smad 4 (Smad4DN) was transfected into RAW 264.7 cells and treated with increasing concentrations of BMP-6. Effect on IL-6 expression was measured using RT-PCR. Transfection process itself modestly increased the baseline expression level of IL-6. Compared with the control, IL-6 induction was suppressed in cells expressing Smad4DN, suggesting that the Smad-dependent pathway is necessary for IL-6 induction in macrophages. C, RAW 264.7 cells were co-transfected with each of the three R-Smads (Smad 1, 5, and 8) and the Co-Smad (Smad 4) along with IL-6-Luc. When treated with BMP-6, cells expressing Smad 1/4 demonstrated the highest level of induction of IL-6 promoter activity. Interestingly, transfection with Smad 8/4 resulted in suppression of IL-6 promoter activity. D, lentiviruses containing shRNA sequences against Smad 1, 5, and 8 were infected into RAW 264.7 cells. Statistically significant knockdown of target gene expression was confirmed using RT-PCR (supplemental Fig. 2). When transfected with IL-6-Luc and treated with BMP-6, knockdown of Smad 1 blocked the induction of luciferase activity whereas that of Smad 8 increased luciferase activity. E, to determine the BMP-6-response element in the IL-6 promoter region, serial deletion constructs of IL-6 promoter were established. Subsequently, the shortened IL-6 promoters were co-transfected with IL-6-Luc into RAW 264.7 cells. In IL-6 promoter, BMP-6-response element was located between −50 and −150 bp 5′ to the transcription initiation site. The results shown are induced luciferase activity by BMP-6. F, ChIP was carried out to determine the presence of interaction between Smads and IL-6 promoter. The −50 to −150 region was amplified in samples immunoprecipitated with Smad 1 and 4 antibodies. As a negative control, the −150 to −300 bp region was targeted. *, statistically significant.
To determine the R-Smad that transduces BMP-6 signal for IL-6 expression, RAW 264.7 cells were co-transfected with each of the R-Smads (Smad 1, 5, and 8) in combination with the Co-Smad (Smad 4) and the IL-6-Luc plasmid. As shown in Fig. 3C, the combination of Smad 1/4 increased IL-6 promoter activity more than any other R-Smad/Co-Smad combination. Interestingly, luciferase activity was consistently lower following the transfection of Smad 8/4. In a reverse experiment, the lentivirus-based shRNA approach was again used (supplemental Fig. 2). As expected, the knockdown of Smad 1 expression led to the abrogation of IL-6 induction by BMP-6 (Fig. 3D) whereas that of Smad 8 led to the induction of IL-6 promoter activity. These results, in sum, demonstrate that Smad 1 is the R-Smad that signals for IL-6 expression when macrophages are stimulated with BMP-6.
To identify the BMP-6-response element within the IL-6 promoter, serial deletion constructs were made using PCR and subcloned. The results demonstrated that the BMP-6-response element is located between −50 and −150 bp 5′ to the transcription initiation site (Fig. 3E). Using the ChIP assay, the −50 to −150 bp region was amplified readily using PCR when immunoprecipitated with either Smad 1 or 4 antibodies (Fig. 3F). As a control, −150 to −300 bp region was targeted.
Activation of p38 Is Required for IL-6 Induction by BMP-6
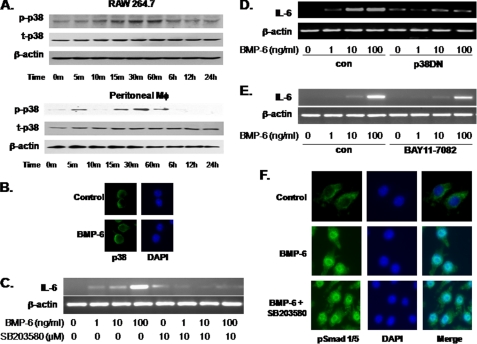
When a software-based analysis of the 100-bp region containing the BMP-6-response element was carried out for potential transcription factor binding sites, surprisingly no consensus Smad-binding element was identified. This suggested that the non-Smad pathway may be involved in BMP-6-mediated IL-6 induction. Thus, the effect of BMP-6 on p38 was investigated. Immunoblot analysis demonstrated that BMP-6 induced the phosphorylation of p38 within 15 min in both RAW 264.7 and murine peritoneal macrophages (Fig. 4A). Immunofluorescence microscopy showed a consistent localization of p38 in the cytosol of RAW 264.7 cells following treatment with BMP-6 (Fig. 4B). When RAW 264.7 was pretreated with the p38 inhibitor SB 203580 (10 μm) for 1 h, IL-6 mRNA expression was no longer induced by BMP-6 (Fig. 4C). Control experiments showed that SB 203580 has no significant effect of Smad phosphorylation up to 10 μm (supplemental Fig. 3). Consistent with this observation, the transfection of dominant negative p38 (p38DN) into RAW 264.7 cells also blocked IL-6 mRNA induction following BMP-6 exposure (Fig. 4D). These results suggest that p38 activation, in addition to the Smad 1, is required for IL-6 induction by BMP-6.
FIGURE 4.
Non-Smad p38 pathway and IL-6 induction in macrophages. A, immunoblot analysis was carried out to determine the effect of BMP-6 on activation of p38. BMP-6 induced the phosphorylation of p38 within 5–15 min in RAW 264.7 and murine peritoneal macrophages. p-p38 = phosphorylated p38; t-p38, total p38. B, immunofluorescence microscopy was used to localize p38 following stimulating with BMP-6 in RAW 264.7. The cytosolic location of p38 did not change with BMP-6 treatment. C, to determine the role of p38 activation on IL-6 induction by BMP-6 in macrophages, RAW 264.7 cells were treated with 10 μm SB 203580, a p38 inhibitor. RT-PCR demonstrated that IL-6 induction was blocked when p38 was inhibited. D, dominant negative p38 (p38DN) was transfected into RAW 264.7 cells, and the effect on IL-6 was measured using RT-PCR. As with cells treated with SB 203580, expression of p38DN blocked the induction of IL-6. E, a classic activator of IL-6 expression is NF-κB. Thus, RAW 264.7 was treated with BAY11-7082 to inhibit the NF-κB pathway. When simultaneously treated with BMP-6, RT-PCR showed no significant effect on IL-6 induction. These results demonstrate that IL-6 induction by BMP-6 does not require the NF-κB pathway. F, to determine the effect of p38 activation on Smad pathway, immunofluorescence microscopy was used. The results revealed that the inhibition of the p38 pathway via SB 203580 had no impact on nuclear translocation of phosphorylated Smad 1/5 (pSmad 1/5).
Because NF-κB is the classic activator of IL-6 promoter, we next studied the potential relationship between BMP-6 signaling and NF-κB in murine peritoneal macrophages using the IKK inhibitor, BAY11-7082. The results demonstrated that the inactivation of NF-κB pathway did not significantly alter IL-6 mRNA expression following exposure to BMP-6 (Fig. 4E). One potential mechanism for the observed requirement of Smad-dependent and p38 pathways is that p38 regulates the nuclear translocation of Smads following BMP-6 treatment. To test this concept, RAW 264.7 cells were again treated with the p38 inhibitor SB 203580, and confocal immunofluorescence microscopy for pSmad 1/5 was carried out. Inactivation of p38 did not alter the nuclear translocation of pSmad 1/5 in response to BMP-6 (Fig. 4F). These results, in total, demonstrate that IL-6 induction by BMP-6 requires the activation of p38 signaling pathway independent of nuclear translocation of Smads.
GATA4 Transcription Factor Is Required for BMP-6-mediated IL-6 Induction
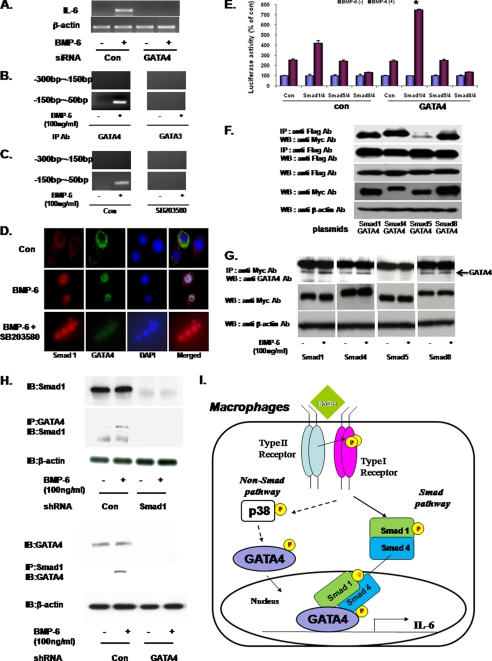
Given that the non-Smad pathway involves p38, transcription factors that signal downstream of p38 were investigated. To this end, GATAs drew our attention because the BMP-6-responsive region in IL-6 promoter contained multiple GATA-binding sites. GATA transcription factors have zinc fingers and play critical roles in cell differentiation and early endoderm development (16). Of the four GATA subtypes identified to date, GATA4 has been reported to interact with Smad 2, 3, and 4 (17). To determine whether GATA4 is involved in BMP-6 signaling in macrophages, siRNA was used. The specificity of GATA4 siRNA (siGATA4) is shown in supplemental Fig. 4. When GATA4 was knocked down in RAW 264.7, BMP-6 no longer induced the expression of IL-6 (Fig. 5A). Consistent with this observation, transfection of dominant negative GATA4 (GATA4DN) also blocked the BMP-6-induced IL-6 expression (supplemental Fig. 5). Next, ChIP assay using the GATA3 and four antibodies revealed that only GATA4 binds to the BMP-6-response element in IL-6 promoter (Fig. 5B). This interaction between GATA4 and the BMP-6-response element was disrupted by the p38 inhibitor SB 203580 (Fig. 5C), demonstrating that GATA4 signals downstream of p38. Next, confocal immunofluorescence microscopy demonstrated a simultaneous nuclear translocation of GATA4 and Smad 1 following BMP-6 treatment (Fig. 5D).
FIGURE 5.
GATA 4 and IL-6 expression in macrophages. A, RAW 264.7 was transfected with GATA4 siRNA (siGATA4), and the effect on IL-6 expression was measured using RT-PCR. When treated with BMP-6, induction of IL-6 was no longer observed when GATA4 was knocked down. B, ChIP was carried out to determine the interaction between GATA4 and the IL-6 promoter. GATA3 was used as a control. Following treatment with BMP-6, the −50 to −150 bp region was amplified in samples immunoprecipitated with GATA4 but not GATA3 antibody. C, when RAW 264.7 cells were treated with SB 203580 and BMP-6, ChIP assay using GATA4 antibody no longer amplified the −50 to −150 bp region. This observation demonstrates that GATA4 signals downstream of p38. D, confocal immunofluorescence microscopy was carried out using antibodies against Smad 1 and GATA 4. BMP-6 treatment induced nuclear translocation of Smad 1 and GATA4 simultaneously. E, RAW 264.7 was transfected with indicated combination of Smad 1, 4, 5, and 8 and GATA4 along with IL-6-Luc. BMP-6 induced a statistically significant level of IL-6 promoter activity when GATA4 was co-transfected with Smad 1/4. However, transfection of GATA4 alone did not induce IL-6 promoter activity (control bar in GATA4 group). F, myc-tagged Smads 1, 4, 5, and 8, along with FLAG-tagged GATA4 were expressed in RAW 264.7. Immunoprecipitation using anti-FLAG antibody followed by immunoblot analysis against myc epitope was performed. The results demonstrated that Smads 1, 4, and 8 but not 5 interacted with GATA4. G, myc-tagged Smads 1, 4, 5, and 8 were expressed in RAW 264.7. Immunoprecipitation using anti-myc antibody followed by immunoblot against endogenous GATA4 was performed. Following treatment with BMP-6, increased levels of GATA4 protein were immunoprecipitated out from cells transfected with Smad 1 or 4. H, interaction between endogenous Smad 1 and GATA4 was investigated using the shRNA approach. When Smad 1 was knocked down and GATA4 was immunoprecipitated, immunoblot for Smad 1 revealed no protein band. Conversely, when GATA4 was knocked down and Smad 1 was immunoprecipitated, immunoblotting for GATA4 demonstrated no protein band. I, proposed mechanism of IL-6 induction by BMP-6 in macrophages. Induction of IL-6 expression by BMP-6 in macrophages requires both Smad and non-Smad pathways. BMP-RII along with ALK2 simultaneously activates Smad 1/4 and p38. Subsequently, p38 activates GATA4. Finally, GATA4 and Smad 1/4 translocate to the nucleus and bind to the BMP-6-response element within IL-6 promoter and induce IL-6 expression. *, statistically significant.
Based on the observation that Smad 1 is required for IL-6 promoter activation by BMP-6 (Fig. 3D), we hypothesized that GATA4 and Smad 1 may complex to activate the transcription of IL-6 in the context of BMP-6 in macrophages. To test this concept, RAW 264.7 cells were transfected with varying combinations of Smads and GATA4 along with IL-6-Luc. The results showed that the overexpression of GATA4 augmented the capacity of Smad 1/4 to induce IL-6 promoter activity (Fig. 5E). However, GATA4 alone did not increase IL-6 promoter activity. To characterize the interaction between Smad 1 and GATA4 further, myc-tagged Smad 1, 4, 5, and 8 along with FLAG-tagged GATA4 were expressed in RAW 264.7 cells. Following the transfection, immunoprecipitation against the FLAG-tagged GATA4 followed by immunoblotting for the myc-tagged Smads demonstrated that Smad 1 interacted with GATA4 (Fig. 5F). In addition, Smad 4 and 8 also complexed with GATA4. Because the observed binding among Smads and GATA4 may be an artifact of overexpression, we next wanted to assay for the interaction of endogenous molecules. However, antibodies that reliably distinguish each of the R-Smads do not exist. Thus, myc-tagged Smads were overexpressed in RAW 264.7 cells and immunoprecipitation against myc-epitope followed by immunoblotting for the endogenous GATA4 was performed. Following treatment with BMP-6, higher levels of GATA4 were immunoprecipitated out in lysates obtained from cells overexpressing Smad 1 and 4 (Fig. 5G). Interestingly, binding of Smad 8 to endogenous GATA4 did not change with the addition of BMP-6. These results along with the observation that overexpression of Smad 1/4 (but not Smad 8) led to an induction of IL-6 expression by BMP-6 (Fig. 3C) demonstrate that GATA4 and Smad 1/4 are part of the transcription machinery that drives the expression of IL-6 in response to BMP-6. To investigate interaction between endogenous Smad 1 and GATA4 further, shRNA knockdown experiments were performed. When Smad1 was knocked down, immunoprecipitation for GATA4 followed by immunoblotting against Smad 1 demonstrated no protein band (Fig. 5H, top panel). Conversely, when GATA4 was knocked down, immunoprecipitation for Smad 1 followed by immunoblotting against GATA4 revealed no positive protein band (Fig. 5H, bottom panel).
Based on these results, we propose a new mechanism of BMP-6 signaling in macrophages (Fig. 5I). In this model, BMP-6 induces macrophages to produce IL-6 expression via a cross-talk between the canonical Smad-dependent and the noncanonical p38 pathways. The actual interaction between the two pathways occurs through GATA4 and Smad 1/4 in which GATA4 and Smad 1/4 cooperate to activate IL-6 promoter in the nucleus of macrophages.
DISCUSSION
In the present study, we have demonstrated that BMP-6 dramatically increased the expression level of IL-6 in a concentration- and time-dependent manner in macrophages. Subsequent studies revealed a novel mechanism of BMP-6 signaling in which a cross-talk between the canonical Smad-dependent and the noncanonical p38 pathways is required for IL-6 induction by BMP-6 in macrophages. Taken together, these observations have uncovered a novel mechanism of BMP-6 signal transduction.
To date, more than 20 subtypes of BMPs have been identified. Based on various knock-out studies, BMPs have differing functions (2), yet, only six BMP receptors, three each for type I and II, have been identified (3). Given that BMP receptors are promiscuous and the same BMP receptor complex binds to multiple BMP subtypes, the mechanism of ligand specificity is unclear. In the present study, IL-6 expression is induced by BMP-6 but not by BMP-2, -4, and -7. Further studies have demonstrated that IL-6 induction by BMP-6 requires the combination of BMP-RII and ALK-2, followed by the phosphorylation of Smad 1 and p38. These observations are consistent with studies that demonstrated a differing affinity among the various BMPs, BMP receptors, and R-Smads (18). Thus, it is likely that the ligand-specific effects of BMPs are mediated in part by the varying optimal ligand/receptors/R-Smad combination.
In the context of type I receptors, ALK-2 was the receptor that induced the expression of IL-6 promoter activity the most. However, ALK-3 also increased IL-6 expression level modestly. This observation suggests that for a given BMP function, there is an optimal receptor. But in the absence of the optimal receptor, the remaining receptor subtype(s) may substitute partly for the missing receptor. This concept requires further investigations for confirmation.
Interestingly, the overexpression of BMP-RII increased whereas that of Act-RIIB decreased the expression of IL-6 upon stimulation with BMP-6. Because different BMP ligands have different binding affinity for each of the type I and type II receptors, it is plausible that the overexpression of Act-RIIB may, in essence, function as a competitive inhibitor of BMP-RII in the context of IL-6 expression. This view that the stoichiometry of the receptors dictates the optimal ligand-receptors interaction is consistent with the report demonstrating that knocking out the expression of BMP-RII attenuated BMP-2 and -4 signaling and augmented BMP-6 and -7 signaling (19). Additional work is necessary to verify this concept.
Despite the varying effects of each BMP subtype, the common theme in BMP signal transduction is the activation of the Smads. Specifically, the canonical BMP signaling pathway involves the R-Smads (Smad 1, 5, and 8) and the Co-Smad (Smad 4). Upon receptor activation by the ligand, the R-Smads are phosphorylated, form heteromeric complexes with Co-Smad, and translocate into the nucleus to regulate gene expression. In addition to this Smad-dependent pathway, the existence of non-Smad pathways has been reported (20). Now, the observation that both p38 and Smad pathways are required for BMP-6 signaling has revealed a novel mechanism of BMP signal transduction in which two signaling pathways interact at the transcriptional level to activate the target gene expression. In this work, we have demonstrated that GATA4, a transcription factor implicated in development and heart formation (16), and Smad 1 cooperate to activate IL-6 promoter. Because the BMP-6-responsive region in IL-6 promoter contains the consensus GATA but not the Smad-binding element, it is likely that GATA4 interacts directly with the promoter whereas Smad 1 is part of the transcription complex that drives IL-6 expression. We cannot rule out though, the possibility of direct binding between Smad 1/4 and IL-6 promoter at nonconsensus Smad-binding element sites. Regardless, this cross-talk between the Smad and p38 pathways adds another point of regulation in BMP signaling in which the host regulates the function of BMPs through the non-Smad pathway.
The observation that a cross-talk between the canonical and the noncanonical pathways is required for BMP-6 signaling may explain, in part, the multifunctional effect of BMPs. Since the initial identification of BMPs, it has been reported that these growth factors have varying effects depending on the cellular context (2). The precise mechanism for these variable effects has been difficult to explain because the activation of Smad-dependent pathway is nearly universal in cells that are responsive to BMPs. Results of the present study revealed the critical role of the noncanonical non-Smad pathway in regulating the expression of certain target genes. Because there are multiple different non-Smad signaling pathways and BMPs do not uniformly activate all the non-Smad pathways (20), the effect of BMPs may be determined by the presence/absence of the noncanonical pathways. In short, the ultimate effect of BMPs may be determined by the non-Smad pathway where the canonical Smad-dependent pathway plays a permissive role.
It should be pointed out that both Smad 4 and 8 also complexed with GATA4. The implications of this interaction are yet to be established. Overexpression of dominant negative Smad 4 completely blocked IL-6 expression. Smad 4 is the lone Co-Smad and likely plays a role in translocation of the R-Smads (Smad 1 in the case of the IL-6 promoter). More recently though, nuclear translocation of R-Smads has been reported to occur independently of Co-Smad (21, 22). Once in the nucleus, Smad 4 has been reported to activate gene transcriptional activity (23). Because the nuclear translocation activity and the transcriptional function of Smad 4 cannot be uncoupled in our experimental setting, the precise biological effect of the GATA4-Smad 4 interaction is uncertain at the present time. Nevertheless, ChIP demonstrated increased binding of Smad 4 to the BMP-6-response element in the IL-6 promoter upon addition of BMP-6. Similarly, increased endogenous GATA4 was seen when immunoprecipitated against Smad 4 following stimulation with BMP-6. These results suggest that Smad 4 is also part of the transcription complex that drives IL-6 expression. With respect to Smad 8, the overexpression of Smad 8 quite surprisingly resulted in consistent suppression of IL-6 expression. In essence, Smad 8 appears to act opposite of Smad 1 with respect to the regulation of the IL-6 promoter. The precise reason for the negative effect of Smad 8 is unclear at the present time. However, ChIP assay against Smad 8 demonstrated that Smad 8 does not bind to the BMP-6-response element within the IL-6 promoter (Fig. 4F). Thus, one potential explanation is that Smad 8 may compete against Smad 1 and prevent the binding of the transcription complex to the target promoter. However, because Smad 8 did not demonstrate increased binding to GATA4 upon stimulation with BMP-6 (Fig. 5G), the possibility that the interaction between Smad 8 and GATA4 may be an artifact of the overexpression study cannot be ruled out. Currently, our laboratory is actively evaluating the interaction between GATA4 and Smad 4/8.
In conclusion, the current study demonstrates that BMP-6 induces IL-6 expression in macrophages. This induction involves a novel mechanism in which a cross-talk between p38 and Smad-dependent signaling pathways via the interaction between GATA4 and Smad 1/4 is necessary for the target gene expression. In the future, the nature of interaction between Smad 1, 4, and 8 and GATA4 as well as the upstream activator(s) of p38 in the context of BMP-6 signaling will be investigated in macrophages.
Supplementary Material
Acknowledgments
We thank Hyun Jung Koo and Han-Eol Fellow from Ewha Womans University, Seoul, Korea, for Fig. 5I and Drs. Joseph Bertino, Robert Dipaola, and Hatem Sabaawy for the invaluable discussions and editing of the manuscript.
This work has been supported in part by grants from the Ruth Estrin Goldberg Memorial Foundation, the Tanzman Foundation, and the Jon Runyan Score for the Cure, and by New Jersey Commission on Cancer Research Grant 09-1975-CCR-EO, Department of Defense Grant PC 080209, and Korea Science and Engineering Foundation Grant R11-2008-014-00001-0 funded by the Korean government Ministry of Education, Science, and Technology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. 1–5.
- BMP
- bone morphogenetic protein
- ALK
- activin-like kinase
- Co-Smad
- common mediator Smad
- DN
- dominant negative
- Luc
- luciferase
- R-Smad
- receptor-activated Smad.
REFERENCES
- 1.Wozney J. M., Rosen V., Celeste A. J., Mitsock L. M., Whitters M. J., Kriz R. W., Hewick R. M., Wang E. A. (1988) Science 242, 1528–1534 [DOI] [PubMed] [Google Scholar]
- 2.Zhao G. Q. (2003) Genesis 35, 43–56 [DOI] [PubMed] [Google Scholar]
- 3.Miyazono K., Maeda S., Imamura T. (2005) Cytokine Growth Factor Rev. 16, 251–263 [DOI] [PubMed] [Google Scholar]
- 4.Liu F., Ventura F., Doody J., Massagué J. (1995) Mol. Cell. Biol. 15, 3479–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kretzschmar M., Massagué J. (1998) Curr. Opin. Genet. Dev. 8, 103–111 [DOI] [PubMed] [Google Scholar]
- 6.Miyazono K., Kamiya Y., Morikawa M. (2010) J. Biochem. 147, 35–51 [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y. E. (2009) Cell Res. 19, 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tokuda H., Hatakeyama D., Shibata T., Akamatsu S., Oiso Y., Kozawa O. (2003) Am. J. Physiol. Endocrinol. Metab. 284, E1202–E1209 [DOI] [PubMed] [Google Scholar]
- 9.Hu M. C., Wasserman D., Hartwig S., Rosenblum N. D. (2004) J. Biol. Chem. 279, 12051–12059 [DOI] [PubMed] [Google Scholar]
- 10.Guicheux J., Lemonnier J., Ghayor C., Suzuki A., Palmer G., Caverzasio J. (2003) J. Bone Miner Res. 18, 2060–2068 [DOI] [PubMed] [Google Scholar]
- 11.Sivertsen E. A., Huse K., Hystad M. E., Kersten C., Smeland E. B., Myklebust J. H. (2007) Eur. J. Immunol. 37, 2937–2948 [DOI] [PubMed] [Google Scholar]
- 12.Kersten C., Sivertsen E. A., Hystad M. E., Forfang L., Smeland E. B., Myklebust J. H. (2005) BMC Immunol. 6, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong J. H., Lee G. T., Lee J. H., Kwon S. J., Park S. H., Kim S. J., Kim I. Y. (2010) Immunology 128, e442–e450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon S. J., Lee G. T., Lee J. H., Kim W. J., Kim I. Y. (2009) Immunology 128, e758–e765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plaisance S., Vanden Berghe W., Boone E., Fiers W., Haegeman G. (1997) Mol. Cell. Biol. 17, 3733–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brewer A., Pizzey J. (2006) Expert Rev. Mol. Med. 8, 1–20 [DOI] [PubMed] [Google Scholar]
- 17.Belaguli N. S., Zhang M., Rigi M., Aftab M., Berger D. H. (2007) Am. J. Physiol. Gastrointest. Liver Physiol. 292, G1520–G1533 [DOI] [PubMed] [Google Scholar]
- 18.Hsu M. Y., Rovinsky S., Penmatcha S., Herlyn M., Muirhead D. (2005) Cancer Metastasis Rev. 24, 251–263 [DOI] [PubMed] [Google Scholar]
- 19.Yu P. B., Beppu H., Kawai N., Li E., Bloch K. D. (2005) J. Biol. Chem. 280, 24443–24450 [DOI] [PubMed] [Google Scholar]
- 20.Derynck R., Zhang Y. E. (2003) Nature 425, 577–584 [DOI] [PubMed] [Google Scholar]
- 21.Dawes L. J., Sleeman M. A., Anderson I. K., Reddan J. R., Wormstone I. M. (2009) Invest Ophthalmol. Vis. Sci. 50, 5318–5327 [DOI] [PubMed] [Google Scholar]
- 22.Wisotzkey R. G., Mehra A., Sutherland D. J., Dobens L. L., Liu X., Dohrmann C., Attisano L., Raftery L. A. (1998) Development 125, 1433–1445 [DOI] [PubMed] [Google Scholar]
- 23.Ross S., Hill C. S. (2008) Int. J. Biochem. Cell Biol. 40, 383–408 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.