Abstract
Plasma membrane compartmentalization spatiotemporally regulates cell-autonomous immune signaling in animal cells. To elucidate immediate early protein dynamics at the plant plasma membrane in response to the bacterial pathogen-associated molecular pattern (PAMP) flagellin (flg22) we employed quantitative mass spectrometric analysis on detergent-resistant membranes (DRMs) of Arabidopsis thaliana suspension cells. This approach revealed rapid and profound changes in DRM protein composition following PAMP treatment, prominently affecting proton ATPases and receptor-like kinases, including the flagellin receptor FLS2. We employed reverse genetics to address a potential contribution of a subset of these proteins in flg22-triggered cellular responses. Mutants of three candidates (DET3, AHA1, FER) exhibited a conspicuous defect in the PAMP-triggered accumulation of reactive oxygen species. In addition, these mutants showed altered mitogen-activated protein kinase (MAPK) activation, a defect in PAMP-triggered stomatal closure as well as altered bacterial infection phenotypes, which revealed three novel players in elicitor-dependent oxidative burst control and innate immunity. Our data provide evidence for dynamic elicitor-induced changes in the membrane compartmentalization of PAMP signaling components.
Keywords: Pathogen-associated Molecular Pattern (PAMP), Plant, Plasma Membrane, Proteomics, Reactive Oxygen Species (ROS), 14N/15N-Metabolic Labeling, Detergent-resistant Membranes, flg22/FlS2, Membrane Rafts, Quantitative Proteomics
Introduction
To cope with the great number of potential pathogens, plants evolved specialized pattern recognition receptors (PRRs)5 through which they detect pathogen-associated molecular patterns (PAMPs) at the cell surface (1). Within seconds to minutes after PAMP perception manifold intracellular responses occur, including ion fluxes across the plasma membrane (PM), increase of cytosolic Ca2+ levels, production of reactive oxygen species (ROS) and protein phosphorylation. At later time points profound transcriptional changes, stomatal closure as well as local cell wall reinforcement take place (2).
The best characterized plant PAMP perception system is the recognition of bacterial flagellin and its elicitor-active epitope, flg22, by the Arabidopsis PRR FLS2 (flagellin sensitive 2; (2)). FLS2 undergoes flg22-induced complex formation with BRl1-associated receptor kinase 1 (BAK1), which precedes and is required for FLS2 endocytosis (2, 3). Indeed, ligand-induced reduction in lateral membrane mobility of FLS2 has been observed in protoplasts (4), which could be explained by either ligand-dependent interactions of FLS2 with e.g. BAK1, the confinement of FLS2 to less mobile membrane compartments, or a combination of both. To ensure adequate perception of PAMPs and tightly regulated downstream signaling, the PM must be spatially highly organized and dynamic. In this context, the recruitment of FLS2 to specialized membrane domains seems crucial to enable ligand-induced endocytosis (5).
During the past years, lateral compartmentalization has become a well-recognized topic in plant membrane research (6). The membrane raft hypothesis provides a plausible explanation for the spatial and temporal organization of biological membranes based on the tight interaction between sterols and sphingolipids. Proteins are believed to associate with membrane rafts in a dynamic manner, allowing stimulus-induced alterations in the raft proteome. Even though the membrane raft hypothesis is still a matter of debate, few researchers doubt the existence of large-scale lateral membrane compartmentalization (7). The most widely used method to study membrane rafts is the isolation of detergent-resistant membranes (DRMs, (8)). The differential solubilization of membrane proteins points to a different membrane environment and/or a differential affinity of proteins to certain lipids. The most meaningful application of DRM extraction is achieved when differences in DRM composition are induced by a biological stimulus (9). DRMs from plant tissues harbor a similar repertoire of proteins as those from animal cells (6, 8).
In this study we applied quantitative proteomics based on 15N/14N-labeled Arabidopsis cells to quantify immediate early responses at the PM following flg22 stimulation. We focused our analysis on changes in the DRM proteome to address the role of induced membrane compartmentalization. This approach revealed 64 proteins that showed significant enrichment in the DRM fraction within 15 min. We employed reverse genetics and pharmacological interference to unravel a potential contribution of these proteins in flg22-induced responses and innate immunity. These experiments identified three novel components that play a role in elicitor-dependent processes and defense against bacterial invasion.
EXPERIMENTAL PROCEDURES
Metabolic Labeling of Suspension Cell Cultures
Full metabolic 15N/14N-labeling of Arabidopsis thaliana (Col-0) suspension cell cultures was carried out as described (10). Briefly, for the 15N-labeled cell cultures the conventional 14N-containing nitrogen source in the medium (K14NO3) was replaced with K15NO3 (Sigma-Aldrich) as the only nitrogen source, yielding a fully 15N-labeled proteome within 2 weeks of growth in the labeling medium.
Experimental Set Up
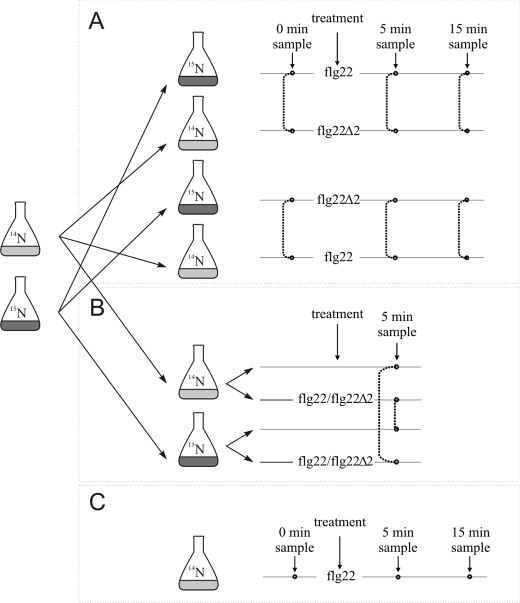
The experimental design is outlined in Fig. 1. 15N- and 14N-labeled parent cell culture suspensions were split up for reciprocal sample pairs and either treated with active flg22 or antagonistic flg22Δ2 (EZBiolab) at 100 nm final concentration. Samples were taken before (0 min) as well as 5 and 15 min after induction. Additionally, cell culture suspensions were treated with flg22 or flg22Δ2 and compared with untreated cells, also in reciprocal pairs. Samples were taken 5 min after peptide addition. After harvesting, equal amounts (gram fresh weight) of labeled and unlabeled cells were pooled for combined protein extraction, DRM preparation, and mass spectrometric analysis.
FIGURE 1.
Schematic representation of the experimental setup. A and B, parental 14N and 15N cultures were split for reciprocal treatment. A, flg22 treatment was compared with flg22Δ2 treatment (in reciprocal pairs). B, flg22 as well as flg22Δ2 treatments were compared with untreated cells (including reciprocal pairs). Samples for DRM extraction and subsequent ratiometric protein quantification were taken before treatment (0 min) as well as 5 and 15 min after peptide addition. Dotted lines indicate reciprocal sample pairs that were extracted and analyzed together. C, label-free quantification.
Plasma Membrane Preparation and Detergent-resistant Membrane Extraction
PM preparation and DRM extraction were performed as described (11). Briefly, PM fractions were isolated by two-phase partitioning, and the protein amount was determined (51). PMs were resuspended in buffer (50 mm Tris-HCl, pH 7.5, 3 mm EDTA) and treated with Triton X-100 at a protein to detergent ratio of 1:13 (final concentration 1%) for 30 min on ice while continuously shaking. Solubilized PM extracts were adjusted to a final concentration of 1.8 m sucrose, overlaid with a sucrose step gradient (1.6, 1.4, and 0.15 m sucrose) and centrifuged at 250,000 × g for 18 h. An opaque ring (DRM fraction; see also Fig. S3) was collected from below the 1.4 m/0.15 m interface. All steps were carried out at 4 °C.
DRM pellets were denatured in 6 m urea and 2 m thiourea and subsequently reduced in 0.5 mm dithiothreitol. Cysteine groups were alkylated in iodoacetamide and proteins were digested with endoproteinase Lys-C (Wako Chemical) and trypsin (Promega). Digested peptides were desalted over C18 STAGE-tips before mass spectrometric analysis (12).
Mass Spectrometry and Protein Identification
Tryptic peptide mixtures were analyzed by LC/MS/MS using nanoflow HPLC (Proxeon Biosystems) and a linear ion trap instrument (LTQ-Orbitrap, Thermo Scientific) as mass analyzer. Peptides were eluted from a 75 μm analytical column (Reprosil C18, Dr. Maisch GmbH) on a linear gradient running from 10–30% acetonitrile in 50 min and sprayed directly into the LTQ-Orbitrap mass spectrometer. Proteins were identified by tandem mass spectrometry (MS/MS) via information-dependent acquisition of fragmentation spectra of multiple-charged peptides. Full scans were obtained at a resolution of FWHM (full width at half-maximum) of 60,000 and CID fragment spectra were acquired in the LTQ. Additional fragmentation through multistage activation was used if peptides displayed a loss of phosphoric acid (neutral loss, 98 Da) upon MS/MS fragmentation. Fragment MS/MS spectra from raw files were extracted as DTA-files and then merged to peak lists using default settings of DTASuperCharge version 1.19 with a tolerance for precursor ion detection of 50 ppm.
Spectra were searched against a non-redundant Arabidopsis protein data base (TAIR8, version 2008–04; 31921 entries) using the Mascot algorithm (version 2.2.0; Matrix Science). The database contained the full Arabidopsis proteome and commonly observed contaminants (human keratin, trypsin, lysyl endopeptidase), thus no taxonomic restrictions were used during automated data base search. The following search parameters were applied: Trypsin as cleaving enzyme, peptide mass tolerance 10 ppm, MS/MS tolerance 0.8 Da, one missed cleavage allowed. Carbamidomethylation of cysteine was set as a fixed modification, and methionine oxidation and phosphorylation of serine, threonine, and tyrosine were chosen as variable modifications. Only peptides with a length of more than five amino acids were considered. MS spectra for protein identification were deposited in the PRoteomics IDEntifications (PRIDE) database, accession numbers 10723–10725).
Quantitative Protein Analysis
For comparative proteomic analysis, reciprocal labeling experiments were set up using 15N-labeled and unlabeled (14N) cell cultures (Fig. 1). The reciprocal labeling setup was chosen rather than an experimental setup using the same 15N-labeled cultures as repeated internal standard to specifically distinguish which proteins are responding to the treatment with flg22 or flg22Δ2 (treatment effects) from those proteins that are a priori different between the 15N-labeled and unlabeled cell cultures (culture effects, Ref. 11).
Intensity ratios of the 15N- to 14N-form of each identified peptide were averaged across all peptides belonging to the same protein within one experimental set. Protein abundance ratios were converted into log2 values and were normalized to the median log2 ratio of all proteins identified in the non-treated sample (0 min). Only those proteins for which intensity ratios were obtained in both of the paired reciprocal experimental sets were considered for further analysis.
The data analysis workflow is based on first determining the variation between cultures based on 15N/14N ratios in independent 1:1 mixtures (Fig. 1) before any treatment is applied. The ratios in two control experiments show normal distribution (supplemental Fig. S2A) and are used to define ratio-dependent standard deviations (11). In a second step, the distances to the diagonal in a graphic display of ratios in reciprocal experiments (supplemental Fig. S2, B and C) were calculated. Proteins, for which the ratios of 15N-form to 14N-form in the two reciprocal experiments lie on a 45° diagonal, are those proteins, which show inherent variation between 15N and 14N cell culture. Proteins with reciprocal behavior, i.e. with high ratios in one of the reciprocal experiments and low ratios in the other, lie away from this diagonal. Using this information, for each data point the p value was determined by a 2-tailed t-distribution (11), and a multiple testing correction was applied to the whole data set using the false discovery rate (FDR) method introduced by Benjamini and Hochberg (13). Reported proteins correspond to a cut-off FDR of 5%. By this method, proteins with clear reciprocal behavior (i.e. high ratios in one experiment and low ratios in the reciprocal experiment) were determined as significant (red symbols in supplemental Fig. S2, B and C).
For label-free protein quantification, separate LC-MS/MS runs were performed for protein extracts treated with flg22 for either 15 or 5 min and respective control cells (either untreated or treated with inactive flg22Δ2). For each protein, ion intensities of all observed charge states of peptides belonging to that protein were extracted from the different LC-MS/MS runs and normalized to total ion intensities per run. Ratios were calculated for each peptide separately based on normalized ion intensities, and peptide ratios were subsequently averaged to obtain protein ratios. In total, two independent cell culture sets were analyzed.
Immunoblot Analysis and Bioassays
Immunoblot analysis of FLS2 in PM-derived DRMs and mutant seedlings, the oxidative burst assay, mitogen-activated protein kinase (MAPK) activity assay, measurement of stomatal aperture, callose (aniline blue) and cell death (trypan blue) staining as well as the bacterial infection assays are described in detail in the supplemental Methods.
RESULTS
Quantification of flg22-triggered Alterations in DRM Composition
To elucidate rapid dynamic changes in PM compartmentalization after flg22 elicitation, we performed quantitative mass spectrometric analyses on PM-derived cell culture DRMs in a time-course experiment. To allow ratiometric quantification of protein levels we employed full 15N/14N metabolic labeling. Since ligand-induced endocytosis of FLS2 takes place as early as 15–20 min after flg22 elicitation (3), PAMP-induced membrane compartmentalization is expected to occur within or even prior to this time frame. Changes in protein abundance due to de novo protein biosynthesis can be largely excluded at these early time points (14, 15). We induced cell cultures with flg22 (100 nm) or control treatment and collected samples after 0, 5, and 15 min (Fig. 1). We previously verified responsiveness of the cell culture to flg22 in an oxidative burst assay (supplemental Fig. S1). In one experimental setup differentially labeled pairs of cell cultures were either induced with flg22 or the antagonistic flg22 derivative, flg22Δ2 (Fig. 1A; Ref. 16). An additional paired sample set with the identical treatment but reciprocal 15N/14N labeling was included. In a second setup, flg22 and flg22Δ2 treatments were each compared with untreated cells (Fig. 1B), again using two pairs of cell cultures with reciprocal 15N/14N labeling. In experiments, PM fractions of pooled 15N- or 14N-labeled treatment and control samples were extracted by two-phase partitioning and subsequently DRMs were isolated by Triton X-100 treatment and sucrose gradient centrifugation. After liquid chromatography coupled to mass spectrometry (LC/MS/MS) analysis proteins were identified based on the fragmentation patterns of peptides using automated data base matching algorithms and ratiometrically quantified (11). With this workflow we efficiently filtered out the between-sample variation and at the same time were able to detect subtle stimulus-induced differences in protein abundance.
Characteristics and Functional Classification of Identified Proteins
Based on the procedure outlined above, we identified 316 unique proteins. These overlapped extensively with the reported inventory of DRM-associated plant proteins (6). 188 of the 316 proteins were present in reciprocal sample pairs and thus met our criteria for quantitative analysis (supplemental Table S1). Histograms of log2-transformed 15N/14N ratios matched Gaussian fits for each tested treatment and time point, indicative of normal distribution of the data sets (supplemental Fig. S2A). Of the 188 proteins suitable for quantification, 34% (n = 64) were significantly enriched in DRMs after flg22 treatment in a least one reciprocal sample pair (p < 0.05; Table 1, and supplemental Fig. S2, B and C, for detailed information see supplemental Table S1). Unaltered abundance of the majority (66%) of the DRM proteins suggests that the PAMP-induced shift in DRM protein abundance is specific and not an artifact caused by a change in phase partitioning behavior after flg22 elicitation. Based on prediction by the ARAMEMNON data base the majority of the 188 proteins (73%) found in the DRM fraction possess at least one transmembrane (TM) domain (56%), a glycosylphosphatidylinositol-anchor (12%) or a lipid modification (5%) (Table 1). Importantly, only 0–2% of the proteins responded in a statistically significant manner in the control samples (either treated with the antagonistic flg22Δ2 peptide or in 1:1 mixtures of untreated cells). We thus conclude that we successfully identified proteins that specifically respond to flg22 exposition. An independent experiment with two replicates using unlabeled cell cultures and quantifying protein abundance based on ion intensities at 0, 5, and 15 min after flg22 or flg22Δ2 treatment corroborated enrichment (with a similar fold change) of the majority of the proteins identified in the reciprocal 15N/14N label experiments (Fig. 1C and supplemental Table S1).
TABLE 1.
Responding proteins
For all proteins present in both samples of a reciprocal pair, quantitation was pursued. Proteins significantly enriched in DRMs after flg22 treatment are indicated in bold (p < 0.05). Functional category (FC); Arabidopsis Genome Initiative code (AGI code); average fold-change (av fold); number of TM domains based on the consensus predicted by ARAMEMNON (TM, (17)); experimental evidence for PM association (PM, (17–20)); transcriptionally co-expressed with FLS2 (46), number indicates rank of co-expressed gene according to ATTED (ATTED); elevated transcript levels in response to flg22 treatment (flg22 up, (14, 21)); phosphorylated after flg22 treatment (P flg22, (15, 22)); (putative) mutants of according genes were analyzed for flg22 responsiveness in this study (MA); flg22-induced ROS (ROS). enriched (enr.), dephosphorylated (de-p), phosphorylation below the significance threshold (✓), not germinated (ng), no ROS = 1, weak ROS = 2, wild-type ROS = 3, ROS higher than wild-type = 4.
| FC AGI code and annotation | flg22 vs flg22Δ2 |
flg22 vs untreated | flg22Δ2 vs untreated | TM | Expression analysis |
MA | ROS | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 min av fold | 5 min av fold | 15 min av fold | 5 min av fold | 5 min av fold | PM | ATTED | flg22 up | P flg22 | ||||
| Signaling receptor-like kinase | ||||||||||||
| AT5G46330 FLS2 (flagellin-sensitive 2) | 1,9 | 1,4 | 1,5 | 1 | 1 | ✓ | fls2 | 1 | ||||
| AT3G17840 RLK902 (receptor-like kinase 902) | 0,8 | 1,9 | 1,7 | 1 | ||||||||
| AT3G51550 FER (feronia) | 1,0 | 1,8 | 1,3 | 1 | enr. | 158 | fer | 4 | ||||
| AT3G02880 LRR transmembrane protein kinase, putative | 0,9 | 1,5 | 1,0 | 1,3 | 1,0 | 1 | enr. | ✓ | ||||
| AT5G16590 LRR transmembrane protein kinase, putative | 1,0 | 1,6 | 1,1 | 1,2 | 1 | enr. | ||||||
| AT2G01820 LRR protein kinase, putative | 1,4 | 1,7 | 2,9 | 1,2 | 1 | N526322 | 3 | |||||
| AT4G36180 LRR family protein | 1,2 | 2,2 | 1 | N800016 | 3 | |||||||
| N800009 | 3 | |||||||||||
| AT1G75640 LRR family protein/protein kinase family protein | 0,6 | 1,4 | 1 | N800023 | 3 | |||||||
| AT3G23750 LRR family protein/protein kinase family protein | 0,9 | 1,6 | 1,3 | 1 | ✓ | |||||||
| AT3G46290 HERK1 (herkules receptor kinase 1) | 0,9 | 1,6 | 1,0 | 1,3 | 1,3 | 1 | 224 | ✓ | N657488 | 2 | ||
| N530215 | 3 | |||||||||||
| AT1G11330 S-locus lectin protein kinase family protein | 2,1 | 1 | N520904 | 3 | ||||||||
| N677232 | 3 | |||||||||||
| Other kinases | ||||||||||||
| AT4G04720 CPK21 (calcium-dependent protein kinase 21) | 1,0 | 1,9 | 1,2 | 0–1/myr enr. | N529412 | 2 | ||||||
| AT5G24010 protein kinase family protein | 0,9 | 1,7 | 1,5 | 1,4 | 1,0 | 1 | ||||||
| Other signalling proteins | ||||||||||||
| AT1G05150 calcium-binding EF hand family protein | 0,9 | 1,4 | 1,1 | 1,4 | 0 | ✓ | ||||||
| Transport Plasma membrane ATPases | ||||||||||||
| AT2G18960 AHA1 (H(+)-ATPase 1) | 0,9 | 1,7 | 1,3 | 1,5 | 1,0 | 10 | ✓ | de-p | ost2-1D | 2 | ||
| N657956 | 3 | |||||||||||
| N658025 | 3 | |||||||||||
| AT4G30190 AHA2 (H(+)-ATPase 2) | 1,0 | 1,8 | 1,2 | 1,4 | 1,0 | 10 | enr. | de-p | ||||
| AT5G57350 AHA3 (H(+)-ATPase 3) | 4,7 | 2,7 | 10 | enr. | N662816 | 2 | ||||||
| AT3G47950 AHA4 (H(+)-ATPase 4) | 2,0 | 1,1 | 1,0 | 10 | ✓ | N657917 | 3 | |||||
| Calcium-transporting ATPases | ||||||||||||
| AT5G57110 ACA8 (autoinhibited Ca2+- ATPase 8) | 1,0 | 1,5 | 1,1 | 1,4 | 1,0 | 8–9 | enr. | |||||
| AT4G29900 ACA10 (autoinhibited Ca2+-ATPase 10) | 1,0 | 1,6 | 1,2 | 1,2 | 9 | ✓ | ✓ | ✓ | ||||
| Vacuolar H(+)-ATPases | ||||||||||||
| AT3G28715 VHA-d2 | 1,2 | 1,5 | 1,5 | 0 | ||||||||
| AT3G28710 VHA-d1 | 1,1 | 1,4 | 1,3 | 1,0 | 0 | ✓ | ||||||
| AT4G39080 VHA-a3 | 1,1 | 1,6 | 1,6 | 1,0 | 6 | |||||||
| AT1G78900 VHA-A | 1,0 | 1,6 | 1,3 | 0–1 | enr. | |||||||
| AT2G21410 VHA-a2 | 1,3 | 1,4 | 1,7 | 1,3 | 6 | |||||||
| AT4G11150 VHA-E1 | 1,0 | 1,8 | 1,3 | 1,3 | 1,0 | 0 | ✓ | |||||
| AT1G76030 VHA.B1 | 0,6 | 1,4 | 1,7 | 0,9 | 0 | ✓ | ||||||
| AT3G58730 VHA-D | 1,0 | 2,0 | 1,4 | 0 | ✓ | |||||||
| AT3G42050 VHA-H | 1,6 | 1,4 | 0 | ✓ | ||||||||
| AT1G12840 VHA-C/DET3 (de-etiolated 3) | 1,6 | 0 | det3 | 2 | ||||||||
| N675700 | 3 | |||||||||||
| N667157 | 3 | |||||||||||
| ABC transporter | ||||||||||||
| AT2G36910 PGP1 (P-glycoprotein 1) | 1,8 | 1,3 | 1,2 | 2,1 | 10 | ✓ | N676004 | 3 | ||||
| AT2G47000 PGP4 (P-glycoprotein 4) | 1,0 | 1,8 | 1,2 | 1,3 | 12 | ✓ | N16269 | 3 | ||||
| N657718 | 3 | |||||||||||
| Other transporter | ||||||||||||
| AT5G50200 WR3 (wound-responsive 3); nitrate transporter | 1,1 | 2,0 | 1,3 | 1,3 | 1,0 | 1 | ||||||
| AT4G13510 AMT1;1 (ammonium transport 1) | 0,9 | 1,8 | 1,5 | 1,3 | 0,9 | 12 | ✓ | ✓ | ✓ | N606389 | 3 | |
| N526874 | 3 | |||||||||||
| AT1G11260 STP1 (sugar transporter 1) | 1,0 | 1,6 | 1,2 | 1,4 | 1,0 | 12 | ✓ | 32 | N654185 | 3 | ||
| N661172 | ng | |||||||||||
| AT3G19930 STP4 (sugar transporter 4) | 1,1 | 1,6 | 1,2 | 1,3 | 1,2 | 12 | ✓ | |||||
| AT4G21120 AAT1 (cationic amino acid transporter 1) | 1,1 | 1,4 | 14 | N655701 | 3 | |||||||
| N668361 | 3 | |||||||||||
| AT5G40780 LHT1 (lysine histidine transporter 1) | 2,4 | 1,1 | 1,5 | 11–12 | ✓ | ✓ | N673254 | 3 | ||||
| AT3G54140 PTR1 (peptide transporter 1) | 0,9 | 1,9 | 1,2 | 1,5 | 0,8 | 11 | ✓ | N859493 | 3 | |||
| N648600 | 3 | |||||||||||
| Cell wall-related | ||||||||||||
| AT1G03870 FLA9 (fasciclin-like arabinogalactan 9) | 1,1 | 1,1 | 1,6 | 0–1 | ✓ | |||||||
| AT4G12420 SKU5 (skewed 5); copper ion binding | 0,7 | 1,6 | 4,6 | 1,7 | 1,1 | 0/GPI | ✓ | |||||
| AT1G05570 CALS1/GSL6 (callose synthase 1) | 1,5 | 16 | 401F09 | 3 | ||||||||
| 867B07 | 3 | |||||||||||
| AT4G03550 GSL05/PMR4 (glucan synthase-like 5)a | 1,0 | 1,8 | 1,5 | 1,4 | 14 | ✓ | (✓) | pmr4-1 | 3 | |||
| Intracellular trafficking | ||||||||||||
| AT3G09740 SYP71 (syntaxin of plants 71) | 1,1 | 2,2 | 1,4 | 1,6 | 1 | ✓ | ||||||
| AT1G32050 SCAMP4 (secretory carrier-associated membrane protein 4) | 1,0 | 1,8 | 1,2 | 1,3 | 1,0 | 4 | ✓ | N25052 | 3 | |||
| N859638 | 3 | |||||||||||
| Metabolism | ||||||||||||
| AT3G16860 COLBL8 (cobra-like protein 8 precursor) | 0,9 | 1,6 | 1,4 | 0/GPI | ✓ | |||||||
| AT3G25290 auxin-responsive family protein | 1,2 | 1,9 | 1,1 | 5 | ✓ | |||||||
| AT4G12980 auxin-responsive protein, putative | 1,0 | 1,7 | 1,5 | 1,2 | 5 | ✓ | N657528 | 3 | ||||
| N668432 | 3 | |||||||||||
| AT3G07570 membrane protein, putative | 1,2 | 1,6 | 1,5 | 5–6 | N660482 | 2 | ||||||
| N659517 | 3 | |||||||||||
| AT1G73650 expressed protein | 1,4 | 2,4 | 1,3 | 1,6 | 4 | ✓ | N664946 | 3 | ||||
| Stress/Redox | ||||||||||||
| AT5G06320 NHL3 (NDR1/HIN1-like 3)b | 0,9 | 1,6 | 1,4 | 1,4 | 1,0 | 1 | ✓ | ✓ | N650318 | 3 | ||
| N535427 | 3 | |||||||||||
| AT1G30360 ERD4 (early-responsive to dehydration 4) | 1,0 | 1,8 | 1,3 | 1,4 | 1,0 | 8–9 | enr. | N658161 | 3 | |||
| N658486 | 2 | |||||||||||
| AT3G54200 expressed protein; similar to harpin-induced 1 | 1,0 | 1,8 | 1,1 | 1 | ✓ | ✓ | N594113 | 3 | ||||
| N676251 | 3 | |||||||||||
| AT1G19110 inter-α-trypsin inhibitor heavy chain-related | 1,1 | 2,1 | 1,6 | 1,5 | 1,0 | 0–1 | N655025 | 3 | ||||
| N655011 | 2 | |||||||||||
| Protein modification | ||||||||||||
| AT3G05560 60S ribosomal protein L22–2 (RPL22B) | 1,1 | 0,6 | 4,5 | 0 | ||||||||
| Other | ||||||||||||
| AT2G45820 REM1.3 (remorin)c | 1,2 | 1,7 | 1,7 | 1,6 | 1,0 | 0 | enr. | ✓ | N670775 | 3 | ||
| AT3G61260 REM1.2 (remorin family protein)c | 0,8 | 1,7 | 1,2 | 1,4 | 1,1 | 0 | enr. | ✓ | N661875 | ng | ||
| AT1G72230 plastocyanin-like domain-containing protein | 1,4 | 1,3 | 1,1 | 1,1 | 0/GPI | |||||||
| Unknown | ||||||||||||
| AT1G32190 expressed protein | 0,9 | 1,9 | 1,5 | 1,4 | 1,0 | 0–1/myr | N655558 | 3 | ||||
| AT3G44150 expressed protein | 1,8 | 1,3 | 1,4 | 1 | N663376 | 3 | ||||||
| AT1G17620 expressed protein | 1,1 | 1,6 | 1,2 | 1,3 | 1,0 | 1 | ✓ | N661308 | 2 | |||
| AT3G01290 band 7 family protein | 1,0 | 1,7 | 1,3 | 1,3 | 0–1/myr | enr. | ✓ | N657289 | 3 | |||
| N668706 | 3 | |||||||||||
| AT1G69840 band 7 family protein | 1,0 | 2,0 | 1,4 | 1,4 | 0,9 | 0 | ✓ | 154 | ✓ | |||
| AT5G62740 band 7 family protein | 1,5 | 2,0 | 1,6 | 1,4 | 0 | ✓ | ||||||
b NHL3 transcript accumulation was specifically observed during the interaction with avirulent Pseudomonas syringae strains (25). NHL3-overexpressing plants are more resistant to Pseudomonas syringae (26). Interestingly, NDR1, one of the founders of the NDR1 /HIN1 -like gene family, was shown to interact with RPM1-interacting protein 4 (RIN4), a negative regulator of plant immunity (27).
Transporters and Receptor-like Kinases (RLKs) Are Most Prominently Enriched in DRMs after flg22 Stimulus
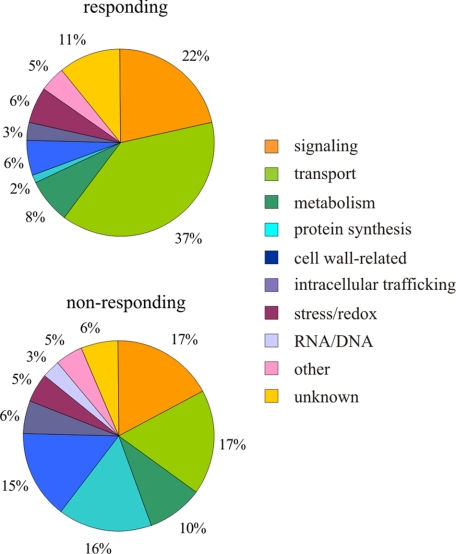
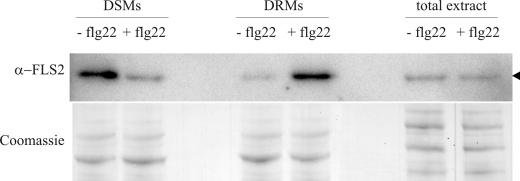
Proteins for which quantification was pursued were classified into MapMan categories (Fig. 2). The apparently large change in the category of “protein synthesis” reflects the successful exclusion of copurifying contaminants (e.g. ribosomal proteins) from the significantly responding group. The largest absolute change occurred in the category of transporters. While comprising 17% among the non-responding proteins, they accounted for 37% of the group of significantly responding proteins. Remarkably, 10 RLKs, including the flagellin receptor FLS2, are significantly enriched in DRMs upon flg22 treatment (Table 1). In fact, FLS2, which undergoes complex formation and endocytosis upon ligand binding (3, 30, 31), is one of the proteins most consistently enriched in DRMs after flg22 elicitation in our quantitative proteome analysis (significantly more abundant in all 15N/14N and label-free sample sets and displaying up to 4–5-fold enrichment). We exemplarily corroborated substantial flg22-triggered enrichment of FLS2 in DRMs by an independent immunoblot experiment (Fig. 3). This also revealed depletion of FLS2 from detergent-soluble membrane (DSM) fractions demonstrating the relocalization within the PM (Fig. 3 and supplemental Fig. S3B). The importance of this pattern recognition receptor in innate immunity is illustrated by the enhanced susceptibility of fls2 mutant plants to the bacterial pathogen Pseudomonas syringae (21). Furthermore, fls2 mutants lack the flg22-induced oxidative burst, callose deposition and stomatal closure (32). Quantitative (15N/14N) proteomic analysis revealed a trend toward depletion of four additional responding proteins and unaltered abundance of several non-responding proteins in the DSM fractions (supplemental Fig. S3). Together, these results strengthen the notion that the observed abundance of the responding proteins in the DRM fractions is the consequence of a dynamic process associated with the concomitant depletion of these proteins from the DSM fractions rather than a change in the overall PM abundance of these proteins. However, further extensive immunoblot analysis would be required to unequivocally rule out the latter possibility.
FIGURE 2.
Classification of proteins exhibiting significant redistribution into detergent-resistant membranes after flg22 elicitation (“responding”) and proteins not responding to flg22 treatment. Functional categories were assigned according to MapMan (47) and manually advanced for some proteins as described in supplemental Methods.
FIGURE 3.
FLS2 immunoblot analysis. Immunoblot showing reduced abundance of FLS2 in DSM and increased abundance of FLS2 in DRM fractions of flg22-treated cells. Cell cultures were treated with flg22 peptide for 10 min as described (+flg22) or remained untreated (−flg22). Subsequently, cell material was homogenized and DRMs were isolated. Total protein extracts of treated and untreated cells were used as a control to demonstrate unaltered overall FLS2 abundance, and Coomassie staining was employed to demonstrate equal loading.
Functional Analysis of Components Identified by the Proteomic Approach
To test whether any of the proteins identified in our proteomic analysis play a role in flg22-induced responses we employed reverse genetics and performed in planta pharmacological interference experiments. We used the occurrence of an oxidative burst, MAPK activity, stomatal closure, and the formation of callose deposits as early and late markers of flg22 responsiveness (2). (Putative) mutant lines for 57 out of the 64 genes encoding proteins enriched in DRMs after flg22 elicitation were selected (Table 1) and screened for the generation of flg22-induced extracellular ROS. Most of the tested lines retained unaltered or weakly reduced responsiveness to flg22 (Table 1).
fer, ost2-1D, and det3 Exhibit Aberrant flg22-triggered Oxidative Burst and MAPK Activity
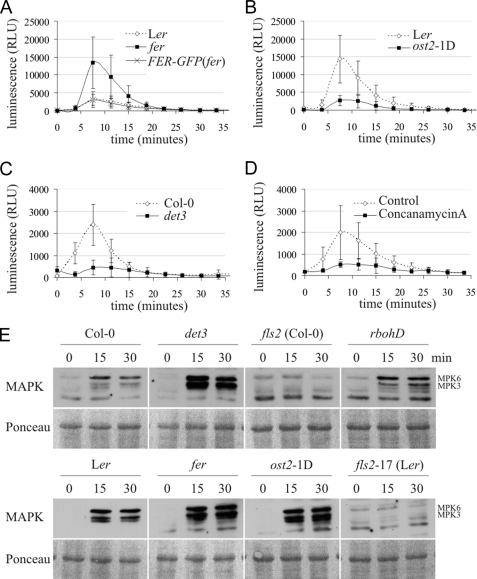
Mutants in genes FER (feronia), OST2 (open stomata 2), and DET3 (de-etiolated 3) showed either a significantly reduced (det3, ost2-1D) or enhanced (fer) accumulation of ROS (Fig. 4, A–C and supplemental Fig. S4, B–D). Immunoblot analysis revealed that these phenotypes are not the consequence of altered FLS2 protein levels (supplemental Fig. S5, A and B). FER codes for a RLK implicated in the female control of pollen tube reception, ost2-1D is a constitutively active mutant of the PM H+-ATPase AHA1 and DET3 encodes a subunit of the Vacuolar H+-ATPase (V-ATPase; (33–35)). All three mutant lines (fer, ost2-1D, and det3) exhibit defects in vegetative development at the adult stage that were, however, less pronounced at the juvenile growth stage used for the majority of our physiological assays (supplemental Fig. S6). Pharmacological interference with V-ATPase function by treatment of wild-type plants with concanamycin A, a specific inhibitor of V-ATPases, phenocopied the effect of the det3 mutant and resulted in strongly reduced generation of ROS (Fig. 4D). A transgenic complementation line expressing GFP-tagged FER in a homozygous fer background (FER-GFP(fer); (33)) restored the aberrant PAMP-induced oxidative burst of this mutant (Fig. 4A). These findings suggest that the observed alterations in flg22-triggered ROS production are genuine effects of the det3 and fer mutations and not due to second-site mutations in these lines. All three mutants (det3, fer, and ost2-1D) revealed enhanced flg22-induced activation of MAPKs (Fig. 4E), further substantiating the notion that flg22 responsiveness is perturbed in these lines.
FIGURE 4.
fer, ost2-1D, and det3 are affected in flg22-induced ROS production and MAPK activity. A–D, oxidative burst in response to 100 nm flg22 in fer (A), ost2-1D (B), det3 (C) and respective wild types was indirectly measured as relative light units (RLU). D, oxidative burst in response to 100 nm flg22 in Col-0 seedlings treated with either 5 μm concanamycin A or with respective amounts of DMSO (Control). Note the different levels of ROS production in wild-type seedlings. Relative changes were similar in all experiments. Error bars represent standard deviation of six (A), ten (B), eleven (C), and six (D) independent samples measured in a single experiment. The experiment was performed four (A) or five (B–D) times with similar results. E, MAPK activity in seedlings in response to 100 nm flg22 was determined in a time course experiment by immunoblot analysis. Ponceau staining served as a loading control. The experiment was repeated once yielding similar results.
fer Shows Aberrant Leaf Cell Death
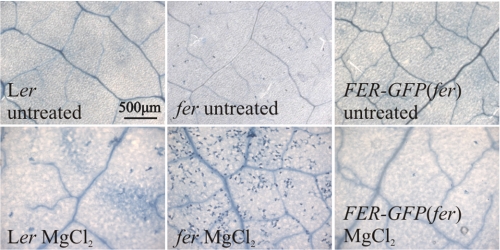
Despite aberrant flg22-induced oxidative burst and MAPK activation, flg22-triggered callose deposition was indistinguishable from wild type in rosette leaves of the det3 and ost2-1D mutants (supplemental Fig. S7). For comparison we included the rbohD (respiratory burst oxidase homolog D) mutant. Notably, although this mutant is fully devoid of any flg22-triggered oxidative burst response (supplemental Fig. S4A; (36)) MAPK activation and callose deposition still occurred under our experimental conditions (Fig. 4E and supplemental Fig. S7). In line with recent publications (e.g. Ref. 37), the extent of flg22-triggered early and late cellular responses was not correlated in our set of tested mutants (det3, ost2-1D, and fer). Rosette leaves of the fer mutant frequently displayed tissue collapse at 24 h after infiltration of flg22, which impeded accurate assessment of callose deposition and prompted us to further study potential anomalous cell death responses in this mutant. Trypan blue staining revealed weak spontaneous cell death and occurrence of pronounced aberrant cell death after infiltration of MgCl2 into fer mutant rosette leaves, but not after infiltration into leaves of control plants, suggesting that fer is hypersensitive to mechanical or osmotic stress (Fig. 5).
FIGURE 5.
fer displays aberrant cell death. fer, FER-GFP(fer), and respective wild-type rosette leaves were infiltrated with 10 mm MgCl2 and cell death was revealed by Trypan blue staining. Representative micrographs of untreated leaves or leaves 24 h after treatment are shown. Bar, 500 μm.
DET3 and FER Are Components of Plant Innate Immunity
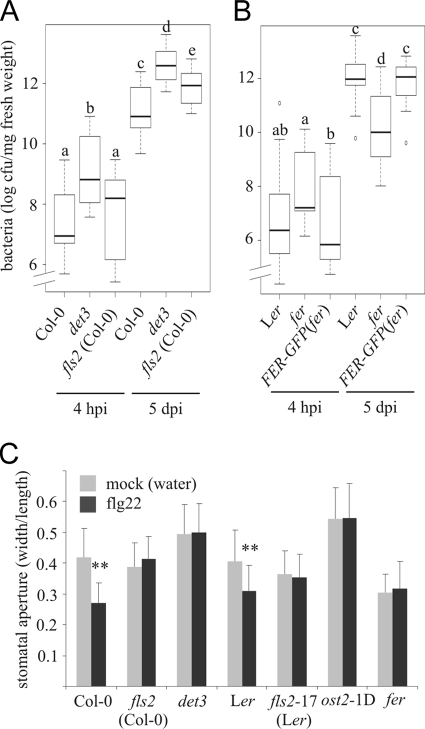
To assess whether altered flg22 responses correlate with an altered immune response, we quantitatively analyzed bacterial infection of the det3, ost2-1D and fer mutants by spray-inoculation with Pseudomonas syringae pv tomato DC3000 (PtoDC3000) ΔavrPtoΔavrPtoB. This virulence-compromised strain proliferates more slowly inside plant tissue and is expected to enable the detection of subtle differences in infection phenotypes (38). While we found no differences in bacterial growth compared with wild-type plants upon spray-inoculation of ost2-1D (supplemental Fig. S8), we observed enhanced bacterial proliferation in det3 as early as 4 hpi (hours postinoculation; Fig. 6A). Because det3 is impaired in stomatal closure upon oxidative stress (39), we hypothesized that it might also be defective in stomatal closure upon biotic stimuli, allowing enhanced stomatal entry of bacteria. We quantitatively assessed stomatal aperture of det3 in response to flg22 and observed compromised flg22-triggered stomatal closure in this mutant (Fig. 6C). Elevated bacterial titers in det3 mutant plants persisted until 5 dpi (days postinoculation) and even exceeded bacterial growth in the fls2 mutant at this time point (Fig. 6A). As expected, the ost2-1D mutant also did not exhibit flg22-triggered stomatal closure (Fig. 6C, Ref. 40). In contrast, the fer mutant had constitutively closed stomata and allowed less bacterial proliferation than wild-type plants at 5 dpi, an effect that was rescued in the FER-GFP(fer) transgenic line (Fig. 6B).
FIGURE 6.
det3 is hypersusceptible to bacterial infection and fer displays reduced bacterial proliferation. Arabidopsis det3 and fls2 (A), fer and FER-GFP(fer) (B) as well as respective wild-type plants were challenged with PtoDC3000ΔavrPtoΔavrPtoB. Depicted are box-plot diagrams representing the statistical distribution of the data. Statistical analysis (ANOVA and subsequent post-hoc test by Tukey's HSD) was done using R software. Thick lines indicate the median, boxes designate the interquartile range, whiskers specify the whole data range, and dots represent outliers. A, experiment was repeated three times, the box plot summarizes three representative data sets. B, experiment was repeated twice, the box plot includes all three data sets. Letters indicate significant differences at the level of p < 0.05. C, stomatal aperture in first true leaves of 2-week-old seedlings was determined following mock or flg22 treatment (3 μm, 2 h) as the ratio of the width to length of 20–63 stomata per genotype. Asterisks indicate a statistically significant difference (p < 0.01) between mock and flg22 treatment (Student's t test). The experiment was repeated once with similar results.
DISCUSSION
To reveal PAMP-induced protein dynamics at the PM of Arabidopsis cells we performed a quantitative proteomics approach aimed at the elucidation of early PM compartmentalization events after flg22 treatment. By ratiometric quantification we showed that PAMP treatment triggered profound changes in the protein composition of DRMs, suggesting that membrane raft association might be an important regulatory mechanism for some PM-resident proteins in this physiological context. We employed a reverse genetic approach to investigate a role of these proteins in PAMP-induced defense responses and discovered three novel components of plant innate immunity. Using a similar quantitative proteomic approach on DRMs, a new player in human B-lymphocyte signaling was previously identified (41). Likewise, stimulus-induced changes in the plant DRM proteome were reported (42–44). However, a functional significance of candidate proteins in the respective biological processes remains to be investigated.
We identified transporters and RLKs as most prominently enriched in DRMs following flg22 stimulation. Among these are FLS2 and FER, the latter originally identified as a key signaling component in the female control of pollen tube reception (33). It has recently been appointed a role in cell elongation during vegetative growth in concert with herkules receptor kinase 1 (HERK1) and theseus 1 (45). Interestingly, we also found HERK1 enriched in DRMs after flg22 treatment (Table 1). In addition to its prominent expression in synergids, FER is expressed throughout the plant (33), suggesting that its gene product might be involved in other processes. Notably, FER and HERK1 are coexpressed with FLS2 throughout a broad range of conditions (46)), and FER becomes rapidly phosphorylated in response to flg22 treatment (15). Our functional data point to a deregulation of otherwise tightly controlled cellular responses such as PAMP-induced oxidative burst, MAPK activity, stomatal aperture, and cell death in the fer mutant. The observed restriction of bacterial growth in fer plants might be caused by a deregulated cell death response that limits bacterial proliferation and/or it could be the consequence of its constitutively closed stomata. These additional mutant phenotypes suggest that FER might have a more general role, e.g. as a potential coreceptor for FLS2 and other RLKs. Its hypothesized function as coreceptor of FLS2 might explain the slightly elevated bacterial titer in fer during early infection, which is reminiscent of fls2 (21). Precedence for such a scenario is the RLK BAK1, which functions in several biological processes, e.g. as a coreceptor for FLS2 and the RLK BRI1 (brassinosteroid insensitive 1) as well as in cell death control (2).
Both H+-ATPases and Ca2+-ATPases have long been proposed to play an essential role in triggering and terminating the PAMP-triggered oxidative burst (2). In the present study we identified four PM H+-ATPases (AHA1, AHA2, AHA3, and AHA4) and two Ca2+-ATPases (autoinhibited Ca2+-ATPase (ACA) 8 and 10) significantly enriched in DRMs upon flg22 elicitation. AHA1, AHA2, and ACA10 were also shown to be rapidly dephosphorylated and phosphorylated, respectively, in response to flg22 treatment (15, 22). In the AHA1 mutant, ost2-1D, a constant hyperpolarization of the PM leads to acidification of the extracellular medium (34). Thus, the reduced production of ROS in this mutant could result from less efficient membrane depolarization leading to perturbed ion fluxes in response to elicitor treatment. Furthermore, ost2-1D is characterized by completely abolished stomatal responses following abscisic acid exposure. Consistently, ost2-1D mutants are more susceptible to PstDC3000 spray inoculation, while they are equally susceptible as wild-type plants to PstDC3000 syringe infiltration, likely because of their defect in PAMP-triggered stomatal closure (40). In our study, we did not observe enhanced susceptibility upon surface inoculation with PtoDC3000ΔavrPtoΔavrPtoB, possibly because of differences in timing, inoculation densities, or bacterial strains used. Our data suggest that the regulation of H+-ATPases besides phosphorylation and dephosphorylation (22) might also involve their recruitment to specialized membrane microdomains. This notion is further corroborated by the fact that immunolocalization studies consistently showed that plant PM H+-ATPases are present in patches at the PM (48).
V-ATPases consist of multiple subunits of which in total we identified 14 in our proteomic analysis and 10 of these underwent relocalization into DRMs after flg22 treatment. It is well accepted that V-ATPases are not only present in vacuolar membranes, but in all types of endomembranes (49) and the PM (50). Their most prominent role is the acidification of endomembrane compartments, but they also act in secretory and endocytic trafficking (49). The recurrent identification of V-ATPases in plant PM-derived DRMs also indicates additional not yet identified functions of V-ATPases at the cell periphery. The det3 mutant, affected in VHA-C, which consequently affects all V-ATPase holoenzyme complexes, lacks stomatal closure in response to extracellular calcium or oxidative stress due to abolished calcium oscillations in guard cells (39). We speculate that PAMP-induced calcium signatures might be perturbed in det3, abolishing subsequent stomatal closure and therefore facilitating bacterial hypercolonization.
In sum, our data suggest that rapid membrane compartmentalization following a PAMP stimulus might be crucial for an appropriate defense response. It remains, however, to be elucidated whether membrane raft association is critical for the function of the identified proteins in general and for their activity in plant-pathogen interactions in particular. It will also be important to unravel the molecular mechanisms underlying the dynamic partitioning of proteins into the various PM compartments.
Supplementary Material
Acknowledgments
We thank Matthew Humphry and Armin Töller for sharing seeds of T-DNA insertion lines and Anja Reinstädler, Sophia Mersmann, Nicolas Frei dit Frey, as well as Thomas Spallek for technical support.
This work was supported in part by grants from the Max Planck Society and the Deutsche Forschungsgemeinschaft (DFG; SFB670) (to R. P. and S. R.). This work was also supported by the University of Zürich and the Swiss National Science Foundation (to U. G.) and the Emmy Noether-Programm (DFG, to W. X. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8, Table S1, and Methods.
- PM
- plasma membrane
- RLK
- receptor-like kinase
- MAPK
- mitogen-activated protein kinase
- PRR
- pattern recognition receptor
- PAMP
- pathogen-associated molecular pattern
- FLS2
- flagellin sensitive 2
- DSM
- detergent-soluble membrane
- ROS
- reactive oxygen species
- DRM
- detergent-resistant membrane.
REFERENCES
- 1.Nicaise V., Roux M., Zipfel C. (2009) Plant Physiol. 150, 1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boller T., Felix G. (2009) Annu. Rev. Plant Biol. 60, 379–406 [DOI] [PubMed] [Google Scholar]
- 3.Robatzek S., Chinchilla D., Boller T. (2006) Genes. Dev. 20, 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ali G. S., Prasad K. V., Day I., Reddy A. S. (2007) Plant Cell Physiol. 48, 1601–1611 [DOI] [PubMed] [Google Scholar]
- 5.Geldner N., Robatzek S. (2008) Plant Physiol. 147, 1565–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zappel N. F., Panstruga R. (2008) Curr. Opin. Plant. Biol. 11, 632–640 [DOI] [PubMed] [Google Scholar]
- 7.van Meer G., Voelker D. R., Feigenson G. W. (2008) Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster L. J., de Hoog C. L., Mann M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lingwood D., Kaiser H. J., Levental I., Simons K. (2009) Biochem. Soc. Trans. 37, 955–960 [DOI] [PubMed] [Google Scholar]
- 10.Engelsberger W., Erban A., Kopka J., Schulze W. (2006) Plant Methods 2, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kierszniowska S., Walther D., Schulze W. X. (2009) Proteomics 9, 1916–1924 [DOI] [PubMed] [Google Scholar]
- 12.Rappsilber J., Ishihama Y., Mann M. (2003) Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 13.Benjamini Y., Hochberg Y. (1995) J. R. Stat. Soc. 57, 289–300 [Google Scholar]
- 14.Navarro L., Zipfel C., Rowland O., Keller I., Robatzek S., Boller T., Jones J. D. G. (2004) Plant Physiol. 135, 1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benschop J. J., Mohammed S., O'Flaherty M., Heck A. J., Slijper M., Menke F. L. H. (2007) Mol. Cell. Proteomics 6, 1198–1214 [DOI] [PubMed] [Google Scholar]
- 16.Bauer Z., Gómez-Gómez L., Boller T., Felix G. (2001) J. Biol. Chem. 276, 45669–45676 [DOI] [PubMed] [Google Scholar]
- 17.Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W. B., Flügge U. I., Kunze R. (2003) Plant Physiol. 131, 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandersson E., Saalbach G., Larsson C., Kjellbom P. (2004) Plant Cell Physiol. 45, 1543–1556 [DOI] [PubMed] [Google Scholar]
- 19.Marmagne A., Rouet M. A., Ferro M., Rolland N., Alcon C., Joyard J., Garin J., Barbier-Brygoo H., Ephritikhine G. (2004) Mol. Cell. Proteomics 3, 675–691 [DOI] [PubMed] [Google Scholar]
- 20.Nelson C. J., Hegeman A. D., Harms A. C., Sussman M. R. (2006) Mol. Cell. Proteomics 5, 1382–1395 [DOI] [PubMed] [Google Scholar]
- 21.Zipfel C., Robatzek S., Navarro L., Oakeley E. J., Jones J. D., Felix G., Boller T. (2004) Nature 428, 764–767 [DOI] [PubMed] [Google Scholar]
- 22.Nühse T. S., Bottrill A. R., Jones A. M., Peck S. C. (2007) Plant J. 51, 931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs A. K., Lipka V., Burton R. A., Panstruga R., Strizhov N., Schulze-Lefert P., Fincher G. B. (2003) Plant Cell 15, 2503–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishimura M. T., Stein M., Hou B. H., Vogel J. P., Edwards H., Somerville S. C. (2003) Science 301, 969–972 [DOI] [PubMed] [Google Scholar]
- 25.Varet A., Parker J., Tornero P., Nass N., Nürnberger T., Dangl J. L., Scheel D., Lee J. (2002) Mol. Plant Microbe Interact. 15, 608–616 [DOI] [PubMed] [Google Scholar]
- 26.Varet A., Hause B., Hause G., Scheel D., Lee J. (2003) Plant Physiol. 132, 2023–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim M., da Cunha L., McFall A. J., Belkhadir Y., DebRoy S., Dangl J. L., Mackey D. (2005) Cell 121, 749–759 [DOI] [PubMed] [Google Scholar]
- 28.Raffaele S., Mongrand S., Gamas P., Niebel A., Ott T. (2007) Plant Physiol. 145, 593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raffaele S., Bayer E., Lafarge D., Cluzet S., German Retana S., Boubekeur T., Leborgne-Castel N., Carde J. P., Lherminier J., Noirot E., Satiat-Jeunemaître B., Laroche-Traineau J., Moreau P., Ott T., Maule A. J., Reymond P., Simon-Plas F., Farmer E. E., Bessoule J. J., Mongrand S. (2009) Plant Cell 21, 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heese A., Hann D. R., Gimenez-Ibanez S., Jones A. M., He K., Li J., Schroeder J. I., Peck S. C., Rathjen J. P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J. D., Felix G., Boller T. (2007) Nature 448, 497–500 [DOI] [PubMed] [Google Scholar]
- 32.Gómez-Gómez L., Felix G., Boller T. (1999) Plant J. 18, 277–284 [DOI] [PubMed] [Google Scholar]
- 33.Escobar-Restrepo J. M., Huck N., Kessler S., Gagliardini V., Gheyselinck J., Yang W. C., Grossniklaus U. (2007) Science 317, 656–660 [DOI] [PubMed] [Google Scholar]
- 34.Merlot S., Leonhardt N., Fenzi F., Valon C., Costa M., Piette L., Vavasseur A., Genty B., Boivin K., Müller A., Giraudat J., Leung J. (2007) EMBO J. 26, 3216–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumacher K., Vafeados D., McCarthy M., Sze H., Wilkins T., Chory J. (1999) Genes Dev. 13, 3259–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torres M. A., Dangl J. L., Jones J. D. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu X., Tintor N., Mentzel T., Kombrink E., Boller T., Robatzek S., Schulze-Lefert P., Saijo Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 22522–22527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Göhre V., Spallek T., Häweker H., Mersmann S., Mentzel T., Boller T., de Torres M., Mansfield J. W., Robatzek S. (2008) Curr. Biol. 18, 1824–1832 [DOI] [PubMed] [Google Scholar]
- 39.Allen G. J., Chu S. P., Schumacher K., Shimazaki C. T., Vafeados D., Kemper A., Hawke S. D., Tallman G., Tsien R. Y., Harper J. F., Chory J., Schroeder J. I. (2000) Science 289, 2338–2342 [DOI] [PubMed] [Google Scholar]
- 40.Liu J., Elmore J. M., Fuglsang A. T., Palmgren M. G., Staskawicz B. J., Coaker G. (2009) PLoS. Biol. 7, e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta N., Wollscheid B., Watts J. D., Scheer B., Aebersold R., DeFranco A. L. (2006) Nat. Immunol. 7, 625–633 [DOI] [PubMed] [Google Scholar]
- 42.Fujiwara M., Hamada S., Hiratsuka M., Fukao Y., Kawasaki T., Shimamoto K. (2009) Plant Cell Physiol. 50, 1191–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minami A., Fujiwara M., Furuto A., Fukao Y., Yamashita T., Kamo M., Kawamura Y., Uemura M. (2009) Plant Cell Physiol. 50, 341–359 [DOI] [PubMed] [Google Scholar]
- 44.Stanislas T., Bouyssie D., Rossignol M., Vesa S., Fromentin J., Morel J., Pichereaux C., Monsarrat B., Simon-Plas F. (2009) Mol. Cell. Proteomics 8, 2186–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo H., Li L., Ye H., Yu X., Algreen A., Yin Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7648–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H. (2007) Nucleic Acids Res. 35, 863–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Usadel B., Nagel A., Thimm O., Redestig H., Blaesing O. E., Palacios-Rojas N., Selbig J., Hannemann J., Piques M. C., Steinhauser D., Scheible W. R., Gibon Y., Morcuende R., Weicht D., Meyer S., Stitt M. (2005) Plant Physiol. 138, 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaxiola R. A., Palmgren M. G., Schumacher K. (2007) FEBS Lett. 581, 2204–2214 [DOI] [PubMed] [Google Scholar]
- 49.Schumacher K. (2006) Curr. Opin. Plant Biol. 9, 595–600 [DOI] [PubMed] [Google Scholar]
- 50.Jefferies K. C., Cipriano D. J., Forgac M. (2008) Arch. Biochem. Biophys. 476, 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bradford M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.