Abstract
The current activation model of the EGF receptor (EGFR) predicts that binding of EGF results in dimerization and oligomerization of the EGFR, leading to the allosteric activation of the intracellular tyrosine kinase. Little is known about the regulatory mechanism of receptor oligomerization. In this study, we have employed FRET between identical fluorophores (homo-FRET) to monitor the dimerization and oligomerization state of the EGFR before and after receptor activation. Our data show that, in the absence of ligand, ∼40% of the EGFR molecules were present as inactive dimers or predimers. The monomer/predimer ratio was not affected by deletion of the intracellular domain. Ligand binding induced the formation of receptor oligomers, which were found in both the plasma membrane and intracellular structures. Ligand-induced oligomerization required tyrosine kinase activity and nine different tyrosine kinase substrate residues. This indicates that the binding of signaling molecules to activated EGFRs results in EGFR oligomerization. Induction of EGFR predimers or pre-oligomers using the EGFR fused to the FK506-binding protein did not affect signaling but was found to enhance EGF-induced receptor internalization. Our data show that EGFR oligomerization is the result of EGFR signaling and enhances EGFR internalization.
Keywords: Fluorescence Resonance Energy Transfer (FRET), Protein Assembly, Protein Phosphorylation, Receptor Endocytosis, Signal Transduction, EGF, EGF Receptor
Introduction
The EGF receptor (EGFR2; ErbB1) has an essential role in the regulation of growth and differentiation of a large range of cell types. The EGFR belongs to the ErbB family, all four members of which have been implicated in the development of different cancers (1). The first step in the signal transduction cascade is the binding of its ligand such as EGF or TGF-α to the ectodomain, which provokes receptor dimerization and oligomerization. Deletion of the dimerization domain, which is present in domain II of the EGFR ectodomain, blocks receptor activation completely, demonstrating that receptor dimerization is critical for the allosteric activation of the tyrosine kinase (2, 3). Activation of the receptor tyrosine kinase results in cross-phosphorylation of the receptors, and the phosphotyrosines in the intracellular domain serve subsequently as docking sites for adaptor proteins such as Grb2 and Shc and enzymes such as phospholipase Cγ, which contain phosphotyrosine-specific SH2 (Src homology 2) or phosphotyrosine-binding domains. Eventually, the active ligand-receptor complex becomes internalized via both clathrin-dependent and clathrin-independent pathways, followed by the intracellular transport to lysosomes, where the ligand-receptor complexes are degraded (4).
Although EGF binding and dimerization seem to be strictly connected, both microscopic and biochemical studies have demonstrated that, in resting cells, the receptor is already found on the cell surface as non-active dimers, the so-called predimers. This phenomenon was initially discovered using electron microscopy and immunogold labeling of the EGFR: in the resting cell, ∼35% of the total receptor population was present as receptor predimers (5). These observations were confirmed by chemical cross-linking and co-immunoprecipitation studies with differentially tagged EGFRs (6–8). More recently, also advanced light microscopic methods have been used to address this question. EGFR predimerization has now been demonstrated using fluorescence correlation spectroscopy, steady-state fluorescence anisotropy, FRET, and single-molecule imaging (9–15). Recent structural data showed that the dimerization of the C-terminal part of the kinase prevents kinase activation and represents a mechanism through which the EGFR tyrosine kinase is inhibited in resting cells (16). Factors controlling EGFR predimer formation, ligand-induced oligomerization, and the function of both phenomena are poorly understood.
In this study, we have used homo-FRET imaging to investigate the regulation of EGFR predimerization and oligomerization. We recently developed a homo-FRET imaging method that allows quantification of the degree of protein clustering on a subcellular level (17). It is based on FRET between identical fluorophores (homo-FRET), meaning that the nanometer proximity between identical reporter fluorophores such as GFP is detected with high sensitivity. Application of this method showed that, in the resting cell, ∼40% of the total EGFR population was already present as predimers (17). A large increase in receptor oligomerization was seen after ligand binding. EGFR predimers were formed independently of kinase activity, whereas EGF-induced receptor clustering was found to require both kinase activity and the substrate tyrosine residues. Inducing predimerization/oligomerization using FK506-binding protein (FKBP) dimerization domains demonstrated that receptor predimer formation enhances EGFR internalization.
EXPERIMENTAL PROCEDURES
Plasmid Construction
To create the FKBP-monomeric GFP (mGFP)-containing constructs, mGFP was first PCR-amplified from pEGFP-N1 using primers 5′-atatactagtatggtgagcaagggcgaggagctgttc-3′ and 5′-atatggatccttacttgtacagctcgtccatgccgagagt-3′, which introduced the flanking restriction sites SpeI and BamHI (underlined). The enhanced GFP PCR product was inserted into the corresponding sites of pC4-Fv1E (ARIAD Pharmaceuticals, Cambridge, MA) to produce pC4-Fv1E-GFP. A monomeric variant of enhanced GFP (mGFP) was constructed by sited-directed mutagenesis using primers 5′-cagtccaagctgagcaaagaccccaacgagaagcgcgatcac-3′ and 5′-gtgatcgcgcttctcgttggggtctttgctcagcttggactg-3′ (with the mutated codons in boldface) as described previously (18), resulting in the FKBP-mGFP plasmid. pcDNA3-EGFR-9YF was made by site-directed mutagenesis (Stratagene mutagenesis kit) of the human EGFR cDNA, resulting in tyrosine-to-phenylalanine transitions at positions 845, 974, 992, 1045, 1068, 1086, 1101, 1148, and 1173. The EGFR constructs were PCR-amplified with primers 5′-atatatcaattgatgcgaccctccgggacggccg-3′ and 5′-atatattctagatgctccaataaattcactgctttgtgg-3′, introducing flanking MunI and XbaI sites (underlined), and inserted into pC4-Fv1E-mGFP. To construct EGFR-mGFP and EGFR-K721A-mGFP, the FKBP domain was removed by digestion with XbaI and SpeI and self-ligation. For cells stably expressing the gene products, the EGFR constructs were subcloned into pcDNA3.1-zeo (Invitrogen). The final constructs were amplified in Escherichia coli, purified using an endotoxin-free plasmid isolation kit, and confirmed by sequencing.
Cell Culture
A431 (ATCC CRL-1555), NIH 3T3 2.2 and Her14 cells were grown in DMEM supplemented with 2 mm l-glutamine and 7.5% fetal calf serum at 37 °C in 5% CO2 under humidified conditions. Transient transfection with all constructs was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells stably expressing EGFR-FKBP-mGFP or its mutants were produced using selective growth conditions (500 μm Zeocin) and FACS. For microscopy, cells were grown on coverslips for 2 days to 50% confluency.
EGFR Internalization and Phosphorylation
The internalization rate constant (Ke) was determined as described previously (19). Briefly, cells were grown on 24-well CellBIND plates to 80% confluency and serum-starved overnight in 0.5% FCS-supplemented DMEM. Cells were incubated for 1 h with ice-cold binding medium (DMEM, 0.1% BSA, and 20 mm HEPES, pH 7.4) supplemented with either 1 μm AP20187 or 0.1% ethanol (mock). EGF (Oxford Biotechnology) was labeled with 125I by the chloramine-T method, yielding a typical specific activity of >400,000 cpm/ng. The EGb4 nanobody (15 μg) was labeled with 1 mCi of 125I in lodogen-coated glass tubes with a typical specific activity of >1500 cpm/ng. Subsequently, 1 ng/ml 125I-EGF or 125I-EGb4 was added in a total volume of 0.5 ml for 3, 6, 9, and 12 min at 37 °C. To collect surface-bound EGF, cells were incubated for 5 min with ice-cold acid wash buffer (for EGF, 150 mm NaCl and 25 mm NaOH/CH3COOH, pH 3.8; and for EGb4, 250 mm NaCl and 100 mm glycine, pH 2.5), and internalized EGF was collected in 1 m NaOH. The ratio of internalized and surface radioactivity was plotted against time, yielding the internalization rate constant (Ke).
NIH 3T3 2.2 cells were transfected using Lipofectamine 2000 with the indicated constructs and incubated overnight in 0.5% FCS-supplemented DMEM, followed by incubation with 1 μm AP20187 for 1 h, with 8 nm EGF for 10 min, or with a combination of both. After washing, cells were lysed in lysis buffer (1% Triton X-100, 100 mm NaCl, 2 mm EDTA, 50 mm Tris-HCl, pH 7.4, and CompleteTM protease inhibitor mixture). The EGFR-FKBP-mGFP constructs were immunoprecipitated using anti-GFP antibody (Roche Applied Science), size-separated by SDS-PAGE, and immunoblotting with the indicated antibodies.
Homo-FRET
Fluorescence anisotropy microscopy was performed essentially as described previously (20). A 473-nm pulsed diode laser (BDL-473, Becker & Hickl) operating at 50 MHz was directly coupled to a modified confocal scan head (C1, Nikon Instruments Europe B. V., Badhoevedorp, The Netherlands). A linear polarizer (Meadowlark, Frederick, CO) was positioned in the laser beam to define the excitation polarization direction. The microscope was equipped with a ×60 (numerical aperture = 1.20) water immersion objective (Plan Apo, Nikon). For GFP, a value of r0 = 0.38 was found instead of the theoretical value of 0.4. The emission light was split into a parallel and perpendicular channel with a broadband polarizing beam splitter cube (OptoSigma, Santa Ana, CA). The two emission channels were coupled to LIMO detection systems (Nikon) (21), equipped with an internal photon-counting photomultiplier tube. All images were recorded in a 160 × 160-pixel mode, covering an area of 50 × 50 μm. The procedures of data analysis, synchronization, and correction for sensitivity differences between the two channels were based on using reference dyes. Anisotropy analysis showed that rinf/rmono values between 1 and 0.87 were found in the monomeric situation, whereas the dimers generated values between 0.87 and 0.77 and between 0.77 and 0, with a mean value of 0.72, corresponding to oligomers. The cluster size images were calculated using anisotropy images. Binning with a factor of 2 was required to obtain sufficient signal to discriminate NAV = 1, NAV = 2, and NAV ≥ 3.
RESULTS
EGFR Forms Predimers in the Plasma Membrane
The dimerization/oligomerization state of the EGFR in the cell was analyzed by homo-FRET imaging, a method that we developed previously (17). For these studies, the EGFR was fused to a monomeric variant of enhanced GFP (A206K), which has a reduced dimerization binding affinity (KD = ∼74 mm) (18). Homo-FRET imaging was done with a confocal time-resolved fluorescence anisotropy imaging microscope (20), which allows for direct quantification of the number of fluorophores in a nanometer scale cluster (17). This method is based upon the fact that GFP monomers have high anisotropy, which decreases with increasing degree of clustering. The measured (steady-state) anisotropy (r) relates directly to cluster size (N), which, in addition, depends on the efficiency of the energy transfer and the relative orientation of the fluorophores (22). This relation is simplified when the limiting anisotropy (rinf) in the time-resolved anisotropy decay is measured instead of the steady-state anisotropy because rinf is not affected by variations in the efficiency of energy transfer (20). The calibration value for the anisotropy of GFP monomers (rmono; theoretically 0.4 but often lower due to high numerical aperture objectives) is determined using a solution of 10 μm GFP in 50:50 glycerol/buffer. At this concentration, GFP can be considered as monomeric because this concentration is well below the affinity value for GFP dimerization (KD = 110 μm) (18). To determine the degree of protein clustering in the cell, we used reference proteins that contain one copy of mGFP and one or two copies of the dimerization domain from FKBP. With these reference proteins, either dimerization or oligomerization can be induced by the addition of the ligand for the FKBP domain: AP20187 (17). Control experiments revealed that our EGFR-FKBP-mGFP constructs were not activated by the addition of AP20187 (supplemental Fig. S1).
Mouse NIH 3T3 fibroblasts, which are devoid of endogenous EGFR (clone 2.2), were stably transfected with a vector encoding mGFP or EGFR-mGFP (for an overview of all EGFR constructs used in this study, see Fig. 1A). FACS was performed to obtain comparable expression levels of the fluorescent constructs, which related to ∼50,000 proteins/cell. Control experiments demonstrated that activation of the EGFR-mGFP construct was indistinguishable from that of the wild-type EGFR, which is in agreement with other studies (15). To determine the anisotropy (rinf), at least five cells per condition were analyzed, and the average value of rinf per cell was used for statistical analysis. The results are presented as the degree of polarization (rinf/rmono). Comparison of cells expressing mGFP or EGFR-mGFP showed that EGFR-mGFP had a significant higher loss of anisotropy compared with cytoplasmic mGFP (Fig. 1B). To prove that the anisotropy loss was due to homo-FRET in receptor predimers, we recorded the average time-resolved anisotropy decay (supplemental Fig. S2). A typical homo-FRET profile was observed: an immediate drop in r (<1 ns) that leveled off to rinf, demonstrating that homo-FRET occurs with high efficiency. This is an important parameter that confirms that the reference constructs can be used. In previous work, we showed that the amount of depolarization due to homo-FRET can be directly related to the degree of clustering (17). For EGFR-mGFP in resting cells, the relative anisotropy value (rinf/rmono) was 0.89. This corresponds to a fraction of clusters of 0.4, which means that 40% of all EGFR molecules are part of a predimer or precluster.
FIGURE 1.
EGFR forms predimers in the cell membrane. A, overview of the constructs used in this study. Full-length EGFR was fused to mGFP, in some constructs preceded by one or two FKBP dimerization domains. ΔICD, intracellular domain deletion. B, mGFP or EGFR-mGFP was expressed in different cell lines: NIH 3T3 2.2 fibroblasts (devoid of endogenous EGFR), Her14 cells (NIH 3T3 2.2 fibroblasts expressing ∼3 × 105 human EGFRs/cell), and A431 cells (expressing ∼2 × 106 EGFRs/cell). Values of rinf were measured and are expressed as fraction of rmono (S.E.). *, p < 0.005.
As a control experiment, we coexpressed EGFR-mGFP in cells with a high level of endogenous EGFRs (3 × 105 and 2 × 106 receptors/cell for Her14 and A431 cells, respectively). This is based upon the idea that heterodimerization of EGFR-mGFP with endogenous EGFR does not result in homo-FRET, resulting in anisotropy values similar to the mGFP control values (10). Anisotropy analysis demonstrated that this was indeed the case: the anisotropy of EGFR-mGFP in these cells was increased in Her14 and A431 cells to the situation observed for mGFP (Fig. 1B). In addition, we analyzed whether this predimer formation was dependent upon the concentration of EGFRs by plotting the intensity values of individual pixels against their anisotropy value. No differences in anisotropy values were apparent in pixels with higher intensities (supplemental Fig. S3), indicating that EGFR predimer formation is concentration-independent. In conclusion, our data show that ∼40% of the total EGFR-mGFP population is predimerized in the plasma membrane of non-stimulated NIH 3T3 2.2 cells.
Ligand-induced Receptor Oligomerization
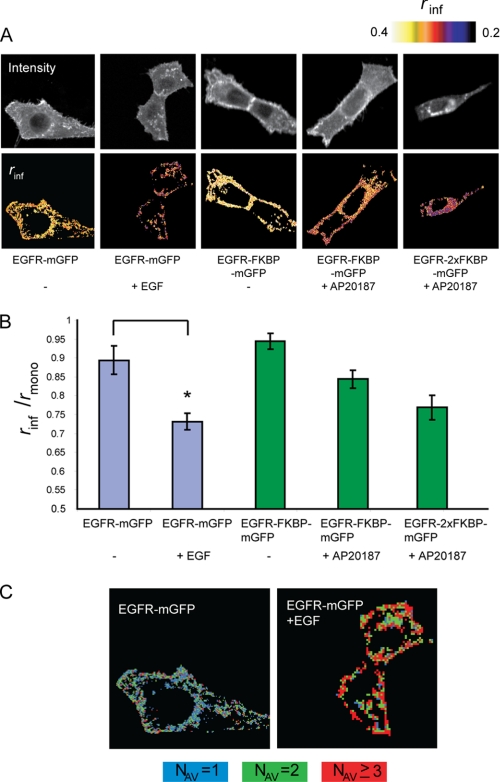
We next investigated the effect of EGF on the oligomerization state of the EGFR using homo-FRET imaging. Cells expressing EGFR-mGFP were treated for 10 min at 37 °C with 8 nm EGF and fixed with 4% formaldehyde. In the resting cell, EGFR-mGFP was located primarily in the plasma membrane, with more intense staining in membrane ruffles (Fig. 2A, upper panels). In EGF-stimulated cells, EGFR-mGFP became increasingly present in intracellular vesicles, reflecting the EGF-induced internalization of the active EGFR (Fig. 2A, upper panels). Effects on anisotropy (rinf) are indicated in false colors: a clear effect of EGF on the anisotropy was observed, reflecting the EGF-induced oligomerization (Fig. 2A, lower panels). Note that also the internalized receptors in the early endosomes were oligomerized. Using the reference rinf values of monomers, dimers, and oligomers, we converted the anisotropy values into a cluster size image (Fig. 2C). In the resting state, the EGFR population was found as a mixture of monomers and dimers. After treatment of the cell with EGF, the majority of EGFRs were found in nanoscale clusters of three or more receptors per cluster (Fig. 2C).
FIGURE 2.
EGFR-mGFP is oligomerized after EGF stimulation. A, cellular distribution of GFP intensities and anisotropy values. NIH 3T3 2.2 cells expressing the indicated EGFR-mGFP constructs were left untreated or were stimulated with 8 nm EGF for 10 min or with 1 μm AP20187 for 2 h. Limiting anisotropy values (rinf) were measured as described under “Experimental Procedures” and are expressed in false colors. B, average anisotropy values expressed as fraction of rmono of the indicated constructs (S.E.). *, p < 0.005. C, representation of the cluster size values of EGFR-mGFP before and after EGF stimulation in false colors. The anisotropy values of the cells shown in A were classified as monomers (NAV = 1 (blue)), dimers (NAV = 2 (green)), and oligomers (NAV ≥ 3 (red)) as described under “Experimental Procedures.”
From a direct comparison of the average depolarization rinf/rmono with reference values (Fig. 2B), we can conclude that EGFR-mGFP forms large clusters upon stimulation with EGF. As described under “Experimental Procedures,” the rinf is calculated on the basis of Gaussian fitting of the anisotropy values of all pixels in the image. To demonstrate the homogeneity in the response, the anisotropy values of two representative cells from both conditions are presented and show a true shift in the mean value of rinf (supplemental Fig. S4). In conclusion, comparison of the calculated average anisotropy values from non-stimulated and EGF-stimulated EGFR-mGFP-expressing cells demonstrated a significant decrease in anisotropy and consequently an increase in EGFR clustering in EGF-stimulated cells.
Receptor Oligomerization Is Kinase-dependent
To analyze the role of EGFR tyrosine kinase activity in the oligomerization of EGFR, a kinase-dead EGFR construct (EGFR-K721A) was fused to mGFP. Cells expressing wild-type EGFR-mGFP or EGFR-K721A-mGFP were incubated for different time periods with 8 nm EGF, fixed, and analyzed by homo-FRET imaging. For EGFR-K721A-mGFP, the distribution pattern was similar at time 0 to that of the wild-type receptor (Fig. 3, A and B). After EGF stimulation, intracellular vesicles appeared after 10–20 min, indicative of receptor internalization (Fig. 3, A and B), which is in agreement with our previous studies (23). In the absence of ligand, the K721A and wild-type receptors displayed similar anisotropy values (Fig. 3C). This indicates that the kinase-dead EGFR forms predimers to a similar extent as the wild-type EGFR; thus, predimer formation is kinase-independent. For the wild-type receptors, the anisotropy values decreased within 5 min to values corresponding to receptor oligomers (Fig. 3, A–C). Ligand-induced EGFR oligomerization was maintained for at least 20 min after stimulation. A gradual increase in anisotropy was seen from 10 to 45 min after activation, which might be caused by receptor dissociation or lysosomal degradation of receptor oligomers. The effect of EGF on the oligomerization of the kinase-dead mutant was analyzed for the same period of time. Remarkably, this mutant did not display any change in anisotropy for the entire observation period of 45 min, indicating that the amount of receptor predimers is not affected and that kinase-dead EGFRs do not oligomerize. On the basis of these results, we conclude that the EGFR predimer formation is kinase-independent, whereas the ligand-induced EGFR oligomerization is kinase-dependent. Consequently, the monomer/predimer ratio in the plasma membrane is not affected by the binding of EGF.
FIGURE 3.
EGFR kinase activity is essential for EGF-induced oligomerization. A, cellular distribution of GFP intensities and anisotropy values of NIH 3T3 2.2 cells expressing EGFR-mGFP, followed in time during stimulation with 8 nm EGF. The limiting anisotropy value was determined as described under “Experimental Procedures.” B, cellular distribution of GFP intensities and anisotropy values of NIH 3T3 2.2 cells expressing EGFR-K721A-mGFP, followed in time during stimulation with 8 nm EGF. The limiting anisotropy value was determined as described under “Experimental Procedures.” C, average anisotropy data of EGFR-mGFP and EGFR-K721A-mGFP, followed in time during stimulation with 8 nm EGF. The limiting anisotropy (rinf) is expressed as fraction of rmono.
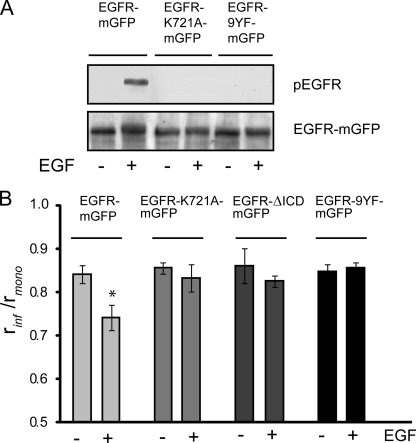
EGFR Oligomerization Requires Receptor Tyrosine Phosphorylation
Because kinase activity was found to be essential for EGFR oligomerization, we wanted to see whether preventing tyrosine phosphorylation of the EGFR had similar effects on receptor oligomerization. To investigate this, we used an EGFR mutant in which nine C-terminal tyrosine residues at positions 845, 974, 992, 1045, 1068, 1086, 1101, 1148, and 1173 were mutated to phenylalanines (EGFR-9YF). Control experiments show that phosphorylation of the wild-type EGFR-FKBP-mGFP protein was induced by EGF, in contrast to the K721A and 9YF mutants (Fig. 4A). NIH 3T3 2.2 cells stably expressing these constructs were left untreated or were stimulated with 8 nm EGF for 10 min and fixed. In the absence of ligand, the anisotropy of the wild-type and mutant K721A and 9YF receptors was similar, reflecting a similar degree of predimer formation (Fig. 4B). As expected, the wild-type receptor showed a decrease in anisotropy after EGF stimulation, reflecting an increase in homo-FRET and consequently in receptor oligomerization. EGF treatment of cells expressing the K721A or 9YF mutant did not result in a change in anisotropy compared with untreated cells. This result demonstrates that the kinase-dependent EGFR oligomerization requires the phosphorylation of tyrosine residues in the intracellular domain of the EGFR.
FIGURE 4.
EGFR tyrosine phosphorylation is essential for EGF-induced oligomerization. A, NIH 3T3 2.2 cells expressing wild-type EGFR-mGFP, kinase-dead EGFR-K721A-mGFP, or EGFR-9YF-mGFP were left untreated or were stimulated with 8 nm EGF for 10 min. Lysates were separated by SDS-PAGE, blotted onto PVDF membrane, and analyzed using antibodies against the activated EGFR (phospho-Tyr1068) or against GFP. B, NIH 3T3 2.2 cells expressing wild-type EGFR-mGFP, EGFR-K721A-mGFP, EGFR-ΔICD-mGFP (intracellular domain deletion), or EGFR-9YF-mGFP were left untreated or were treated with 8 nm EGF for 10 min. The limiting anisotropy value (rinf) was determined as described under “Experimental Procedures” and is expressed as a fraction of rmono (S.E.). *, p < 0.005.
EGFR Clustering Stimulates Receptor Internalization
An important question concerns the possible function of both receptor predimer formation and receptor oligomerization. Modeling of EGFR activation via monomers and predimers predicts that activation via the predimers would result in 100-fold faster activation or phosphorylation of the receptor (13). Consequently, the predimerization or even pre-oligomerization would result in faster signaling and higher sensitivity of the cell for EGF. To test a role for EGFR predimer/oligomer formation in signaling, we expressed EGFR fused to one or two copies of FKBP (Fig. 1A). Cells were either left untreated or were pretreated with 1 μm AP20187 for 1 h to induce predimer/oligomer formation, followed by incubation with 8 nm EGF for 9 min at 37 °C. Anisotropy analysis already showed that incubation of the cells expressing this construct led to a reduction in anisotropy, reflecting the increase in EGFR predimers (Fig. 2B). However, combined treatment with AP20187 and EGF did not, at least under the conditions used, enhance receptor activation compared with EGF alone (Fig. 5A and supplemental Fig. S5). Also, downstream targets of EGFR signaling such as Akt and MAPK were not affected, indicating that no function for receptor predimerization in the EGFR activation process could be discerned (Fig. 5A and supplemental Fig. S5). Moreover, similar results were obtained with an EGFR construct with two copies of FKBP, which became pre-oligomerized (Fig. 5A and supplemental Fig. S5). Further refinement of this analysis with additional time intervals (3–12 min) and lower ligand concentrations (0.25–8 nm EGF) yielded similar results.
FIGURE 5.
EGFR predimerization stimulates receptor internalization but not signaling. A, NIH 3T3 2.2 cells expressing EGFR-FKBP-mGFP were left untreated or were incubated with 1 μm AP20187 for 2 h at 4 °C, with 8 nm EGF for 9 min at 37 °C, or with both. Cell lysates were analyzed by immunoblotting using antibodies against the activated EGFR (phospho-Tyr1068), against GFP to detect EGFR-FKBP-mGFP, and against the indicated signaling proteins (Akt and MAPK); representative blots are shown. Quantification of signaling is shown in supplemental Fig. S5. B, NIH 3T3 2.2 cells expressing the indicated wild-type EGFR-FKBP constructs were incubated for 2 h with 1 μm AP20187 at 4 °C. Then, 1 ng/ml radiolabeled nanobody against EGFR (125I-EGb4) was added at 37 °C for different time periods (3, 6, 9, and 12 min) in the presence and absence of 8 nm EGF, and finally, the internalization rate constant (Ke) was determined. C, NIH 3T3 cells expressing EGFR-FKBP-mGFP were incubated with 1 μm AP20187 (AP) for 2 h, with 8 nm EGF for 10 min, or both. Cells were analyzed by confocal immunofluorescence microscopy. D, NIH 3T3 2.2 cells expressing the wild-type EGFR-FKBP and kinase-dead EGFR-K721A-FKBP constructs were incubated for 2 h with 1 μm AP20187 at 4 °C. Then, 1 ng/ml radiolabeled EGF (125I-EGF) was added for different time periods (3, 6, 9, and 12 min), and finally, the internalization rate constant (Ke) was determined.
This approach was subsequently used to investigate a possible role for EGFR clustering in receptor internalization. The internalization rate constant (Ke) was determined using a single-domain llama antibody, or nanobody, that does not activate the receptor or compete for EGF binding (24). Cells stably expressing the EGFR-FKBP-mGFP or EGFR-2×FKBP-mGFP construct were either left untreated or pretreated with AP20187 to induce EGFR predimerization or pre-oligomerization, respectively, and then incubated with radiolabeled anti-EGFR nanobody (125I-EGb4) in the absence of EGF or in the presence of a high concentration EGF (8 nm EGF). The data show that, in the control situation, the monovalent EGb4 nanobody was internalized at a low rate, reflecting fluid-phase endocytosis (Fig. 5B). The addition of EGF stimulated the internalization rate constant significantly, demonstrating the dominant role for kinase activity in the internalization process. However, internalization rate constants were dramatically increased after predimerization of the EGFR prior to EGF treatment. Remarkably, pre-oligomerization of the receptor did not further increase this Ke value, indicating that predimerization is already sufficient to stimulate internalization.
To further substantiate the role for EGFR predimer formation in the internalization process, we made use of the fact that kinase-dead EGFR does not oligomerize upon EGF binding and is internalized upon binding of EGF, albeit at a low rate. Internalization rate constants were determined for wild-type and kinase-dead EGFRs and compared with rate constants from the same receptors that were predimerized by preincubation with 1 μm AP20187 for 1 h, performed at 4 °C to prevent receptor internalization. The cells were incubated with a high dose of EGF (8 nm) for confocal microscopy and with a low dose of 125I-EGF (1 ng/ml) to determine the internalization rate constant (Ke). Although untreated EGFR-FKBP-mGFP was present predominantly in the plasma membrane, the AP20187-treated cells show internalized EGFR as judged by the presence of intracellular endocytic vesicles (Fig. 5C). Also at a low dose, the addition of EGF to cells with predimerized EGFR induced a prominent internalization of the EGFR. At this low EGF dose, the Ke of wild-type EGFR internalization was higher than at a high EGF dose (Fig. 5C), which is in agreement with previous work (25). As expected, the kinase-dead EGFR-FKBP-mGFP construct was endocytosed at a low rate (Ke(KD) = 0.10 min−1), which was much slower than the internalization of the wild-type EGFR (Ke(WT) = 0.22 min−1). Increasing the amount of EGFR predimers using AP20187 resulted in an ∼30% increase in the Ke of the wild-type receptor (Ke = 0.29 min−1). The Ke of the K721A construct was even doubled from 0.10 min−1 in the absence of AP20187 to 0.20 min−1 in its presence. In summary, these results show that predimerization of the EGFR increases the internalization rate of the EGFR, independent of ligand concentration or kinase activity.
DISCUSSION
To monitor the dimerization and oligomerization of the EGFR before and during receptor activation, we applied a novel non-invasive homo-FRET-based technique that allows the imaging of receptor dimers and oligomers in its cellular context. A time-resolved anisotropy analysis was chosen because data obtained with this method are more accurate in the determination of protein clustering compared with steady-state measurements (20). Analysis of EGFR clustering was done using reference EGFR constructs that allow for controlled dimerization or oligomerization by induced binding (17). For both the reference and EGFR-mGFP constructs, a high homo-FRET efficiency was observed, which means that the GFP fluorophores are in close proximity and their relative orientation is very similar. This is the major prerequisite for using the reference anisotropy values for monomer, dimers, and oligomers. The reporter protein that we used in this study is the monomeric version of enhanced GFP, GFP-A206K, or mGFP. This mutant was found to have a reduced homo-association constant compared with wild-type GFP and was therefore considered to be strictly monomeric (18). The reliability of this determination is high despite the fact that, in an ideal case, membrane-bound GFP would be preferred. In the plasma membrane, however, artifacts can be introduced when membrane anchoring induces clustering (18).
The homo-FRET data demonstrated that, in resting cells, ∼40% of the total population of EGFRs was present as predimers. Evidence for the presence of EGFR predimers was based upon different arguments. First, the anisotropy of EGFR-mGFP was significantly lower than for the GFP monomer as well as for cytosolic mGFP, and the time-resolved anisotropy decay showed a typical homo-FRET profile. Second, EGFR predimer formation was demonstrated by diluting mGFP-tagged EGFR with untagged EGFR in EGFR-expressing cells. Assuming that EGFR-mGFP associates with equal preference with EGFR and EGFR-mGFP, the formation of hetero- or homodimers is proportional to the relative expression levels of both receptor types. As the amount of non-fluorescent receptor increased, a decrease in apparent cluster size was found, which approximated the level of cytosolic mGFP. Similar data were provided by Lidke et al. (10), who used CHO cells instead of 3T3 fibroblasts. Third, the clustering was concentration-independent, indicating that predimer formation is not the result of overexpression of the receptor.
Our homo-FRET results are in perfect agreement with both light and electron microscopic data obtained from A431 cells (5, 15). Recently, the AP-2-binding region in the C-terminal part of the tyrosine kinase domain has been shown to interact with its counterpart in the predimer, resulting in an auto-inhibited receptor dimer (16). Mutations in this “electrostatic hook” were found to activate the tyrosine kinase. Thus, predimerization of the EGFR is a mechanism to inhibit the EGFR in the resting cell (16). An interesting question is the mechanism that controls EGFR predimer formation. First of all, predimerization is not the result of basal receptor activity in the absence of EGF. This can be concluded from the observation that, for the kinase-dead EGFR, no significant differences in anisotropy values were obtained compared with the non-stimulated wild-type EGFR (Fig. 3). Recently, cholesterol levels were found to regulate the number of predimers (11). We have previously demonstrated that the EGFR colocalizes with the lipid raft marker GM1 (24). On the basis of these data, we hypothesize that the partitioning of the EGFR in lipid raft domains might play a role in the stabilization of the EGFR predimer.
EGFR activation modeling studies have suggested that signaling via the predimer might occur 2 orders of magnitude faster than via the monomeric receptor simply because the time to find a binding partner is not required (13). To study the functional importance of EGFR predimerization, we increased the amount of receptor predimers using the FKBP domain. Importantly, our EGFR-FKBP-mGFP construct was not activated by FKBP-mediated predimerization, which is in contrast to previous studies (26). This may be caused by the fusion with mGFP or by differences in the orientation of the dimerization domain. Thus, our FKBP-mediated predimerization system generates functional non-active EGFR predimers. The amount of EGFR predimers was enhanced by treatment of the cells with the FKBP ligand, which did not affect EGF-mediated signaling. Also, further refinement of the assay by checking at shorter time intervals or different EGF concentrations did not reveal any difference. However, Chung et al. (27) have shown recently that differences in EGFR phosphorylation are noticeable at very short time intervals (14.6 s). Although dimer formation itself is essential for receptor activation, an increase in the amount of receptor predimers does not affect signaling at longer time intervals.
The FKBP system was subsequently used to analyze a possible function for EGFR clustering in the internalization process of the EGFR. To directly monitor receptor internalization, we used a VHH (VH domain of heavy chain-only) antibody or nanobody recognizing the ectodomain of the EGFR that does not block EGF binding or activate the receptor (24). Although the induction of receptor dimerization prior to EGF binding clearly stimulated EGFR internalization, receptor oligomerization did not stimulate this process any further. These results show that predimerization of the EGFR enhances the ligand-induced internalization of the EGFR. Receptor oligomerization did not stimulate this process any further, indicating that predimer formation is already sufficient to prime the EGFR for internalization. This might be mediated by the two clathrin-binding motifs that are present in the intracellular domain of the EGFR: a double-leucine motif at positions 1010 and 1011 and a tyrosine-based motif at position 974 (26, 28). We suggest that receptor oligomerization contributes to the internalization process by enhancing the binding of EGFRs to the AP-2 complexes of the internalization machinery. This would result in the binding of the EGFR to already preformed clathrin-coated pits, which is in line with recent observations (29). In conclusion, although the formation of receptor dimers is essential for signaling, the number of EGFR predimers is not, but is involved in the onset of EGFR internalization for both kinase-dead and wild-type EGFRs.
Ever since the report of Yarden and Schlessinger (30), ligand-induced aggregation of the EGFR was considered as the activation step of the tyrosine kinase. In contrast, our data show that receptor oligomerization after EGFR activation is the result of receptor tyrosine kinase activity rather than the cause of it. The results obtained with the 9YF mutant suggest that the kinase-induced oligomerization is exerted via the phosphotyrosines located in the intracellular domain of the EGFR. Based upon our data, the following sequence of events during the process of EGFR activation is proposed. The receptor is present both as monomers and predimers in the plasma membrane. Factors involved in the regulation of this system may include the lipid composition of the membrane. Predimers are auto-inhibited and keep receptors in an inactive state (16). Ligand binding induces a conformational change in the ectodomain, leading to the reorientation of the intracellular kinase domains, resulting in the activation of the asymmetric kinase dimer. As signaling proceeds, activated receptors will bind to phosphotyrosine-binding proteins such as actin, Cbl, and Grb2, resulting in the oligomerization of the EGFR. This process occurs more efficiently with increasing amounts of predimers. Receptor oligomerization enhances EGF-induced endocytosis via either the clathrin-dependent or clathrin-independent pathway, finally resulting in the down-regulation of the EGFR.
Supplementary Material
Acknowledgments
We thank Hein Sprong for helpful discussions, Ger Arkesteijn for support with FACS, and ARIAD Pharmaceuticals for supplying the ARGENT regulated homodimerization kit.
This work was supported by Dutch Organization for Scientific Research (NWO) Grant 805.47.084 (to E. G. H. and A. N. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- EGFR
- EGF receptor
- FKBP
- FK506-binding protein
- mGFP
- monomeric GFP.
REFERENCES
- 1.Ullrich A., Coussens L., Hayflick J. S., Dull T. J., Gray A., Tam A. W., Lee J., Yarden Y., Libermann T. A., Schlessinger J. (1984) Nature 309, 418–425 [DOI] [PubMed] [Google Scholar]
- 2.Garrett T. P., McKern N. M., Lou M., Elleman T. C., Adams T. E., Lovrecz G. O., Zhu H. J., Walker F., Frenkel M. J., Hoyne P. A., Jorissen R. N., Nice E. C., Burgess A. W., Ward C. W. (2002) Cell 110, 763–773 [DOI] [PubMed] [Google Scholar]
- 3.Zhang X., Gureasko J., Shen K., Cole P. A., Kuriyan J. (2006) Cell 125, 1137–1149 [DOI] [PubMed] [Google Scholar]
- 4.Sigismund S., Woelk T., Puri C., Maspero E., Tacchetti C., Transidico P., Di Fiore P. P., Polo S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Belzen N., Rijken P. J., Hage W. J., de Laat S. W., Verkleij A. J., Boonstra J. (1988) J. Cell. Physiol. 134, 413–420 [DOI] [PubMed] [Google Scholar]
- 6.Moriki T., Maruyama H., Maruyama I. N. (2001) J. Mol. Biol. 311, 1011–1026 [DOI] [PubMed] [Google Scholar]
- 7.Yu X., Sharma K. D., Takahashi T., Iwamoto R., Mekada E. (2002) Mol. Biol. Cell 13, 2547–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu H. J., Iaria J., Orchard S., Walker F., Burgess A. W. (2003) Growth Factors 21, 15–30 [DOI] [PubMed] [Google Scholar]
- 9.Gadella T. W., Jr., Jovin T. M. (1995) J. Cell Biol. 129, 1543–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lidke D. S., Nagy P., Barisas B. G., Heintzmann R., Post J. N., Lidke K. A., Clayton A. H., Arndt-Jovin D. J., Jovin T. M. (2003) Biochem. Soc. Trans. 31, 1020–1027 [DOI] [PubMed] [Google Scholar]
- 11.Saffarian S., Li Y., Elson E. L., Pike L. J. (2007) Biophys. J. 93, 1021–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sako Y., Minoghchi S., Yanagida T. (2000) Nat. Cell Biol. 2, 168–172 [DOI] [PubMed] [Google Scholar]
- 13.Teramura Y., Ichinose J., Takagi H., Nishida K., Yanagida T., Sako Y. (2006) EMBO J. 25, 4215–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin-Fernandez M., Clarke D. T., Tobin M. J., Jones S. V., Jones G. R. (2002) Biophys. J. 82, 2415–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clayton A. H., Walker F., Orchard S. G., Henderson C., Fuchs D., Rothacker J., Nice E. C., Burgess A. W. (2005) J. Biol. Chem. 280, 30392–30399 [DOI] [PubMed] [Google Scholar]
- 16.Jura N., Endres N. F., Engel K., Deindl S., Das R., Lamers M. H., Wemmer D. E., Zhang X., Kuriyan J. (2009) Cell 137, 1293–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bader A. N., Hofman E. G., Voortman J., en Henegouwen P. M., Gerritsen H. C. (2009) Biophys. J. 97, 2613–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. (2002) Science 296, 913–916 [DOI] [PubMed] [Google Scholar]
- 19.Fallon L., Bélanger C. M., Corera A. T., Kontogiannea M., Regan-Klapisz E., Moreau F., Voortman J., Haber M., Rouleau G., Thorarinsdottir T., Brice A., van Bergen En Henegouwen P. M., Fon E. A. (2006) Nat. Cell Biol. 8, 834–842 [DOI] [PubMed] [Google Scholar]
- 20.Bader A. N., Hofman E. G., van Bergen en Henegouwen P. M., Gerritsen H. C. (2007) Optics Express 15, 6934–6945 [DOI] [PubMed] [Google Scholar]
- 21.de Grauw C. J., Gerritsen H. C. (2001) Appl. Spectrosc. 55, 670–678 [Google Scholar]
- 22.Runnels L. W., Scarlata S. F. (1995) Biophys. J. 69, 1569–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoorvogel W., Kerstens S., Fritzsche I., den Hartigh J. C., Oud R., van der Heyden M. A., Voortman J., van Bergen en Henegouwen P. M. (2004) J. Biol. Chem. 279, 11562–11569 [DOI] [PubMed] [Google Scholar]
- 24.Hofman E. G., Ruonala M. O., Bader A. N., van den Heuvel D., Voortman J., Roovers R. C., Verkleij A. J., Gerritsen H. C., van Bergen en Henegouwen P. M. (2008) J. Cell Sci. 121, 2519–2528 [DOI] [PubMed] [Google Scholar]
- 25.Sigismund S., Argenzio E., Tosoni D., Cavallaro E., Polo S., Di Fiore P. P. (2008) Dev. Cell 15, 209–219 [DOI] [PubMed] [Google Scholar]
- 26.Wang Q., Zhu F., Wang Z. (2007) Exp. Cell Res. 313, 3349–3363 [DOI] [PubMed] [Google Scholar]
- 27.Chung I., Akita R., Vandlen R., Toomre D., Schlessinger J., Mellman I. (2010) Nature 464, 783–787 [DOI] [PubMed] [Google Scholar]
- 28.Goh L. K., Huang F., Kim W., Gygi S., Sorkin A. (2010) J. Cell Biol. 189, 871–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rappoport J. Z., Simon S. M. (2009) J. Cell Sci. 122, 1301–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yarden Y., Schlessinger J. (1987) Biochemistry 26, 1434–1442 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.