Abstract
To investigate the mechanisms by which O-linked β-N-acetylglucosamine modification of nucleocytoplasmic proteins (O-GlcNAc) confers stress tolerance to multiple forms of cellular injury, we explored the role(s) of O-GlcNAc in the regulation of heat shock protein (HSP) expression. Using a cell line in which deletion of the O-GlcNAc transferase (OGT; the enzyme that adds O-GlcNAc) can be induced by 4-hydroxytamoxifen, we screened the expression of 84 HSPs using quantitative reverse transcriptase PCR. In OGT null cells the stress-induced expression of 18 molecular chaperones, including HSP72, were reduced. GSK-3β promotes apoptosis through numerous pathways, including phosphorylation of heat shock factor 1 (HSF1) at Ser303 (Ser(P)303 HSF1), which inactivates HSF1 and inhibits HSP expression. In OGT null cells we observed increased Ser(P)303 HSF1; conversely, in cells in which O-GlcNAc levels had been elevated, reduced Ser(P)303 HSF1 was detected. These data, combined with those showing that inhibition of GSK-3β in OGT null cells recovers HSP72 expression, suggests that O-GlcNAc regulates the activity of GSK-3β. In OGT null cells, stress-induced inactivation of GSK-3β by phosphorylation at Ser9 was ablated providing a molecular basis for these findings. Together, these data suggest that stress-induced GlcNAcylation increases HSP expression through inhibition of GSK-3β.
Keywords: Chaperone Chaperonin, Glycogen Synthase Kinase 3, Glycosylation, Heat Shock Protein, Signal Transduction, O-GlcNAc, Intracellular Glycosylation
Introduction
Dynamic modification of Ser/Thr residues by monosaccharides of O-linked β-N-acetylglucosamine (O-GlcNAc) has been implicated in the regulation of over 500 intracellular proteins (1–3). Unlike prototypical O-linked glycosylation, O-GlcNAc occurs on nuclear, cytoplasmic (1, 2), and mitochondrial proteins (4, 5); and with the exception of plant nuclear pore proteins (6, 7), O-GlcNAc is not extended into more complex structures. The addition and removal of O-GlcNAc is catalyzed by the uridine diphospho-N-acetylglucosamine:peptide β-N-acetylglucosaminyl transferase (OGT)6 (EC 2.4.1.-) (8, 9) and a neutral O-GlcNAc-specific hexosaminidase (O-GlcNAcase; EC 3.2.1.52) (10–13), respectively. Like the O-GlcNAc modification, these enzymes are also localized to the mitochondria (4, 5, 14), nucleus, and cytoplasm. Deletion of OGT is lethal in animals, tissues, and single cells, highlighting the key role of O-GlcNAc in regulating cellular function (15, 16).
O-GlcNAc is thought to regulate protein function in a manner analogous to protein phosphorylation (1, 3, 17). One mechanism by which O-GlcNAc may mediate these activities is by altering the phosphorylation status of proteins (18–21). Notably, O-GlcNAc and O-phosphate have been mapped to the same Ser/Thr residue on key cellular proteins, such as c-Myc (22) and RNA polymerase II (23, 24). Both OGT and O-GlcNAcase have been found in complexes with phosphatases and kinases, suggesting that the exchange between phosphorylation and GlcNAcylation is rapid and dynamic (21, 25–27). Similar to phosphorylation, O-GlcNAc is dynamically added and removed in response to numerous extracellular signals, including: extracellular glucose concentration (28–30), cell cycle (31), development (32, 33), hormones such as insulin (34, 35), phorbol esters (36), and the focus of this study, cellular stress (37).
In response to injury, cells remodel their metabolic and signaling pathways to promote survival, the so-called cellular stress response (38, 39). GlcNAcylation of intracellular proteins appears to be one target of the cellular stress response (1, 17, 25, 37, 40–55). O-GlcNAc levels increase in a dose-dependent manner in response to numerous forms of cellular injury (37), and decline in some cell models of apoptosis (48). These data, combined with those demonstrating that elevating O-GlcNAc levels before (1, 40–45, 47, 48, 50, 51, 54), or immediately after (46), cellular injury improves survival in numerous models (heat stress, oxidative stress, hypoxia, ischemia reperfusion injury, and trauma hemorrhage injury) has led to the conclusion that stress-induced GlcNAcylation is a novel regulator of the cellular stress response. Interestingly, in some models of cellular injury, for example, ischemia reperfusion injury and trauma hemorrhage, the levels of O-GlcNAc decrease (45, 47, 48, 57–59). Here, it appears that simply blocking the decrease in O-GlcNAc levels is protective. It is unclear if cells/tissues that die during ischemia reperfusion injury and trauma hemorrhage are undergoing necrosis/apoptosis for similar reasons as the OGT null (16).
Modulating O-GlcNAc appears to impact several pathways in a manner consistent with increased survival. These include: 1) improving protein solubility in models of heat stress (25, 60); 2) capacitative calcium entry (41, 46, 61); 3) reducing calpain activation (46); 4) p38 MAP kinase phosphorylation (43, 47); 5) modulation of circulating IL-6 and TNF-α levels (57, 62); 6) maintenance of mitochondrial membrane potential (48); 7) BCL2 translocation (63); and finally 8) regulating the expression of HSP72 (37).
HSP72 belongs to a large family of ATP-dependent molecular chaperones, expressed constitutively or inducibly in most subcellular compartments (38, 39, 64–66). HSPs act to prevent cell death via numerous mechanisms, including: 1) refolding proteins; 2) preventing protein aggregation; 3) altering transcription; and 4) inhibiting apoptotic signaling pathways (38, 39, 64–66). Of the nine HSP70s in mammalian cells, the predominant stress-induced nuclear and cytosolic HSP70 is the result of transcription from two genes: Hsp70.1a and Hsp70.1b (64). The products of these genes are 98% identical and are often referred to collectively as HSP72, which also delineates them from other HSP70 family members (64). HSP72 is a key regulator of cell survival and deletion of HSP72 genes renders cells sensitive to environmental and physiological stress (67–70).
HSP72 expression is predominantly regulated at the level of transcription, which is mediated by the transcription factor, heat shock factor 1 (HSF1) (71, 72). HSF1 is regulated at numerous points; in unstressed cells HSF1 is maintained as an inert monomer in a complex with HSP90 and other proteins in the cytoplasm. Upon stress, HSF1 disassociates from the HSP90, exposing a trimerization and nuclear localization domain. Once in the nucleus, HSF1 can bind DNA but is not fully active until phosphorylated at over 10 sites (71, 72). HSF1 is also negatively regulated by phosphorylation (72–76) and acetylation (77). Phosphorylation by GSK-3β at Ser303 maintains HSF1 in an inactive state in unstressed cells, and is involved in deactivation of HSF1 post-stress promoting reduced DNA binding, nuclear export, and cytoplasmic sequestration by 14-3-3ϵ (73–76). Recently, it has been reported that HSF1 is acetylated and that this appears to peak post-stress enhancing HSF1 deactivation (77).
Previously, we have shown that O-GlcNAc appears to regulate the expression of HSP72 and HSP40 (37). To confirm these data and to determine whether O-GlcNAc regulates the expression of other molecular chaperones we have performed an RT-PCR array directed against 84 molecular chaperones. We have shown that O-GlcNAc appears to regulate the expression of at least 18 chaperones, including HSP72. To determine the mechanism(s) by which this occurs we have focused on the mechanism by which O-GlcNAc regulates the expression of HSP72. Two other groups have suggested that O-GlcNAc regulates HSP72 expression by either: 1) stabilizing the housekeeping transcription factor Sp1 to elevated temperatures, thus leading to enhanced HSP72 expression (60); or 2) by modulating the localization of HSF1 (78). However, in this study we present an alternative model in which O-GlcNAc regulates HSP expression by modulating the phosphorylation and inactivation of GSK-3β. As GSK-3β promotes apoptosis through numerous pathways (79–82), these studies highlight additional pathways through which O-GlcNAc may regulate cell survival, implicating O-GlcNAc in diseases/conditions in which the activity of GSK-3β is mis-regulated, such as Alzheimer disease and numerous forms of cancer (79, 81, 83, 84).
EXPERIMENTAL PROCEDURES
Reagents
Reagents were purchased from Sigma, unless otherwise noted. LiCl was used at 10 mm in 0.5 m HEPES, pH 7.5, 1 h prior to heat stress. O-(2-Acetamido-2-deoxy-d-glucopyranosylidene)-amino N-phenylcarbamate (PUGANc; Toronto Research Chemicals Inc., ON, Canada) was used at 50 μm in water for 8 or 18 h. 1,2-Dideoxy-2′-methyl-α-d-glucopyranoso[2,1-d]-δ2′-thiazoline (GlcNAc-thiazaline or GT; a gift from G. W. Hart) was used at 10 μm in water for 8 or 18 h unless otherwise noted. 4-Hydroxytomoxifen (4HT) was used at 0.5 μm in ethanol for 24 h. The GSK-3β inhibitor AR-A014418 (in DMSO) was used at 100 nm, 1 h prior to heat stress. A complete list of antibodies can be found under supplemental “Experimental Procedures”.
Cell Culture
COS-7 cells or mouse embryonic fibroblasts (MEFs) were cultured in Dulbecco's modified Eagle's medium (DMEM; 1 g/liter glucose; Mediatech, Manassas, VA), 10% (v/v) fetal bovine serum (FBS), 1% (v/v) penicillin/streptomycin at 37 °C in a water-jacketed, humidified CO2 (5%) incubator. Typically, cells were plated at 5 × 105 cells/100-mm plate or 1 × 06 cells/150-mm plate (Corning Inc., Corning, NY), 46 h prior to the initiation of experiments.
Generation of an Inducible OGT Knock-out Cell Line
MEF (OGTF/Y) (16) were stably transduced with lentivirus encoding either green fluorescent protein (GFP) or mutated estrogen receptor (mER)-Cre-2A-GFP. Transfected cells fluoresce green and these were sorted from untransfected cells at The Johns Hopkins University Flow Cytometry facility on a FACSVantage SE (BDIS). Unless otherwise noted, Cre-recombinase was activated using 0.5 μm 4HT 8 h post-plating. 4HT was removed at 24 h and experiments were initiated 46 h post-plating, ∼38 post-4HT treatment.
Stress Treatments
Cells 46 h post-plating, and ∼38 h post-4HT, were stressed at 45 °C for up to 1 h in a humidified water-jacketed CO2 incubator and returned to 37 °C for the indicated lengths of time. For cells treated ±4HT, medium containing the drug was removed 14 h prior to the experiment, the cells were washed with phosphate-buffered saline, pH 7.4, (PBS; 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, pH 7.4) to remove residual 4HT, and replaced with complete medium (DMEM 1 g/liter glucose, 10% FBS, penicillin/streptomycin).
Growth Assay
Typically, cells were plated at 2 × 103 cells/well in a 96-well plate. At 24 h 4HT (0.5 μm) was added and then was removed at 48 h. Subsequently medium was replaced every 24 h. Cell growth was measured using the CellTiter 96® AQ reagent (Promega, Madison, WI) according to the manufacturers' instructions, and absorbance was read at 492 nm on a BioTek SynergyTM HT 96-well plate reader.
Quantitative Reverse Transcriptase-PCR (qRT-PCR)
Cells were washed with ice-cold PBS, harvested, snap frozen, and stored at −80 °C. Total RNA was extracted using the RNeasy Midi Kit according to the manufacturer's instructions (Qiagen, Valencia, CA). cDNA was generated using an RT2 First Strand Kit (SABiosciences Corp., Frederick, MD). Probes for 84 chaperone genes were arrayed in a 96-well plate, along with 4 control proteins. Using SYBR Green PCR master mix, RT-PCR was performed on a MX300PTM System (Stratagene, La Jolla, CA) according to the manufacturer's instructions (SABiosciences). Each time point was analyzed independently three times. Expression was normalized to three control genes: β-actin (ActB), β-glucuronidase (GusB), and hypoxanthine guanine phosphoribosyltransferase 1 (hrpt1). Heat shock protein α90 kDa (cytosolic) and glyceraldehyde-3-phosphate dehydrogenase were also supplied as controls, but we observed stress-induced changes in expression of these proteins (supplemental Table S2) and as a result these transcripts were excluded as controls.
Protein Expression
Cells were washed with ice-cold PBS, harvested, and stored at −80 °C. Total protein was extracted with sonication (5S, Setting 3, Fisher 550 Sonic dismembrator) in extraction buffer (1% (v/v) Nonidet P-40 in TBS (50 mm Tris-HCl, pH 7.5, 150 mm NaCl) with 0.5 mm PMSF, PIC1, PIC2, 2 μm O-(2-acetamido-2-deoxy-d-glucopyranosylidene)-amino N-phenylcarbamate, 5 mm KF, and 5 mm β-glycerophosphate). Cellular debris were pelleted at 18,000 × g (30 min at 4 °C). Protein concentration was estimated using Coomassie Plus Protein Assay Reagent (Pierce Biotechnology). Equal protein (typically between 20 and 30 μg) was separated on either Bio-Rad Tris Glycine SDS-PAGE gels (7.5 or 4–15%; Criterion, Bio-Rad) or Invitrogen BisTris SDS-PAGE gels (8 or 4–12%). Proteins were transferred to nitrocellulose or PVDF and blocked in either 3% (w/v) milk in TBST or 3% (w/v) BSA in TBST. HRP was detected using Immobilon Western Chemiluminescent substrate (Millipore Corporation, Billerica, MA) and detected on ECL-Hyperfilm (GE Healthcare).
Nuclear and Cytoplasmic Extracts
Cells were washed with ice-cold PBS, harvested, snap frozen, and stored at −80 °C until required. Nuclear and cytoplasmic extracts were performed as previously reported by Qing and co-workers (85) in the presence of protease, phosphatase, and O-GlcNAcase inhibitors.
Densitometry
Densitometry was performed using non-saturated chemiluminescent-exposed films and quantitated using a MacBAS bio-imaging analyzer (version 2.5, Fuji Photo Film Co.). Typically, multiple exposures from the same experiment were used to confirm that the signal was within the linear range. Levels of HSF1, Actin, or Ser(P)303 HSF1 were normalized to that in OGT wild-type unstressed cells. In all instances, data are averaged from independent experiments (n > 3). Statistics are the result of a Student's t test or a two-way analysis of variance as indicated.
Immunoprecipitation
OGT wild-type and null cells were treated with heat shock (45 °C) for either 20 or 40 min, or with the protein phosphatase inhibitor calyculin A (100 nm, 30 min). Cells were extracted with sonication (5S, Setting 3, Fisher 550 Sonic dismembrator) in buffer (20 mm HEPES, 150 mm NaCl, 1 mm EDTA, 0.5% (v/v) Nonidet P-40, pH 7.5) in the presence of protease, phosphatase, and O-GlcNAcase inhibitors. Cellular debris was pelleted by centrifugation at 18,000 × g (30 min at 4 °C). Extracts (2.5 mg) were immunoprecipitated with anti-HSF1 antibody (Assay Designs; 10 μg) or control rabbit immunoglobulin (Santa Cruz Biotechnology; 10 μg); which were covalently coupled to tosyl-activated Dynabeads (Invitrogen) according to the manufacturer's instructions. Extracts were incubated with beads overnight at 4 °C, with rotation, washed 4 times in extraction buffer, and resuspended in SDS-PAGE sample buffer. Immunoprecipitates were separated on 8% BisTris gels and O-GlcNAc, HSF1, Ser(P)303 HSF1, and actin were detected by immunoblot.
RESULTS
Generation of an Inducible OGT Knock-out Cell Line
Previously, we have shown that O-GlcNAc levels can be reduced by overexpression of Cre-recombinase in MEFs carrying an Ogt exon flanked by loxP recombination sites (MEFs (OGTF/Y)). Activation of these recombination sites deletes amino acids 206–232, the consequent addition of 27 amino acids and a new translational termination site, effectively deleting OGT. Overexpression of Cre-recombinase using retrovirus results in OGT null cells ∼5–7 days after the initial infection (16, 37). This is an effective method for reducing OGT levels, but is plagued by three issues: 1) viral infection can stimulate stress response pathways (86); 2) the efficiency and timing of the knockout varies; and 3) retrovirus only infects dividing cells. To overcome these issues, we have stably transfected MEFs (OGTF/Y) cells with either GFP or mER-Cre-2A-GFP (a gift from A. Michaelis and M. K. Meffert). Using the GFP tag we sorted cells to select a polyclonal cell population that expresses the Cre-recombinase chimera (mER-Cre-2A-GFP-MEFs (OGTF/Y)) and a control cell line that expresses only GFP (GFP-MEFs (OGTF/Y); data not shown).
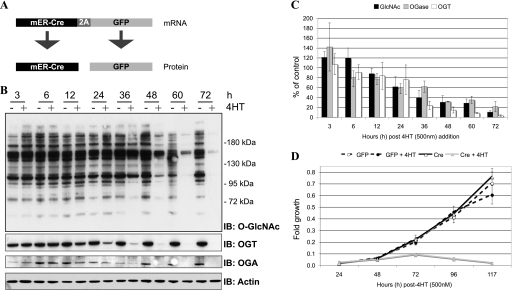
The mER-Cre-2A-GFP plasmid contains a 2A sequence between the mER-Cre and GFP, and as a result, whereas one mRNA is transcribed, two proteins are translated: mER-Cre and GFP (87) (Fig. 1A). mER-Cre encodes a mER fusion protein that has a higher affinity for 4HT than estrogen (88). In the absence of ligand, the mER-Cre chimera is retained in the cytoplasm. In the presence of ligand, the mER-Cre chimera can dimerize and translocate to the nucleus recombining the loxP sites, resulting in an Ogt null allele (Ogt is X-linked). As expected, treatment of the mER-Cre-GFP-2A-MEFs (OGTF/Y) cells with 0.5 μm 4HT results in reduced OGT protein expression at 24 h. A >80% loss of OGT and O-GlcNAc was not achieved until ∼48 h (Fig. 1, B and C). A small amount of OGT persists, which is either a result of inefficiency of the Cre-recombinase, or a small number of untransduced cells. Notably, O-GlcNAcase expression is also reduced in OGT null cells (Fig. 1, B and C, discussed below).
FIGURE 1.
Characterization of the inducible OGT knockout. A, MEFs (OGTF/Y) were stably transfected with mER-Cre-2A-GFP. This construct encodes one transcript that is translated as two proteins, a Cre-recombinase estrogen receptor fusion protein and GFP. Cells transfected with mER-Cre-2A-GFP were treated with and without 0.5 μm 4HT for 24 h. B, cells were harvested at regular intervals (as indicated) and the expression of O-GlcNAc, OGT, and O-GlcNAcase were determined by immunoblot (IB). Actin is shown as a loading control (n = 3; representative data shown). C, densitometry of O-GlcNAc (black), OGT (white), and O-GlcNAcase (gray) in OGT null cells relative to wild-type cells are shown. Error bars represent one standard deviation. D, cell growth was assessed every 24 h using the CellTiter 96® AQ reagent (n = 3, 12 replicates per experiment). MEFs (OGTF/Y) transfected with GFP are depicted by a dashed line, +4HT (●) and −4HT (○). Whereas, MEFs (OGTF/Y) transfected with mER-Cre-2A-GFP are depicted by a solid line, + 4HT (▴) and −4HT (△). Cells that are null for OGT are in gray. Error bars represent one standard deviation.
Previously, we have reported that deletion of OGT in cell culture is lethal, occurring after 2 rounds of cell division and an apparent period of senescence (16). Consistent with previous data, OGT null cells appear to grow normally for 48 h, cell growth then slows, and cell death ensues between days 4 and 5 (Fig. 1D). Consistent with these data, 48 h after the addition of 4HT we do not observe an increase in LDH release from OGT null cells when compared with wild-type cells (supplemental Fig. S1A). Moreover, OGT null cells do not have elevated levels of caspase-3 activity or demonstrate cleavage of either caspase-3 or poly(ADP-ribose) polymerase (supplemental Fig. S1, B and C) 48 h post-4HT treatment. With the exception of experiments examining the kinetics of OGT deletion, all experiments using the mER-Cre-2A-GFP-MEFs (OGTF/Y) cells were initiated and completed before 48 h.
The Expression of Numerous Molecular Chaperones Is Regulated by O-GlcNAc
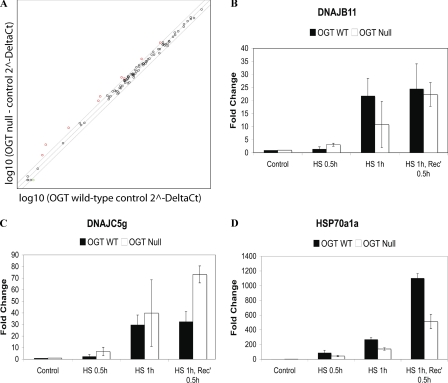
The expression of 84 molecular chaperones was assessed simultaneously by qRT-PCR using a RT2 ProfilerTM PCR array (SABiosciences) in OGT wild-type and null cells that had been stressed for varying lengths of time. Of the chaperones examined, the basal mRNA expression of 8 chaperones was affected more than 1.5-fold (Table 1, supplemental Fig. S1, and Fig. 2), and the stress-induced expression of 21 chaperones was affected, by deletion of OGT/O-GlcNAc (Table 2, supplemental Fig. S2, and Fig. 2). 18 molecular chaperones had a reduction in their mRNA expression (Tables 1 and 2, supplemental Tables S1 and S2, and Fig. 2), whereas 3 chaperones had enhanced expression (Dnajc5g, Dnajc5B, and Dnajb8; Table 2 and supplemental Table S2). Notably, the basal expression of Dnajb8 also appears up-regulated, although the data are not statistically significant (Fig. 2, green dot, and supplemental Table S1). Interestingly, only one chaperone whose expression is induced by stress was unaffected by deletion of OGT: Dnajb11 (Fig. 2).
TABLE 1.
Molecular chaperones whose basal expression was affected by deletion of OGT
Complete data are shown under supplemental Table 1.
| Symbol | -Fold change | p value |
|---|---|---|
| Caba1 | 2.29 | 0.004 |
| Cct6b | 3.6 | 0.003 |
| Dnajb9 | 2.06 | 0.005 |
| Dnajc12 | 2.36 | 0.002 |
| Dnajc9 | 1.86 | 0.033 |
| Hsap1aa,b | 1.97 | 0.01 |
| Hspa1ba,b | 1.51 | 0.023 |
| Hspb1a | 1.61 | 0.011 |
a Stress induced expression is also affected.
b The proteins translated from these genes are known as HSP72.
FIGURE 2.
Deletion of OGT alters the expression of 21 molecular chaperones, including HSP72. OGT wild-type and null cells (38 h post-OGT deletion) were heat stressed at 45 °C for 30, 60, or 60 min with 30 min recovery at 37 °C. The last time point in the experiment was harvested 39.5 h post-OGT deletion. mRNA was quantified using RT-PCR (n = 3). A, a comparison of basal mRNA expression in OGT wild-type (control group) and OGT null (group 1) cells. The bars represent a 1.5-fold change in expression. Transcripts down-regulated in OGT null cells are indicated in red, whereas transcripts up-regulated in OGT null cells are highlighted in green. B, expression of Dnajb11, a protein whose stress-induced expression is not altered in OGT null cells. C, expression of Dnajc5G, a protein whose stress-induced expression is up-regulated in OGT null cells. D, expression of Hsp70a1a, a protein whose stress-induced expression is down-regulated in OGT null cells. Error bars represent one standard deviation.
TABLE 2.
Molecular chaperones whose stress-induced expression was affected by deletion of OGT
Complete data are shown under supplemental Table 2. -Fold change as compared to control is reported at each time point.
| Protein | OGT wild-type |
OGT null |
||||
|---|---|---|---|---|---|---|
| 0.5 h at 45 °C | 1 h at 45 °C | 1 h at 45 °C, 0.5 h at 37 °C | 0.5 h at 45 °C | 1 h at 45 °C | 1 h at 45 °C, 0.5 h at 37 °C | |
| Proteins up-regulated in OGT null cells | ||||||
| Dnajb8 | 0.58 | 1.16 | 1.18 | 1.17a | 2.46 | 5.54 |
| Dnajc5b | 0.88 | 1.18 | 0.87 | 0.97 | 1.26 | 1.62 |
| Dnajc5g | 2.52 | 29.83 | 32.45 | 6.64 | 39.84 | 73.20b |
| Proteins down-regulated in OGT null cells | ||||||
| Bag2 | 1.52 | 1.52 | 1.56 | 1.08c | 0.98c | 1.12b |
| Bag3 | 2.59 | 3.85 | 7.97 | 1.94 | 2.44b | 3.68b |
| Cct4 | 1.56 | 2.07 | 1.50 | 0.94 | 0.99b | 0.86a |
| Cryab | 1.22 | 1.33 | 1.43 | 0.98a | 0.95 | 1.01b |
| Dnaja1 | 2.17 | 2.02 | 2.81 | 1.34 | 1.61 | 1.88c |
| Dnaja2 | 1.18 | 1.28 | 1.33 | 0.88c | 0.78 | 0.97b |
| Dnaja3 | 1.25 | 1.36 | 1.22 | 0.88b | 0.86a | 0.92c |
| Dnaja4 | 4.56 | 6.81 | 36.60 | 3.85 | 6.18 | 16.63 |
| Dnajb1 | 7.31 | 11.91 | 27.03 | 6.67 | 9.31 | 16.09b |
| Dnajb7 | 3.49 | 5.63 | 4.78 | 2.32c | 3.37b | 3.03 |
| Hsph1 | 3.81 | 6.28 | 7.00 | 1.86 | 2.96 | 3.76 |
| Hsp90aa1 | 1.40 | 2.23 | 2.28 | 1.11c | 1.36 | 1.79c |
| Hspa1ad | 87.48 | 270.16 | 1101.90 | 43.39 | 139.06a | 513.61b |
| Hspa1bd | 49.26 | 179.84 | 368.97 | 43.26 | 107.72c | 248.50a |
| Hspb1 | 7.30 | 15.73 | 22 | 4.29c | 7.91 | 11.18b |
| Hspb8 | 1.55 | 1.99 | 2.75 | 1.26c | 1.54c | 1.81c |
| Hspd1 | 1.77 | 1.87 | 1.98 | 0.92 | 1.10 | 1.19a |
| 1.51 | 2.27 | 3.04 | 1.56 | 1.51 | 1.91 | |
a p value <0.01. p values derived from t test comparing chaperone expression in wild-type and OGT null cells are each time point.
b p value <0.05 derived from t test comparing chaperone expression in wild-type and OGT null cells are each time point.
c p value <0.1 derived from t test comparing chaperone expression in wild-type and OGT null cells are each time point.
d The proteins transcribed from these genes are known as HSP72.
To confirm these data, we examined the expression of a subset of these molecular chaperones by immunoblot (Fig. 3). The stress-induced expressions of HSP72 and HSP90α were reduced in OGT null cells. However, expression of the closely related chaperones GRP75 and HSC70 were not altered, suggesting that the effect of deleting OGT on the expression of HSP72 (and other chaperones) was specific rather than a global effect on transcription or translation. To ensure that the effect of HSP expression was due to OGT deletion, rather than treatment of cells with 4HT, we performed these experiments in the OGTf/y MEFs that were stably transduced with GFP. Treatment of cells with 4HT, in the presence of OGT, did not alter the expression of HSPs (supplemental Fig. S2).
FIGURE 3.
Deletion of OGT abrogates the expression of HSP72, HSP90, and GRP78, but not the closely related chaperones GRP75 and HSC70. OGT wild-type and null cells (38 h post-OGT deletion) were heat stressed at 45 °C for 60 min, and recovered at 37 °C for 2, 4, or 8 h. Cells were harvested 41, 43, and 47 h post-OGT deletion. The expression of OGT, O-GlcNAc, and the HSPs HSP72, HSC70, GRP78, and GRP75 were assessed by immunoblot (IB) (n = 5). Actin is shown as a loading control.
Interestingly, the expression of GRP78 was only affected at the protein level. As we only observe GRP78 protein levels at 8 h, it is unclear if this is as GRP78 expression is regulated through a different mechanism or alternatively the transcription of GRP78 is induced after our last time point (HS 60 min, recovery 30 min).
De Novo O-GlcNAcylation Is Required for HSP Expression
To determine whether stress-induced GlcNAcylation is required for HSP expression, we showed that incubating cells with GlcNAc-thiazaline at the onset of Cre-activation (addition of 4HT) could maintain O-GlcNAc levels for 24 h, but not 48 h (supplemental Fig. S3). As such, we examined the expression of HSP72 in cells 30 h post-4HT, incubated with and without GlcNAc-thiazaline (20 μm). We observed no augmentation of HSP72 expression in OGT null cells (Fig. 4), suggesting that stress-induced O-GlcNAcylation is important for the induction of HSP72.
FIGURE 4.
Stress-induced O-GlcNAcylation is required for appropriate HSP72 expression. The O-GlcNAcase inhibitor GlcNAc-thiazaline (GT; 20 μm) was incubated with OGT wild-type and null cells concurrent with 4HT treatment (time = 0 h post-OGT deletion). GlcNAc-thiazaline (20 μm) was replaced 18 h prior to the induction of the experiment (time = 12 h post-OGT deletion). OGT wild-type and null cells with and without GlcNAc-thiazaline were heat stressed (45 °C, 1 h, time = 30 h post-OGT deletion) and returned to 37 °C for 1, 2, and 4 h (time = 32, 33, and 34 h post-OGT deletion). O-GlcNAc, OGT, O-GlcNAcase, HSP72, and Actin were determined in total cell lysates by immunoblot (IB) (n = 4).
HSF1 Does Not Appear to be O-GlcNAc Modified
HSF1 is regulated by its trimerization status, localization, and by both activating and inhibitory phosphorylation events (71, 72). Deletion of OGT did not alter stress-induced hyper-phosphorylation of HSF1 (supplemental Fig. S4).
Previously, it has been reported that glutamine acts to regulate the localization of HSF1 in an O-GlcNAc-dependent manner (78). Unlike the study of Singleton and Wischmeyer (78) deletion of OGT did not appear to alter nuclear localization of HSF1 (supplemental Fig. S5); and glutamine was able to induce HSP70 in the OGT null (supplemental Fig. S6). Consistent with these observations, our data suggests that HSF1 is not directly GlcNAcylated in either the MEFs (supplemental Fig. S7A) or COS-7 cells used in this study (supplemental Fig. S7B) before or after heat stress. In addition to CTD110.6, we failed to detect O-GlcNAc on HSF1 with either RL2 or sWGA (data not shown). O-GlcNAc may regulate HSF1 although several mechanisms and this may be cell type dependent. Notably, Hatsell and colleagues (89) have previously shown that Plakoglobin is O-GlcNAc modified in some tissues, and not in others.
Phosphorylation of Ser303, an Inhibitory Phosphorylation Site of HSF1, Is Reduced in OGT Null Cells
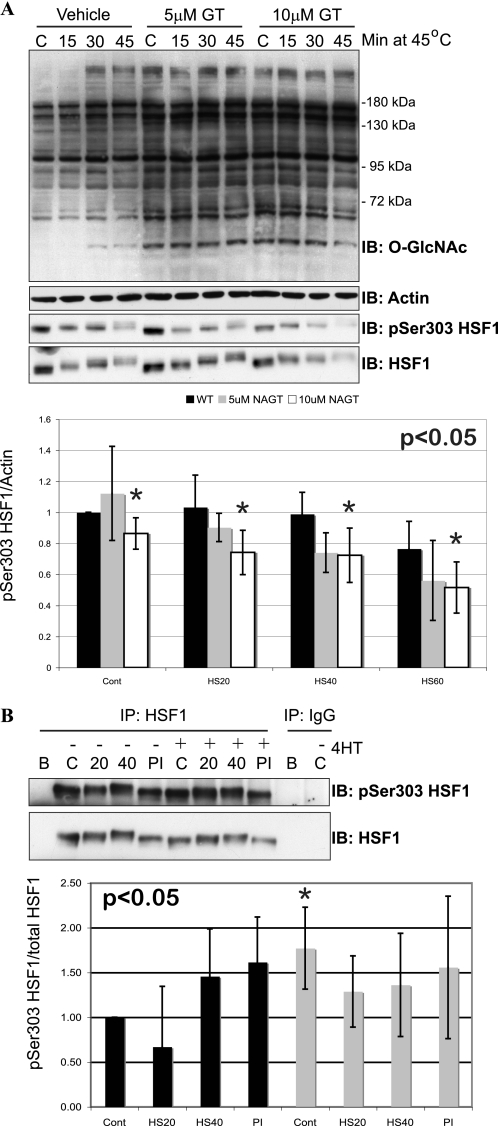
HSF1 can be inhibited by phosphorylation at Ser303, which is catalyzed by GSK-3β (73, 74). Consistent with this observation, in control cells we observe a decrease in Ser(P)303 HSF1 from cells harvested during the heat stress, and Ser(P)303 HSF1 increases again once cells are returned to 37 °C (supplemental Fig. S7B). In COS-7 cells treated with GlcNAc-thiazaline (elevates O-GlcNAc levels), we observe an enhanced rate of Ser(P)303 dephosphorylation (Fig. 5A). To confirm these data, HSF1 was immunoprecipitated from OGT wild-type and null cells. Notably, the Ser(P)303 HSF1 antibody was raised against human HSF1 and recognizes mouse HSF1 poorly in total cell lysates, but does recognize mouse HSF1 in immunoprecipitates. We observed higher basal levels of Ser(P)303 in OGT null cells; Ser(P)303 HSF1 decreases initially (HS20) in response to stress and this is dulled in OGT null cells (Fig. 5b). Consistent with these signals being dependent on phosphorylation, treatment of cells with Calyculin A (PI in figure), a protein phosphatase 1 and 2 inhibitor, also elevated the Ser(P)303 signal. To confirm this mild trend, we also overexpressed O-GlcNAcase in COS-7 cells and observed elevated levels of Ser(P)303 phosphorylation on HSF1 (supplemental Fig. S8). As we are unable to detect O-GlcNAc on HSF1, it is unlikely these data result from direct competition of O-GlcNAc and O-phosphate for the Ser303 hydroxyl group. As such, we examined the activation/inactivation of GSK-3β in response to heat stress.
FIGURE 5.
Modulating O-GlcNAc levels alters phosphorylation at Ser303. A, COS-7 cells were treated with 5 or 10 μm GlcNAc-thiazaline for 8 h. Cells were heat stressed at 45 °C for 20, 40, or 60 min. Expression of O-GlcNAc, Actin, HSF1, and Ser(P)303 HSF1 was determined in total cell lysates by immunoblot (n = 3). Densitometry representing the ratio of Ser(P)303 HSF1/Actin is reported for three experiments. p values are derived from a two-way analysis of variance comparing Ser(P)303 HSF1 phosphorylation between OGT wild-type and null cells. Error bars represent 1 standard deviation. An asterisk represents a p-value <0.05. B, HSF1 was immunoprecipitated from OGT wild-type and null cells (38h post-OGT deletion). Ser(P)303 HSF1 was determined on HSF1 immunoprecipitates (IP) by immunoblot (IB) with anti-Ser(P)303 HSF1 (n = 6). PI denotes cells treated with Calyculin A. Densitometry was performed as above, OGT wild-type cells are represented by black bars and OGT null cells in gray bars.
Phosphorylation at Ser9 of GSK-3β Is Mis-regulated in OGT Null Cells
We examined stress-induced phosphorylation at Ser9 of GSK-3β in cytosolic and nuclear extracts of OGT wild-type and null cells. Phosphorylation and inhibition of GSK-3β in cytosolic extracts was unaffected by deletion of OGT. However, in nuclear extracts of OGT null cells where GSK-3β acts on HSF1, phosphorylation at Ser9 was delayed ∼50 min (Fig. 6).
FIGURE 6.
Stress induced inactivation of GSK-3β is defective in the OGT knockout. OGT wild-type and null cells (38 h post-OGT deletion) were heat stressed (45 °C) and harvested every 10 min. The last time point was harvested 39 h post-OGT deletion. GSK-3β, Ser(P)9 GSK-3β, OGT, Total AKT/PKB, p308 AKT/PKB, p473 AKT/PKB, and Actin were detected in cytoplasmic fractions (A) or nuclear fractions (B) by immunoblot (IB). The quality of the cytoplasmic and nuclear fractions was assessed by immunoblotting for Lamin and Tubulin (n = 3). C, densitometry representing the ratio of Ser(P)9 GSK-3 β/total GSK-3β in nuclear fractions is reported for three experiments. p values are derived from a Student's t test (Paired, two-tailed) comparing Ser(P)9 GSK-3 β phosphorylation between OGT wild-type and null cells. Error bars represent 1 standard deviation. An asterisk represents a p-value <0.1, a double asterisk represents a p-value of <0.05.
Numerous kinases are known to phosphorylate GSK-3β at Ser9, including AKT/PKB, ERK, MAPK, AMPK, and mTOR (79–83, 90). Of these, the PI 3-kinase-AKT/PKB signaling pathway appears to phosphorylate GSK-3β in models of heat stress (91). Consistent with this observation, inhibition of PI 3-kinase blocks stress-induced phosphorylation of GSK-3β in the MEFs used in this study. Based on these data, and on published data showing a role for O-GlcNAc in regulating the PI 3-kinase-AKT/PKB signaling pathway (92–95), we explored a role for O-GlcNAc in mediating the stress-induced activation of AKT/PKB. We observed a mild defect in nuclear translocation of AKT/PKB (Fig. 6 and supplemental Fig. S9), which may result in the disruption of GSK-3β phosphorylation observed in OGT null cells.
Inhibition of GSK-3β in OGT Null Cells Recovers HSP72 Expression
Consistent with elevated GSK-3β activity in OGT null cells resulting in reduced HSP72 levels, inhibition of GSK-3β with either LiCl or AR-A014418 resulted in the recovery of HSP72 expression in the OGT knockout (Fig. 7).
FIGURE 7.
Inhibition of GSK-3β recovers expression of HSP72 in the OGT null. OGT wild-type and null cells (38 h post-OGT deletion) were pre-treated (1 h) with GSK-3β inhibitors LiCl2 (10 mm) (A) or AR-A014418 (100 nm) (B). Cells were stressed (45 °C, 60 min; 39 h post-OGT deletion) and recovered at 37 °C (2 h; 41 h post-OGT deletion)). HSP72 and Actin were detected in whole cell lysate by immunoblot (IB) (n = 3). Densitometry representing HSP72 expression in heat-stressed cells is reported. Densitometry from OGT null cells is depicted in gray. C, model of the mechanism by which O-GlcNAc regulates HSP expression. Error bars represent one standard deviation.
O-GlcNAcase Expression Is Reduced in OGT Null Cells
Previously, we and others have reported that modulating O-GlcNAc levels (pharmacologically or genetically) results in cells altering the expression of either OGT or O-GlcNAcase to return GlcNAcylation to baseline (31). As shown in supplemental Fig. S3, an increasing concentration of GlcNAc-thiazaline (raises O-GlcNAc levels) is accompanied by elevation of O-GlcNAcase expression. Interestingly, when GlcNAc-thiazaline-treated cells are heat stressed, and where GlcNAcylation should be elevated, the increase in O-GlcNAcase expression declines (Fig. 4). These data are consistent with the idea that the “cell can sense its state and adjust GlcNAcylation appropriately.” Based on these data, we examined the expression of O-GlcNAcase in OGT null cells. In response to 4HT treatment, the expression of O-GlcNAcase is reduced with similar kinetics to that of OGT (Fig. 1, B and C), even though the genes encoding each enzyme reside on different chromosomes. Interestingly, if cells incubated with 4HT also receive GlcNAc-thiazaline (20 μm), O-GlcNAc levels and O-GlcNAcase expression are rescued (supplemental Fig. S3 and Fig. 4). Together, these data suggest that: 1) the reduction of O-GlcNAcase is not an off-target effect of 4HT; 2) the deletion of OGT does not compromise the O-GlcNAcase gene; and 3) that the cell senses declining GlcNAcylation, a result of reduced OGT expression, and lowers O-GlcNAcase expression in an attempt to correct O-GlcNAc levels. Recently, it was reported that OGT and O-GlcNAcase interact directly (27). Raising O-GlcNAc levels with an inhibitor of O-GlcNAcase (GT; Fig. 4) stabilizes O-GlcNAcase expression in the absence of OGT, suggesting that the interaction between OGT and O-GlcNAcase is not necessary for this phenomenon.
DISCUSSION
In this paper we have investigated the mechanisms by which O-GlcNAc regulates the expression of HSPs, to gain insight into the role(s) of O-GlcNAc in mediating cell survival. To facilitate these studies, we have generated a novel cell line in which OGT deletion can be induced by the addition of 4HT.
Notably, the OGT knockout is essentially also an O-GlcNAcase null (Fig. 1). These data suggest that conclusions drawn from OGT knockouts will be complicated, as the resulting phenotype may also be the result of reduced O-GlcNAcase expression. Interestingly, the cells appear to contain a mechanism by which they can match the cellular state to GlcNAcylation levels. This is evidenced by 1) the maintenance of O-GlcNAcase expression in OGT null cells treated GlcNAc-thiazaline; and 2) that O-GlcNAcase expression is reduced in GlcNAc-thiazaline-treated cells that have been heat stressed.
Using the inducible OGT null cell line, we have screened the expression of 84 molecular chaperones and have shown that 18 proteins belonging to the HSP72 superfamily have reduced mRNA expression in the absence of OGT/O-GlcNAcase/O-GlcNAc. These data confirm our previous studies showing that O-GlcNAc regulates the expression of HSP72 (37), and implicates O-GlcNAc in the regulation of other key chaperones including: DNAJ, HSP90α, Serpin H, HSP105, GRP78, and BAG3. Many of these proteins, if not all, have been shown to promote cell survival through numerous mechanisms in response to various forms of cellular injury.
Given the key role of HSF1 in transactivating these genes (96, 97), we explored a role for O-GlcNAc in the regulation of this transcription factor. We found little evidence of HSF1 being modified by O-GlcNAc, but we did observe elevated levels of Ser(P)303 on HSF1 in OGT null cells. Phosphorylation at this site is catalyzed by GSK-3β, and inhibits HSF1 by reducing DNA binding, increasing nuclear export, and elevating binding to 14-3-3ϵ (73–75). Consistent with stressed OGT null cells having elevated GSK-3β activity, inhibition of GSK-3β recovered HSP72 expression. Underlying this observation, we observed reduced inhibition by phosphorylation at Ser9 of GSK-3β in the nuclei of OGT null cells. These data have led to a model (Fig. 7C) in which O-GlcNAc promotes phosphorylation and inhibition of GSK-3β, leading to a release of the inhibition of HSF1 and enhanced HSP synthesis. Consistent with previously published studies, the AKT/PKB-PI 3-kinase signaling pathway is required for stress-induced phosphorylation of GSK-3β at Ser9. We observed a small defect in the nuclear translocation AKT/PKB (Fig. 6 and supplemental Fig. S9), which may result in reduction in GSK-3β inactivation. Other alternatives may include altered subnuclear localization of AKT/PKB and GSK-3β, or differential association with scaffold proteins required for efficient phosphorylation of AKT/PKB substrates.
Glutamine, a key metabolite in the synthesis of UDP-GlcNAc, is also known to regulate the expression of HSP72 (78, 98–103). Previously, it was suggested that O-GlcNAcylation of HSF1, or a HSF1-binding protein, was one mechanism by which glutamine elicits an effect on HSP72 expression (78). However, in the cells used in these studies we observed no evidence of the O-GlcNAc modification on HSF1 and no defect in glutamine-induced HSP70 expression.
We, and others, have shown that O-GlcNAc appears to be one target of the cellular stress response. In this paper we have reported that O-GlcNAc regulates the expression of numerous molecular chaperones, including HSP72. Our findings that O-GlcNAc regulates the stress-induced inactivation of GSK-3β may have far reaching implications in models such as ischemic reperfusion injury. In addition to reducing heat shock protein expression through inactivation of HSF1 (discussed in this study), GSK-3β promotes the intrinsic pathway of apoptosis through numerous mechanisms: 1) GSK-3β binds to and promotes the acetylation of p53, which in turn activates proapoptotic changes in gene expression (104); 2) phosphorylation of myeloid cell leukemia sequence-1 by GSK-3β induces ubiquitination and subsequent degradation of this anti-apoptotic BCL-2 protein; 3) GSK-3β phosphorylates and inactivates the translation initiation factor eIF2B; 4) GSK-3β phosphorylates Bax at Ser163, promoting mitochondiral localization; 5) GSK-3β is required for the stress-induced expression of BIM; and 6) GSK-3β promotes collapse of mitochondrial membrane potential through opening of the mitochondrial permeability transition pore (56, 79, 104–107). The latter appears to be regulated by a number of events including direct phosphorylation by GSK-3β of VDAC, BCL-2, and p53, as well as binding of phospho-GSK-3β (inactive) to the adenine nucleotide translocase, which suppresses association with cyclophilin D reducing the sensitivity of the mitochondrial permeability transition pore to Ca2+ induced opening (105, 106). Notably, elevating O-GlcNAc levels has been shown to reduce opening of the mitochondrial permeability transition pore and to promote the maintenance of mitochondrial membrane potential during ischemia reperfusion injury (48, 50, 53, 54), as such the data presented in this study may provide molecular insight into the mechanisms underlying this phenomena. These data also suggest additional GSK-3β-dependent mechanisms by which dynamic stress-induced changes in GlcNAcylation may regulate cellular survival in response to injury. Finally, as GSK-3β plays a key role regulating the pathogenesis of Alzheimer disease, cardiac hypertrophy, as well as of many types of cancer, these data may suggest a role for O-GlcNAc in the etiology of these diseases.
Supplementary Material
Acknowledgments
We thank G. W. Hart (Department of Biological Chemistry, Johns Hopkins University School of Medicine) for the gift of antibodies, Anca Michaelis and M. K. Meffert (Department of Biological Chemistry, Johns Hopkins University School of Medicine) for the gift of constructs, Shivang Doshi and Srona Sengupta for technical help, and comments of our colleagues Dr. G. W. Hart, Dr. C. Slawson, Dr. D. Raben, and Dr. M. Wolfgang.
This work was supported by American Heart Association Grant SDG 0930162N (to N. E. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S9, and Tables S1 and S2.
- OGT
- uridine diphospho-N-acetylglucosamine:peptide β-N-acetylglucosaminyl transferase
- 4HT
- 4-hydroxytamoxifen
- GRP
- glucose-regulated protein
- GSK
- glycogen synthase kinase
- HSF1
- heat shock factor 1
- HSP
- heat shock protein
- MEF
- mouse embryonic fibroblast
- GlcNAc-thiazaline or GT
- 1,2-dideoxy-2′-methyl-α-d-glucopyranoso[2,1-d]-δ2′-thiazoline
- O-GlcNAc
- O-linked β-N-acetylglucosamine
- O-GlcNAcase
- O-GlcNAc-specific hexosaminidase
- mER
- mutated estrogen receptor
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Zachara N. E., Hart G. W. (2006) Biochim. Biophys. Acta 1761, 599–617 [DOI] [PubMed] [Google Scholar]
- 2.Zachara N. E., Hart G. W. (2004) Trends Cell Biol. 14, 218–221 [DOI] [PubMed] [Google Scholar]
- 3.Wang Z., Gucek M., Hart G. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13793–13798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanover J. A., Yu S., Lubas W. B., Shin S. H., Ragano-Caracciola M., Kochran J., Love D. C. (2003) Arch. Biochem. Biophys. 409, 287–297 [DOI] [PubMed] [Google Scholar]
- 5.Love D. C., Kochan J., Cathey R. L., Shin S. H., Hanover J. A., Kochran J. (2003) J. Cell Sci. 116, 647–654 [DOI] [PubMed] [Google Scholar]
- 6.Heese-Peck A., Raikhel N. V. (1998) Plant Cell 10, 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heese-Peck A., Cole R. N., Borkhsenious O. N., Hart G. W., Raikhel N. V. (1995) Plant Cell 7, 1459–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreppel L. K., Blomberg M. A., Hart G. W. (1997) J. Biol. Chem. 272, 9308–9315 [DOI] [PubMed] [Google Scholar]
- 9.Lubas W. A., Frank D. W., Krause M., Hanover J. A. (1997) J. Biol. Chem. 272, 9316–9324 [DOI] [PubMed] [Google Scholar]
- 10.Dong D. L., Hart G. W. (1994) J. Biol. Chem. 269, 19321–19330 [PubMed] [Google Scholar]
- 11.Gao Y., Wells L., Comer F. I., Parker G. J., Hart G. W. (2001) J. Biol. Chem. 276, 9838–9845 [DOI] [PubMed] [Google Scholar]
- 12.Wells L., Gao Y., Mahoney J. A., Vosseller K., Chen C., Rosen A., Hart G. W. (2002) J. Biol. Chem. 277, 1755–1761 [DOI] [PubMed] [Google Scholar]
- 13.Farook V. S., Bogardus C., Prochazka M. (2002) Mol. Genet. Metab. 77, 189–193 [DOI] [PubMed] [Google Scholar]
- 14.Hu Y., Suarez J., Fricovsky E., Wang H., Scott B. T., Trauger S. A., Han W., Hu Y., Oyeleye M. O., Dillmann W. H. (2009) J. Biol. Chem. 284, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shafi R., Iyer S. P., Ellies L. G., O'Donnell N., Marek K. W., Chui D., Hart G. W., Marth J. D. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Donnell N., Zachara N. E., Hart G. W., Marth J. D. (2004) Mol. Cell. Biol. 24, 1680–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zachara N. E., Hart G. W. (2004) Biochim. Biophys. Acta 1673, 13–28 [DOI] [PubMed] [Google Scholar]
- 18.Hart G. W., Chou T. Y., Jiang M. S., Greis K. D., Cole R. N., Comer F. I., Arnold C. S., Matsuoka T., Snow D. M., Hayes B. K., Kreppel L. K., Earles B. J. (1995) NIH Catalyst 3, 8–9 [Google Scholar]
- 19.Griffith L. S., Schmitz B. (1999) Eur. J. Biochem. 262, 824–831 [DOI] [PubMed] [Google Scholar]
- 20.Lefebvre T., Alonso C., Mahboub S., Dupire M. J., Zanetta J. P., Caillet-Boudin M. L., Michalski J. C. (1999) Biochim. Biophys. Acta 1472, 71–81 [DOI] [PubMed] [Google Scholar]
- 21.Wells L., Kreppel L. K., Comer F. I., Wadzinski B. E., Hart G. W. (2004) J. Biol. Chem. 279, 38466–38470 [DOI] [PubMed] [Google Scholar]
- 22.Chou T. Y., Hart G. W., Dang C. V. (1995) J. Biol. Chem. 270, 18961–18965 [DOI] [PubMed] [Google Scholar]
- 23.Kelly W. G., Dahmus M. E., Hart G. W. (1993) J. Biol. Chem. 268, 10416–10424 [PubMed] [Google Scholar]
- 24.Comer F. I., Hart G. W. (2001) Biochemistry 40, 7845–7852 [DOI] [PubMed] [Google Scholar]
- 25.Cheung W. D., Hart G. W. (2008) J. Biol. Chem. 283, 13009–13020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slawson C., Lakshmanan T., Knapp S., Hart G. W. (2008) Mol. Biol. Cell 19, 4130–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung W. D., Sakabe K., Housley M. P., Dias W. B., Hart G. W. (2008) J. Biol. Chem. 283, 33935–33941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han I., Oh E. S., Kudlow J. E. (2000) Biochem. J. 350, 109–114 [PMC free article] [PubMed] [Google Scholar]
- 29.Konrad R. J., Kudlow J. E. (2002) Int. J. Mol. Med. 10, 535–539 [PubMed] [Google Scholar]
- 30.Walgren J. L., Vincent T. S., Schey K. L., Buse M. G. (2003) Am. J. Physiol. Endocrinol. Metab 284, E424–434 [DOI] [PubMed] [Google Scholar]
- 31.Slawson C., Zachara N. E., Vosseller K., Cheung W. D., Lane M. D., Hart G. W. (2005) J. Biol. Chem. 280, 32944–32956 [DOI] [PubMed] [Google Scholar]
- 32.Slawson C., Shafii S., Amburgey J., Potter R. (2002) Biochim. Biophys. Acta 1573, 121–129 [DOI] [PubMed] [Google Scholar]
- 33.Lefebvre T., Baert F., Bodart J. F., Flament S., Michalski J. C., Vilain J. P. (2004) J. Cell. Biochem. 93, 999–1010 [DOI] [PubMed] [Google Scholar]
- 34.Majumdar G., Harmon A., Candelaria R., Martinez-Hernandez A., Raghow R., Solomon S. S. (2003) Am. J. Physiol. Endocrinol. Metab 285, E584-E591 [DOI] [PubMed] [Google Scholar]
- 35.Gewinner C., Hart G., Zachara N., Cole R., Beisenherz-Huss C., Groner B. (2004) J. Biol. Chem. 279, 3563–3572 [DOI] [PubMed] [Google Scholar]
- 36.Kearse K. P., Hart G. W. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 1701–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zachara N. E., O'Donnell N., Cheung W. D., Mercer J. J., Marth J. D., Hart G. W. (2004) J. Biol. Chem. 279, 30133–30142 [DOI] [PubMed] [Google Scholar]
- 38.Lindquist S. (1986) Annu. Rev. Biochem. 55, 1151–1191 [DOI] [PubMed] [Google Scholar]
- 39.Nollen E. A., Morimoto R. I. (2002) J. Cell Sci. 115, 2809–2816 [DOI] [PubMed] [Google Scholar]
- 40.Sohn K. C., Lee K. Y., Park J. E., Do S. I. (2004) Biochem. Biophys. Res. Commun. 322, 1045–1051 [DOI] [PubMed] [Google Scholar]
- 41.Liu J., Pang Y., Chang T., Bounelis P., Chatham J. C., Marchase R. B. (2006) J. Mol. Cell Cardiol. 40, 303–312 [DOI] [PubMed] [Google Scholar]
- 42.Yang S., Zou L. Y., Bounelis P., Chaudry I., Chatham J. C., Marchase R. B. (2006) Shock 25, 600–607 [DOI] [PubMed] [Google Scholar]
- 43.Fulop N., Zhang Z., Marchase R. B., Chatham J. C. (2007) Am. J. Physiol. Heart Circ. Physiol. 292, H2227–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fülöp N., Marchase R. B., Chatham J. C. (2007) Cardiovasc. Res. 73, 288–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Champattanachai V., Marchase R. B., Chatham J. C. (2007) Am. J. Physiol. Cell Physiol. 292, C178–187 [DOI] [PubMed] [Google Scholar]
- 46.Liu J., Marchase R. B., Chatham J. C. (2007) Am. J. Physiol. Heart Circ. Physiol. 293, H1391–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fülöp N., Zhang Z., Marchase R. B., Chatham J. C. (2007) Am. J. Physiol. Heart Circ. Physiol. 292, H2227–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones S. P., Zachara N. E., Ngoh G. A., Hill B. G., Teshima Y., Bhatnagar A., Hart G. W., Marbán E. (2008) Circulation 117, 1172–1182 [DOI] [PubMed] [Google Scholar]
- 49.Ohn T., Kedersha N., Hickman T., Tisdale S., Anderson P. (2008) Nat. Cell Biol. 10, 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ngoh G. A., Watson L. J., Facundo H. T., Dillmann W., Jones S. P. (2008) J. Mol. Cell Cardiol. 45, 313–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chatham J. C., Nöt L. G., Fülöp N., Marchase R. B. (2008) Shock 29, 431–440 [DOI] [PubMed] [Google Scholar]
- 52.Guinez C., Mir A. M., Dehennaut V., Cacan R., Harduin-Lepers A., Michalski J. C., Lefebvre T. (2008) FASEB J. 22, 2901–2911 [DOI] [PubMed] [Google Scholar]
- 53.Ngoh G. A., Jones S. P. (2008) J. Pharmacol. Exp. Ther. 327, 602–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ngoh G. A., Facundo H. T., Hamid T., Dillmann W., Zachara N. E., Jones S. P. (2009) Circ. Res. 104, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laczy B., Hill B. G., Wang K., Paterson A. J., White C. R., Xing D., Chen Y. F., Darley-Usmar V., Oparil S., Chatham J. C. (2009) Am. J. Physiol. Heart Circ. Physiol. 296, H13–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dugo L., Collin M., Thiemermann C. (2007) Shock 27, 113–123 [DOI] [PubMed] [Google Scholar]
- 57.Zou L., Yang S., Champattanachai V., Hu S., Chaudry I. H., Marchase R. B., Chatham J. C. (2009) Am. J. Physiol. Heart Circ. Physiol. 296, H515–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laczy B., Marsh S. A., Brocks C. A., Wittmann I., Chatham J. C. (September10, 2010) Am. J. Physiol. Heart Circ. Physiol. 299, H1715–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nöt L. G., Brocks C. A., Vámhidy L., Marchase R. B., Chatham J. C. (2010) Crit. Care Med. 38, 562–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lim K. H., Chang H. I. (2006) FEBS Lett. 580, 4645–4652 [DOI] [PubMed] [Google Scholar]
- 61.Nagy T., Champattanachai V., Marchase R. B., Chatham J. C. (2006) Am. J. Physiol. Cell Physiol. 290, C57–65 [DOI] [PubMed] [Google Scholar]
- 62.Huang J. B., Clark A. J., Petty H. R. (2007) Cell. Immunol. 245, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Champattanachai V., Marchase R. B., Chatham J. C. (2008) Am. J. Physiol. Cell Physiol. 294, C1509–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daugaard M., Rohde M., Jäättelä M. (2007) FEBS Lett. 581, 3702–3710 [DOI] [PubMed] [Google Scholar]
- 65.Kregel K. C. (2002) J. Appl. Physiol. 92, 2177–2186 [DOI] [PubMed] [Google Scholar]
- 66.Becker J., Craig E. A. (1994) Eur. J. Biochem. 219, 11–23 [DOI] [PubMed] [Google Scholar]
- 67.Oh K. H., Kim J. Y., Kim D., Lee E. M., Oh H. Y., Seo J. S., Han J. S., Kim S., Lee J. S., Ahn C. (2004) Transplant. Immunol. 13, 273–281 [DOI] [PubMed] [Google Scholar]
- 68.Shim E. H., Kim J. I., Bang E. S., Heo J. S., Lee J. S., Kim E. Y., Lee J. E., Park W. Y., Kim S. H., Kim H. S., Smithies O., Jang J. J., Jin D. I., Seo J. S. (2002) EMBO Rep. 3, 857–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee S. H., Kim M., Yoon B. W., Kim Y. J., Ma S. J., Roh J. K., Lee J. S., Seo J. S. (2001) Stroke 32, 2905–2912 [DOI] [PubMed] [Google Scholar]
- 70.Huang L., Mivechi N. F., Moskophidis D. (2001) Mol. Cell. Biol. 21, 8575–8591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Voellmy R. (2004) Cell Stress Chaperones 9, 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holmberg C. I., Tran S. E., Eriksson J. E., Sistonen L. (2002) Trends Biochem. Sci. 27, 619–627 [DOI] [PubMed] [Google Scholar]
- 73.Xavier I. J., Mercier P. A., McLoughlin C. M., Ali A., Woodgett J. R., Ovsenek N. (2000) J. Biol. Chem. 275, 29147–29152 [DOI] [PubMed] [Google Scholar]
- 74.Wang X., Grammatikakis N., Siganou A., Calderwood S. K. (2003) Mol. Cell. Biol. 23, 6013–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang X., Grammatikakis N., Siganou A., Stevenson M. A., Calderwood S. K. (2004) J. Biol. Chem. 279, 49460–49469 [DOI] [PubMed] [Google Scholar]
- 76.Wang X., Khaleque M. A., Zhao M. J., Zhong R., Gaestel M., Calderwood S. K. (2006) J. Biol. Chem. 281, 782–791 [DOI] [PubMed] [Google Scholar]
- 77.Westerheide S. D., Anckar J., Stevens S. M., Jr., Sistonen L., Morimoto R. I. (2009) Science 323, 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Singleton K. D., Wischmeyer P. E. (2008) JPEN J. Parenter. Enteral. Nutr. 32, 371–376 [DOI] [PubMed] [Google Scholar]
- 79.Kannoji A., Phukan S., Sudher Babu V., Balaji V. N. (2008) Expert Opin. Ther. Targets 12, 1443–1455 [DOI] [PubMed] [Google Scholar]
- 80.Beurel E., Jope R. S. (2006) Prog. Neurobiol. 79, 173–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jope R. S., Johnson G. V. (2004) Trends Biochem. Sci. 29, 95–102 [DOI] [PubMed] [Google Scholar]
- 82.Frame S., Cohen P. (2001) Biochem. J. 359, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cohen P., Goedert M. (2004) Nat. Rev. Drug Discov. 3, 479–487 [DOI] [PubMed] [Google Scholar]
- 84.Imahori K., Uchida T. (1997) J. Biochem. 121, 179–188 [PubMed] [Google Scholar]
- 85.Qing G., Yan P., Xiao G. (2006) Cell Res. 16, 895–901 [DOI] [PubMed] [Google Scholar]
- 86.Santoro M. G. (1996) EXS 77, 337–357 [DOI] [PubMed] [Google Scholar]
- 87.de Felipe P., Ryan M. D. (2004) Traffic 5, 616–626 [DOI] [PubMed] [Google Scholar]
- 88.Tannour-Louet M., Porteu A., Vaulont S., Kahn A., Vasseur-Cognet M. (2002) Hepatology 35, 1072–1081 [DOI] [PubMed] [Google Scholar]
- 89.Hatsell S., Medina L., Merola J., Haltiwanger R., Cowin P. (2003) J. Biol. Chem. 278, 37745–37752 [DOI] [PubMed] [Google Scholar]
- 90.Hartigan J. A., Xiong W. C., Johnson G. V. W. (2001) Biochem. Biophys. Res. Commun. 284, 485–489 [DOI] [PubMed] [Google Scholar]
- 91.Dajani R., Fraser E., Roe S. M., Young N., Good V., Dale T. C., Pearl L. H. (2001) Cell 105, 721–732 [DOI] [PubMed] [Google Scholar]
- 92.Soesanto Y. A., Luo B., Jones D., Taylor R., Gabrielsen J. S., Parker G., McClain D. A. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E974–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park S. Y., Ryu J., Lee W. (2005) Exp. Mol. Med. 37, 220–229 [DOI] [PubMed] [Google Scholar]
- 94.Musicki B., Kramer M. F., Becker R. E., Burnett A. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11870–11875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vosseller K., Wells L., Lane M. D., Hart G. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5313–5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Y., Huang L., Zhang J., Moskophidis D., Mivechi N. F. (2002) J. Cell. Biochem. 86, 376–393 [DOI] [PubMed] [Google Scholar]
- 97.McMillan D. R., Xiao X., Shao L., Graves K., Benjamin I. J. (1998) J. Biol. Chem. 273, 7523–7528 [DOI] [PubMed] [Google Scholar]
- 98.Hayashida N., Inouye S., Fujimoto M., Tanaka Y., Izu H., Takaki E., Ichikawa H., Rho J., Nakai A. (2006) EMBO J. 25, 4773–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peng Z. Y., Hamiel C. R., Banerjee A., Wischmeyer P. E., Friese R. S., Wischmeyer P. (2006) JPEN J. Parenter. Enteral. Nutr. 30, 400–406 [DOI] [PubMed] [Google Scholar]
- 100.Peng Z. Y., Serkova N. J., Kominsky D. J., Brown J. L., Wischmeyer P. E. (2006) JPEN J. Parenter. Enteral. Nutr. 30, 373–378 [DOI] [PubMed] [Google Scholar]
- 101.Morrison A. L., Dinges M., Singleton K. D., Odoms K., Wong H. R., Wischmeyer P. E. (2006) Am. J. Physiol. Cell Physiol. 290, C1625–1632 [DOI] [PubMed] [Google Scholar]
- 102.Eliasen M. M., Brabec M., Gerner C., Pollheimer J., Auer H., Zellner M., Weingartmann G., Garo F., Roth E., Oehler R. (2006) J. Mol. Med. 84, 147–158 [DOI] [PubMed] [Google Scholar]
- 103.Curi R., Lagranha C. J., Doi S. Q., Sellitti D. F., Procopio J., Pithon-Curi T. C., Corless M., Newsholme P. (2005) J. Cell. Physiol. 204, 392–401 [DOI] [PubMed] [Google Scholar]
- 104.Eom T. Y., Jope R. S. (2009) Mol. Cancer 8, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xi J., Wang H., Mueller R. A., Norfleet E. A., Xu Z. (2009) Eur. J. Pharmacol. 604, 111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miura T., Miki T. (2009) Circ. J. 73, 1184–1192 [DOI] [PubMed] [Google Scholar]
- 107.Forde J. E., Dale T. C. (2007) Cell Mol. Life Sci. 64, 1930–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.