Abstract
PTPD1, a cytosolic non-receptor protein-tyrosine phosphatase, stimulates the Src-EGF transduction pathway. Localization of PTPD1 at actin cytoskeleton and adhesion sites is required for cell scattering and migration. Here, we show that during EGF stimulation, PTPD1 is rapidly recruited to endocytic vesicles containing the EGF receptor. Endosomal localization of PTPD1 is mediated by interaction with KIF16B, an endosomal kinesin that modulates receptor recycling at the plasma membrane. Silencing of PTPD1 promotes degradation of EGF receptor and inhibits downstream ERK signaling. We also found that PTPD1 is markedly increased in bladder cancer tissue samples. PTPD1 levels positively correlated with the grading and invasiveness potential of these tumors. Transgenic expression of an inactive PTPD1 mutant or genetic knockdown of the endogenous PTPD1 severely inhibited both growth and motility of human bladder cancer cells. These findings identify PTPD1 as a novel component of the endocytic machinery that impacts on EGF receptor stability and on growth and motility of bladder cancer cells.
Keywords: Phosphatase, Receptor-tyrosine Kinase, Signal Transduction, Tumor, Tumor Marker
Introduction
PTPD1 is a cytosolic non-receptor tyrosine phosphatase that associates with and activates Src tyrosine kinase. PTPD1 up-regulates Src in response to growth factor stimulation and stimulates the EGF transduction pathway (1–6). The 1174-amino acid PTPD1 protein carries an N-terminal sequence homologous to the FERM (four point one ezrin-radixin-moesin) domain protein family, which includes PTPH1 and PTPMEG1. PTPD1 localizes along actin filaments and at adhesion plaques through interaction with actin and focal adhesion kinase, respectively. By recruiting Src to its targets at adhesion sites and actin filaments, PTPD1 exerts major effects on cell adhesion, scattering, and migration (7). PTPD1-Src complex also associates with AKAP121, a protein kinase A-anchoring protein that attaches to the outer mitochondrial membrane. The PTPD1-Src-AKAP121 complex is required for efficient maintenance of mitochondrial membrane potential and ATP oxidative synthesis (8–10). When not in complex with AKAP121, PTPD1 directs EGF/Src signaling to the nucleus, activating ERK1/2- and Elk1-dependent gene transcription (3). These activities of PTPD1 exemplify a mechanism by which tyrosine kinase signaling may be dynamically routed to different intracellular organelles by molecular platforms assembled at distinct subcellular locations.
Endocytosis is a biological process by which membrane receptors, ligands, nutrients, fluids, and membrane lipids are internalized by a cell into endosomal vesicles that traffic to distinct intracellular compartments (11, 12). This constitutes an essential mechanism to control the interaction between the cell and the extracellular environment. Generation and directional transport of endosomes require dynamic interaction of the vesicles with cytoskeletal elements and coordinated activation of enzymes, scaffolds, and adapter molecules (13, 14). Endocytic vesicles containing activated receptors fuse with early endosomes through a mechanism involving the small GTP-binding proteins of the Ras superfamily, namely Rabs (15–17). Rab5- and EEA1 (early endocytic antigen 1)-enriched early endosomes may rapidly recycle to the cell membrane by a Rab4-dependent mechanism or traffic to the recycling compartment that is enriched in Rab11. Maturation from early to late endosomes is characterized by accumulation of Rab7. Late endosomes cluster in proximity to the nucleus as multivesicular bodies that eventually fuse with lysosomes (18). Trafficking from early endosomes to the lysosomal degradative pathway is a control mechanism to attenuate receptor signaling (19).
In this paper, we show that PTPD1 is a novel component of the endocytic pathway that supports EGF receptor stability and mitogenic signaling. PTPD1 is required for growth and motility of urothelial cancer cells in vitro, and its expression in human bladder cancer tissue positively correlates with tumor stage and invasive potential. Thus, PTPD1 represents a novel marker and possible therapeutic target for bladder cancer.
EXPERIMENTAL PROCEDURES
Cell Lines
The human embryonic kidney cell line (HEK293), human bladder cells (J82, RT4, 5637, and HT; purchased from ATTC), human neuroblastoma cells (SK), human bladder papilloma cells (RT4), human bladder cancer cells (J82), human urinary bladder cancer cells (5637, and human bladder carcinoma cells (HT-1376) were cultured and propagated in Dulbecco's modified Eagle's medium containing 10% fetal calf serum in an atmosphere of 5% CO2.
Antibodies and Chemicals
Antibodies against the following proteins or epitopes were used: phospho-ERK (Tyr204) and ERK2 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), FLAG (Sigma), EGFR2 (Upstate Biotechnology, Inc.), Myc tag (Sigma), phosphotyrosine (Upstate), tubulin (Sigma), pancytokeratins (Sigma), Rab7 (Sigma), Rab11 (Santa Cruz Biotechnology, Inc.), LAMP-1 (Developmental Studies Hybridoma Bank, University of Iowa), EEA1 (BD Biosciences). Where indicated, we used a monoclonal anti-EGFR (anti-EGFR 108) that binds to the EGFR without affecting the interaction with its ligand (EGF). Anti-EGFR 108 was a kind gift of Dr. Oreste Segatto (Regina Elena Cancer Institute, Rome). Fluorescein- or rhodamine-tagged anti-rabbit and anti-mouse IgG secondary antibodies were purchased from Technogenetics. A polyclonal antibody directed against human PTPD1 was raised as previously described (ab2) (7). We also used an anti-PTPD1 antibody raised against the peptide sequence 618QEVSEPLTAARHAQ631 of human PTPD1 previously described (ab1) (2). Both anti-PTPD1 antibodies gave identical immunostaining patterns. The following chemicals were used: forskolin and cAMP (Sigma), H-89 (Calbiochem), and EGF (Upstate).
Plasmids and Transfection
For the hemagglutinin (HA) epitope, PTPD1 cDNA was excised from pBKS and subcloned into the pcDNA 3.1 vector (Invitrogen). HA epitope was placed at the extreme N terminus of PTPD1 or its inactive mutant PTPD1C1108S by PCR as described previously (3). The vector encoding for FLAG-tagged human PTPD1 was purchased from GeneCopoeia; FLAG-PTPD1Δ1–325 was generated by PCR using specific oligonucleotide and subcloned in pcDNA3 vector; KIF16B vectors (either wild type and mutant) were kindly provided by Dr. Hong Wan Jin (BMSI, Singapore); the vector encoding for Src (wild type and inactive mutant) was previously described (3). siGENOME duplex siRNAs of four distinct segments of human PTPD1 (siRNAPTPD1) were purchased from Dharmacon. We used three distinct siRNAPTPD1 mixtures: (a) siRNAPTPD1 SMARTpool, containing equimolar concentrations of all four duplex siRNAs; (b) siRNAPTPD1 1, containing equimolar concentrations of two duplex siRNAs (LQ-009379-01 and LQ-009379-02); and (c) siRNAPTPD1 2, containing equimolar concentrations of two duplex siRNAs (LQ-009379-03 and LQ-009379-04). siRNA duplex composed of non-targeting sequence (siRNAc) was used as control. siRNAs were transiently transfected using Lipofectamine 2000 (Invitrogen) at a final concentration of 250 pmol/ml culture medium. Plasmid transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Immunofluorescence Analysis
Cells were rinsed with PBS and fixed in 3% paraformaldehyde for 20 min. After permeabilization with 0.5% Triton X-100 in PBS for 5 min, the cells were incubated with 1× PBS, 0.1 mg/ml bovine serum albumin for 60 min at room temperature. Double immunofluorescence was carried out with the following antibodies: anti-EEA1 (Sigma) and anti-PTPD1 rabbit polyclonal. Fluorescein- or rhodamine-tagged anti-rabbit and anti-mouse IgG (Technogenetics) secondary antibodies were used. EGFR labeling studies were performed as follows. Before fixation, cells were incubated with fluorescein-labeled EGF, washed with phosphate-buffered saline, and formalin-fixed. Coverslips were analyzed by confocal microscopy. Where indicated, J82 cells were subjected to hypo-osmotic shock (lysis-squirting), as described previously (25). Coverslips were then rapidly fixed with 4% paraformaldehyde in PIPES, pH 7.0, and subjected to immunofluorescence analysis.
Immunoprecipitation and Immunoblot Analyses
Tissue samples and cells were homogenized and sonicated in lysis buffer (20 mm Tris-HCl, pH 7.4, 0.15 m NaCl, 10 mm EDTA, 1% Triton X-100) containing aprotinin (5 μg/ml), leupeptin (10 μg/ml), pepstatin (2 μg/ml), 0.5 mm PMSF, 2 mm orthovanadate, and 10 mm NaF. The lysates were cleared by centrifugation at 15,000 × g for 15 min. Cell lysates were resolved by SDS-PAGE, transferred to PROTRAN membrane, and immunoblotted with specific antibodies. ECL signals were quantified by scanning densitometry (Amersham Biosciences).
Cell Lysate Fractionation
Cells were harvested and resuspended in buffer A (250 mm mannitol, 1 mm EGTA, 25 mm Hepes, pH 7.2, 1.5 mm MgCl2) containing aprotinin (5 μg/ml), leupeptin (10 μg/ml), pepstatin (2 μg/ml), 0.5 mm PMSF, 2 mm orthovanadate, and 10 mm NaF. After centrifugation at 3,000 × g for 10 min, the supernatant was isolated and cleared by centrifugation at 9,000 × g for 10 min. Further purification was achieved by applying the new supernatant fraction to a discontinuous gradient of sucrose (0.5/1.5 m) and centrifuged at 110,000 × g for 40 min. The pellet (endosome-enriched fraction) and supernatant fractions were isolated and further analyzed.
Cell Migration Assays
Migration was assayed in a standard Transwell kit assay (Costar). 1 × 105 cells were suspended in migration medium and added to the upper chamber. At selected time points, the cells were washed with PBS, fixed in 3.7% formaldehyde for 20 min, and stained with crystal violet. Migrated cells were counted and scored. Cells that migrated into the membrane were stained with crystal violet and counted.
CFSE Analysis and DNA Synthesis
J82 cells were labeled with the fluorescent dye CFSE (Molecular Probes, Eugene, OR) used at 1 μg/ml. Flow cytometric analysis of CFSE dilution was performed by a FACSCalibur apparatus (BD Biosciences) and analyzed by Cell Quest software (BD Biosciences). DNA synthesis was monitored by thymidine incorporation. J82 cells (15,000 cells/well) were incubated with [3H]thymidine (Amersham Biosciences) (0.5 μCi/well) and harvested 6 h later from labeling. Incorporated radioactivity was measured with a β-cell plate scintillation counter, according to the manufacturer's instructions (Wallac, Gaithersburg, MD).
Human Bladder Samples
Tissue samples were isolated from patients affected by urothelial hyperplasia or benign or malignant urothelial neoplasia. Normal mucosa surrounding the bladder lesion of the same patient was used as control. Tissues were retrieved from the files of the Department of Biomorphological and Functional Sciences, Pathology Section, and Department of Urology, University “Federico II” of Naples, Italy. The risk grade was assessed based upon the categorization provided by the World Health Organization 2004 bladder grading. The scoring was performed by a single pathologist.
Immunohistochemistry
Formalin-fixed, paraffin-embedded tissues from patients subjected to cystoscopic biopsy were selected. Two polyclonal antibodies raised against distinct domains (residues 751–910 and 618–631) of human PTPD1 protein were used. The specificity of both antibodies was described previously (2, 7). Both anti-PTPD1 antibodies gave an identical immunostaining pattern. Representative sections were incubated with the listed primary antibodies overnight at 4 °C. Subsequently, the slides were incubated with biotinylated secondary antibodies, peroxidase-labeled streptavidin (LSAB kit HRP, DAKO (Carpinteria, CA)), and chromogenic substrate diaminobenzidine (Vector Laboratories, Burlingame, CA) for the development of the peroxidase activity. Omission of primary antibody and substitution with phosphate-buffered saline were used as negative controls. Section analysis was performed by two pathologists blind to the histological typing and to the follow-up data of the single cases of bladder carcinoma. Only cells with a definite membrane and cytoplasmic staining were judged as positive for each antibody.
RESULTS
PTPD1 Is a Component of the Endocytic Pathway
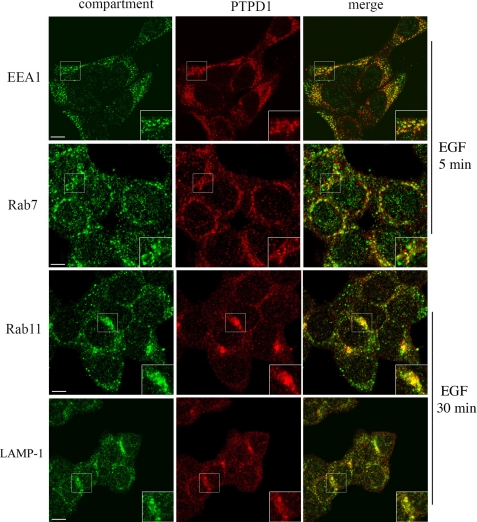
To probe the mechanism of PTPD1 action, we first determined its intracellular distribution and then asked if its expression and localization affected downstream mitogenic signaling. Immunostaining of HEK293 cells revealed PTTD1 localization at the cell periphery and cytoplasm and partly within the nucleus. Cytoplasmic PTPD1 staining showed a vesicular-like pattern, resembling that of endosomes (Fig. 1). To show that PTPD1, in fact, localizes within or is associated with endosomes, we stimulated cells with EGF and performed immunostaining analysis with antibodies directed against different endosome components. Fig. 1 shows that, following EGF stimulation (5 min), vesicles enriched for EEA1, an antigen exclusively targeted to early endosomes (20), were mostly dispersed throughout the cytoplasm. Some of these vesicles partially co-localized with those staining for PTPD1. PTPD1 staining also co-distributed with Rab7, a protein that accumulates during maturation from early to late endosomes. At a later time point (30 min) from EGF stimulation, the presence of PTPD1 in late endosomes and its selective accumulation in the multivesicular bodies was confirmed by double labeling with Rab11 (Fig. 1). Moreover, co-localization of PTPD1 with LAMP-1-enriched vesicles in proximity to the perinuclear compartment strongly supports the notion that a relevant fraction of the phosphatase traffics from late endosome/multivesicular bodies to the lysosome degradative pathway.
FIGURE 1.
PTPD1 is a component of endosomes. HEK293 cells were subjected to double immunostaining for PTPD1 and EEA1, Rab7, Rab11, or LAMP-1. Before fixation, cells were serum-deprived overnight and stimulated with EGF for 5 min (EEA1 and Rab7) and 30 min (Rab11 and LAMP-1). Fluorescence images were collected and analyzed by a confocal microscope. A merged composite for each immunostaining is shown. Magnification of selected areas is shown (insets). Bar, 10 μm.
KIF16B Anchors PTPD1 to Endosomes
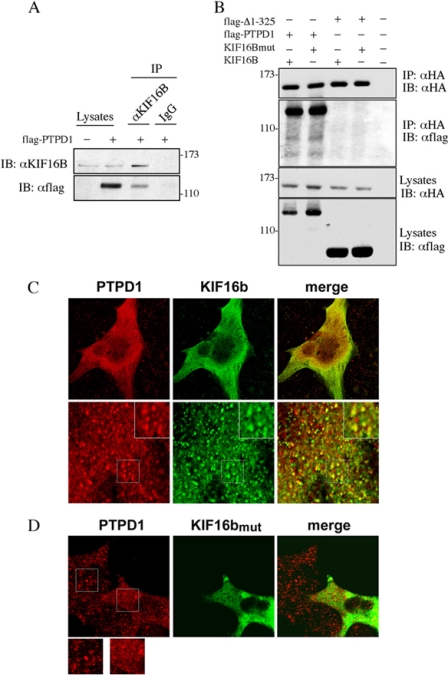
To identify the molecular determinants mediating the interaction of PTPD1 with endosomes, we performed co-imunoprecipitation assays using antibodies directed against distinct residents of endocytic vesicles. In this way, we identified endogenous KIF16B as an interacting partner of PTPD1 (Fig. 2A). KIF16B is a kinesin-3 family member that attaches to the membrane of early endosomes, regulates the motility of these organelles (21, 22), and is required for efficient EGF receptor recycling from early endosomes to cell membrane (21). Binding of KIF16B to endosomes is mediated by strong interaction between its PH domain and endosomal membrane lipids. Mutations of two PH domain residues (L1248A/F1249A) abrogate KIF16B binding to early endosomes (22). Nevertheless, the mutant KIF16B still bound PTPD1 (Fig. 2B). Interaction with KIF16B requires the FERM domain; a PTPD1 mutant lacking residues 1–325 (PTPD1Δ1–325) failed to co-precipitate with KIF16B (Fig. 2B).
FIGURE 2.
PTPD1 interacts and colocalizes with KIF16B through its FERM domain. A, lysates from HEK293 cells transiently transfected with FLAG-PTPD1 were immunoprecipitated (IP) with anti-KIF16B antibody or control IgG and immunoblotted (IB) with anti-FLAG and anti-KIF16B antibodies. B, lysates from HEK293 cells transiently co-transfected with FLAG-PTPD1 or FLAG-PTPD1Δ1–325 and HA-KIF16B or HA-L1248A/F1249A mutant were immunoprecipitated with anti-HA and immunoblotted with anti-FLAG antibody. As a control, we used a lysate from untransfected cells. C and D, cells were transiently transfected with vector encoding Myc-tagged KIF16B, either wild type (C) or L1248A/F1249A mutant (D). Where indicated, a GFP vector was included in the transfection mixture (D). Twenty-four h after transfection, cells were serum-deprived overnight and then stimulated with EGF (100 ng/ml) for 30 min. Cells were subjected to double immunostaining with anti-Myc and anti-PTPD1 antibodies. Images were collected by confocal microscopy. A merged composite and magnified images are shown.
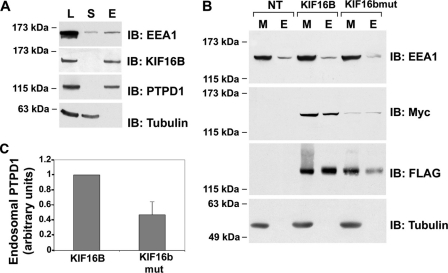
We then analyzed localization of endogenous PTPD1 in cells transfected with KIF16B vector. Fig. 2C shows partial co-localization of PTPD1 and KIF16B. To demonstrate if KIF16B targets PTPD1 to the endosomes, we transfected mutant KIF16B (L1248A/F1249A) and analyzed the intracellular distribution of endogenous PTPD1. To identify transfected cells, a vector encoding for green fluorescent protein (GFP) was included in the transfection mixture. As shown in Fig. 2D, cells expressing mutant KIF16B (GFP-positive) present a more diffuse PTPD1 staining, compared with surrounded untransfected (GFP-negative) cells. Fractionation assays confirmed that endogenous PTPD1 and KIF16B partly co-purified with EEA1 (Fig. 3A), as FLAG-PTPD1 and Myc-KIF16B (Fig. 3B). As expected, the amount of KIF16B mutant associated with membrane (M) and endosomal (E) fractions was significantly reduced (Fig. 3B). Accordingly, expression of mutant KIF16B reduced the levels of PTPD1 recovered in the endosome-enriched fraction (Fig. 3, B and C), supporting the notion that KIF16B anchors PTPD1 to endosomes.
FIGURE 3.
KIF16B anchors PTPD1 to endosomes. A, lysates (L), supernatant (S), and endosomal (E) fractions were isolated from HEK293 lysates and immunoblotted (IB) for PTPD1, EEA1, KIF16B, and tubulin. B, total membranes (M) and endosomal (E) fractions were purified from HEK293 cells transiently transfected with Myc-KIF16B (either wild type or mutant) and FLAG-PTPD1 vectors. Protein fractions were immunoblotted with the indicated antibodies. C, quantitative analysis of the experiments shown in B. A mean value ± S.E. (error bars) of five independent experiments is shown.
Taken together, these findings indicate that a significant fraction of PTPD1 is retained by endosomes through physical interaction with KIF16B. The other PTPD1 vesicles may be associated with the Golgi or endoplasmic reticulum because PTPD1 is known to interact with KIFIC, a component of these organelles (23).
PTPD1 Regulates EGFR Stability
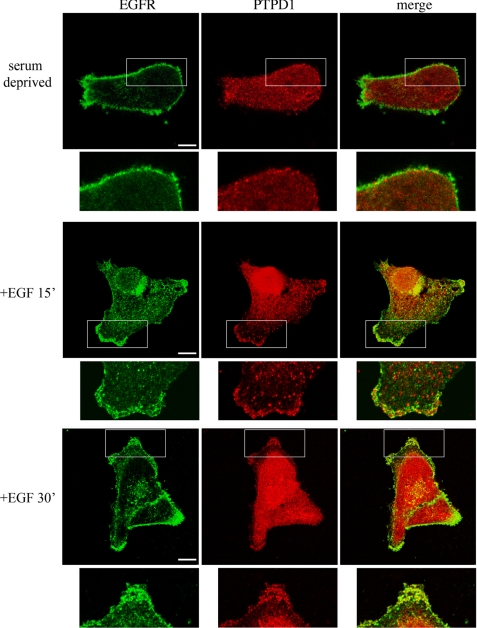
In mammalian epithelial cells, ligand binding induces EGFR phosphorylation and activation. Activated EGFR routes through the endocytic pathway, where it phosphorylates and modulates the activity of a large number of substrates and effector molecules (24, 25). Sorting of the receptor from early endosomes to the lysosomal degradative pathway attenuates EGF signaling and promotes receptor desensitization. However, a significant fraction of activated EGFR is retained within the early endocytic vesicles. This fraction evades proteolysis and is recycled and redirected to the plasma membrane (19, 26). KIF16B mediates EGFR recycling to the membrane and attenuates receptor turnover (21). We showed above that PTPD1 is targeted to endosomes through interaction with KIF16B. Given the positive role of the phosphatase in sustaining EGF signaling (3), we suspected that the endosomal fraction of PTPD1 might control EGFR recycling and stability. To test this notion, we first determined the intracellular distribution of EGFR and PTPD1 in quiescent or EGF-treated J82 urothelial tumor cells. Fig. 4 shows that EGFR was mostly distributed along the plasma membrane in serum-deprived cells, whereas PTPD1 staining was dispersed throughout the cytoplasm and within the nucleus. EGF stimulation induced vesicular clustering of EGFR at the plasma membrane. After 15 or 30 min of EGF stimulation, the PTPD1 signal partially clustered in vesicles, some of which overlapped with EGFR. These findings indicate that EGF stimulation recruits a fraction of PTPD1 to the same membrane compartment where EGFR accumulates. Co-localization of PTPD1 and EGFR was not due to a direct physical interaction between the two proteins. Co-immunoprecipitation experiments revealed no association between PTPD1 and EGFR in lysates from serum-deprived or EGF-treated cells (data not shown).
FIGURE 4.
PTPD1 co-localizes with EGF receptor. J82 cells were serum-deprived overnight (top) and stimulated with EGF (100 ng/ml) for 15 min (middle) and 30 min (bottom). Before fixation, cells were pulsed with mouse anti-EGFR antibody (see “Experimental Procedures”), washed with phosphate-buffered saline, and formalin-fixed. Immunostaining was performed using anti-PTPD1 antibody.
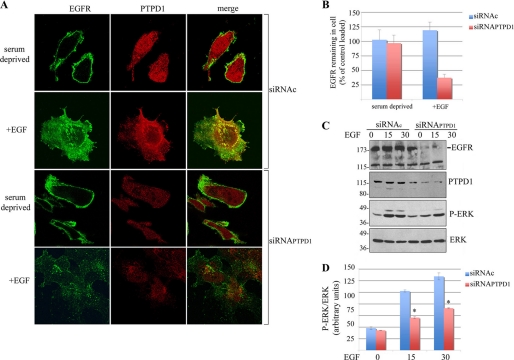
To ask if PTPD1 regulates EGFR routing and stability, we analyzed the distribution of the receptor in cells where PTPD1 expression was silenced. J82 cells were transiently transfected with siRNA duplexes targeting endogenous PTPD1 (siRNAPTPD1) or with nonspecific siRNAs as control. Twenty-four h after transfection, cells were serum-deprived and then stimulated with EGF for 30 min. As shown above, EGF stimulation induced co-localization of PTPD1 and EGFR (Fig. 5A). As expected, transfection with siRNAPTPD1 decreased PTPD1 levels, both in basal and EGF-stimulated cells. In serum-deprived control cells, EGFR staining was distributed along the plasma membrane. However, in contrast to the control cells, EGF stimulation in PTPD1-deficient cells markedly dispersed and significantly reduced total EGFR staining (Fig. 5, A and B). Silencing of PTPD1 and concomitant down-regulation of EGFR was confirmed by immunoblot analysis (Fig. 5C, upper panels). Accordingly, EGF-induced phosphorylation of the downstream effector kinase ERK was inhibited by PTPD1 silencing, compared with controls (Fig. 5, C (lower panels) and D). This suggests that PTPD1 is, indeed, required for EGFR recycling and stability and for downstream mitogenic signaling.
FIGURE 5.
PTPD1 regulates EGFR stability. A, J82 cells were transiently transfected with siRNAPTPD1 or siRNAc. Twenty-four h after transfection, cells were serum-deprived overnight and then stimulated with EGF (100 ng/ml) for 30 min. Before fixation, cells were pulsed with mouse anti-EGFR, washed with phosphate-buffered saline, and formalin-fixed. Cells were subjected to immunostaining for PTPD1. Images were collected by confocal microscopy. A merged composite is shown on the right. B, quantification of fluorescence signals in cells subjected to pulse-chase experiments as described in A. Fluorescence values ± S.E. (error bars) were normalized to the value of control, serum-deprived cells. C, total lysates from J82 transfected cells (as in A) were subjected to immunoblot analysis with the indicated antibodies. A representative set of autoradiograms is shown. D, quantitative analysis of the experiments shown in C. A mean value ± S.E. of four independent experiments is shown. *, p < 0.01 versus control (siRNAc). P-ERK, phospho-ERK.
PTPD1 Is Required for Motility and Growth of Urothelial Tumor Cells
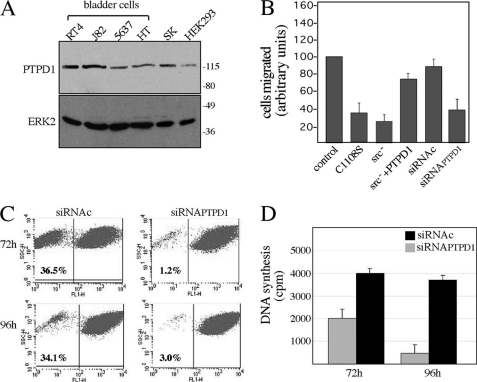
We showed previously that PTPD1 is required for proper cell adhesion and migration (7). PTPD1 mRNA is up-regulated in several forms of cancer (27, 28). Up-regulated EGFR signaling has been mechanistically linked to the malignant behavior of several epithelial human tumors, including bladder cancer. This led us to ask if and how PTPD1 contributes to the phenotype of urothelial cancer cells. First, we measured PTPD1 expression in four human bladder tumor cell lines (J82, RT4, 5637, and HT-1376). Fig. 6A shows that PTPD1 is expressed at moderate (5637 and HT1376) to high levels (J82 and RT4) in these lines. PTPD1 was also detected in human kidney (HEK293) and neuroblastoma (SK) cell lines.
FIGURE 6.
PTPD1 is required for growth and motility of bladder cancer cells. A, immunoblot analysis for PTPD1 on total lysates from human bladder cells (RT4, J82, 5637, and HT), human neuroblastoma (SK), and human embryonic kidney (HEK293) cell lines. B, motility assays from J82 cells transiently transfected with vectors expressing the following transgenes: CMV (control), HA-PTPD1C1108S, dominant negative Src (Src−), HA-PTPD1, siRNAPTPD1, and siRNAc. Twenty-four h after transfection, cells were plated on a Transwell apparatus and incubated for an additional 24 h. Migrated cells were fixed, stained, and counted. Cumulative data are presented as mean ± S.E. (error bars) of 3–5 independent experiments. Values from control (CMV) cells were set as 100. C, fluorescence-activated cell sorter analysis on J82 transiently transfected with siRNAPTPD1 or siRNAc. Cells were harvested and analyzed by FACS at 72 h (upper panels) and 96 h (lower panels) after transfection. The experiment shown is representative of four independent experiments that gave similar results. D, J82 cells (15,000 cells/well) were transiently transfected with siRNAPTPD1 or siRNAc. DNA synthesis was monitored by thymidine (0.5 μCi/well) incorporation at 72 and 96 h after transfection. Data are expressed as mean ± S.E.
To causally link PTPD1 to urothelial cancer cell malignancy, we varied the levels and activity of PTPD1 and monitored the growth and motility of bladder cells in vitro. J82 cells were transfected with a catalytically inactive PTPD1 mutant (C1108S) and assayed for motility. Fig. 6B shows that expression of C1108S markedly inhibited cell motility. Motility was also inhibited by expression of an inactive Src mutant (Src−). This is probably due to titration of PTPD1 by the Src mutant because motility was restored when PTPD1 was co-expressed with the mutant. Additional evidence for the role of PTPD1 in bladder cell motility was obtained by reducing PTPD1 levels with siRNAs. Thus, PTPD1 silencing also markedly inhibited cell motility (Fig. 6B). In addition, PTPD1 down-regulation significantly inhibited cell growth, as demonstrated by fluorescence-activated cell sorter analysis performed at various times following transfection (Fig. 6C). A requirement for PTPD1 for robust growth was confirmed by monitoring DNA synthesis in siRNAPTPD1-transfected cells by [3H]thymidine incorporation. Down-regulation of PTPD1 reduced DNA synthesis 3-fold (Fig. 6D).
PTPD1 Is Overexpressed in Urothelial Carcinoma
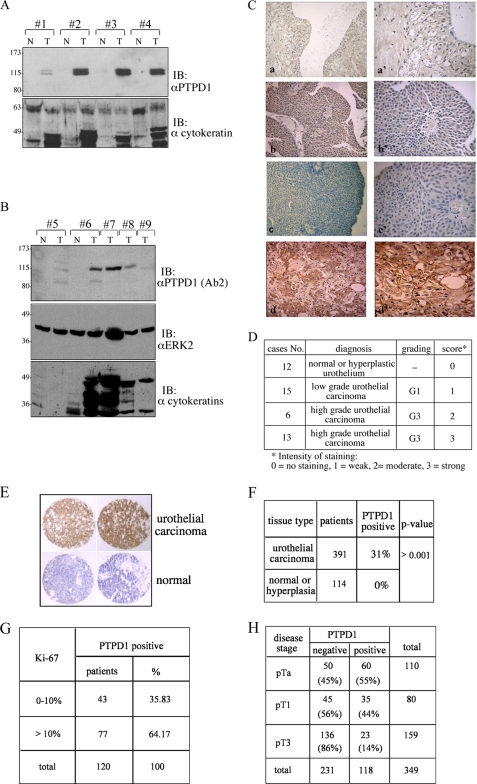
Our findings indicate that PTPD1 regulates critical aspects of cell-cell communication, growth and motility of urothelial cancer cells. This led us to investigate the expression profile of PTPD1 in human bladder cancer tissues, comparing tumors with different recurrence rates and metastatic potential. Specimens with a diagnosis of urothelial hyperplasia, urothelial papilloma, and low or high grade urothelial carcinoma were obtained from 46 patients. Tissue samples were homogenized, and protein lysates were immunoblotted with anti-PTPD1 antibody. Fig. 7A shows that PTPD1 was nearly undetectable in normal bladder tissue, hyperplastic urothelium, and urothelial papilloma, whereas low levels were visible in low grade urothelial carcinoma. Notably, elevated PTPD1 concentrations were seen in samples derived from high grade urothelial carcinomas. These tumors express high levels of cytokeratins, which are typical molecular markers of epithelial bladder cancer (29, 30). Similar findings were obtained using an anti-PTPD1 antibody raised against a different polypeptide epitope (Fig. 7B).
FIGURE 7.
PTPD1 is highly expressed in bladder carcinomas. A and B, tumor samples (T) were isolated from patients affected by high grade (lanes 2, 3, 4, 6, 7, and 8) or low grade (lanes 1, 5, and 9) urothelial carcinoma. Normal tissue (N) surrounding each neoplastic lesion was also isolated. Tissue samples were lysed, resolved on 8% SDS-polyacrylamide gels, and immunoblotted (IB) with the following antibody: anti-peptide PTPD1 (ab1) (A) or anti-polypeptide PTPD1 (ab2) (B), anti-ERK2, and anti-cytokeratins. C, tissue sections from normal bladder (a), hyperplastic bladder (b and c), and high grade (d) of urothelial carcinoma were immunostained with anti-PTPD1 antibody and analyzed by light microscopy. Higher resolution panels (a′, b′, c′, and d′) of each set of images are shown on the right. D, bladder lesions were grouped into three subcategories: normal/hyperplastic, low grade urothelial carcinoma, and high grade urothelial carcinoma. Cumulative data and relative abundance of PTPD1 in each category are shown. E, a tissue microarray of 505 bladder samples ranging from normal tissue to benign lesions and urothelial carcinomas was immunostained with anti-PTPD1 polyclonal antibody. Shown is an enlarged section of representative biopsies of normal and cancer lesions immunostained with anti-PTPD1 antibody. F, cumulative data are expressed as the percentage of PTPD1-positive samples within the two main categories (normal/hyperplastic lesions and urothelial carcinomas). p value is indicated on the right. G, PTPD1-positive urothelial carcinomas were scored for Ki-67 positivity. The cut-off value represents the percentage of Ki-67-positive cells versus total cells scored. H, inverse correlation between bladder stage disease (pTa, pT1, and pT3) and PTPD1 signal. The analysis was carried out on a total of 349 patients with urothelial carcinoma.
To further address this issue, we performed immunohistochemistry analysis on bladder tissue derived from the same specimens described in Fig. 1. The results shown in Fig. 7C are consistent with the immunoblotting data. PTPD1 accumulated in high grade urothelial carcinoma, whereas PTPD1 levels were low to nearly absent in other tissue samples (normal urothelium, urothelial hyperplasia, urothelial papilloma, and low grade urothelial carcinoma). The results of this analysis, which was performed on a total of 46 patients, are summarized in Fig. 7D. A score of 0–3 was given. We also evaluated PTPD1 expression in a large number of human bladder cancers by tissue microarray analysis. We used a preexisting bladder tissue microarray with clinical follow-up data containing 391 formalin-fixed bladder cancer tissues and 114 normal bladder mucosa. Fig. 7E shows overexpression of PTPD1 in two representative urothelial carcinomas, compared with normal bladder tissues. Immunohistochemical analysis (Fig. 7F) shows that PTPD1 was overexpressed in 120 bladder tumors (31%), whereas a low or undetectable immunoreactive signal was obtained in other samples, including normal or hyperplastic urothelium. Ki-67 is a proliferative marker, and its cut-off value of 10% is commonly used as predictive parameter of bladder cancer recurrence and progression (31, 32). As shown in Fig. 7G, a greater number of PTPD1-positive sections were evident in malignant lesions with a Ki-67 cut-off value of >10%, compared with those lesions with a Ki-67 value of <10% (64% versus 36%, respectively).
Next, we assessed whether PTPD1 immunoreactivity correlates with the staging of bladder disease. As shown in Fig. 7H, urothelial bladder cancers in an early developmental stage (pTa) include more PTPD1-positive cells (60%) compared with cancers in intermediate (pT1) (35%) or advanced (pT3) (23%) disease stages. The inverse correlation between PTPD1 expression and disease progression might reflect a requirement of PTPD1 in an early step of tumor progression, when cells first acquire a high proliferative rate and a more invasive behavior.
DISCUSSION
In the present study, we show that PTPD1, an activator of Src tyrosine kinase, is a newly identified component of the endocytic pathway. PTPD1 silencing enhanced ligand-induced degradation of EGFR and slowed the growth and motility of bladder cancer cells, consistent with a role of PTPD1 in tumorigenesis. Accordingly, we found that PTPD1 is strongly up-regulated in high grade bladder cancer lesions.
Several steps of the EGFR pathway are positively regulated by Src. Functional interference with Src activity profoundly inhibits downstream propagation of EGF signals (33). By activating Src, PTPD1 up-regulates the EGF-dependent mitogenic pathway in a wide variety of normal and cancer cells. PTPD1 is also a component of the multivalent scaffold complex nucleated by FAK at specific intracellular sites. By modulating Src-FAK signaling at adhesion sites, PTPD1 promotes cytoskeleton events required for cell adhesion and migration (7). In this paper, we have provided a novel insight into the role of PTPD1 in specialized cell functions. Our data indicate that EGF stimulation promotes the recruitment of a significant fraction of PTPD1 into endosomes through interaction with KIF16B, a component of the endocytic pathway. KIF16B is a kinesin family motor protein that regulates intracellular transport of early endosomes along microtubules, controlling the stability and signaling of membrane receptors. KIF16B overexpression enhances recycling of internalized receptors to the cell membrane, whereas down-regulation of KIF16B impairs endocytic cargo movement and promotes receptor degradation (21). Similarly, recruitment of PTPD1 to endosomes is functionally linked to receptor stability. Thus, genetic knockdown of endogenous PTPD1 dramatically decreased the levels of the EGFR and impaired downstream phosphorylation of mitogenic kinases, deeply impacting on cell growth and motility. This implies that PTPD1 may have a synergistic role with KIF16B in favoring recycling of internalized EGFR through the endocytic pathway. PTPD1 also interacts with KIF1C, an evolutionarily related KIF family member that is potentially involved in retrograde transport of vesicles from the Golgi to the endoplasmic reticulum (23). These observations strongly support a more general role of PTPD1 as a cargo-associated protein that controls the route and recycling of vesicles between cell surface and specialized intracellular compartments. Whether PTPD1 dephosphorylates endosomal targets or acts as scaffold protein for signaling enzymes/adapter molecules remains to be experimentally addressed.
We analyzed PTPD1 levels in human bladder lesions. We found that transition from benign lesions to low and high grade malignant tumors, as determined by Ki-67 levels, is marked by sustained accumulation of PTPD1. Bladder cancer usually begins as a superficial protrusion that can be located at the mucosa (Ta tumors) or the submucosa (invasive T1 tumors). The tumors can be flat, with features of high grade dysplasia (carcinoma in situ). Ta and T1 tumors are often recurrent (50–70%). Carcinoma in situ lesions commonly represent the precursor stage of aggressive carcinoma (29, 34, 35). EGF signaling and EGFR are implicated in different aspects of bladder cancer. Increased EGF signaling is associated with progression of superficial bladder lesions carrying p53 or Rb mutations to an invasive phenotype (36). Development of invasiveness entails sequential activation of distinct biochemical pathways that lead to disruption of the extracellular matrix and increased cell motility (37, 38). The role of EFGR in this pathway is unclear. We found that the percentage of PTPD1-positive cancer tissues inversely correlated with tumor expansion. PTPD1 is more abundant in bladder lesions at the early stage (pTa) of disease progression than in the late advanced (pT3) stages, when there is muscle invasion. Based on this data, we hypothesize that PTPD1 is a positive regulator of membrane receptor recycling that underlies transition from low to high grade cancer lesions. PTPD1 may act in the early steps of tumor development when EGF signaling is necessary for cell growth, matrix invasion, and spreading to the surrounding tissue. Consistent with the above model, silencing of PTPD1 down-regulated EGFR levels and downstream signaling, inhibiting the growth and motility of bladder cancer cells.
Taken together, these findings point to PTPD1 as a novel component of the endocytic pathway that controls the strength and duration of receptor signaling. Up-regulation of PTPD1 in human bladder cancer may contribute to increased receptor recycling and downstream signaling events, promoting progression of the disease. Ultimately, PTPD1 may serve as a novel biomarker for bladder tumor invasiveness and a potential target for cancer therapy.
Acknowledgments
We thank Dr. Oreste Segatto for kindly providing the mouse anti-EGFR antibody and Dr. Annalisa Morano for generating PTPD1 mutants.
This work was supported by a grant from Associazione Italiana per la Ricerca sul Cancro and Italian Ministry of University and Research MIUR 2008–2009 Grant 2007KS47FW_002, and Union for International Cancer Control Study Grant YY1/09/012 (to A. F.).
- EGFR
- epidermal growth factor receptor; 5,6-carboxyfluorescein diacetate succinimidyl ester
- siRNAc
- control siRNA duplex composed of non-targeting sequence.
REFERENCES
- 1.Andersen J. N., Jansen P. G., Echwald S. M., Mortensen O. H., Fukada T., Del Vecchio R., Tonks N. K., Møller N. P. (2004) FASEB J. 18, 8–30 [DOI] [PubMed] [Google Scholar]
- 2.Møller N. P., Møller K. B., Lammers R., Kharitonenkov A., Sures I., Ullrich A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7477–7481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardone L., Carlucci A., Affaitati A., Livigni A., DeCristofaro T., Garbi C., Varrone S., Ullrich A., Gottesman M. E., Avvedimento E. V., Feliciello A. (2004) Mol. Cell. Biol. 24, 4613–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jui H. Y., Tseng R. J., Wen X., Fang H. I., Huang L. M., Chen K. Y., Kung H. J., Ann D. K., Shih H. M. (2000) J. Biol. Chem. 275, 41124–41132 [DOI] [PubMed] [Google Scholar]
- 5.Ostman A., Hellberg C., Böhmer F. D. (2006) Nat. Rev. Cancer 6, 307–320 [DOI] [PubMed] [Google Scholar]
- 6.Barr A. J., Ugochukwu E., Lee W. H., King O. N., Filippakopoulos P., Alfano I., Savitsky P., Burgess-Brown N. A., Müller S., Knapp S. (2009) Cell 136, 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlucci A., Gedressi C., Lignitto L., Nezi L., Villa-Moruzzi E., Avvedimento E. V., Gottesman M., Garbi C., Feliciello A. (2008) J. Biol. Chem. 283, 10919–10929 [DOI] [PubMed] [Google Scholar]
- 8.Livigni A., Scorziello A., Agnese S., Adornetto A., Carlucci A., Garbi C., Castaldo I., Annunziato L., Avvedimento E. V., Feliciello A. (2006) Mol. Biol. Cell 17, 263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlucci A., Lignitto L., Feliciello A. (2008) Trends Cell Biol. 18, 604–613 [DOI] [PubMed] [Google Scholar]
- 10.Carlucci A., Adornetto A., Scorziello A., Viggiano D., Foca M., Cuomo O., Annunziato L., Gottesman M., Feliciello A. (2008) EMBO J. 27, 1073–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorkin A., von Zastrow M. (2009) Nat. Rev. Mol. Cell Biol. 10, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doherty G. J., McMahon H. T. (2009) Annu. Rev. Biochem. 78, 857–902 [DOI] [PubMed] [Google Scholar]
- 13.Sandilands E., Frame M. C. (2008) Trends Cell Biol. 18, 322–329 [DOI] [PubMed] [Google Scholar]
- 14.Simonsen A., Tooze S. A. (2009) J. Cell Biol. 186, 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sann S., Wang Z., Brown H., Jin Y. (2009) Trends Cell Biol. 19, 317–324 [DOI] [PubMed] [Google Scholar]
- 16.Zwang Y., Yarden Y. (2009) Traffic 10, 349–363 [DOI] [PubMed] [Google Scholar]
- 17.Mosesson Y., Mills G. B., Yarden Y. (2008) Nat. Rev. Cancer 8, 835–850 [DOI] [PubMed] [Google Scholar]
- 18.Murphy J. E., Padilla B. E., Hasdemir B., Cottrell G. S., Bunnett N. W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17615–17622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorkin A., Goh L. K. (2009) Exp. Cell Res. 315, 683–696 [DOI] [PubMed] [Google Scholar]
- 20.Mu F. T., Callaghan J. M., Steele-Mortimer O., Stenmark H., Parton R. G., Campbell P. L., McCluskey J., Yeo J. P., Tock E. P., Toh B. H. (1995) J. Biol. Chem. 270, 13503–13511 [DOI] [PubMed] [Google Scholar]
- 21.Hoepfner S., Severin F., Cabezas A., Habermann B., Runge A., Gillooly D., Stenmark H., Zerial M. (2005) Cell 121, 437–450 [DOI] [PubMed] [Google Scholar]
- 22.Blatner N. R., Wilson M. I., Lei C., Hong W., Murray D., Williams R. L., Cho W. (2007) EMBO J. 26, 3709–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorner C., Ciossek T., Müller S., Møller P. H., Ullrich A., Lammers R. (1998) J. Biol. Chem. 273, 20267–20275 [DOI] [PubMed] [Google Scholar]
- 24.Hynes N. E., MacDonald G. (2009) Curr. Opin. Cell Biol. 21, 177–184 [DOI] [PubMed] [Google Scholar]
- 25.Bogdan S., Klämbt C. (2001) Curr. Biol. 11, R292–R295 [DOI] [PubMed] [Google Scholar]
- 26.Eden E. R., White I. J., Futter C. E. (2009) Biochem. Soc. Trans. 37, 173–177 [DOI] [PubMed] [Google Scholar]
- 27.Dunican D. S., McWilliam P., Tighe O., Parle-McDermott A., Croke D. T. (2002) Oncogene 21, 3253–3257 [DOI] [PubMed] [Google Scholar]
- 28.Dyrskjøt L., Kruhøffer M., Thykjaer T., Marcussen N., Jensen J. L., Møller K., Ørntoft T. F. (2004) Cancer Res. 64, 4040–4048 [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Carbayo M., Socci N. D., Lozano J., Saint F., Cordon-Cardo C. (2006) J. Clin. Oncol. 24, 778–789 [DOI] [PubMed] [Google Scholar]
- 30.Sanchez-Carbayo M., Socci N. D., Lozano J. J., Haab B. B., Cordon-Cardo C. (2006) Am. J. Pathol. 168, 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanchet P., Droupy S., Eschwege P., Viellefond A., Paradis V., Pichon M. F., Jardin A., Benoit G. (2001) Eur. Urol. 40, 169–175 [DOI] [PubMed] [Google Scholar]
- 32.Liedberg F., Anderson H., Chebil G., Gudjonsson S., Höglund M., Lindgren D., Lundberg L. M., Lövgren K., Fernö M., Månsson W. (2008) Urol. Oncol. 26, 17–24 [DOI] [PubMed] [Google Scholar]
- 33.Kim L. C., Song L., Haura E. B. (2009) Nat. Rev. Clin. Oncol. 6, 587–595 [DOI] [PubMed] [Google Scholar]
- 34.Blaveri E., Simko J. P., Korkola J. E., Brewer J. L., Baehner F., Mehta K., Devries S., Koppie T., Pejavar S., Carroll P., Waldman F. M. (2005) Clin. Cancer Res. 11, 4044–4055 [DOI] [PubMed] [Google Scholar]
- 35.Kaufman D. S., Shipley W. U., Feldman A. S. (2009) Lancet 374, 239–249 [DOI] [PubMed] [Google Scholar]
- 36.Baffa R., Letko J., McClung C., LeNoir J., Vecchione A., Gomella L. G. (2006) J. Exp. Clin. Cancer Res. 25, 145–160 [PubMed] [Google Scholar]
- 37.Wallerand H., Reiter R. R., Ravaud A. (2008) Curr. Opin. Urol. 18, 524–532 [DOI] [PubMed] [Google Scholar]
- 38.Black P. C., Dinney C. P. (2008) Curr. Urol. Rep. 9, 55–61 [DOI] [PubMed] [Google Scholar]