Abstract
PA700, the 19 S regulatory subcomplex of the 26 S proteasome, contains a heterohexameric ring of AAA subunits (Rpt1 to -6) that forms the binding interface with a heteroheptameric ring of α subunits (α1 to -7) of the 20 S proteasome. Binding of these subcomplexes is mediated by interactions of C termini of certain Rpt subunits with cognate binding sites on the 20 S proteasome. Binding of two Rpt subunits (Rpt2 and Rpt5) depends on their last three residues, which share an HbYX motif (where Hb is a hydrophobic amino acid) and open substrate access gates in the center of the α ring. The relative roles of other Rpt subunits for proteasome binding and activation remain poorly understood. Here we demonstrate that the C-terminal HbYX motif of Rpt3 binds to the 20 S proteasome but does not promote proteasome gating. Binding requires the last three residues and occurs at a dedicated site on the proteasome. A C-terminal peptide of Rpt3 blocked ATP-dependent in vitro assembly of 26 S proteasome from PA700 and 20 S proteasome. In HEK293 cells, wild-type Rpt3, but not Rpt3 lacking the HbYX motif was incorporated into 26 S proteasome. These results indicate that the C terminus of Rpt3 was required for cellular assembly of this subunit into 26 S proteasome. Mutant Rpt3 was assembled into intact PA700. This result indicates that intact PA700 can be assembled independently of association with 20 S proteasome and thus may be a direct precursor for 26 S proteasome assembly under normal conditions. These results provide new insights to the non-equivalent roles of Rpt subunits in 26 S proteasome function and identify specific roles for Rpt3.
Keywords: ATP-dependent Protease, Proteasome, Protein Assembly, Protein Degradation, Ubiquitin
Introduction
ATP-dependent protease complexes commonly comprise two distinct subcomplexes: a cylinder-shaped protease with internally sequestered catalytic sites and an ATPase regulatory module required for delivery of substrates to those sites (1–3). The eukaryotic 26 S proteasome represents the most structurally and functionally elaborate example of such complexes (4, 5). Its protease subcomplex, the 20 S proteasome, contains two copies each of 14 different gene products arranged as four axially stacked heteroheptameric rings (6, 7). Each identical outer ring contains seven different α-type subunits (α1–α7), and each identical inner ring contains seven different β-type subunits (β1–β7). Three of the seven β-type subunits feature N-terminal threonine residues that serve as catalytic nucleophiles and line an interior chamber in the center of the barrel-shaped structure (8, 9). The regulatory subcomplex of 26 S proteasome, known as PA700 or 19 S regulator, contains about 20 different gene products, including six distinct ATPases associated with various activities (AAA)2 subunits (Rpt1 to -6) (4, 10). The Rpt subunits are arranged in a hexameric ring that forms the binding interface of PA700 with the α rings of the 20 S proteasome (11–14). Binding of PA700 to the 20 S proteasome results in repositioning of interlaced N-terminal peptides of α subunits that normally occlude a narrow pore in the center of the α ring (15–19). This conformational rearrangement opens a route for substrates to reach the otherwise inaccessible catalytic sites in the interior of the proteasome. Although short peptides and some unstructured proteins pass the opened pore by simple diffusion, most physiological substrates of the 26 S proteasome are folded proteins covalently modified with a polyubiquitin chain (20–22). Polyubiquitin serves as the principal method of targeting protein substrates to the proteasome via polyubiquitin-binding subunits of PA700, but its client substrates require additional processing by PA700 for delivery to the sites of proteolysis (23, 24). Substrate processing includes unfolding, detachment from the polyubiquitin chain by resident deubiquitylating subunits, and translocation through the open pore. These coordinated activities appear mechanistically linked to one another and to Rpt-catalyzed ATP hydrolysis (21, 25, 26). Although molecular details of this coordination and linkage remain poorly understood, the Rpt subunits of PA700 are topologically situated and functionally suited to play a central role in proteasome function.
In addition to the obligatory role of ATP for 26 S proteasome degradation of polyubiquitylated proteins, ATP also is necessary for PA700 binding to and activation of the 20 S proteasome (27, 28). However, unlike the former process, the latter requires ATP binding but not hydrolysis (21, 22). Thus, the ATP-bound state of one or more Rpt subunits probably promotes a conformation in the Rpt subunit ring that optimizes its interaction with cognate binding sites on the α subunit ring of the 20 S proteasome. Considerable insight into the molecular details of binding and consequent proteasome activation has been achieved from studies of 20 S proteasome-ATPase regulatory complexes in archaea. This structurally simpler system features a 20 S proteasome composed of a homoheptameric α ring and an ATPase regulator, proteasome-activating nucleotidase (PAN), composed of a homohexameric AAA subunit ring lacking additional non-ATPase subunits (3, 29, 30). These properties have facilitated imaging and crystallographic analysis of the resulting complex, revealing that residues at the extreme C terminus of PAN bind to pockets between adjacent α subunits and induce proteasome gate opening (18, 19, 31, 32) Remarkably, a seven-residue peptide corresponding to the C terminus of PAN is sufficient for both binding and activation of archaeal as well as eukaryotic proteasomes (33). The carboxyl group of the C-terminal arginine of PAN makes an essential interaction with an ϵ-amino side chain of a lysine residue on one α subunit while the hydroxyl of the penultimate tyrosine residue interacts with residues on the adjacent α subunit. Although conflicting data have been presented about the exact identity of these latter contacts, there is general agreement that the interactions stabilize a proline-containing reverse loop in an open gate conformation of the proteasome (18, 19, 32). This general mechanism explains how the C terminus of PAN participates in proteasome binding and activation. Notably, tyrosine also is the penultimate residue in four of the six distinct Rpt subunits in eukaryotic PA700; three of the Rpt subunits share with PAN an HbYX motif (where Hb is a hydrophobic amino acid) at the last three residues. Previous work by us and others showed that the different Rpt subunits of eukaryotic PA700 have at least some non-equivalent roles with respect to proteasome binding and activation. For example, enzymatic removal of the HbYX motifs from only two (Rpt2 and Rpt5) of the six Rpt subunits of PA700 completely inhibited PA700 binding to and activation of the proteasome (33, 34). Moreover, as with PAN, peptides corresponding to the C terminus of Rpt2 and Rpt5 were each sufficient to bind to and activate the 20 S proteasome in a manner that depended on an intact HbYX motif. Finally, binding of the Rpt2 and Rpt5 peptides occurred at distinct and dedicated sites on the fixed order heteromeric α ring, as judged by chemical cross-linking (34). However, a C-terminal peptide of another HbYX motif-containing subunit, Rpt3, as well as C-terminal peptides of the three non-HbYX-containing subunits, had no demonstrable proteasome-activating activity. The lack of activating function of non-activating peptides could reflect either their lack of proteasome binding or their inability to induce conformational changes required for gating after binding. The purpose of this work was to explore roles for non-activating Rpt subunits of 26 S proteasome.
EXPERIMENTAL PROCEDURES
Proteins
PA700, PA700 subassemblies (PS-1, PS-2, and PS-3), 20 S proteasome, and 26 S proteasome were purified from bovine red blood cells as described previously (21, 28, 34–37). SUMO-Rpt peptide fusion proteins were generated by amplification of the whole pET28a-SUMO cassette with primers containing nucleotides appropriate for amino acid sequences of the desired peptides. The resulting His-tagged recombinant SUMO-Rpt peptide fusion proteins were expressed in Escherichia coli BL21 (DE3) cells at 15 °C overnight and purified by affinity chromatography utilizing nickel-nitrilotriacetic acid beads (Qiagen). SUMO-Rpt-chimeric peptide fusion proteins were produced by generating two point mutations in the HbYX motif of a SUMO-Rpt peptide fusion protein sequence. The SUMO-chimeric peptide fusion proteins were expressed and purified as described for the SUMO-Rpt peptide fusion proteins.
Peptide Synthesis
Peptides corresponding to the sequences (or variants thereof) of C termini of Rpt subunits of PA700 were synthesized using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry and purified using HPLC by the Protein Core Facility at the University of Texas Southwestern Medical Center. Sequences of all peptides were verified by mass spectrometry. The sequences of these peptides from N to C termini are as follows: Rpt1, SATPRYMTYN; Rpt2, QEGTPEGLYL; Rpt3, KDEQEHEFYK; Rpt4, LESKLDYKPV; Rpt5, KKKANLQYYA; Rpt6, KNMSIKKLWK; Rpt3(−3C), KDEQEHE; Rpt3-Rpt1, KDEQEHETYN; Rpt1-Rpt3, SATPRYMFYK; Rpt5-Rpt1, KKKANLQTYN; Rpt1-Rpt5, SATPRYMYYA; Rpt5-Rpt3, KKKANLQFYK; Rpt3-Rpt5, KDEQEHEYYA. For cross-linking studies, peptides containing 3,4-dihydroxyphenylalanine (DOPA) and either biotin or fluorescein were synthesized and had the following sequences: biotin-DOPA-Rpt3, biotin-DOPA-GSKDEQEHEFYK; biotin-DOPA-Rpt3(−3C), biotin-DOPA-GSKDEQEHE; fluorescein-DOPA-Rpt3, fluorescein-GGG-DOPA-GSKDEQEHEFYK; fluorescein-DOPA-Rpt3(−3C), fluorescein-GGG-DOPA-GSKDEQEHE.
Proteasome Activity and Activation Assays
Proteasome activity was measured by determining rates of enzymatic cleavage of 7-amino-4-methylcourmarin (AMC) from peptide substrates Suc-LLVY-AMC, Suc-LLE-AMC, and benzyloxycarbonyl-VLR-AMC, as described previously (21). Standard assay conditions included 45 mm Tris-HCl, pH 8.0, 5 mm β-mercaptoethanol, 15 nm latent 20 S proteasome, and 200 μm substrate in a volume of 50 μl. Incubations were carried out at 37 °C for 21 min in a Biotek FL600 fluorescence plate reader with filters at 380-nm excitation/460-nm emission. AMC fluorescence was monitored once per min during the assay, and progress curves were analyzed with kinetic software. Proteasome activation by Rpt peptides and SUMO-Rpt peptide fusion proteins was determined similarly but included preincubation of 20 S proteasome with peptides or SUMO proteins for 15 min at 37 °C (34). Proteasome activity is expressed as arbitrary fluorescent units produced/min. Routine control assays included reactions without proteasome. Proteasome activity against a protein substrate, [methyl-14C]casein, was determined as described previously (37). Other details of individual experiments are provided in the appropriate figure legends. In some experiments, semiquantitative measures of proteasome activity were obtained by overlay of peptide substrates in situ on proteins separated in native 4% polyacrylamide gels, as described previously (38). After incubation at 37 °C for 10–30 min, AMC at the position of the protease in the gel responsible for its production was visualized by UV light.
26 S Proteasome Assembly Assay
Assembly of 26 S proteasome from purified 20 S proteasome and PA700 was conducted by preincubating 20 S proteasome with PA700 in 45 mm Tris-HCl, pH 8.0, 5.6 mm DTT, 10 mm MgCl2, and 100 μm ATP at 37 °C for 30 min (35). 20 S proteasome and PA700 concentrations for given experiments are provided in the appropriate figure legends. Samples were either assayed directly for proteasome activity or subjected to native PAGE, after which gels were stained for protein and assayed for in situ proteasome activity, as described above.
Rpt Peptide-20 S Proteasome Binding Assays
Binding of Rpt peptides to 20 S proteasome was determined by pull-down assays utilizing purified bovine 20 S proteasome and recombinant His-tagged SUMO-Rpt peptide fusion proteins. In typical assays, 100 μm His-tagged SUMO fusion protein was incubated with 214 nm 20 S proteasome at 37 °C for 15 min in 50 mm Tris-HCl, pH 7.6, and 1 mm β-mercaptoethanol in a volume of 100 μl. Twenty-five μl of nickel beads were added and mixed for 2 h at 4 °C. The beads were washed with 50 mm Tris-HCl, pH 7.6, 1 mm β-mercaptoethanol and eluted with 300 mm imidazole. Eluted proteins were separated by SDS-PAGE and Western blotted with antibodies against 20 S proteasome.
Chemical Cross-linking
Chemical cross-linking of Rpt peptides was conducted by methods similar to those described previously (34). Biotin- or fluorescein-containing DOPA peptides described above were incubated for 10 min at room temperature with 720 nm 20 S proteasome, 20 mm Tris-HCl, pH 7.6, 20 mm NaCl, 1 mm EDTA, and 10% glycerol (Buffer H) in a final volume of 20 μl. Cross-linking was initiated by the addition of 10 mm sodium periodate and quenched after 30 s by 50 mm β-mercaptoethanol. Cross-linked products were detected by either fluorescence spectrometry (488-nm excitation/523-nm emission) using a Typhoon 9410 scanning imager (GE Healthcare) or Western blotting with HRP-linked neutravidin or infrared dye-labeled streptavidin (see below) after SDS-PAGE. In some experiments, samples were detected by these methods after two-dimensional gel electrophoresis. In other experiments, samples of cross-linked proteins were purified by HPLC using a Jupiter C4 5-ml reverse phase column (Phenomenex). Samples were applied to the column in 0.05% TFA and eluted with a gradient of acetonitrile at a flow rate of 1 ml/min. In preliminary experiments, we determined that fluorescent subunits representing the cross-linked products eluted between 45 and 50% acetonitrile. Therefore, the gradient was developed from 0 to 45% acetonitrile in 15 min and from 45 to 50% in 30 min. Column fractions of 1.0 ml were collected, dried by vacuum, and redissolved in either SDS-sample buffer or isoelectric focusing sample buffer (7 m urea, 2 m thiourea, 4% CHAPS, 65 mm dithiothreitol, Pharmolytes® (pH 3–10), and bromphenol blue). Isoelectric focusing was conducted using a ReadystripTM pH 3–10 support (Bio-Rad).
Samples subjected to cross-linking with biotin-containing peptides were enriched for cross-linked product by binding to monomeric avidin beads after exposure to denaturing conditions. Cross-linking was performed as described above. Non-cross-linked Rpt3 peptides were removed by multiple washes through a Microcon-YM100 centrifugal filter in Buffer H with 0.5 m NaCl and 0.05% Tween 20. The samples were exposed to 7 m guanidine HCl for 30 min at room temperature. The samples were diluted to decrease the guanidine concentration to 1 m and mixed with monomeric avidin beads in Buffer H containing 0.5 m NaCl and 0.05% Tween 20. Beads were washed with the same buffer, and retained proteins were eluted in either SDS sample buffer or isoelectric sample buffer. Samples were separated by either one- or two-dimensional gels and Western blotted with either infrared dye-labeled streptavidin or with antibodies against selected 20 S proteasome subunit and the respective infrared dye-labeled secondary antibody to visualize the proteins of interest utilizing an Odyssey infrared imaging system (Li-Cor).
Transient Expression of FLAG-Rpt3 in HEK293 Cells
HEK293 cell lines were maintained in Dulbecco's modified Eagle's medium (Invitrogen) containing high glucose and glutamine, supplemented with 10% fetal bovine serum in the presence of 5% CO2 at 37 °C. HEK293 cells were transfected at ∼60% confluence with cDNA for either FLAG-human Rpt3 or FLAG-human Rpt3 lacking the last three C-terminal residues subcloned into the pIRESpuro3 vector (Clontech) using FuGene 6 reagent (Roche Applied Science). Forty-eight h after transfection, cells were washed with phosphate-buffered saline and harvested with buffer consisting of 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 1 mm β-mercaptoethanol, 1 mm ATP, and protease inhibitor mixture (Roche Applied Science). Whole cell extracts were prepared by 15 passages through a 27-gauge needle and centrifuged for 20 min. Expression of Rpt proteins was determined by Western blot analysis using anti-FLAG M2 antibody (Sigma) and anti-Rpt3 antibody (Boston Biochem).
Glycerol Density Gradient Centrifugation
Glycerol density gradient centrifugation was conducted as described previously (39) using 10–40% linear glycerol gradients.
Affinity Purification of FLAG-Rpt Proteins from HEK293 Cell Extracts
Approximately 7 mg of a whole cell extract was mixed gently for 2 h at 4 °C with 100 μl of anti-FLAG M2-agarose beads (Sigma) in 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1 mm β-mercaptoethanol, 1 mm ATP, 5 mm MgCl2, 10% glycerol, and 0.1% Nonidet P-40. The beads were harvested by centrifugation and washed three times with the same buffer. Bound proteins were eluted overnight at 4 °C with 2 bed volumes of binding buffer containing 200 μg/ml FLAG peptide (Sigma).
RESULTS
The C Terminus of Rpt3 Binds to but Does Not Activate the 20 S Proteasome
We previously discovered that isolated peptides corresponding to the C termini of PA700 subunits Rpt2 and Rpt5, but not those corresponding to the C termini of the other four Rpt subunits of this regulatory complex, stimulated 20 S proteasome-catalyzed hydrolysis of model substrates by a mechanism that involves enhanced gating of the substrate entry channel (34). The differential effects of the various Rpt C-terminal peptides could reflect their differential binding to the proteasome or their differential ability to promote gate opening and substrate passage after binding. To distinguish between these possibilities, we directly tested the relative binding of the Rpt peptides to the proteasome. We expressed and purified recombinant proteins in which the C-terminal 10 residues of each Rpt subunit were appended to the C terminus of His-tagged SUMO-1, a protein that otherwise does not interact detectably with the proteasome. After incubation, 20 S proteasome bound by these fusion proteins was isolated by pull-down assays on Ni2+ beads and detected by Western blotting. As expected, the 20 S proteasome bound to SUMO proteins containing the C terminus of Rpt2 and of Rpt5 (Fig. 1A, lanes 3 and 6). Surprisingly, however, 20 S proteasome also bound to SUMO containing the C terminus of Rpt3 (Fig. 1A, lane 4), a subunit whose C terminus does not enhance proteasome activity. The Rpt3-containing SUMO protein consistently pulled down more 20 S proteasome than did the Rpt2 and Rpt5-containing proteins, suggesting that it bound with greater affinity to the proteasome than did the Rpt2- and Rpt5-containing proteins. The proteasome failed to bind detectably to SUMO proteins with C termini of the remaining non-activating Rpt subunits (Rpt1, Rpt4, and Rpt6). Binding of the 20 S proteasome to SUMO-Rpt3 was blocked by excess free Rpt3 C-terminal peptide but not by excess free Rpt5 C-terminal peptide (Fig. 1B, lanes 2–4) or by excess free Rpt2 C-terminal peptide (Fig. 1C). Likewise, the Rpt3 peptide did not block binding of the SUMO-Rpt5 to the proteasome (Fig. 1B, lanes 5 and 6). These results indicate that binding of the Rpt3 peptide was specific and probably occurred at a site unique from those bound by Rpt2 and Rpt5. As with the binding of Rpt2 and Rpt5, binding of the SUMO-Rpt3 protein depended on the presence of the last three residues. Thus, proteins lacking the last two or three residues had no detectable proteasome binding, whereas a protein lacking the only the last residue displayed detectable but greatly diminished binding (Fig. 1D). Binding of SUMO-Rpt3 to the 20 S proteasome was demonstrated independently in a gel shift assay (Fig. 1E). Thus, the SUMO-Rpt3 fusion protein with an intact C-terminal peptide, but not that without the last three residues, retarded the migration of 20 S proteasome during native gel electrophoresis (Fig. 1E, lanes 1–4). Similar results were obtained with SUMO-Rpt5 proteins (Fig. 1E, lanes 5–7). In-gel proteasome activity assays also reflected the differential ability of the C termini of Rpt5 and Rpt3 to activate proteasome function. Collectively, these results confirm the differential ability of the C termini of different Rpt subunits to bind to the proteasome and identify Rpt3 as a PA700 subunit that binds to the proteasome but does not directly activate proteasome hydrolysis of model peptide substrates.
FIGURE 1.
The C terminus of Rpt3 binds to the 20 S proteasome. Fusion proteins of SUMO and the C termini of the indicated Rpt proteins were expressed, purified, and used for pull-down assays of purified 20 S proteasome as described under “Experimental Procedures.” A, pull-down assays with the indicated His-tagged SUMO-Rpt peptide fusion peptides (lanes 2–7) were performed as described under “Experimental Procedures.” Lane 1 shows an assay with 20 S proteasome but no SUMO-Rpt protein. Relative intensities of the bands in lanes 1–8, as determined by densitometry are 0, 0, 1.0, 4.32, 0, 1.49, 0, and 0.96, respectively. B, pull-down assays of 20 S proteasome with indicated SUMO-Rpt peptide fusion proteins were conducted in the absence (lanes 2 and 5) or presence (lanes 3, 4, 6, and 7) of the indicated Rpt C-terminal peptides. 20 S proteasome, SUMO-Rpt protein, and Rpt peptides were present in relative concentrations of 240 nm, 100 μm, and 1 mm, respectively. Relative intensities of the bands in lanes 1–8, as determined by densitometry, are 0, 1.0, 0.01, 0.80, 0.16, 0.19, 0.03, and 0.30, respectively. C, pull-down assays of 20 S proteasome with the indicated SUMO-Rpt peptide fusion proteins and Rpt C-terminal peptides as in B. Relative intensities of the bands in lanes 1–5, as determined by densitometry, are 0, 1.0, 1.1, 0.01, and 1.06, respectively. D, pull-down assays were conducted with the indicated His-tagged SUMO-Rpt3 C-terminal peptide fusion proteins. −3C, −2C, and −1C denote deletions of the last 3, 2, and 1 C-terminal Rpt3 residues, respectively. Relative intensities of the bands in lanes 1–6, as determined by densitometry, are 0, 1.0, 0.01, 0, 0.44, and 1.93, respectively. E, the indicated SUMO-Rpt C-terminal peptide fusion proteins (300 μm) were preincubated with 650 nm 20 S proteasome for 15 min at 37 °C, subjected to native PAGE, and visualized after overlay of Suc-LLVY-AMC fluorescent peptide substrate. Similar results for experiments in all panels were obtained in at least four separate experiments. WB, Western blot.
The C Terminus of Rpt3 Binds to a Dedicated Site on the 20 S Proteasome
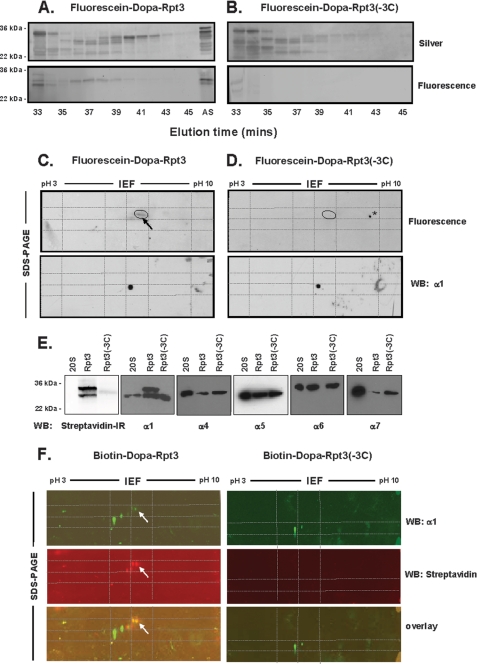
Previously, we showed that activating peptides from the C termini of Rpt2 and Rpt5 chemically cross-link to distinct, dedicated, and identifiable subunits of the 20 S proteasome (34). Consistent with the results presented above, these findings indicate that the fixed order heterohexameric ring of Rpt subunits of PA700 binds to the fixed order heteroheptameric ring of 20 S α subunits with an invariant interring subunit registration. This model predicts that Rpt3 also should bind at a unique and dedicated site on the α ring of the 20 S proteasome and cross-link to a specific subunit. To test this hypothesis, we applied the same general chemical cross-linking strategy to Rpt3 employed previously for Rpt2 and Rpt5. We synthesized DOPA-containing peptides corresponding to the C terminus of Rpt3 that either contained or lacked the last three residues and included either biotin or fluorescein for detection of cross-linked products. Cross-linking of the intact C-terminal peptide with 20 S proteasome produced one major product, which was similar by each detection method after SDS-PAGE (Figs. 2, A and B). In some experiments, a second band, whose intensity varied among independent cross-linking reactions, also was detected. No cross-linked product was detected with an Rpt3 peptide lacking the last three amino acids, indicating that the cross-linking was specific for conditions required for peptide binding. We exploited respective characteristics of the fluorescein and biotin tags of Rpt peptides for independent identification of the Rpt cross-linked subunit of 20 S proteasome. For ease of detection of the cross-linked product, we continued subsequent analysis with the fluorescein-tagged peptide. For more facile enrichment of the cross-linked product, we exploited the biotin moiety. With the fluorescein-labeled peptide, a single cross-linked product also was detected by two-dimensional gel electrophoresis; in some experiments, such as that shown in Fig. 2C, the product appeared as two or three closely separated spots. However, the subunit complexity of 20 S proteasome from the unenriched sample and low protein content on these gels prevented us from further identification of the cross-linked product at this stage. Therefore, we subjected cross-linked 20 S proteasome to reverse phase HPLC to enrich and purify the modified subunit. SDS-PAGE of gradient fractions showed that this method separated most proteasome subunits from one another and from the major fluorescently labeled band (Fig. 3A). We subjected this band to two-dimensional PAGE, which like the unenriched sample, usually appeared as two or three closely separated spots, whose position did not correspond to that of any unmodified proteasome subunit. The fluorescent spot was extracted, digested with trypsin, and subjected to mass spectrometry, which identified peptides of only one 20 S proteasome subunit, α1 (PSMA6), in repeated independent experiments (supplemental Table 1); in one experiment, peptides of the α7 (PSMA3) subunit were detected in addition to α1. No proteasome peptides were identified when an equivalent area of the gel was analyzed from a 20 S proteasome sample subjected to identical treatment with an Rpt3 peptide lacking the last three residues (data not shown). We attempted to confirm the identity of α1 as the cross-linked protein by Western blotting, but the only available antibodies were insufficiently sensitive to detect the low protein content at the position corresponding to the fluorescent spot. No antibody to other 20 S proteasome subunits produced a detectable signal at this position (data not shown). Non-fluorescent α1 subunit was identified by Western blotting at its expected position in the gel; it was similar for the intact and truncated cross-linking peptide and presumably represents the non-cross-linked portion of the α1 protein.
FIGURE 2.
DOPA-Rpt3 C-terminal peptide cross-links to a specific α subunit of 20 S proteasome. The indicated concentrations of biotin-DOPA-Rpt3 or biotin-DOPA-Rpt3(−3C) (A) or fluorescein-DOPA-Rpt3 or fluorescein-DOPA-Rpt3(−3C) (B) peptides were used for cross-linking assays with 20 S proteasome as described under “Experimental Procedures.” Proteasome subunits were separated by SDS-polyacrylamide gel and detected by Coomassie Blue staining, as indicated, Western blotting with α-neutravidin (A), or fluorescence scanning (B). C, cross-linking was conducted as above with fluorescein-DOPA-Rpt3. The sample was divided and subjected to either SDS-PAGE (left) or two-dimensional PAGE (right), as described under “Experimental Procedures.” Cross-linked product was detected by fluorescence scanning. D, 20 S proteasome was subjected to two-dimensional PAGE and stained with Coomassie Blue. Similar results for experiments in all panels were obtained in at least four separate experiments. IEF, isoelectric focusing.
FIGURE 3.
Identification of the 20 S proteasome subunit cross-linked to the C-terminal peptide of Rpt3. Chemical cross-linking of 20 S proteasome with fluorescein-DOPA-Rpt3 (A) or fluorescein-DOPA-Rpt3(−3C) (B) and subsequent HPLC purification were conducted as described under “Experimental Procedures.” HPLC fractions were subjected to SDS-PAGE and either stained for protein with silver (top) or scanned for fluorescence (bottom). AS, sample applied to the column. Fractions containing the major fluorescent (36–39 min) (left) and equivalent fractions from unlabeled control samples (right) were pooled, concentrated, and subjected to two-dimensional PAGE (C and D, respectively). The area of the gel containing the fluorescent spot (denoted by an oval, upper left) and the corresponding area of the non-fluorescent control gel (upper right) were excised and processed for mass spectrometric identification of proteins, as described under “Experimental Procedures.” Gels were Western blotted for proteasome subunit α1 (lower right and left). Grids indicate relative registration of gels based on the position of common markers. The spot marked by the asterisk is an imaging artifact unique to this individual experiment. Similar results for experiments in all panels were obtained in at least four separate experiments. E and F, chemical cross-linking of 20 S was performed with biotin-DOPA-Rpt3 or biotin-DOPA-Rpt3(−3C), and the samples were enriched on monoavidin beads as described under “Experimental Procedures.” Enriched samples were subjected to Western blotting after SDS-PAGE (E) or two-dimensional PAGE (F) using either Streptavidin-IR or antibodies against individual 20 S proteasome subunits, as indicated. Untreated 20 S proteasome (20S) was used as a control for each blot (E). Two-dimensional gels of the indicated samples were blotted for both the α1 subunit (F, top, green channel) and biotin (F, middle, red channel). F, bottom, merged image (yellow) for the two blots. The arrows point to cross-linked products. IEF, isoelectric focusing; WB, Western blot.
To confirm the identification of α1 as the cross-linked product of the Rpt3 peptide, we utilized the biotin-containing peptide to enrich the resulting cross-linked product on monomeric avidin beads under denaturing conditions, as described under “Experimental Procedures.” In contrast to the analysis described above, this enrichment method permitted isolation of sufficient product for subsequent analysis by Western blotting after one- and two-dimensional PAGE. As shown in Fig. 3E, cross-linking with the intact peptide but not with the C-terminally truncated peptide, produced a biotin-labeled band that migrated indistinguishably from a modified α1 band, detected by Western blotting on SDS-PAGE. Likewise, after two-dimensional PAGE, Western blotting for the α1 subunit revealed modified spots in positions similar to those observed for the fluorescently cross-linked protein in samples cross-linked with the intact peptide but not with the truncated peptide. Moreover, biotin was detected only in the modified spots, which coincided precisely with those detected by the anti-α1 antibody (Fig. 3F). Control experiments with antibodies against several other α subunits failed to detect modified proteins as a consequence of cross-linking in Western blots of either the one- or two-dimensional gels (Fig. 3E) (data not shown). The spots detected coincidentally by infrared dye-labeled streptavidin (for biotin) and the α1 antibody were extracted, digested with trypsin, and subjected to mass spectrometry. As with the analogous experiment described above, peptides of the α1 subunit were selectively identified (supplemental Table 1). Collectively, these results indicate that α1 is the probable cross-linked product of the Rpt3 C-terminal peptide. This subunit is distinct from subunits identified previously as cross-linked products of C-terminal peptides of Rpt2 (α7) and Rpt5 (α4). These results support the general model for a fixed and distinct registration between subunits of the interacting heteromeric Rpt subunits and α-subunit rings (see “Discussion”).
Binding of Rpt3 Does Not Affect Proteasome Activation by Rpt2 or Rpt5
C-terminal peptides of Rpt2 and Rpt5 activate substrate hydrolysis by the 20 S proteasome, and their effects are either additive (with short peptide substrates) or synergistic (with longer protein substrates). Such results are consistent with the binding of these peptides to distinct sites on the proteasome and support a model in which the substrate access pore can be gated to variable degrees by multiple independent binding events (40). Therefore, we tested whether Rpt3 could modulate the gating effects of Rpt2 and/or Rpt5, despite its inability to induce gating independently. Rpt3 had no proteasome-activating activity by itself or in combination with Rpt2 and/or Rpt5, regardless of the substrate tested (Fig. 4). Thus, binding of Rpt3 C-terminal peptide neither opens the substrate access pore directly nor modulates the effect of other Rpt C-terminal peptides that do so. These results, however, monitor the relative roles of physically separated binding molecules and may not reflect the roles and effects of these peptides when they function in the context of an intact PA700 complex (see “Discussion”).
FIGURE 4.
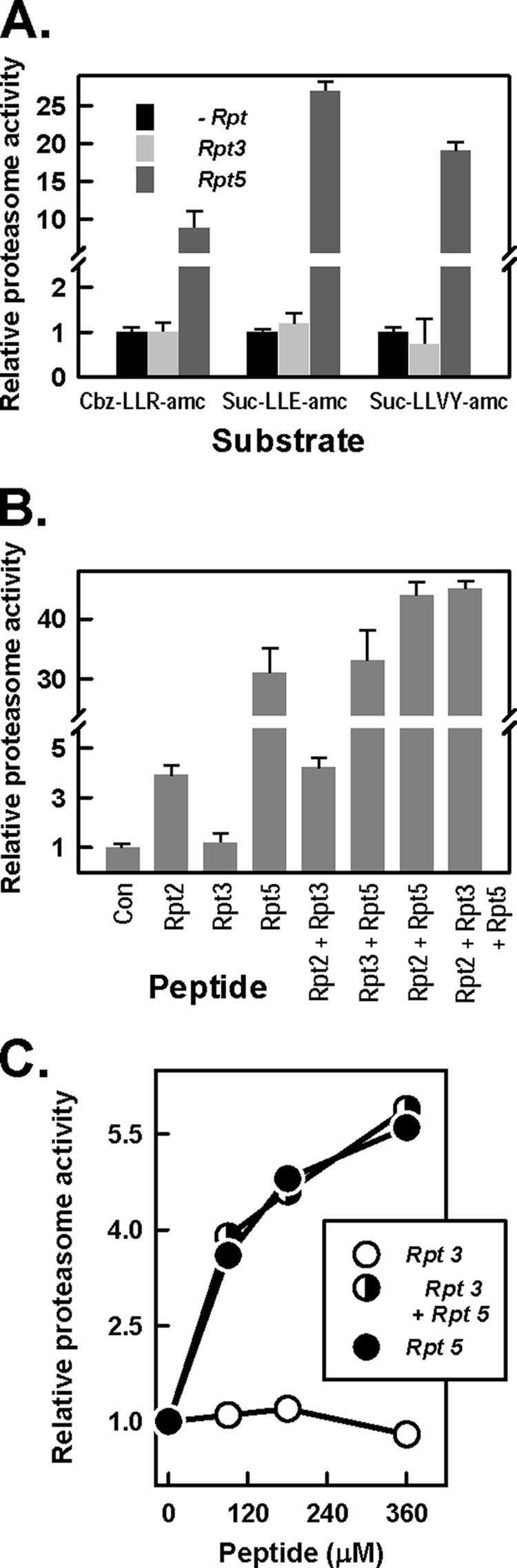
The C terminus of Rpt3 does not influence 20 S proeasome activation. A, 20 S proteasome (12 nm) activity against the indicated peptide substrates was determined in the presence and absence of peptides corresponding to the last 10 residues of Rpt3 (400 μm) or Rpt5 (200 μm). B, 20 S proteasome (12 nm) activity against Suc-LLVY-AMC substrate was determined in the absence (Con) and presence of the indicated C-terminal Rpt peptides (400 μm). C, 20 S proteasome (30 nm) activity against [methyl-14C]casein substrate was determined in the presence of the indicated Rpt peptides. For each experiment, activity in the absence of Rpt peptides was assigned a value of 1.0, and other activities are expressed as relative values. Data represent mean values ± S.D. of triplicate assays. Similar results were obtained in at least four separate experiments.
Features of Proteasome Binding and Activation Are Determined by Both the HbYX Motifs and Adjacent Residues of the Rpt C Termini
Previous work by us and others has established and emphasized the important role of the last three amino acids of C-terminal Rpt peptides in proteasome binding and activation by Rpt2 and Rpt5 (33, 34). These residues (LYL and YYA for Rpt2 and Rpt5, respectively) conform to a motif of HbYX. This motif also is present in Rpt3 (FYK) and, in an imperfect form, in Rpt1 (TYN). Thus, various HbYX motif-containing peptides display distinct functional properties with respect to proteasome binding and activation. Although such disparity probably reflects differences among the HbYX motifs and their cognate binding sites on the α ring of the 20 S proteasome, features of Rpt C-terminal peptides other than the HbYX motif may provide additional determinants of proteasome binding and/or activation. To explore this possibility, we synthesized “chimeric” peptides containing the HbYX motif (hereafter denoted as the “tail”) of given Rpt subunits and the adjacent N-terminal seven residues (hereafter denoted as the “head”) of other Rpt subunits. We also produced recombinant fusion proteins of SUMO and the corresponding chimeric peptides. We selected for this analysis examples of C-terminal peptides that (i) both bind to and activate the proteasome (e.g. Rpt5); (ii) bind to but do not activate the proteasome (e.g. Rpt3); and (iii) neither bind to nor activate the proteasome (e.g. Rpt1). First, we determined the ability of various SUMO-Rpt chimeric peptide fusion proteins to bind to the 20 S proteasome in pull-down assays analogous to those used with their wild-type counterparts (Fig. 5). Neither SUMO-Rpt3 head-Rpt1 tail (B, lane 3) nor SUMO-Rpt5 head-Rpt1 tail (B, lane 6) displayed appreciable proteasome binding, thereby supporting a critical role of the HbYX motif of wild-type Rpt3 and Rpt5 C-terminal peptides for their respective proteasome binding. Surprisingly, however, chimeric peptides consisting of an Rpt3 or Rpt5 tail with an Rpt1 head displayed proteasome binding properties suggestive of an important influence of the Rpt1 head. Thus, the Rpt1 head reduced the proteasome binding expected of the Rpt3 tail (compare lanes 2 and 4) but increased the proteasome binding expected of the Rpt5 tail (compare lanes 5 and 7). The influence of head region on proteasome binding also was demonstrated by the lack of binding of the chimeric peptide consisting of an Rpt3 head and an Rpt5 tail (i.e. a peptide containing both a head and a tail of binding peptides; compare lanes 4 and 8). In contrast, a peptide consisting of an Rpt5 head and an Rpt3 tail featured a proteasome binding affinity similar to that of Rpt3. These results further highlight the obligatory role of specific HbYX motifs for proteasome binding but demonstrate the influence of additional elements of the C terminus on this process.
FIGURE 5.
Structural determinants of Rpt C-terminal peptides on proteasome binding and activation. A, structures of C-terminal Rpt peptides used for proteasome binding and activation assays. B, His-tagged SUMO proteins containing the C-terminal fusions of the indicated peptides were expressed, purified, and used for pull-down assays with 20 S proteasome, as described under “Experimental Procedures.” Lane 1, assay containing 20 S proteasome but no SUMO-Rpt protein. Relative intensities of the bands in lanes 1–10, as determined by densitometry, are 0, 1.0, 0.01, 0.13, 0.29, 0.02, 1.14, 0.89, 0.01, and 0.88, respectively. C, 20 S proteasome activity against Suc-LLVY-AMC substrate was measured in the presence of the indicated SUMO-Rpt C-terminal peptide fusion proteins (100 μm). Proteasome activity in the absence of SUMO-Rpt protein was assigned a value of 1.0, and all other activities are expressed as relative values. Data represent mean values of triplicate assays ± S.D. Similar results were obtained in at least three separate experiments. D, peptides were synthesized corresponding to either the C-terminal 10 residues of each indicated Rpt subunit (RptX peptide; black bars) or the C-terminal three residues of each indicated Rpt subunit (RptX) and the adjacent seven residues of Rpt5 (Rpt5 head-RptX tail; gray bars). 20 S proteasome (12 nm) activity was assayed in the absence (Control) or presence of these peptides (400 μm) using Suc-LLVY-AMC substrate. Activity in the absence of Rpt peptides was assigned a value of 1.0, and all other activities are expressed as relative values. Data represent mean values of triplicate assays ± S.D. (error bars). Similar results were obtained in at least four separate experiments. WB, Western blot.
To explore the relationship between binding of the HbYX motif and proteasome activation, we compared the effects of the various SUMO-chimeric peptide fusion proteins on proteasome activation with those of their wild-type counterparts and with the binding features of these chimeric peptides. As expected, no non-binding chimeric peptide activated 20 S proteasome catalysis. Moreover, chimeric peptides containing both a head and a tail of non-activating Rpt subunits (e.g. Rpt1 and Rpt3), regardless of their binding capacity, did not activate the proteasome. Instead, proteasome activation by chimeric proteins required the tail (HbYX motif) of a normally activating Rpt peptide. Thus, neither SUMO-Rpt5 head-Rpt1 tail, nor SUMO-Rpt5 head-Rpt3 tail activated the proteasome, although each could bind. In contrast, SUMO-Rpt1head-Rpt5 tail activated the proteasome to a greater extent than did SUMO-Rpt5, an effect that mirrored the relative proteasome binding of these proteins. These results indicate that proteasome binding is influenced by features of both the HbYX motif and the adjacent residues of specific Rpt subunits but that proteasome activation is restricted to the binding of a normally activating HbYX motif (e.g. Rpt5). To test this further, we synthesized a series of peptides consisting of an Rpt5 head and the tail of each of the six Rpt subunits. Only chimeric peptides containing the activating HbYX tails Rpt2 and Rpt5 stimulated 20 S proteasome activity (Fig. 5D). Interestingly, the Rpt5 head-Rpt2 tail peptide stimulated the proteasome to a greater extent than the wild-type Rpt2 peptide but to a lesser extent than the wild type Rpt5 (Fig. 5D). These results provide additional evidence for the influence of the head region on the function of gating-competent HbYX motifs.
Rpt3 C-terminal Peptide Attenuates 26 S Proteasome Assembly in Vitro
The data presented above identify the C terminus of Rpt3 as an important binding element of intact PA700 to the proteasome. To test the role of Rpt3 in binding of intact PA700 to the 20 S proteasome, we examined the effect of a C-terminal peptide of Rpt3 on the ATP-dependent in vitro reconstitution of 26 S proteasome from purified PA700 and 20 S proteasome. Both Rpt3 peptide (Fig. 6A) and the SUMO-Rpt3 fusion protein (data not shown) inhibited the PA700-dependent activation of the 20 S proteasome, an indirect monitor of 26 S proteasome assembly. The inhibitory effect was dependent on peptide concentration and required the last three residues. Inhibition of assembly of activated 26 S proteasome activation by the intact Rpt3 C-terminal peptide also was demonstrated by native PAGE (Fig. 6B). The Rpt3 peptide had no effect on the activity of intact purified 26 S proteasome, indicating that the peptide did not exert its effect in the assembly assay by inhibiting the activity of assembled 26 S proteasome or by promoting 26 S proteasome disassembly (Fig. 6, C and D). These results suggest that the isolated Rpt3 peptide functions as a dominant negative inhibitor of 26 S proteasome assembly by competitively blocking binding of intact PA700 to the 20 S proteasome. Remarkably, this effect is manifested despite the presence of at least two other PA700 subunits (Rpt2 and Rpt5) with the capacity to bind 20 S proteasome (see “Discussion”).
FIGURE 6.
The C-terminal peptide of Rpt3 inhibits 26 S proteasome assembly and activation in vitro. A and B, in vitro 26 S proteasome assembly and activation from purified 20 S proteasome and PA700 was conducted as described under “Experimental Procedures.” A, 20 S proteasome (15 nm) and PA700 (75 nm) were preincubated in the presence or absence of the indicated Rpt3 C-terminal peptide or Rpt3(−3C) C-terminal peptide. After 30 min, proteasome activity was measured using Suc-LLVY-AMC substrate. 20 S proteasome activity in the absence of PA700 was assigned a value of 1.0, and all other activities are expressed as relative values. B, 20 S proteasome (75 nm) and PA700 (200 nm) were preincubated in the presence or absence of Rpt3 C-terminal peptide or Rpt3(−3C) C-terminal peptide, as indicated. After 30 min, samples were subjected to native PAGE. The gel was incubated with a solution of Suc-Leu-Leu-Val-Tyr-AMC, incubated for 15 min at 37 °C, and exposed to UV light. The arrows indicate established migration positions of 26 S proteasome and 20 S proteasome, respectively. Relative fluorescence intensities of the bands in lanes 1–4 were quantified as 0/1.0, 1.0/0.57, 0.40/0.89, and 1.01/0.34, respectively, for 26 S/20 S proteasome bands, respectively. Similar results were obtained in four separate experiments. C, purified 26 S proteasome (10 nm) was assayed against Suc-LLVY-AMC substrate in the presence of the indicated concentrations of Rpt3 C-terminal peptide. Data points represent mean values of triplicate assays. Similar results were obtained in three separate experiments. D, purified 26 S proteasome (20 nm) was preincubated with indicated concentrations of Rpt3 C-terminal peptide for 30 min. Samples were then subjected to native PAGE and then assayed for in-gel proteasome activity by incubation with Suc-LLVY-AMC, as in B, or stained for protein with Coomassie Blue. Similar results were obtained in two separate experiments. E, 20 S proteasome (3 nm) was incubated in the absence (−) or presence (+) of 2 μg each of PA700 subcomplexes PS1, PS2, and PS3 and either 1 mm Rpt3 or Rpt3 (−3C) peptide, as indicated, or no peptide (−). Samples were incubated for 30 min at 37 °C in 45 mm Tris-HCl, pH 8.0, 5.6 mm DTT, 10 mm MgCl2, and 100 μm of ATP in a volume of 50 μl. Proteasome activity was assayed with 200 μm Suc-LLVY-AMC substrate peptide, as described above. Proteasome activity in the absence of subassemblies and Rpt peptide was assigned a value of 1.0, and all other activities are expressed as relative values. Data represent mean values of triplicate assays ± S.D. (error bars). Similar results were obtained in three separate experiments. AFU, arbitrary fluorescent units.
Previously, we showed that 26 S proteasome also could be assembled in vitro from 20 S proteasome and three subcomplexes that collectively form intact PA700 (35). The C-terminal Rpt3 peptide but not the peptide lacking the last three residues blocked assembly of 26 S proteasome from these PA700 subassemblies (Fig. 6E).
The C Terminus of Rpt3 Is Essential for Assembly of 26 S Proteasome in Intact Cells
To evaluate the relative role and importance of the C terminus of Rpt3 to 26 S proteasome assembly in intact cells, we transfected HEK293 cells with expression vectors for either FLAG-tagged wild-type Rpt3 or FLAG-tagged Rpt3 lacking the last three C-terminal residues. We analyzed cells in which expressions of these proteins were approximately equal to one another and equal to or less than that of endogenous Rpt3 (Fig. 7A). The two Rpt3-expressing cell types were indistinguishable from one another and from non-transfected HEK293 cells by general morphological features and by rates of growth (data not shown). They also had similar overall proteasome activity (Fig. 7A). In non-transfected control cells, endogenous Rpt3 displayed a trimodal distribution when soluble extracts were subjected to glycerol density gradient centrifugation. Most of the Rpt3 protein sedimented in fractions characteristic of the 26 S proteasome. Smaller amounts were found in slower sedimenting fractions corresponding to free PA700 and other lower molecular weight complexes (Fig. 7B, top). FLAG-tagged wild-type Rpt3 displayed a distribution pattern that was qualitatively similar to that of endogenous Rpt3, although proportionally more exogenous protein was distributed to the slowest sedimenting complexes (Fig. 7B, middle). The reasons for and significance of this quantitative distinction are unclear. Nevertheless, an appreciable portion of expressed wild-type FLAG-Rpt3 was assembled normally into 26 S proteasome as judged by its sedimentation position in the glycerol gradient and by anti-FLAG immunoprecipitation of proteins with structural and functional features of 26 S proteasome (Fig. 7, B and C; see below). In contrast, little or no detectable FLAG-tagged Rpt3 protein lacking C-terminal residues was present in gradient fractions corresponding to the 26 S proteasome and instead accumulated in slower sedimenting fractions corresponding to those characteristic of PA700 and smaller subcomplexes (Fig. 7B, bottom). Moreover, although immunoprecipitation was equally efficient for the wild-type and mutant Rpt3 proteins, the resulting immunoprecipitates differed significantly in other features. For example, FLAG immunoprecipitation from extracts of cells expressing wild-type Rpt3 isolated a protein with features characteristic of intact 26 S proteasome as judged by Western blotting of representative component subunits (Fig. 7C, left) (data not shown), migration on native PAGE (Fig. 7C, middle), and proteasome activity (Fig. 7C, middle and right). In contrast, FLAG immunoprecipitation from extracts of cells expressing mutant Rpt3 isolated a protein with subunits characteristic of PA700 but without 20 S proteasome subunits or proteasome activity. Collectively, these results show that lack of an intact C terminus prevented Rpt3 from incorporation into 26 S proteasome and that the contributions of intact binding elements of Rpt2 and Rpt5 were not sufficient to overcome this deficiency. These results are consistent with the ability of isolated C-terminal Rpt3 peptide to attenuate binding of intact PA700 to the proteasome and highlight an important role of Rpt3 binding in 26 S proteasome assembly.
FIGURE 7.
The C terminus of Rpt3 is required for assembly of 26 S proteasome. HEK293 cells were transfected with expression vectors without insert (Mock) or with inserts for either FLAG-tagged wild-type Rpt3 (Rpt3) or FLAG-tagged Rpt3 lacking the last three C-terminal residues (Rpt3−3C), as described under “Experimental Procedures.” A, whole cell extracts were Western blotted for the indicated proteins (top) and assayed for hydrolysis of Suc-LLVY-AMC (bottom). Activity assays were normalized for total extract protein content and represent mean values of triplicate assays ± S.D. B, extracts from non-transfected cells (Control) and from indicated Rpt3-expressing cells were subjected to glycerol density gradient centrifugation as described under “Experimental Procedures.” Fractions were Western blotted for the indicated proteins. The arrows indicate the normal peak of sedimentation profile for purified PA700 and 26 S proteasome (data not shown). C, extracts of the indicated cells were subjected to immunoprecipitation with anti-FLAG beads as described under “Experimental Procedures.” Immunoprecipitates were subjected to the following: Western blotting (WB) for the indicated antigens, including FLAG, β5 subunit of 20 S proteasome, Rpt2, Rpt5, and Rpn12 (left); native PAGE, followed by silver staining (middle; arrows indicate known migration positions of purified singly and doubly capped 26 S proteasome); and proteasome activity assays using Suc-LLVY-AMC as substrate (right). Data represent mean values of triplicate assays ± S.D. (error bars) and were normalized for FLAG content. Similar results for data in each panel were obtained in three separate experiments.
DISCUSSION
The results presented here reveal new details about structural and functional interactions between the 20 S proteasome and PA700, two multisubunit subcomplexes that compose the 26 S proteasome. PA700 and 20 S proteasome bind at an axial interface of two different heteromeric rings. PA700 contributes a heterohexameric ring of AAA subunits (Rpt1 to -6), whereas the 20 S proteasome contributes a heteroheptameric ring of α-type subunits (α1 to -7). Thus, the interaction between subcomplexes of the eukaryotic 26 S proteasome has greater structural complexity than that between subcomplexes of archaeal proteasome complexes featuring interacting rings of homomeric proteins. Previous work has established an important role for the C termini of certain Rpt subunits for binding of PA700 to the proteasome and consequent proteasome gating (33, 34). However, the relative roles of the different Rpt subunits in these processes remain uncertain. The clearest examples of Rpt C termini that interact directly with the proteasome are those of Rpt2 and Rpt5. Each features an HbYX motif that binds to pockets between specific adjacent α subunits of the 20 S proteasome ring. Although the structure of intact 26 S proteasome has not been solved at atomic resolution, possible explanations for the non-equivalent roles of the C termini of different Rpt subunits in proteasome binding and activation have been provided by structural studies of heterologous, artificially engineered, and simpler archaeal model systems (18, 19, 31, 32, 41). For example, the significance of the HbYX motif has been illustrated by showing the atomic details of how these residues from PAN and certain Rpt subunits interact with proteasome subunits to promote gate opening (19, 31, 32). Despite the considerable insight gained by these studies, a comprehensive molecular understanding of the relative structure-function relationships of the Rpt subunits for proteasome binding and activation of authentic 26 S proteasome remains elusive. Nevertheless, the non-equivalent capacity of different Rpt subunits to bind to and activate the proteasome must depend on both the features of individual Rpt C-terminal residues and the specific α subunits that create their respective binding pockets. Thus, whereas Rpt2 and Rpt5 bind to gating-competent sites on the proteasome, Rpt3 probably binds to a site that cannot directly promote gating. The lack of proteasome activation by an isolated peptide, however, does not exclude its role in proteasome gating when it is part of the intact PA700 complex.
The current data provide initial information about the influence of residues upstream of the HbYX motif in proteasome binding and activation. Goldberg and colleagues (33) previously established a seven-residue minimum length requirement for proteasome binding and activation of features of the HbYX motif peptide of PAN. Multiple substitutions for residues N-terminal to the HbYX motif had little effect on binding and activation, suggesting that they did not make identity-specific contributions to these processes. In contrast, the results presented here show that the identity of residues adjacent to the HbYX motif of given Rpt peptides can have appreciable influence on proteasome binding and/or activation. Although an HbYX motif was always necessary for binding, alterations to adjacent residues could either diminish or enhance the apparent affinity of this effect. Likewise, residues adjacent to the HbYX motif had significant effects on proteasome activation. It is unclear from the current data whether these various effects reflect general structural features of the substituted peptides or features specific to the normal function of given Rpt subunits. Information about the exact binding sites of chimeric peptides and their relationship to the normal binding sites of each component will be required for complete interpretation of these data.
The identification of α1 as the proteasome subunit to which Rpt3 specifically cross-links extends our previous results that identified α7 and α4 as the cross-linked products of Rpt2 and Rpt5 peptides, respectively (34). These collective results support other data indicating that different Rpt C termini bind to different and dedicated sites on the 20 S proteasome. However, these cross-linked products do not necessarily represent the subunits to which their respective HbYX motif residues directly bind because the cross-linking peptides' reactive DOPA residue is located up to 10 amino acids away from this site. Thus, it is not certain that these data can be used to fix the registration of interacting 20 S proteasome and PA700 rings, each of which is composed of subunits with invariant order (12, 42). Our attempts to cross-link Rpt peptides in which the DOPA residue was located closer to the HbYX motif were unsuccessful. This could have many causes but may reflect the importance of the identity of residues adjacent to the HbYX motif for proper binding.
The current results demonstrate that the isolated C-terminal peptide of Rpt3 peptide blocks the in vitro assembly of 26 S proteasome from intact PA700 and 20 S proteasome. This effect most likely results from competition of the Rpt3 peptide with the C terminus of the intact Rpt3 subunits in PA700 and indicates that binding contributions of Rpt2 and Rpt5 in intact PA700 are not sufficient to overcome this inhibition. Thus, diminished binding of only one of several competent binding elements of intact PA700 can severely impair overall 26 S proteasome assembly. Although our current studies have focused on Rpt3, it is likely that analogous results would be achieved by interference with the binding of Rpt2 and Rpt5. In fact, results compatible with this prediction have been obtained in previous independent studies in which enzymatic modification of the C terminus of either Rpt2 or Rpt5 was sufficient to inhibit 26 S proteasome assembly in vitro (34).
Additional evidence for a critical role of the HbYX motif of Rpt3 in 26 S proteasome assembly was obtained in intact cells. Unlike wild-type Rpt3, mutant Rpt3 lacking this motif was excluded from 26 S proteasome. Consistent with the biochemical data noted above, this result indicates that other binding-competent Rpt subunits (i.e. Rpt2 and Rpt5) are unable to overcome the binding deficiency of mutant Rpt3 with respect to its incorporation into 26 S proteasome. We suspect that HbYX deletion mutants of Rpt2 and Rpt5 will be similarly defective in their cellular incorporation into 26 S proteasome. Our results on the role of Rpt3 in 26 S proteasome assembly and activation appear to conflict with those of others obtained using a different experimental design and system. For example, a point mutation in the penultimate tyrosine residue or the deletion of the C-terminal lysine residue of Rpt3 each diminished the activity but not the cellular assembly of yeast 26 S proteasome (33). In a similar study, yeast expressing Rpt3 lacking a single C-terminal residue showed reduced but not abolished assembly and activation of 26 S proteasome (33, 43). Reduced assembly of the single-residue deletion mirrors the effect observed for this same modification in our pull-down assays. In general, these more limited perturbations of the HbYX motif may produce less severe effects on these processes than does complete truncation.
In both previous and current work, we have established that 26 S proteasome can be assembled in vitro by ATP-dependent reconstitution from purified 20 S proteasome and PA700 (27, 28). It is unclear, however, whether this process mimics the physiological pathway of 26 S proteasome assembly. In fact, several recent reports provide evidence that intact PA700 may not be a direct intermediate of the cellular 26 S proteasome assembly pathway but rather that 26 S proteasome is formed by sequential binding of multiple subassemblies of PA700 to 20 S proteasome, which would serve as a template for PA700 formation (43, 44). Notably, three described subassemblies in these studies each contained two of the six different Rpt subunits including one HbYX motif subunit (Rpt2, Rpt3, and Rpt5) and one non-binding subunit (Rpt1, Rpt6, and Rpt4), respectively (45–48). Independently, we purified three subassemblies of PA700 that collectively account for all component PA700 subunits (35). Each had the same content of Rpt subunits found in several of the aforementioned cellular studies, and two were identical in overall composition to cellular assembly intermediates found by others (46, 49). Although we have not investigated the physiological significance of these subassemblies in detail, we note that they can be reconstituted in vitro into functional PA700 in the absence of 20 S proteasome and into 26 S proteasome in the presence of 20 S proteasome. The former result indicates that the 20 S proteasome is not an obligatory template for PA700 formation. Moreover, the cellular studies described here indicate that the C-terminal mutant Rpt3 protein defective in 26 S proteasome assembly accumulated as intact PA700. Thus, the C-terminal mutation of Rpt3 prevented only assembly into 26 S proteasome and not into intact PA700. Although this effect could reflect a direct decrease in binding affinity of the truncated protein for the proteasome, it also could be mediated by indirect mechanisms. For example, recent work has identified multiple Rpt-binding proteins that serve as 26 S proteasome assembly chaperones (45–50). Each of these chaperones binds to a unique Rpt subunit prior to 26 S proteasome assembly but is released during assembly. One such protein, p28 (also known as gankyrin or, in yeast, Nas6) binds to a C-terminal domain of Rpt3 (51). Thus, structural alterations of the Rpt3 C terminus might block 26 S proteasome assembly by attenuating an otherwise required dissociation of Nas6 from Rpt3. In fact, previous work in yeast has shown that Rpt3 lacking a single C-terminal residue failed to release Nas6, resulting in defective association with the 20 S proteasome (43). Additional work will be required to determine the precise mechanism for the defective assembly of mammalian Rpt3 with a larger truncation studied here.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Gillette for assistance with the Li-Cor imaging system and the Protein Core Facility at University of Texas Southwestern for assistance with mass spectrometry.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK46181. This work was also supported by Welch Foundation Grant I-500 (to G. N. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- AAA
- ATPases associated with a variety of cellular activities
- AMC
- 7-amino-4-methylcourmarin
- PAN
- proteasome-activating nucleotidase
- SUMO
- small ubiquitin-like modifier
- DOPA
- 3,4-dihydroxyphenylalanine
- Suc
- succinyl.
REFERENCES
- 1.Pickart C. M., Cohen R. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 177–187 [DOI] [PubMed] [Google Scholar]
- 2.Smith D. M., Benaroudj N., Goldberg A. L. (2006) J. Struct. Biol. 156, 72–83 [DOI] [PubMed] [Google Scholar]
- 3.Groll M., Bochtler M., Brandstetter H., Clausen T., Huber R. (2005) Chembiochem 6, 222–256 [DOI] [PubMed] [Google Scholar]
- 4.Voges D., Zwickl P., Baumeister W. (1999) Annu. Rev. Biochem. 68, 1015–1068 [DOI] [PubMed] [Google Scholar]
- 5.Bedford L., Paine S., Sheppard P. W., Mayer R. J., Roelofs J. (2010) Trends Cell Biol. 20, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumeister W., Walz J., Zühl F., Seemüller E. (1998) Cell 92, 367–380 [DOI] [PubMed] [Google Scholar]
- 7.Bochtler M., Ditzel L., Groll M., Hartmann C., Huber R. (1999) Annu. Rev. Biophys. Biomol. Struct. 28, 295–317 [DOI] [PubMed] [Google Scholar]
- 8.Groll M., Clausen T. (2003) Curr. Opin. Struct. Biol. 13, 665–673 [DOI] [PubMed] [Google Scholar]
- 9.Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. (1997) Nature 386, 463–471 [DOI] [PubMed] [Google Scholar]
- 10.DeMartino G. N., Gillette T. G. (2007) Cell 129, 659–662 [DOI] [PubMed] [Google Scholar]
- 11.Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V. A., Finley D. (1998) Cell 94, 615–623 [DOI] [PubMed] [Google Scholar]
- 12.Tomko R. J., Jr., Funakoshi M., Schneider K., Wang J., Hochstrasser M. (2010) Mol. Cell 38, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nickell S., Beck F., Scheres S. H., Korinek A., Förster F., Lasker K., Mihalache O., Sun N., Nagy I., Sali A., Plitzko J. M., Carazo J. M., Mann M., Baumeister W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11943–11947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nickell S., Mihalache O., Beck F., Hegerl R., Korinek A., Baumeister W. (2007) Biochem. Biophys. Res. Commun. 353, 115–120 [DOI] [PubMed] [Google Scholar]
- 15.Groll M., Bajorek M., Kohler A., Moroder L., Rubin D. M., Huber R., Glickman M. H., Finley D. (2000) Nat. Struct. Biol. 11, 1062–1067 [DOI] [PubMed] [Google Scholar]
- 16.da Fonseca P. C., Morris E. P. (2008) J. Biol. Chem. 283, 23305–23314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Köhler A., Cascio P., Leggett D. S., Woo K. M., Goldberg A. L., Finley D. (2001) Mol. Cell 7, 1143–1152 [DOI] [PubMed] [Google Scholar]
- 18.Förster A., Masters E. I., Whitby F. G., Robinson H., Hill C. P. (2005) Mol. Cell 18, 589–599 [DOI] [PubMed] [Google Scholar]
- 19.Stadtmueller B. M., Ferrell K., Whitby F. G., Heroux A., Robinson H., Myszka D. G., Hill C. P. (2010) J. Biol. Chem. 285, 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C. W., Corboy M. J., DeMartino G. N., Thomas P. J. (2003) Science 299, 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C. W., Li X., Thompson D., Wooding K., Chang T. L., Tang Z., Yu H., Thomas P. J., DeMartino G. N. (2006) Mol. Cell 24, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith D. M., Kafri G., Cheng Y., Ng D., Walz T., Goldberg A. L. (2005) Mol. Cell 20, 687–698 [DOI] [PubMed] [Google Scholar]
- 23.Finley D. (2009) Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C., Schwartz M. P., Prakash S., Iwakura M., Matouschek A. (2001) Mol. Cell 7, 627–737 [DOI] [PubMed] [Google Scholar]
- 26.Yao T., Cohen R. E. (2002) Nature 419, 403–407 [DOI] [PubMed] [Google Scholar]
- 27.Adams G. M., Crotchett B., Slaughter C. A., DeMartino G. N., Gogol E. P. (1998) Biochemistry 37, 12927–12932 [DOI] [PubMed] [Google Scholar]
- 28.DeMartino G. N., Moomaw C. R., Zagnitko O. P., Proske R. J., Chu-Ping M., Afendis S. J., Swaffield J. C., Slaughter C. A. (1994) J. Biol. Chem. 269, 20878–20884 [PubMed] [Google Scholar]
- 29.Zwickl P., Ng D., Woo K. M., Klenk H. P., Goldberg A. L. (1999) J. Biol. Chem. 274, 26008–26014 [DOI] [PubMed] [Google Scholar]
- 30.Pühler G., Weinkauf S., Bachmann L., Müller S., Engel A., Hegerl R., Baumeister W. (1992) EMBO J. 11, 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabl J., Smith D. M., Yu Y., Chang S. C., Goldberg A. L., Cheng Y. (2008) Mol. Cell 30, 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Y., Smith D. M., Kim H. M., Rodriguez V., Goldberg A. L., Cheng Y. (2010) EMBO J. 29, 692–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith D. M., Chang S. C., Park S., Finley D., Cheng Y., Goldberg A. L. (2007) Mol. Cell 27, 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillette T. G., Kumar B., Thompson D., Slaughter C. A., DeMartino G. N. (2008) J. Biol. Chem. 283, 31813–31822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson D., Hakala K., DeMartino G. N. (2009) J. Biol. Chem. 284, 24891–24903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeMartino G. N. (2005) Methods Enzymol. 398, 295–306 [DOI] [PubMed] [Google Scholar]
- 37.McGuire M. J., McCullough M. L., Croall D. E., DeMartino G. N. (1989) Biochim. Biophys. Acta 995, 181–186 [DOI] [PubMed] [Google Scholar]
- 38.Elsasser S., Schmidt M., Finley D. (2005) Methods Enzymol. 398, 353–363 [DOI] [PubMed] [Google Scholar]
- 39.Koulich E., Li X., DeMartino G. N. (2008) Mol. Biol. Cell 19, 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X., DeMartino G. N. (2009) Biochem. J. 421, 397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadre-Bazzaz K., Whitby F. G., Robinson H., Formosa T., Hill C. P. (2010) Mol. Cell 37, 728–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Förster F., Lasker K., Nickell S., Sali A., Baumeister W. (2010) Mol. Cell Proteomics 9, 1666–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park S., Roelofs J., Kim W., Robert J., Schmidt M., Gygi S. P., Finley D. (2009) Nature 459, 866–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hendil K. B., Kriegenburg F., Tanaka K., Murata S., Lauridsen A. M., Johnsen A. H., Hartmann-Petersen R. (2009) J. Mol. Biol. 394, 320–328 [DOI] [PubMed] [Google Scholar]
- 45.Kaneko T., Hamazaki J., Iemura S., Sasaki K., Furuyama K., Natsume T., Tanaka K., Murata S. (2009) Cell 137, 914–925 [DOI] [PubMed] [Google Scholar]
- 46.Funakoshi M., Tomko R. J., Jr., Kobayashi H., Hochstrasser M. (2009) Cell 137, 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saeki Y., Toh-E A., Kudo T., Kawamura H., Tanaka K. (2009) Cell 137, 900–913 [DOI] [PubMed] [Google Scholar]
- 48.Le Tallec B., Barrault M. B., Guérois R., Carré T., Peyroche A. (2009) Mol. Cell 33, 389–399 [DOI] [PubMed] [Google Scholar]
- 49.Isono E., Nishihara K., Saeki Y., Yashiroda H., Kamata N., Ge L., Ueda T., Kikuchi Y., Tanaka K., Nakano A., Toh-e A. (2007) Mol. Biol. Cell 18, 569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roelofs J., Park S., Haas W., Tian G., McAllister F. E., Huo Y., Lee B. H., Zhang F., Shi Y., Gygi S. P., Finley D. (2009) Nature 459, 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakamura Y., Nakano K., Umehara T., Kimura M., Hayashizaki Y., Tanaka A., Horikoshi M., Padmanabhan B., Yokoyama S. (2007) Structure 15, 179–189 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.