Abstract
The Hippo pathway has emerged as a conserved signaling pathway that is essential for the proper regulation of organ growth in Drosophila and vertebrates. Although the mechanisms of signal transduction of the core kinases Hippo/Mst and Warts/Lats are relatively well understood, less is known about the upstream inputs of the pathway and about the downstream cellular and developmental outputs. Here, we review recently discovered mechanisms that contribute to the dynamic regulation of Hippo signaling during Drosophila and vertebrate development. We also discuss the expanding diversity of Hippo signaling functions during development, discoveries that shed light on a complex regulatory system and provide exciting new insights into the elusive mechanisms that regulate organ growth and regeneration.
Keywords: Cell signaling, Drosophila, Mouse development, Organ growth control, Tumor suppressor genes
Introduction
Although growth is fundamental to animal development, surprisingly little is known about the mechanisms that control organ size (Neto-Silva et al., 2009; Stanger, 2008). How, for example, do cells know when to stop dividing after an organ has reached its correct size? How do injured organs regenerate damaged parts, and how do cells sense that part of an organ is missing? Over the past two decades, much progress has been made in deciphering the mechanisms that are responsible for tissue patterning, while the mechanisms that control organ size have remained largely a mystery.
A puzzling but fascinating aspect of growth control in many animal species is that the number of divisions that a cell can undertake is often not programmed into organ progenitor cells, which may thus exhibit a variable number of cell divisions. This feature of growing tissues makes compensation for the loss of cells during development possible, a phenomenon referred to as regulative development. For example, mouse embryos that have been reduced to 10% of their normal size by administration of mitomycin C just before the onset of organogenesis engage a compensatory growth program, resulting in nearly normal sized embryos 48 hours later (Snow and Tam, 1979). Compensatory proliferation also occurs in Drosophila imaginal discs. For example, larvae in which over 60% of imaginal disc cells have been ablated by X-rays produce normal sized adults (Haynie and Bryant, 1977). Even more remarkably, some adult animals have the ability to regenerate missing parts: salamanders and crickets, for example, can regenerate entire limbs after amputation (Bryant et al., 1977; French et al., 1976). A major conclusion from such compensation and regeneration experiments is that cell-to-cell signaling is required to regulate cell proliferation as a function of organ size. The identity of these signals, however, remains largely unknown (Affolter and Basler, 2007; Neto-Silva et al., 2009; Stanger, 2008).
The recent discovery of the Hippo pathway as a key regulator of organ growth in Drosophila has generated much excitement, as it provides novel insight into the mechanisms that control organ size. In addition, the Hippo pathway is deregulated in many different types of cancers (for reviews, see Chan et al., 2010; Fernandez and Kenney, 2010; Zeng and Hong, 2008; Zhao et al., 2010a). The Hippo pathway transduces signals from the plasma membrane to the nucleus, where it then regulates gene expression (for reviews, see Badouel et al., 2009b; Reddy and Irvine, 2008; Zhao et al., 2010a). Although much progress has been made in understanding the molecular mechanisms of signal transduction between the core components of this cascade, other aspects of the Hippo pathway have been less well explored. Key unanswered questions include: What are the upstream regulators and biological roles of Hippo pathway regulation? How does the Hippo pathway interface with other global signals that control organ size and compensatory proliferation? What are the developmental outputs of Hippo signaling? And, what role does the pathway play in adult homeostasis and abnormal growth? In this review, we discuss recent findings from both Drosophila and vertebrate studies that shed new light on some of these questions.
The core Hippo pathway
The Hippo pathway is composed of a highly conserved core kinase cascade that is regulated by multiple upstream inputs and has multiple transcriptional outputs. Here, we briefly review the components of the core kinase cascade and the consequences of mis-regulated Hippo signaling in Drosophila and mammalian systems.
Hippo signaling regulates growth in Drosophila
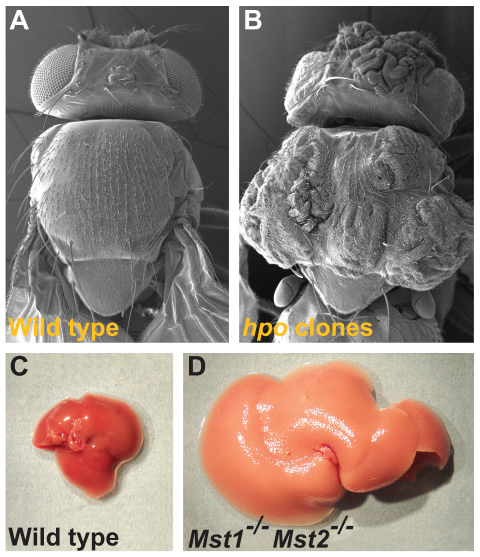
Many of the known Hippo pathway components were discovered through genetic screens in Drosophila (Badouel et al., 2009b; Reddy and Irvine, 2008). Loss of function of Hippo (Hpo) or of Warts (Wts), two kinases that lie at the center of the Hippo pathway, results in dramatic overgrowth of imaginal discs and of corresponding adult structures (Fig. 1A,B) (Harvey et al., 2003; Jia et al., 2003; Justice et al., 1995; Pantalacci et al., 2003; Udan et al., 2003; Wu et al., 2003; Xu et al., 1995). Animals with hpo mutant eye discs, for example, produce adults with severely overgrown eyes and heads that are folded and darker than normal. The hpo gene was thus named after its mutant adult head phenotype, which resembles the hide of the hippopotamus (Udan et al., 2003). The identification of the Hpo kinase and the realization that Hpo forms a signaling module together with the scaffold adaptor protein Salvador (Sav) and with Wts marked the beginning of our understanding of the so-called `Hippo pathway' in Drosophila (Fig. 2A). In Drosophila, loss of Hippo signaling in imaginal discs causes overgrowth because mutant cells proliferate faster than normal cells. They also continue to proliferate beyond normal disc size and are resistant to the pro-apoptotic signals that would normally eliminate extra cells (Harvey et al., 2003; Jia et al., 2003; Kango-Singh et al., 2002; Pantalacci et al., 2003; Tapon et al., 2002; Udan et al., 2003; Wu et al., 2003). Thus, the Hippo pathway restricts cell proliferation and promotes the apoptosis of excess cells. Importantly, in imaginal discs, the Hippo pathway primarily affects the number of cells produced and has only minor effects on tissue patterning. Thus, the Hippo pathway is a key regulator of organ growth and tissue size in Drosophila.
Fig. 1.
Hippo mutant phenotypes in flies and mice. (A,B) Scanning electron micrographs of (A) a wild-type fly and (B) a fly with clones of cells homozygous mutant for hippo that exhibit overgrowth of the adult cuticle (Udan et al., 2003). (C) A mouse liver at 2 months of age from a wild-type animal and (D) a liver at 2 months of age from a mouse mutant in which both Mst1 and Mst2 (Stk3 and Stk4), two mammalian Hippo homologs, have been conditionally inactivated in the developing liver (Lee et al., 2010; Lu et al., 2010; Song et al., 2010; Zhou et al., 2009). The double null Mst1/2 mutant liver is overgrown owing to an increase in cell numbers.
Fig. 2.
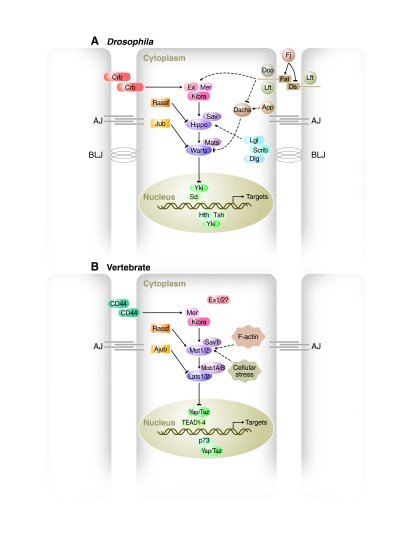
Schematics of the Hippo pathway in flies and mice. Cells (outlined in grey, nuclei in green) are shown with adherens junctions (AJ) and basolateral junctions (BLJ). (A,B) Hippo pathway components in (A) Drosophila and (B) vertebrate are shown in various colors, with pointed and blunt arrowheads indicating activating and inhibitory interactions, respectively. Continuous lines indicate direct interactions, whereas dashed lines indicate unknown mechanisms. See text for further details. Abbreviations: Ajub, Ajuba; App, Approximated; Crb, Crumbs; Dco, Discs overgrown; Dlg, Discs large; Ds, Dachsous; Ex, Expanded; Fj, Four-jointed; Hth, Homothorax; Jub, Drosophila Ajuba; Lats, Large tumor suppressor; Lft, Lowfat; Lgl, Lethal giant larvae; Mer, Merlin; Mats, Mob as a tumor suppressor; Mob1A/B, Mps1 binder; Mst, Mammalian sterile 20 like; Rassf, Ras-associated factor; Sav, Salvador; Scrib, Scribble; Sd, Scalloped; Taz, transcriptional co-activator with PDZ-binding motif; TEAD, TEA domain protein; Tsh, Teashirt; Yap, Yes associated protein; Yki, Yorkie.
Several other genes have since been added to the Hippo pathway in Drosophila and other systems, and a complex signaling pathway with positive and negative regulators has emerged (Fig. 2, Table 1). At the center of the Hippo pathway are two kinases, the Ste20-like kinase Hpo and the nuclear Dbf2-related (NDR) family kinase Wts, that form a kinase cascade (Harvey et al., 2003; Jia et al., 2003; Justice et al., 1995; Pantalacci et al., 2003; Udan et al., 2003; Wu et al., 2003; Xu et al., 1995). The Hpo and Wts kinases, together with their co-factors Sav (Salvador) and Mats (Mob as a tumor suppressor) (Kango-Singh et al., 2002; Lai et al., 2005; Tapon et al., 2002) and the Yorkie (Yki) transcriptional co-activator (Huang et al., 2005), form the `core' of the Hippo pathway. The mechanisms of Hpo and Wts action in this core are relatively well understood (Oh and Irvine, 2010; Reddy and Irvine, 2008; Zhao et al., 2010a). When active, Hpo in complex with Sav phosphorylates Wts and its co-factor Mats, thereby activating Wts kinase activity (Wei et al., 2007; Wu et al., 2003); the pathway is now considered to be in an activated state (Fig. 2A). Hpo can be deactivated by dephosphorylation of its activation loop by the protein phosphatase 2A (PP2A) complex Striatin-interacting phosphatase and kinase (STRIPAK) (Ribeiro et al., 2010). Activated Wts/Mats phosphorylates Yki at three separate sites (S111, S168 and S250), thereby inhibiting its function (Dong et al., 2007; Huang et al., 2005; Oh and Irvine, 2008; Oh and Irvine, 2009; Zhao et al., 2007). Although phosphorylation at each of these sites contributes to the regulation of Yki, S168 is a particularly important site as its phosphorylation results in the binding of Yki to 14-3-3 phosphopeptide binding proteins, resulting in the retention of Yki in the cytoplasm thereby suppressing its transcriptional activity (Dong et al., 2007; Oh and Irvine, 2008; Oh and Irvine, 2009; Ren et al., 2009). Yki is also regulated by phosphorylation-independent mechanisms; direct interaction with Hpo, Wts and the upstream regulator Expanded (Ex) can suppress Yki activity, possibly by cytoplasmic retention (Badouel et al., 2009a; Oh et al., 2009; Ren et al., 2009), although the physiological relevance of this mechanism is not known as these findings relied mostly on in vitro studies. When Yki is not inhibited, it localizes to the nucleus, complexes with transcription factors and induces the expression of target genes, such as the cell cycle regulator cyclin E, the inhibitor of apoptosis diap1, the growth promoter Myc and the growth and cell survival-promoting miRNA bantam, which drive cell proliferation and cell survival (Goulev et al., 2008; Harvey et al., 2003; Huang et al., 2005; Jia et al., 2003; Kango-Singh et al., 2002; Neto-Silva et al., 2010; Nolo et al., 2006; Pantalacci et al., 2003; Peng et al., 2009; Tapon et al., 2002; Thompson and Cohen, 2006; Udan et al., 2003; Wu et al., 2003; Wu et al., 2008; Zhang et al., 2008b; Ziosi et al., 2010). Yki is thus a growth promoter, whereas Hpo, Sav, Wts and Mats act as tumor suppressors by suppressing the growth-promoting activity of Yki.
Table 1.
Components of the Hippo pathway in flies and mice
Hippo signaling in mammalian organ size determination
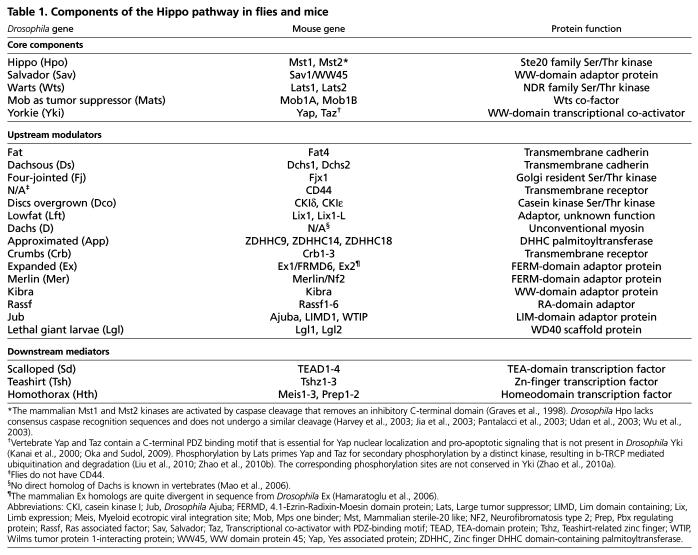
All of the core components of the Drosophila Hippo pathway have direct homologs (orthologs) in mammals (Table 1). These include Mst1/2 (Stk3 and Stk4 – Mouse Genome Informatics; Hpo homologs) (Creasy and Chernoff, 1995a; Creasy and Chernoff, 1995b), Sav1 (the Sav homolog) (Tapon et al., 2002), Lats1/2 (Wts homologs) (Tao et al., 1999; Yabuta et al., 2000), Mob1A/B (Mats homologs) (Bichsel et al., 2004; Chow et al., 2010; Stavridi et al., 2003), and Yap and Taz (Yki homologs) (Kanai et al., 2000; Sudol, 1994). Analogous to the core module in Drosophila, Mst1/2 and Lats1/2 form a kinase cascade, are regulated by Sav1 and Mob1A/B, and phosphorylate Yap and Taz (Fig. 2B) (Chan et al., 2005; Dong et al., 2007; Hao et al., 2008; Hirabayashi et al., 2008; Lei et al., 2008; Oka et al., 2008; Praskova et al., 2008; Zhang et al., 2008a; Zhao et al., 2010b; Zhao et al., 2007). Thus, the core components of the mammalian Hippo pathway act together in a signaling module as they do in Drosophila (Badouel et al., 2009b; Oh and Irvine, 2010; Zhao et al., 2010a). However, although the evolutionary conservation of Hippo signaling is extensive, there are some differences in pathway components between flies and mammals, and the Hippo pathway appears to be more complex in mammals (see Table 1 footnotes).
In mammals, the first studies that connected Hippo signaling to organ size control employed a Yap overexpression strategy that mimics pathway inactivation and showed that the induction of Yap expression in the adult mouse liver leads to a dramatic three- to fourfold increase in liver mass as a result of increased cell numbers (Camargo et al., 2007; Dong et al., 2007). Remarkably, when Yap overexpression is terminated, the liver rapidly reverts to its normal size, suggesting that intrinsic size control mechanisms are then activated, presumably to reduce cell numbers through an apoptotic process (Dong et al., 2007). These studies infer that Hippo signaling is crucial for regulating the size of the mammalian liver, and that it acts through a mechanism that integrates global organ size control signals with the regulation of Yap. However, although the effect of Yap overexpression on liver size is dramatic, whether Hippo signaling is required to maintain liver size was not addressed by these studies. More recently, it has been reported that Mst1/2 and Sav1 function to maintain hepatocyte quiescence and to restrict liver growth postnatally (Fig. 1C,D) (Lee et al., 2010; Lu et al., 2010; Song et al., 2010; Zhou et al., 2009). Yap phosphorylation is significantly reduced in mst1/2 double mutant livers, suggesting that these kinases exert their effects through downstream core Hippo signaling components (Lee et al., 2010; Lu et al., 2010; Song et al., 2010; Zhou et al., 2009). However, an unknown kinase may be responsible for phosphorylation and activation of Lats1/2 in liver cells and a kinase other than Lats1/2 may phosphorylate Yap in MEFs (Zhou et al., 2009). Thus, the majority of genetic and biochemical data show that the core components of the mammalian Hippo pathway act together in a signaling module as they do in Drosophila; however, there are also indications that the pathway may be more complex in mammals than it is in Drosophila.
Although these findings show that Hippo signaling functions in regulating the size of the liver, it appears that Hippo signaling, at least that mediated by the Mst1 and Mst2 kinases, does not regulate the size or growth of other mammalian tissues to the same degree (Song et al., 2010) (discussed in detail in Box 1). One possibility is that Hippo signaling is required to control growth in some contexts, whereas in others its primary role may be to regulate cell cycle exit, as has been shown in the case of the Drosophila optic neuroepithelia (Reddy et al., 2010). Further studies will be required to fully address the role of Hippo signaling in mammals, including tissue-specific deletion of core Hippo signaling components in multiple organ systems and at different times during development and in the adult.
Box 1. Hippo signaling in mammals: growth control or cell cycle exit?
Overexpression of Yap (Camargo et al., 2007; Dong et al., 2007) and targeted deletion of Mst1/2 in the mouse liver (Lu et al., 2010; Song et al., 2010; Zhou et al., 2009) provide evidence that Hippo signaling has a pivotal role in regulating organ size, suggesting that Hippo signaling is a universal growth regulatory mechanism. However, other studies question whether Hippo signaling regulates growth in all mammalian tissues. Clearly, some tissues do not require Hippo signaling to regulate organ size. For example, the targeted deletion of Mst1/2 (Stk3/4) in mouse limb bud tissues has a relatively mild effect on the growth plate, but does not result in enlarged limbs (Song et al., 2010). Furthermore, overexpression of activated Yap in the mouse small intestine leads to Notch-dependent hyperplasia and loss of terminally differentiated cell types, but does not appreciably increase the overall size of the organ (Camargo et al., 2007). Likewise, targeted deletion of the adaptor protein Sav1 (Salvador homolog 1) in various mouse epithelial tissues leads to hyperplasia and the loss of terminal differentiated phenotypes, including in the small intestine and skin, but does not result in markedly enhanced organ size (Lee et al., 2008). These studies suggest that an important function of Hippo signaling in mammals might be to modulate cell cycle exit and terminal differentiation, as has been suggested in Drosophila neural development (Reddy et al., 2010). How this is achieved at a mechanistic level is poorly understood. Additional studies in which the conditional deletion of core Hippo signaling components is carried out in a range of tissues will be required to resolve this issue.
Hippo pathway regulation by multiple inputs
In systems that show regulative growth control, global organ size information must be transmitted to single cells, which then decide whether to continue or to stop proliferating. The phenotypes of Hippo pathway mutants demonstrate that this pathway has a key role in growth control, raising the possibility that its activity is dynamically regulated by global growth control signals. If this is the case, what are these signals and how are these translated into decisions made by single cells?
Recent studies have shed light on the dynamic nature of the regulation of the Hippo pathway, which can respond to specific developmental cues and to a variety of stress signals (Densham et al., 2009; Nishioka et al., 2009; Oka et al., 2008; Rogulja et al., 2008; Taylor et al., 1996; Willecke et al., 2008; Zecca and Struhl, 2010). As we discuss below, these findings show that multiple inputs feed into the core of the Hippo pathway, and these inputs can act in both a coordinated and independent fashion. These inputs can also act at various levels within the cascade, and some are associated with the plasma membrane and might thus relay information from the extracellular milieu or from cell-cell contacts, revealing a complex network of regulatory mechanisms (Fig. 2A,B).
Although Hippo pathway activity can be perturbed by genetic mutations in Drosophila, it remains unclear whether the activity of the pathway is temporally or spatially patterned. In imaginal discs, for example, the core components of the Hippo pathway are ubiquitously required and Yki localization is largely uniform (Dong et al., 2007; Harvey et al., 2003; Huang et al., 2005; Jia et al., 2003; Kango-Singh et al., 2002; Oh and Irvine, 2008; Pantalacci et al., 2003; Tapon et al., 2002; Udan et al., 2003; Wu et al., 2003). Thus, it appears that all cells in imaginal discs require Hippo signaling for normal growth regulation, and that Hippo pathway activity is uniformly distributed. Several negative-feedback loops exist in the pathway that may contribute to a balanced level of its activity (Genevet et al., 2010; Hamaratoglu et al., 2006). It is thus unknown to what extent the Hippo pathway is constitutively active and is simply permissive for growth regulation, or whether it is regulated and transduces instructive growth controlling signals. However, several Hippo pathway inputs are modulated by external signals and may therefore convey information about imaginal disc size to core Hippo signaling components. These various inputs are discussed below.
The Fat branch of the Hippo pathway
Fat is a large atypical cadherin that regulates growth and planar cell polarity (PCP) (Bryant et al., 1988; Mahoney et al., 1991) (reviewed by Lawrence et al., 2008; Reddy and Irvine, 2008; Sopko and McNeill, 2009). Fat functions upstream of core Hippo pathway components to regulate growth (Fig. 2A), but it modulates planar cell polarity independently of Hippo signaling (Bennett and Harvey, 2006; Cho et al., 2006; Silva et al., 2006; Tyler and Baker, 2007; Willecke et al., 2006). Fat localizes to the sub-apical region of the plasma membrane by a mechanism that involves the conserved protein Lowfat, which directly interacts with the Fat intracellular domain (Mao et al., 2009). The binding of Fat to its ligand Dachsous (Ds), another large atypical cadherin related to Fat (Clark et al., 1995), promotes phosphorylation of the Fat intracellular domain by the Casein Kinase 1ε homolog Discs overgrown (Dco) (Feng and Irvine, 2009; Sopko et al., 2009). The intracellular domain of Fat does not contain informative protein motifs and the molecular mechanism of subsequent signaling from Fat to the Hippo pathway is poorly understood. However, Fat signaling is known to require the myosin Dachs (Cho et al., 2006; Cho and Irvine, 2004; Mao et al., 2006; Rogulja et al., 2008), which accumulates at the membrane when Fat is inactive and which also requires the palmitoyltransferase Approximated (App) for its membrane localization (Matakatsu and Blair, 2008). Dachs binds to Wts and influences its protein levels (Cho et al., 2006), and Fat/Dachs signaling also affects the levels of membrane-localized Ex, thereby regulating the activity of the Hippo pathway (Bennett and Harvey, 2006; Feng and Irvine, 2007; Silva et al., 2006; Willecke et al., 2006).
Fat activity is regulated by interaction with Ds, and this interaction is modulated by phosphorylation of the extracellular domains of Fat and Ds by the kinase Four-jointed (Fj), which localizes to the Golgi (Bennett and Harvey, 2006; Brittle et al., 2010; Casal et al., 2006; Cho et al., 2006; Cho and Irvine, 2004; Ishikawa et al., 2008; Lawrence et al., 2007; Ma et al., 2003; Matakatsu and Blair, 2004; Matakatsu and Blair, 2006; Silva et al., 2006; Simon et al., 2010; Strutt and Strutt, 2002; Willecke et al., 2006; Yang et al., 2002). Interestingly, Ds and Fj are expressed in complementary gradients in imaginal discs, such as the eye and wing discs (Clark et al., 1995; Villano and Katz, 1995). The vector or direction of these gradients is important for the orientation of PCP in eye discs but may act redundantly with other PCP orienting systems in other tissues such as wing discs (Adler et al., 1998; Casal et al., 2002; Matakatsu and Blair, 2004; Rawls et al., 2002; Simon, 2004; Strutt and Strutt, 2002; Yang et al., 2002; Zeidler et al., 1999; Zeidler et al., 2000). The regulation of Fat by Ds and Fj in the Hippo pathway, by contrast, is dependent on the steepness of their gradients (Rogulja et al., 2008; Willecke et al., 2008). Intriguingly, Hippo pathway activity is suppressed in a cell when its neighbors express a different amount of either Ds or Fj. Thus, juxtaposing cells with different Ds or Fj levels cause the induction of Hippo pathway target genes, leading to cell proliferation on either side of the Ds/Fj expression boundary. This `boundary effect' is unusual, in that the response of a cell is not proportional to the amount of Ds present, but rather depends on whether neighboring cells have either more or less Ds activity. The mechanism of this effect is not known but may involve the intracellular polarization of Dachs localization, which then modulates the core of the Hippo pathway (Mao et al., 2006; Reddy and Irvine, 2008; Rogulja et al., 2008).
Fat signaling: mechanisms of tissue size regulation
The above findings suggest that the gradients of Ds and Fj are important for normal growth. Indeed, flies with uniform Ds and Fj expression have small wings and are smaller than normal (Rogulja et al., 2008; Willecke et al., 2008). Two models, the polar coordinate model and the feed-forward model, have been used to explain how Ds and Fj control growth (Rogulja et al., 2008; Willecke et al., 2008; Zecca and Struhl, 2010).
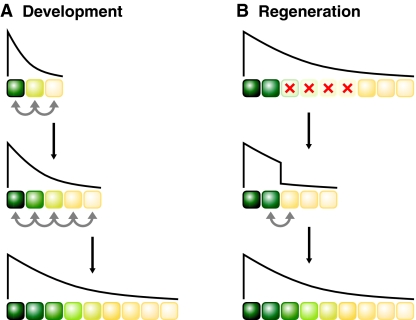
According to the polar coordinate model (Fig. 3), tissue growth depends on the shape of positional information in a field of cells (Bryant et al., 1977; French et al., 1976; Lawrence et al., 2008). During development, cells acquire positional information, e.g. by reading the concentration of a morphogen gradient, thereby establishing a gradient of positional information across a field. Neighboring cells then compare their positional information, and, if the values are too different, cells are stimulated to proliferate. New cells intercalate between existing cells, thereby decreasing the disparity of positional values between neighboring cells until the differential falls below a certain threshold, when cells stop proliferating. The polar coordinate model can thus explain how organs stop growing at the appropriate size: while the gradient of positional information is steep initially, its slope decreases owing to expansion of the tissue until the gradient can no longer drive growth. The polar coordinate model thus proposes the existence of signals that cells use to compare their positional information and that regulate cell proliferation. The Ds/Fj/Fat signaling system fits these criteria (see Box 2 for details): Ds and Fj are expressed in gradients reflecting positional information and they regulate growth depending on how different expression levels are between neighboring cells (Clark et al., 1995; Rogulja et al., 2008; Villano and Katz, 1995; Willecke et al., 2008).
Fig. 3.
The polar coordinate model and Hippo signaling during organ growth. (A,B) Schematics representing the polar coordinate model to explain growth during (A) normal development and (B) regeneration. The schematics depict cells in growing tissues; the graded colors and the gradient lines indicate gradients of positional information or Dachsous (Ds) activity. When the difference of positional information between two neighboring cells exceeds a certain threshold, this triggers cell proliferation at the position of discontinuity (gray arrows) followed by intercalation of intermediate values. This `boundary effect' will drive proliferation until the gradient of positional information or Ds activity is smooth and differences between neighboring cells are below the threshold. (A) During development, gradients of positional information are steep initially, triggering intercalary proliferation until the tissue has reached its proper size and gradient of positional information. (B) During regeneration, after some cells are ablated (red crosses), cells at the wound site are juxtaposed to cells with inappropriate positional values (gray arrow), which triggers intercalary proliferation until the normal situation is restored. Ds activity is graded in developing Drosophila imaginal discs, reflecting positional information, and differences in Ds activity between neighboring cells can suppress the Hippo pathway and trigger proliferation (Rogulja et al., 2008; Willecke et al., 2008).
Box 2. Experimental evidence for Fat signaling and the polar coordinate model of growth control
The polar coordinate model predicts that increasing the steepness of the gradients, or the presence of juxtaposing cells with disparate expression levels, should induce growth, whereas flattening the gradients can reduce growth. Indeed, local ectopic expression of Ds or Fj in imaginal discs induces proliferation at expression boundaries while uniform expression of Ds and Fj leads to smaller flies that have smaller wings (Rogulja et al., 2008; Willecke et al., 2008). Thus, the Ds and Fj gradients provide essential growth control information. However, artificial uniform expression of Ds and Fj does not completely eliminate growth, indicating that other, unknown mechanisms act in addition to Fat signaling to control tissue size (Rogulja et al., 2008; Willecke et al., 2008). The polar coordinate model was initially developed to explain limb regeneration in insects and amphibians (Bryant et al., 1977; French et al., 1976; Lawrence et al., 2008). Consistent with this mechanism, the Ds/Fj/Fat signaling module is essential for leg regeneration in the cricket Gryllus bimaculatus (Bando et al., 2009). Crickets can regenerate amputated legs during their nymphal stages, when they grow through molting. Amputated legs precisely regenerate the missing piece, indicating that cells of the regenerating blastema are aware of their positional identity and stop proliferation when the missing part is regenerated. Cricket Ds and Fj are expressed in complementary gradients in each of the developing leg segments and are re-expressed in regenerating legs to recapitulate their embryonic expression patterns. Importantly, treatment of regenerating legs with RNAi against Fat or Ds results in loss of regenerative growth (Bando et al., 2009). Cricket leg regeneration thus provides an exciting opportunity to explore the regulation of the Hippo pathway in response to organ size control and regeneration.
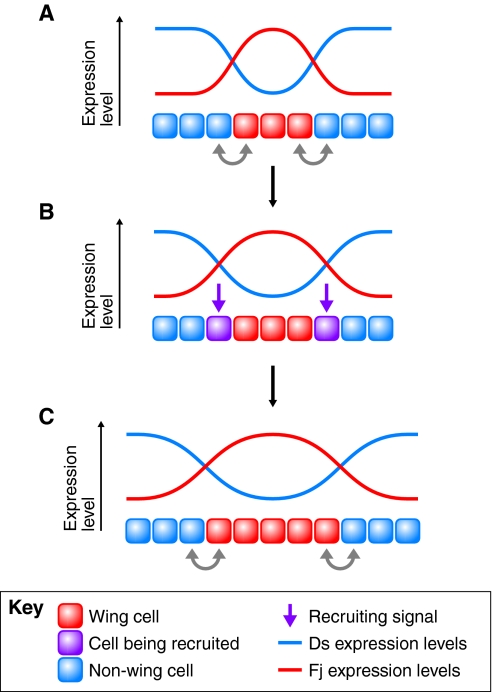
The Ds/Fj/Fat signaling system may also control the size of the Drosophila wing through the local recruitment of cells into the developing wing field by a feed-forward signaling process (Fig. 4) (Zecca and Struhl, 2010). In this model, Fat activity is suppressed in cells just outside of the developing wing field where the Fj and Ds gradients show maximal steepness. Suppression of Fat causes suppression of the Hippo pathway and activation of Yki, which re-programs these cells to become wing cells, thereby expanding the wing field. The newly recruited cells then upregulate Fj expression and downregulate Ds expression according to their new fate, which moves the expression patterns of Fj and Ds, thus initiating another round of cell recruitment.
Fig. 4.
Recruitment of cells into the developing Drosophila wing by a Ds/Fj-dependent feed-forward signal. Schematics showing Dachsous (Ds) and Four-jointed (Fj) expression levels in the wing pouch area of a Drosophila wing imaginal disc. (A) Cells of the wing pouch (red) are specified by the expression of the wing selector gene vestigial (vg), which induces high levels of Fj expression, while cells surrounding the wing pouch (blue) express high levels of Ds. The gradients of Ds and Fj are thus steepest at the periphery of the vg-expressing wing pouch, which leads to suppression of Hippo signaling through the Ds boundary effect (gray arrows). (B) Suppression of Hippo signaling then leads to the induction of Hippo target genes, one of which is vg. Ds/Fj signaling thus results in the expansion of the vg expression domain (purple) and the Ds boundary effect acts as a recruitment signal (purple arrow) (Zecca and Struhl, 2010). (C) Vg in these newly recruited pouch cells induces Fj expression and causes suppression of Ds expression, which moves the Ds and Fj expression gradients outwards and leads to another cycle of signaling and recruitment of more neighboring cells, thereby further expanding the territory of the wing pouch (Zecca and Struhl, 2010). This growth control model is specific for the Drosophila wing and it can partially explain the control of growth of the developing wing. Other unknown mechanisms must act in addition, as ds,fj double mutant flies can still produce small wings (Rogulja et al., 2008; Willecke et al., 2008).
The polar coordinate model and the feed-forward model are not mutually exclusive and both mechanisms appear to be important for growth control. However, replacing the endogenous Ds and Fj gradients with uniform expression does not completely eliminate growth during Drosophila development (Rogulja et al., 2008; Willecke et al., 2008). Thus, other, currently unknown, mechanisms must exist that control growth in addition to the Ds/Fj/Fat signaling system. Whether these also act through the Hippo pathway is not known.
In mammals there are four Fat-related atypical cadherins (Fat1-4), with Fat4 being the ortholog of Drosophila Fat, two Ds homologs (Dchs1-2) and one Fj homolog (Fjx1) (Ashery-Padan et al., 1999; Nakajima et al., 2001; Rock et al., 2005; Tanoue and Takeichi, 2005). Although there is evidence that the Fat1 cadherin plays a role in regulating focal adhesions and actin dynamics (Ishiuchi et al., 2009; Moeller et al., 2004; Tanoue and Takeichi, 2004), and that Fat4 regulates planar cell polarity during kidney development (Saburi et al., 2008), a connection between mammalian Fat- or Ds-related cadherins and the core mammalian Hippo pathway has not been explored. However, the observation by Skouloudaki et al. (Skouloudaki et al., 2009) that depletion of zebrafish Yap suppresses the effect of Fat1 knockdown in pronephric development is consistent with a role for Fat-related cadherins in modulating Hippo signaling in vertebrate systems. Additional experiments are, however, necessary to determine whether this effect is mediated by core Hippo signaling components such as Mst1/2 and Lats1/2.
The Expanded, Merlin, Kibra complex
Another key upstream regulatory module that feeds into the Drosophila Hippo pathway is the Expanded/Merlin/Kibra (Ex/Mer/Kibra) complex (Fig. 2A) (Baumgartner et al., 2010; Cho et al., 2006; Genevet et al., 2010; Hamaratoglu et al., 2006; Pellock et al., 2007; Tyler and Baker, 2007; Yu et al., 2010). Ex, Mer and Kibra are adaptor proteins that localize to the sub-apical region of epithelial cells (Baumgartner et al., 2010; Boedigheimer and Laughon, 1993; Boedigheimer et al., 1997; Genevet et al., 2010; McCartney et al., 2000; Yu et al., 2010). The imaginal disc phenotypes of flies carrying mutations in these genes are relatively weak compared with those of flies with mutations affecting downstream components of the pathway, such as hpo and wts. However, double mutant combinations of ex, mer and kibra show synergistic phenotypes that are more severe than those of the single mutants and that resemble those of hpo mutants (Baumgartner et al., 2010; Genevet et al., 2010; Hamaratoglu et al., 2006; Maitra et al., 2006; Yu et al., 2010). Ex, Mer and Kibra act genetically upstream of Hpo, form a complex in cultured cells, and colocalize in vivo (Baumgartner et al., 2010; Genevet et al., 2010; McCartney et al., 2000; Yu et al., 2010). They connect directly to Hpo and Sav via multiple interactions between Ex and Hpo, Mer and Sav, and Kibra and Sav (Yu et al., 2010); however, the way in which they regulate Hpo activity is not known. In addition, Ex can directly interact with Yki, thereby sequestering Yki from the nucleus and downregulating its activity (Badouel et al., 2009a; Oh et al., 2009). Ex, Mer and Kibra thus function together as an apical scaffold that promotes Hippo pathway activity.
Mer and Kibra are conserved in mammals and Ex has divergent homologs, although to date only Mer has been directly implicated in having a role in the mammalian Hippo pathway (Striedinger et al., 2008; Yokoyama et al., 2008; Zhang et al., 2010; Zhao et al., 2007). Mer is required for contact inhibition in cultured mammalian cells (Curto et al., 2007; Lallemand et al., 2003; Striedinger et al., 2008), it promotes nuclear export of Yap in response to contact inhibition (Striedinger et al., 2008; Zhao et al., 2007), and it is required for Hippo pathway activity and to restrict Yap activity in the mouse liver (Zhang et al., 2010). Mer/NF2 depletion in the mouse liver thus causes overgrowth phenotypes that are similar to those of mst1/2 double mutant mice (Benhamouche et al., 2010; Zhang et al., 2010). Mammalian Kibra binds to Mer, and their co-expression results in synergistic phosphorylation of Lats1/2 in vitro (Yu et al., 2010; Zhang et al., 2010). Thus, mammalian Kibra and Mer may function together to regulate the Hippo pathway as they do in Drosophila.
Input from the cell polarity determinant Crumbs
The Drosophila Crumbs (Crb) protein was recently added to the Hippo pathway as a potential new receptor (Chen et al., 2010; Ling et al., 2010; Robinson et al., 2010). Crb is a large single-pass transmembrane protein with a short intracellular domain of 37 residues (for a review, see Bazellieres et al., 2009). Crb localizes to the sub-apical region of epithelial cells and is part of the machinery that establishes and maintains apical-basal cell polarity. In contrast to its role in embryonic epithelia, Crb is not required for cell polarity in imaginal discs. However, discs that are mutant for crb overgrow and produce adult structures that are larger than normal (Chen et al., 2010; Ling et al., 2010; Richardson and Pichaud, 2010; Robinson et al., 2010). Loss of Crb causes upregulation of Hippo target genes, which requires Yki. Crb mutant cells lose Ex from the sub-apical membrane, and Crb and Ex directly interact in cultured cells (Chen et al., 2010; Ling et al., 2010; Robinson et al., 2010). Thus, Crb regulates growth of imaginal discs through the Hippo pathway by recruiting Ex to the sub-apical region of the plasma membrane. Notably, different motifs in the Crb intracellular domain are required to maintain cell polarity and to bind Ex (Chen et al., 2010; Ling et al., 2010; Robinson et al., 2010).
Several observations suggest that Crb may function to transduce extracellular cues to the Hippo pathway. First, Crb is required for proper Crb localization on neighboring cells, indicating that it acts as a homophilic adhesion receptor (Izaddoost et al., 2002; Tanentzapf et al., 2000). Second, Crb recruits Ex to the membrane autonomously through its intracellular domain and nonautonomously through the binding of Crb on neighboring cells (Chen et al., 2010). Hence, the membrane localization of Ex requires Crb-mediated cell-cell contact and is thus `regulated' by Crb-dependent cell-cell signaling. These findings indicate that Crb acts as a cell-contact-dependent receptor that regulates growth by modulating Hippo pathway activity.
Vertebrates have three Crb homologs, all of which share some conservation with the intracellular domain of Drosophila Crb and which function in apical-basal polarity (Bazellieres et al., 2009). Crb3 has been implicated as a tumor suppressor in selected immortal mouse kidney epithelial cells (Karp et al., 2008), but it is not known whether any of the vertebrate Crb homologs act through the Hippo pathway.
Regulation by the tumor suppressor Lethal giant larvae
The Lethal giant larvae (Lgl) cell polarity factor is part of the Discs large (Dlg) module of cell polarity determinants that also includes Scribble (Scrib) (reviewed in Humbert et al., 2008). All three are adaptor proteins that localize to the basolateral membrane of epithelial cells and function interdependently to organize apical-basal cell polarity. Mutations in lgl, dlg and scrib cause loss of basolateral markers and the expansion of apical markers, leading to defects in apical-basal cell polarity, and also result in massively overgrown imaginal discs. As such, they are referred to as `neoplastic tumor suppressors'. However, the pathways through which they regulate growth are poorly understood.
Lgl has recently been linked to the Hippo pathway (Grzeschik et al., 2010); Lgl mutant clones in eye discs cause extra cell proliferation, reduced apoptosis, relocalization of Yki from the cytoplasm to the nucleus and up-regulation of Hippo target genes. These effects are significant, although not as strong as those observed for mutations in core Hippo pathway components. Increased cell proliferation in lgl eye disc clones occurs without loss of apical-basal polarity, indicating separable roles for Lgl in controlling cell proliferation and cell polarity (Grzeschik et al., 2007). Notably, Ex is localized normally in lgl mutant cells but Hpo and Ras associated family protein (Rassf), an adaptor protein that binds to Hpo and modulates its activity in flies and vertebrates (Guo et al., 2007; Ikeda et al., 2009; Khokhlatchev et al., 2002; Polesello et al., 2006; Praskova et al., 2004), are mis-localized (Grzeschik et al., 2010). The molecular mechanisms that link Lgl to Hpo and Rassf localization are not known. Thus, although Lgl is a cell polarity determinant like Crb, it exerts its effect on the Hippo pathway in a manner that is different from that of Crb. Collectively, these observations suggest that multiple pathways modulate Hippo pathway activity in response to signals emanating from polarity complexes.
Whether other components of the Dlg complex affect the Hippo pathway in imaginal discs is unclear (Grzeschik et al., 2010). Loss of Lgl, Scrib or Dlg causes upregulation of the Hippo targets cyclin E and diap1 in the ovarian follicular epithelium, although it is not known whether these effects require increased Yki activity (Zhao et al., 2008b). Additional evidence that Dlg complex components regulate Hippo signaling comes from studies in zebrafish. A zebrafish homolog of Scrib genetically interacts with Yap during embryonic kidney development and suppresses Yap activity in cultured cells in a manner similar to Lats2, indicating that the link between cell polarity and the Hippo pathway may be conserved in vertebrates (Skouloudaki et al., 2009).
Regulation by Ajuba
The mammalian Ajuba LIM proteins (Ajuba, LIMD1 and WTIP) and the single Drosophila ortholog Jub have recently been shown to be inhibitory regulators of Hippo signaling (Das Thakur et al., 2010). The mammalian and Drosophila Ajuba proteins physically interact with Lats/Wts and with Sav1/Sav, and Ajuba inhibits Yap phosphorylation in response to Mst1 or Lats1/2 expression in cultured mammalian cells (Das Thakur et al., 2010). Drosophila jub mutant imaginal disc cells grow poorly and show increased apoptosis, phenotypes reminiscent of yki mutant cells, and Jub acts genetically downstream of Hpo but upstream of Wts (Das Thakur et al., 2010). The phenotype of jub mutant cells can thus be explained by the action of Jub on the Hippo pathway. By contrast, mammalian Ajuba proteins may have additional functions as they are able to interact with other proteins such as the Snail/Slug transcriptional repressors (Langer et al., 2008).
Ajuba proteins are adaptors that localize to adherens junctions and are thought to link cell adhesive properties with nuclear responses (Langer et al., 2008; Marie et al., 2003). Thus, Ajuba proteins may transduce signals that emanate at the plasma membrane to the Hippo pathway. Interestingly, Ajuba proteins require adherens junctions for their localization, and the formation of cell-cell contacts in confluent cultures promotes the recruitment of Ajuba to the membrane (Marie et al., 2003). If membrane localization regulates the activity of Ajuba, then recruitment of Ajuba to adherens junctions may contribute to the contact dependent inhibition of cell proliferation by disabling the action of Ajuba on Wts/Lats, thereby allowing the Hippo pathway to repress Yki/Yap and cell proliferation. Analysis of how the activity of Ajuba is regulated may thus give new insights into the regulation of the Hippo pathway by extracellular signals.
Tissue-specific differences in upstream regulation
Strikingly, the requirement for Hippo pathway components differs between different Drosophila epithelia, such as imaginal discs and ovarian follicle cells (Meignin et al., 2007; Polesello and Tapon, 2007; Yu et al., 2008). Although mutations in core pathway components (such as sav, hpo, wts and mats) show strong phenotypes in both tissues, the loss-of-function phenotypes of upstream regulators differ. In posterior follicle cells, loss of Mer and Kibra produces phenotypes that are as severe as those of hpo mutants, while their imaginal disc phenotypes are much weaker than those of hpo mutants (Baumgartner et al., 2010; Genevet et al., 2010; Hamaratoglu et al., 2006; MacDougall et al., 2001; Meignin et al., 2007; Milton et al., 2010; Pellock et al., 2007; Polesello and Tapon, 2007; Yu et al., 2008; Yu et al., 2010). Conversely, loss of Ex produces only mild phenotypes in the ovary and Fat is entirely dispensable, but both show relatively strong phenotypes in imaginal discs (Bryant et al., 1988; Hamaratoglu et al., 2006; Meignin et al., 2007; Milton et al., 2010; Pellock et al., 2007; Polesello and Tapon, 2007; Tyler and Baker, 2007; Yu et al., 2008). Similarly, Fat and Ex play only minor roles in the pupal eye, unlike the larval eye disc (Milton et al., 2010). In addition, upstream components such as Fat, Ex, Mer and Kibra can act partially redundantly in different tissues at different stages, which adds further complexity to the regulation of the Hippo pathway (Baumgartner et al., 2010; Genevet et al., 2010; Hamaratoglu et al., 2006; Maitra et al., 2006; Silva et al., 2006; Willecke et al., 2006; Yu et al., 2010). Thus, the strength and identity of upstream inputs into the Hippo pathway differ between different tissues and developmental stages.
Multiple inputs in mammalian systems
Several other inputs into the mammalian Hippo pathway are known, in addition to those discussed above (Fig. 2B). The mammalian Hpo kinase orthologs Mst1 and Mst2 were originally identified by virtue of their homology to yeast sterile 20 kinase (Creasy and Chernoff, 1995a; Creasy and Chernoff, 1995b), and the first physiological function attributed to either Mst1 or Mst2 (also known as Krs1 or Krs2) was increased kinase activity in response to pro-apoptotic stimuli, such as staurosporine (Taylor et al., 1996). Later, it was shown that oncogenic stress signals, such as those provided by activated Ras, also induced Mst1 activity, leading to apoptosis through an interaction with Rassf-like co-factors (Khokhlatchev et al., 2002) that relieve Mst1 inhibition by Raf-1 (O'Neill et al., 2004). Other inputs reported to modulate mammalian Hippo signaling include DNA damage (Hamilton et al., 2009), contact inhibition (Ota and Sasaki, 2008; Zhao et al., 2007), F-actin depolymerization (Densham et al., 2009) and CD44, a cell-surface hyaluronan receptor (Xu et al., 2010). Taken together, these observations demonstrate that mammalian Hippo signaling can be regulated in cultured cells, and further suggest that the pathway might likewise be modulated in vivo, although direct evidence for stress- or oncogene-induced pathway activation in vivo is currently lacking.
Unlike in Drosophila imaginal discs, mammalian Hippo signaling activity appears to be non-uniform, at least in some tissues. Mechanisms employed affect the level of localized expression, nuclear accumulation and stability of the downstream effector Yap. For example, in the mouse embryo and in several adult tissues, Yap protein expression is spatially restricted, often exhibiting highest levels in stem and progenitor cells, as seen in the skin and intestine (for more on the role of Hippo signaling in stem and progenitor cells, see Box 3) (Camargo et al., 2007; Lee et al., 2008; Ota and Sasaki, 2008). The mechanisms that control Yap levels in vivo are not known and may involve a combination of transcriptional and post-transcriptional mechanisms. Post-translational modifications affect Yap protein stability (Zhao et al., 2010b) and these may modulate Yap levels in vivo. Hence, differences in the levels of Yap expression may render cells more or less sensitive to alterations in the activity of upstream pathway components and can serve as an effective method of Hippo pathway modulation in vivo.
Box 3. Hippo signaling in stem cells and progenitor populations
Several lines of evidence suggest that Hippo signaling regulates the balance between pluripotent stem cells and lineage-restricted progenitor pools in mammals. First, Yap protein expression and nuclear localization are found to a greater degree in the intestinal crypts and the basal layer of the skin, where stem and progenitor cells reside, in both adult humans and mice (Camargo et al., 2007; Lam-Himlin et al., 2006). Second, in both mouse adult skin and intestinal epithelia, the attenuation of Hippo signaling expands the progenitor pool at the expense of differentiated cells (Camargo et al., 2007; Lee et al., 2008). Hippo signaling has also been reported to modulate stem cell proliferation in the regenerating Drosophila intestinal epithelium (Staley and Irvine, 2010). Third, overexpression of Yap or inhibition of Mst1/2 in the developing neuroepithelium of chick embryos expands the pool of neural progenitors in the spinal cord (Cao et al., 2008). Fourth, inactivation of either Sav1 or Mst1/2 in murine hepatocytes leads to activation of oval cells, a facultative progenitor population that is thought to derive from resident liver stem cells (Lee et al., 2010; Lu et al., 2010). Likewise, conditional deletion of Mer/Nf2 in the mouse liver results in oval cell expansion and an increased liver to body weight ratio (Benhamouche et al., 2010; Zhang et al., 2010). Finally, in mouse embryonic stem cells, Yap promotes pluripotency and inhibits differentiation in a Lats1/2-dependent manner (Lian et al., 2010). These observations reflect a role for Hippo signaling in maintaining the balance between stem, progenitor and differentiated cells although the way in which it does this remains to be determined.
An example of Yap regulation by the Hippo pathway is seen in the pre-implantation mouse embryo when the outer trophectoderm (TE, which gives rise to the extra-embryonic lineages) and the inner cell mass (ICM, which gives rise to the embryo proper) are specified. A key regulatory factor in TE development is the homeodomain transcription factor Cdx2, which is expressed in trophoblast cells and is both necessary and sufficient for TE development. Cdx2 expression depends on selective nuclear localization of Yap in presumptive TE cells, and this localization is mediated by core Hippo signaling components (Nishioka et al., 2009). Yap localization also depends on the position of cells in the embryo: in cells exposed to the surface, Yap is in both the cytoplasm and the nucleus; in internally located cells, nuclear Yap is absent. When outer cells are manipulated to an internal location, nuclear accumulation of Yap is lost. Similarly, when inner cells are exposed to the embryo surface, Yap accumulates in the nucleus. Hence, cell position within the embryo, via an unknown mechanism, regulates Yap localization by blocking Hippo signaling activity. One possible signal is E-cadherin-mediated cell adhesion, which is required for patterned Yap localization. These studies are an excellent example of how Hippo signaling can function instructively rather than simply being a permissive pathway, the activity of which is constitutively required.
Multiple outputs of Hippo signaling
Depending on the context, Hippo signaling can suppress growth, mediate stress-induced apoptosis or regulate cell fate decisions. These diverse biological outcomes result from tissue and cell-type specific pathway outputs of Hippo signaling. At least two main mechanisms contribute to the different outcomes of Hippo signaling: first, Yki and its mammalian orthologs Yap and Taz have multiple binding partners; and second, the transcriptional output of Hippo signaling is cell-type dependent.
Roles of Scalloped and Yorkie in Drosophila
In Drosophila imaginal discs, the TEAD family transcription factor Scalloped (Sd) primarily mediates the function of Yki, which itself does not bind to DNA (Goulev et al., 2008; Wu et al., 2008; Zhang et al., 2008b; Zhao et al., 2008a). Yki and Sd form a complex that directly binds to Hippo response elements in target genes, such as Drosophila inhibitor of apoptosis 1 (diap1) (Wu et al., 2008; Zhang et al., 2008b). Co-expression of Sd with Yki in imaginal discs enhances overgrowth caused by Yki overexpression, and Sd depletion suppresses it (Goulev et al., 2008; Wu et al., 2008; Zhang et al., 2008b; Zhao et al., 2008a). These and other findings led to a model in which Sd recruits Yki to Hippo pathway target genes, with Yki providing the co-activator function necessary for target gene expression. However, in reality the situation may be more complex than Sd simply being the dedicated Hippo pathway transcription factor.
For example, the phenotypes of sd and yki loss-of-function mutants are distinct. Sd is expressed at high levels in the developing wing pouch but not in dividing eye progenitor cells, and sd-null mutant cell clones grow poorly only in the developing wing pouch (Campbell et al., 1992; Liu et al., 2000). By contrast, Yki is required in all imaginal discs for cell proliferation (Huang et al., 2005). Thus, if Yki requires Sd to regulate Hippo pathway target genes, why does Sd not show the same requirement as Yki? One possible answer may be that Sd acts as a repressor without Yki. In this scenario, loss of Sd would result in a medium level of expression of target genes, whereas loss of Yki would result in suppression of target gene expression. Loss of Yki may thus produce a stronger phenotype than would loss of Sd. Alternatively, Yki may interact with additional transcription factors to regulate gene expression. Indeed, Yki also interacts with the Homothorax (Hth) transcription factor to regulate cell proliferation and expression of the bantam miRNA in the developing eye (Peng et al., 2009) and of Myc in the developing wing hinge (Ziosi et al., 2010). However, Hth regulates only a subset of Yki targets in the eye, and other factors or mechanisms may thus exist to explain the actions of Yki.
Sd also has functions that are independent of Yki. In the developing wing, Sd forms a complex with the Vestigial (Vg) co-activator that acts as a wing selector gene by inducing the expression of genes important for wing development (Halder et al., 1998; Paumard-Rigal et al., 1998; Simmonds et al., 1998). Notably, the sets of genes regulated by Yki and Vg are different, and whereas Sd/Vg regulates wing specification, Sd/Yki regulates growth. How, then, does Sd complexed with Vg regulate a different set of target genes than Sd complexed with Yki? It is possible that the two complexes bind to different target genes or that each complex requires other factors that bind DNA independently to regulate target gene expression. Notably, the interaction of Sd with Vg changes the DNA-binding specificity of Sd; thus, interaction of Sd with different co-factors may influence target gene selection (Halder and Carroll, 2001). Clearly, additional work is required to solve this puzzle.
Roles of Yap and Taz in mammals
In mammals, an additional level of complexity is achieved by the existence of two Yki-related co-activators, Yap and Taz, which bind to the TEAD/TEF family of sequence-specific DNA-binding proteins, TEAD 1-4 (Chan et al., 2009; Vassilev et al., 2001; Zhang et al., 2009; Zhao et al., 2008b). The TEAD proteins are required for many activities of Yap and Taz, including contact inhibition, epithelial-mesenchymal transition, transformation, apoptosis inhibition and trophectoderm development (Nishioka et al., 2009; Ota and Sasaki, 2008; Zhang et al., 2009; Zhao et al., 2008b). In addition to TEAD, Yap can interact with other transcription factors (Zhao et al., 2010a). For example, the pro-apoptotic effects of Yap are reported to occur via the binding of Yap to the tumor suppressor p73 (Strano et al., 2005; Strano et al., 2001). The Yap/p73 complex localizes to the nucleus where it regulates known p73 targets, such as puma and p53AIP1, that stimulate programmed cell death. Intriguingly, both the pro- and anti-apoptotic effects of Yap have been shown to depend on upstream core components of the Hippo signaling pathway (Hamilton et al., 2009; Oka et al., 2008). How Yap selectively induces cell death by interacting with p73 in some contexts while repressing apoptosis in other situations through TEAD binding is unclear. One possibility is that context-dependent post-translational modification of Yap allows for selective choice of interaction partners.
Taz also has specific binding partners, some of which are shared with Yap (reviewed in Wang et al., 2009). For example, Taz promotes osteoblast differentiation of mesenchymal stem cells by binding to and potentiating Runx2 activity, while simultaneously inhibiting adipocyte differentiation by complexing to and inhibiting Pparγ-mediated transcription (Hong et al., 2005). Yap can also bind to Runx2 (Zaidi et al., 2004), but is unable to bind to Pparγ. Whether upstream core Hippo signaling components modulate Taz activity in the context of mesenchymal stem cell differentiation has not been explored, but these findings suggest that Taz interacts with a variety of sequence-specific binding proteins, some in common with Yap, and some distinct. This probably contributes to different Hippo signaling outputs in different contexts.
Tissue-specific transcriptional responses
Transcriptional targets of Yap/TEAD or Taz/TEAD have been identified in mammalian cells by microarray profiling of fibroblasts, mammary epithelial cells and hepatocytes (Dong et al., 2007; Hao et al., 2008; Lu et al., 2010; Overholtzer et al., 2006). Interestingly, although there are common Yap/Taz/TEAD targets among the genes identified, most appear to be cell-type specific. Similarly, in Drosophila, Hippo pathway output shows cell type variation. For example, in eye and wing imaginal discs, Hippo signaling regulates genes involved in proliferation and apoptosis, such as cyclin E, diap1 and bantam (Harvey et al., 2003; Jia et al., 2003; Kango-Singh et al., 2002; Nolo et al., 2006; Pantalacci et al., 2003; Tapon et al., 2002; Thompson and Cohen, 2006; Udan et al., 2003; Wu et al., 2003) but in wing discs it also regulates vg and wg expression (Cho and Irvine, 2004; Zecca and Struhl, 2010), and in differentiating photoreceptor cells, Hippo signaling regulates the expression of specific opsin genes (Mikeladze-Dvali et al., 2005). In addition to growth, Hippo signaling also regulates cell morphology in imaginal discs, where it regulates the size of the apical domain (Genevet et al., 2009; Hamaratoglu et al., 2009). Hence, in both mammals and flies, the biological outputs of Hippo signaling can vary depending on when and where the pathway is deployed.
Conclusions
The emerging picture of the Hippo pathway is that of a pleiotropic pathway that is used for multiple aspects of signaling during development. When activity of this pathway is attenuated pathological phenotypes, such as cancer, can arise. Thus, the Hippo pathway has context-dependent roles and regulates different sets of downstream target genes in different cell types. In addition to Hippo signaling having pleiotropic effects, it has been shown that Hippo pathway components are dedicated not only to the Hippo pathway but that they also mediate crosstalk with other pathways. Recent examples include the Wnt, TGFβ and BMP pathways. Taz can inhibit Wnt signal transduction by binding to the Wnt pathway component Dvl (Varelas et al., 2010), while Taz binding to the TGFβ signal transducing Smad2/3 complex couples it to the transcriptional machinery (Varelas et al., 2008). In addition, binding of Yap to Smad1 enhances BMP signaling (Alarcon et al., 2009). Given its contextual dependency, Hippo signaling is not strictly involved in regulating cell proliferation, cell survival and growth alone, but is also involved in the regulation of cell type specification and differentiation. Clearly, much work needs to be done to understand the multiple roles of the Hippo pathway during development and disease. One of the most pressing unknown issues is still the question of upstream regulatory mechanisms and signals: do global organ growth controlling signals act via the Hippo pathway to regulate growth and regeneration? Given the pace of discoveries in the Hippo field, we can expect many new insights into this pathway in the near future.
Note added in proof
Two recent papers reveal further details about the function of the Hippo pathway during Drosophila midgut regeneration (Shaw et al., 2010; Karpowicz et al., 2010). Together with the report by Staley and Irvine (Staley and Irvine, 2010), these papers show that Jun N-terminal kinase signaling activates Yki in differentiated midgut cells in response to tissue damage. Yki then drives the expression of cytokines, which activate Janus kinase/Signal transducer and activator of transcription signaling in nearby intestinal stem cells, stimulating their proliferation. In addition, Yki plays a cell-autonomous role in intestinal stem cells, where it is activated upon tissue damage and required for the regenerative proliferation of intestinal stem cells (Shaw et al., 2010; Karpowicz et al., 2010). These reports thus reveal cell-autonomous and non cell-autonomous functions of the Hippo pathway in regulating intestinal stem cell proliferation during regeneration, and also provide an example of regulated Hippo activity.
Acknowledgments
We thank L. McCord for help with art production, and members of the Halder laboratory for comments on the manuscript. Research in G.H.'s laboratory is supported by the NIH and CPRIT (Cancer Prevention Research Institute of Texas) and in R.L.J.'s laboratory by the NIH. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- Adler P. N., Charlton J., Liu J. (1998). Mutations in the cadherin superfamily member gene dachsous cause a tissue polarity phenotype by altering frizzled signaling. Development 125, 959-968 [DOI] [PubMed] [Google Scholar]
- Affolter M., Basler K. (2007). The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat. Rev. Genet. 8, 663-674 [DOI] [PubMed] [Google Scholar]
- Alarcon C., Zaromytidou A. I., Xi Q., Gao S., Yu J., Fujisawa S., Barlas A., Miller A. N., Manova-Todorova K., Macias M. J., et al. (2009). Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell 139, 757-769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashery-Padan R., Alvarez-Bolado G., Klamt B., Gessler M., Gruss P. (1999). Fjx1, the murine homologue of the Drosophila four-jointed gene, codes for a putative secreted protein expressed in restricted domains of the developing and adult brain. Mech. Dev. 80, 213-217 [DOI] [PubMed] [Google Scholar]
- Badouel C., Gardano L., Amin N., Garg A., Rosenfeld R., Le Bihan T., McNeill H. (2009a). The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev. Cell 16, 411-420 [DOI] [PubMed] [Google Scholar]
- Badouel C., Garg A., McNeill H. (2009b). Herding Hippos: regulating growth in flies and man. Curr. Opin. Cell Biol. 21, 837-843 [DOI] [PubMed] [Google Scholar]
- Bando T., Mito T., Maeda Y., Nakamura T., Ito F., Watanabe T., Ohuchi H., Noji S. (2009). Regulation of leg size and shape by the Dachsous/Fat signalling pathway during regeneration. Development 136, 2235-2245 [DOI] [PubMed] [Google Scholar]
- Baumgartner R., Poernbacher I., Buser N., Hafen E., Stocker H. (2010). The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev. Cell 18, 309-316 [DOI] [PubMed] [Google Scholar]
- Bazellieres E., Assemat E., Arsanto J. P., Le Bivic A., Massey-Harroche D. (2009). Crumbs proteins in epithelial morphogenesis. Front. Biosci. 14, 2149-2169 [DOI] [PubMed] [Google Scholar]
- Benhamouche S., Curto M., Saotome I., Gladden A. B., Liu C. H., Giovannini M., McClatchey A. I. (2010). Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 24, 1718-1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett F. C., Harvey K. F. (2006). Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr. Biol. 16, 2101-2110 [DOI] [PubMed] [Google Scholar]
- Bichsel S. J., Tamaskovic R., Stegert M. R., Hemmings B. A. (2004). Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein. J. Biol. Chem. 279, 35228-35235 [DOI] [PubMed] [Google Scholar]
- Boedigheimer M., Laughon A. (1993). Expanded: a gene involved in the control of cell proliferation in imaginal discs. Development 118, 1291-1301 [DOI] [PubMed] [Google Scholar]
- Boedigheimer M. J., Nguyen K. P., Bryant P. J. (1997). Expanded functions in the apical cell domain to regulate the growth rate of imaginal discs. Dev. Genet. 20, 103-110 [DOI] [PubMed] [Google Scholar]
- Brittle A. L., Repiso A., Casal J., Lawrence P. A., Strutt D. (2010). Four-jointed modulates growth and planar polarity by reducing the affinity of dachsous for fat. Curr. Biol. 20, 803-810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant P. J., Bryant S. V., French V. (1977). Biological regeneration and pattern formation. Sci. Am. 237, 66-76, 81 [DOI] [PubMed] [Google Scholar]
- Bryant P. J., Huettner B., Held L. I., Jr, Ryerse J., Szidonya J. (1988). Mutations at the fat locus interfere with cell proliferation control and epithelial morphogenesis in Drosophila. Dev. Biol. 129, 541-554 [DOI] [PubMed] [Google Scholar]
- Camargo F. D., Gokhale S., Johnnidis J. B., Fu D., Bell G. W., Jaenisch R., Brummelkamp T. R. (2007). YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17, 2054-2060 [DOI] [PubMed] [Google Scholar]
- Campbell S., Inamdar M., Rodrigues V., Raghavan V., Palazzolo M., Chovnick A. (1992). The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes Dev. 6, 367-379 [DOI] [PubMed] [Google Scholar]
- Cao X., Pfaff S. L., Gage F. H. (2008). YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 22, 3320-3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J., Struhl G., Lawrence P. A. (2002). Developmental compartments and planar polarity in Drosophila. Curr. Biol. 12, 1189-1198 [DOI] [PubMed] [Google Scholar]
- Casal J., Lawrence P. A., Struhl G. (2006). Two separate molecular systems, Dachsous/Fat and Starry night/Frizzled, act independently to confer planar cell polarity. Development 133, 4561-4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E. H., Nousiainen M., Chalamalasetty R. B., Schafer A., Nigg E. A., Sillje H. H. (2005). The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24, 2076-2086 [DOI] [PubMed] [Google Scholar]
- Chan S. W., Lim C. J., Loo L. S., Chong Y. F., Huang C., Hong W. (2009). TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J. Biol. Chem. 284, 14347-14358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. W., Lim C. J., Chen L., Chong Y. F., Huang C., Song H., Hong W. (2010). The hippo pathway in biological control and cancer development. J. Cell Physiol. (in press). [DOI] [PubMed] [Google Scholar]
- Chen C. L., Gajewski K. M., Hamaratoglu F., Bossuyt W., Sansores-Garcia L., Tao C., Halder G. (2010). The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc. Natl. Acad. Sci. USA 107, 15810-15815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E., Irvine K. D. (2004). Action of fat, four-jointed, dachsous and dachs in distal-to-proximal wing signaling. Development 131, 4489-4500 [DOI] [PubMed] [Google Scholar]
- Cho E., Feng Y., Rauskolb C., Maitra S., Fehon R., Irvine K. D. (2006). Delineation of a Fat tumor suppressor pathway. Nat. Genet. 38, 1142-1150 [DOI] [PubMed] [Google Scholar]
- Chow A., Hao Y., Yang X. (2010). Molecular characterization of human homologs of yeast MOB1. Int. J. Cancer 126, 2079-2089 [DOI] [PubMed] [Google Scholar]
- Clark H. F., Brentrup D., Schneitz K., Bieber A., Goodman C., Noll M. (1995). Dachsous encodes a member of the cadherin superfamily that controls imaginal disc morphogenesis in Drosophila. Genes Dev. 9, 1530-1542 [DOI] [PubMed] [Google Scholar]
- Creasy C. L., Chernoff J. (1995a). Cloning and characterization of a human protein kinase with homology to Ste20. J. Biol. Chem. 270, 21695-21700 [DOI] [PubMed] [Google Scholar]
- Creasy C. L., Chernoff J. (1995b). Cloning and characterization of a member of the MST subfamily of Ste20-like kinases. Gene 167, 303-306 [DOI] [PubMed] [Google Scholar]
- Curto M., Cole B. K., Lallemand D., Liu C. H., McClatchey A. I. (2007). Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. J. Cell Biol. 177, 893-903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur M., Feng Y., Jagannathan R., Seppa M. J., Skeath J. B., Longmore G. D. (2010). Ajuba LIM proteins are negative regulators of the hippo signaling pathway. Curr. Biol. 20, 657-662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Densham R. M., O'Neill E., Munro J., Konig I., Anderson K., Kolch W., Olson M. F. (2009). MST kinases monitor actin cytoskeletal integrity and signal via c-Jun N-terminal kinase stress-activated kinase to regulate p21Waf1/Cip1 stability. Mol. Cell. Biol. 29, 6380-6390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A., Pan D. (2007). Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120-1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Irvine K. D. (2007). Fat and expanded act in parallel to regulate growth through warts. Proc. Natl. Acad. Sci. USA 104, 20362-20367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Irvine K. D. (2009). Processing and phosphorylation of the Fat receptor. Proc. Natl. Acad. Sci. USA 106, 11989-11994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L. A., Kenney A. M. (2010). The Hippo in the room: a new look at a key pathway in cell growth and transformation. Cell Cycle 9, 2292-2299 [DOI] [PubMed] [Google Scholar]
- French V., Bryant P. J., Bryant S. V. (1976). Pattern regulation in epimorphic fields. Science 193, 969-981 [DOI] [PubMed] [Google Scholar]
- Genevet A., Polesello C., Blight K., Robertson F., Collinson L. M., Pichaud F., Tapon N. (2009). The Hippo pathway regulates apical-domain size independently of its growth-control function. J. Cell Sci. 122, 2360-2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevet A., Wehr M. C., Brain R., Thompson B. J., Tapon N. (2010). Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell 18, 300-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulev Y., Fauny J. D., Gonzalez-Marti B., Flagiello D., Silber J., Zider A. (2008). SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 18, 435-441 [DOI] [PubMed] [Google Scholar]
- Graves J. D., Gotoh Y., Draves K. E., Ambrose D., Han D. K., Wright M., Chernoff J., Clark E. A., Krebs E. G. (1998). Caspase-mediated activation and induction of apoptosis by the mammalian Ste20-like kinase Mst1. EMBO J. 17, 2224-2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik N. A., Amin N., Secombe J., Brumby A. M., Richardson H. E. (2007). Abnormalities in cell proliferation and apico-basal cell polarity are separable in Drosophila lgl mutant clones in the developing eye. Dev. Biol. 311, 106-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik N. A., Parsons L. M., Allott M. L., Harvey K. F., Richardson H. E. (2010). Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr. Biol. 20, 573-581 [DOI] [PubMed] [Google Scholar]
- Guo C., Tommasi S., Liu L., Yee J. K., Dammann R., Pfeifer G. P. (2007). RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr. Biol. 17, 700-705 [DOI] [PubMed] [Google Scholar]
- Halder G., Carroll S. B. (2001). Binding of the Vestigial co-factor switches the DNA-target selectivity of the Scalloped selector protein. Development 128, 3295-3305 [DOI] [PubMed] [Google Scholar]
- Halder G., Polaczyk P., Kraus M. E., Hudson A., Kim J., Laughon A., Carroll S. (1998). The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in Drosophila. Genes Dev. 12, 3900-3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H., Halder G. (2006). The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8, 27-36 [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F., Gajewski K., Sansores-Garcia L., Morrison C., Tao C., Halder G. (2009). The Hippo tumor-suppressor pathway regulates apical-domain size in parallel to tissue growth. J. Cell Sci. 122, 2351-2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton G., Yee K. S., Scrace S., O'Neill E. (2009). ATM regulates a RASSF1A-dependent DNA damage response. Curr. Biol. 19, 2020-2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Chun A., Cheung K., Rashidi B., Yang X. (2008). Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 283, 5496-5509 [DOI] [PubMed] [Google Scholar]
- Harvey K. F., Pfleger C. M., Hariharan I. K. (2003). The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457-467 [DOI] [PubMed] [Google Scholar]
- Haynie J. L., Bryant P. J. (1977). The effects of X-rays on the proliferation dynamics of cells in the imaginal wing disc of Drosophila melanogaster. Roux Arch. Dev. Biol. 183, 85-100 [DOI] [PubMed] [Google Scholar]
- Hirabayashi S., Nakagawa K., Sumita K., Hidaka S., Kawai T., Ikeda M., Kawata A., Ohno K., Hata Y. (2008). Threonine 74 of MOB1 is a putative key phosphorylation site by MST2 to form the scaffold to activate nuclear Dbf2-related kinase 1. Oncogene 27, 4281-4292 [DOI] [PubMed] [Google Scholar]
- Hong J. H., Hwang E. S., McManus M. T., Amsterdam A., Tian Y., Kalmukova R., Mueller E., Benjamin T., Spiegelman B. M., Sharp P. A., et al. (2005). TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 309, 1074-1078 [DOI] [PubMed] [Google Scholar]
- Huang J., Wu S., Barrera J., Matthews K., Pan D. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421-434 [DOI] [PubMed] [Google Scholar]
- Humbert P. O., Grzeschik N. A., Brumby A. M., Galea R., Elsum I., Richardson H. E. (2008). Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene 27, 6888-6907 [DOI] [PubMed] [Google Scholar]
- Ikeda M., Kawata A., Nishikawa M., Tateishi Y., Yamaguchi M., Nakagawa K., Hirabayashi S., Bao Y., Hidaka S., Hirata Y., et al. (2009). Hippo pathway-dependent and -independent roles of RASSF6. Sci Signal 2, ra59 [DOI] [PubMed] [Google Scholar]
- Ishikawa H. O., Takeuchi H., Haltiwanger R. S., Irvine K. D. (2008). Four-jointed is a Golgi kinase that phosphorylates a subset of cadherin domains. Science 321, 401-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T., Misaki K., Yonemura S., Takeichi M., Tanoue T. (2009). Mammalian Fat and Dachsous cadherins regulate apical membrane organization in the embryonic cerebral cortex. J. Cell Biol. 185, 959-967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaddoost S., Nam S. C., Bhat M. A., Bellen H. J., Choi K. W. (2002). Drosophila Crumbs is a positional cue in photoreceptor adherens junctions and rhabdomeres. Nature 416, 178-183 [DOI] [PubMed] [Google Scholar]
- Jia J., Zhang W., Wang B., Trinko R., Jiang J. (2003). The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 17, 2514-2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice R. W., Zilian O., Woods D. F., Noll M., Bryant P. J. (1995). The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 9, 534-546 [DOI] [PubMed] [Google Scholar]
- Kanai F., Marignani P. A., Sarbassova D., Yagi R., Hall R. A., Donowitz M., Hisaminato A., Fujiwara T., Ito Y., Cantley L. C., et al. (2000). TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 19, 6778-6791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kango-Singh M., Nolo R., Tao C., Verstreken P., Hiesinger P. R., Bellen H. J., Halder G. (2002). Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development 129, 5719-5730 [DOI] [PubMed] [Google Scholar]
- Karp C. M., Tan T. T., Mathew R., Nelson D., Mukherjee C., Degenhardt K., Karantza-Wadsworth V., White E. (2008). Role of the polarity determinant crumbs in suppressing mammalian epithelial tumor progression. Cancer Res. 68, 4105-4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz P., Perez J., Perrimon N. (2010). The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development 137, 4135-4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokhlatchev A., Rabizadeh S., Xavier R., Nedwidek M., Chen T., Zhang X. F., Seed B., Avruch J. (2002). Identification of a novel Ras-regulated proapoptotic pathway. Curr. Biol. 12, 253-265 [DOI] [PubMed] [Google Scholar]
- Lai Z. C., Wei X., Shimizu T., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L. L., Li Y. (2005). Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell 120, 675-685 [DOI] [PubMed] [Google Scholar]
- Lallemand D., Curto M., Saotome I., Giovannini M., McClatchey A. I. (2003). NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes Dev. 17, 1090-1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam-Himlin D. M., Daniels J. A., Gayyed M. F., Dong J., Maitra A., Pan D., Montgomery E. A., Anders R. A. (2006). The hippo pathway in human upper gastrointestinal dysplasia and carcinoma: a novel oncogenic pathway. Int. J. Gastrointest. Cancer 37, 103-109 [DOI] [PubMed] [Google Scholar]
- Langer E. M., Feng Y., Zhaoyuan H., Rauscher F. J., 3rd, Kroll K. L., Longmore G. D. (2008). Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev. Cell 14, 424-436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence P. A., Struhl G., Casal J. (2007). Planar cell polarity: one or two pathways? Nat. Rev. Genet. 8, 555-563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence P. A., Struhl G., Casal J. (2008). Do the protocadherins Fat and Dachsous link up to determine both planar cell polarity and the dimensions of organs? Nat. Cell Biol. 10, 1379-1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Kim T. S., Yang T. H., Koo B. K., Oh S. P., Lee K. P., Oh H. J., Lee S. H., Kong Y. Y., Kim J. M., et al. (2008). A crucial role of WW45 in developing epithelial tissues in the mouse. EMBO J. 27, 1231-1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. P., Lee J. H., Kim T. S., Kim T. H., Park H. D., Byun J. S., Kim M. C., Jeong W. I., Calvisi D. F., Kim J. M., et al. (2010). The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc. Natl. Acad. Sci. USA 107, 8248-8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Q. Y., Zhang H., Zhao B., Zha Z. Y., Bai F., Pei X. H., Zhao S., Xiong Y., Guan K. L. (2008). TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell. Biol. 28, 2426-2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian I., Kim J., Okazawa H., Zhao J., Zhao B., Yu J., Chinnaiyan A., Israel M. A., Goldstein L. S., Abujarour R., et al. (2010). The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 24, 1106-1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C., Zheng Y., Yin F., Yu J., Huang J., Hong Y., Wu S., Pan D. (2010). The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc. Natl. Acad. Sci. USA 107, 10532-10537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. Y., Zha Z. Y., Zhou X., Zhang H., Huang W., Zhao D., Li T., Chan S. W., Lim C. J., Hong W., et al. (2010). The Hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCFbeta-TrCP E3 ligase. J. Biol. Chem. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Grammont M., Irvine K. D. (2000). Roles for scalloped and vestigial in regulating cell affinity and interactions between the wing blade and the wing hinge. Dev. Biol. 228, 287-303 [DOI] [PubMed] [Google Scholar]
- Lu L., Li Y., Kim S. M., Bossuyt W., Liu P., Qiu Q., Wang Y., Halder G., Finegold M. J., Lee J. S., et al. (2010). Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. USA 107, 1437-1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Yang C. H., McNeill H., Simon M. A., Axelrod J. D. (2003). Fidelity in planar cell polarity signalling. Nature 421, 543-547 [DOI] [PubMed] [Google Scholar]
- MacDougall N., Lad Y., Wilkie G. S., Francis-Lang H., Sullivan W., Davis I. (2001). Merlin, the Drosophila homologue of neurofibromatosis-2, is specifically required in posterior follicle cells for axis formation in the oocyte. Development 128, 665-673 [DOI] [PubMed] [Google Scholar]
- Mahoney P. A., Weber U., Onofrechuk P., Biessmann H., Bryant P. J., Goodman C. S. (1991). The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell 67, 853-868 [DOI] [PubMed] [Google Scholar]
- Maitra S., Kulikauskas R. M., Gavilan H., Fehon R. G. (2006). The tumor suppressors Merlin and expanded function cooperatively to modulate receptor endocytosis and signaling. Curr. Biol. 16, 702-709 [DOI] [PubMed] [Google Scholar]
- Mao Y., Rauskolb C., Cho E., Hu W. L., Hayter H., Minihan G., Katz F. N., Irvine K. D. (2006). Dachs: an unconventional myosin that functions downstream of Fat to regulate growth, affinity and gene expression in Drosophila. Development 133, 2539-2551 [DOI] [PubMed] [Google Scholar]
- Mao Y., Kucuk B., Irvine K. D. (2009). Drosophila lowfat, a novel modulator of Fat signaling. Development 136, 3223-3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie H., Pratt S. J., Betson M., Epple H., Kittler J. T., Meek L., Moss S. J., Troyanovsky S., Attwell D., Longmore G. D., et al. (2003). The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J. Biol. Chem. 278, 1220-1228 [DOI] [PubMed] [Google Scholar]
- Matakatsu H., Blair S. S. (2004). Interactions between Fat and Dachsous and the regulation of planar cell polarity in the Drosophila wing. Development 131, 3785-3794 [DOI] [PubMed] [Google Scholar]
- Matakatsu H., Blair S. S. (2006). Separating the adhesive and signaling functions of the Fat and Dachsous protocadherins. Development 133, 2315-2324 [DOI] [PubMed] [Google Scholar]
- Matakatsu H., Blair S. S. (2008). The DHHC palmitoyltransferase approximated regulates Fat signaling and Dachs localization and activity. Curr. Biol. 18, 1390-1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney B. M., Kulikauskas R. M., LaJeunesse D. R., Fehon R. G. (2000). The Neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development 127, 1315-1324 [DOI] [PubMed] [Google Scholar]
- Meignin C., Alvarez-Garcia I., Davis I., Palacios I. M. (2007). The salvador-warts-hippo pathway is required for epithelial proliferation and axis specification in Drosophila. Curr. Biol. 17, 1871-1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeladze-Dvali T., Wernet M. F., Pistillo D., Mazzoni E. O., Teleman A. A., Chen Y. W., Cohen S., Desplan C. (2005). The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell 122, 775-787 [DOI] [PubMed] [Google Scholar]
- Milton C. C., Zhang X., Albanese N. O., Harvey K. F. (2010). Differential requirement of Salvador-Warts-Hippo pathway members for organ size control in Drosophila melanogaster. Development 137, 735-743 [DOI] [PubMed] [Google Scholar]
- Moeller M. J., Soofi A., Braun G. S., Li X., Watzl C., Kriz W., Holzman L. B. (2004). Protocadherin FAT1 binds Ena/VASP proteins and is necessary for actin dynamics and cell polarization. EMBO J. 23, 3769-3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima D., Nakayama M., Kikuno R., Hirosawa M., Nagase T., Ohara O. (2001). Identification of three novel non-classical cadherin genes through comprehensive analysis of large cDNAs. Brain Res. Mol. Brain Res. 94, 85-95 [DOI] [PubMed] [Google Scholar]
- Neto-Silva R. M., Wells B. S., Johnston L. A. (2009). Mechanisms of growth and homeostasis in the Drosophila wing. Annu. Rev. Cell Dev. Biol. 25, 197-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto-Silva R. M., de Beco S., Johnston L. A. (2010). Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev. Cell 19, 507-520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka N., Inoue K., Adachi K., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R. O., Ogonuki N., et al. (2009). The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398-410 [DOI] [PubMed] [Google Scholar]
- Nolo R., Morrison C. M., Tao C., Zhang X., Halder G. (2006). The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 16, 1895-1904 [DOI] [PubMed] [Google Scholar]
- Oh H., Irvine K. D. (2008). In vivo regulation of Yorkie phosphorylation and localization. Development 135, 1081-1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Irvine K. D. (2009). In vivo analysis of Yorkie phosphorylation sites. Oncogene 28, 1916-1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Irvine K. D. (2010). Yorkie: the final destination of Hippo signaling. Trends Cell Biol. 20, 410-417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Reddy B. V., Irvine K. D. (2009). Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev. Biol. 335, 188-197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Sudol M. (2009). Nuclear localization and pro-apoptotic signaling of YAP2 require intact PDZ-binding motif. Genes Cells 14, 607-615 [DOI] [PubMed] [Google Scholar]
- Oka T., Mazack V., Sudol M. (2008). Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP). J. Biol. Chem. 283, 27534-27546 [DOI] [PubMed] [Google Scholar]
- O'Neill E., Rushworth L., Baccarini M., Kolch W. (2004). Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1. Science 306, 2267-2270 [DOI] [PubMed] [Google Scholar]
- Ota M., Sasaki H. (2008). Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development 135, 4059-4069 [DOI] [PubMed] [Google Scholar]
- Overholtzer M., Zhang J., Smolen G. A., Muir B., Li W., Sgroi D. C., Deng C. X., Brugge J. S., Haber D. A. (2006). Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl. Acad. Sci. USA 103, 12405-12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S., Tapon N., Leopold P. (2003). The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell. Biol. 5, 921-927 [DOI] [PubMed] [Google Scholar]
- Paumard-Rigal S., Zider A., Vaudin P., Silber J. (1998). Specific interactions between vestigial and scalloped are required to promote wing tissue proliferation in Drosophila melanogaster. Dev. Genes Evol. 208, 440-446 [DOI] [PubMed] [Google Scholar]
- Pellock B. J., Buff E., White K., Hariharan I. K. (2007). The Drosophila tumor suppressors Expanded and Merlin differentially regulate cell cycle exit, apoptosis, and Wingless signaling. Dev. Biol. 304, 102-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H. W., Slattery M., Mann R. S. (2009). Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 23, 2307-2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesello C., Tapon N. (2007). Salvador-warts-hippo signaling promotes Drosophila posterior follicle cell maturation downstream of notch. Curr. Biol. 17, 1864-1870 [DOI] [PubMed] [Google Scholar]
- Polesello C., Huelsmann S., Brown N. H., Tapon N. (2006). The Drosophila RASSF homolog antagonizes the hippo pathway. Curr. Biol. 16, 2459-2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praskova M., Khoklatchev A., Ortiz-Vega S., Avruch J. (2004). Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem. J. 381, 453-462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praskova M., Xia F., Avruch J. (2008). MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 18, 311-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls A. S., Guinto J. B., Wolff T. (2002). The cadherins fat and dachsous regulate dorsal/ventral signaling in the Drosophila eye. Curr. Biol. 12, 1021-1026 [DOI] [PubMed] [Google Scholar]
- Reddy B. V., Irvine K. D. (2008). The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development 135, 2827-2838 [DOI] [PubMed] [Google Scholar]
- Reddy B. V., Rauskolb C., Irvine K. D. (2010). Influence of fat-hippo and notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development 137, 2397-2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F., Zhang L., Jiang J. (2009). Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev. Biol. 337, 303-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro P. S., Josue F., Wepf A., Wehr M. C., Rinner O., Kelly G., Tapon N., Gstaiger M. (2010). Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell 39, 521-534 [DOI] [PubMed] [Google Scholar]
- Richardson E. C., Pichaud F. (2010). Crumbs is required to achieve proper organ size control during Drosophila head development. Development 137, 641-650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson B. S., Huang J., Hong Y., Moberg K. H. (2010). Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein expanded. Curr. Biol. 20, 582-590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock R., Schrauth S., Gessler M. (2005). Expression of mouse dchs1, fjx1, and fat-j suggests conservation of the planar cell polarity pathway identified in Drosophila. Dev. Dyn. 234, 747-755 [DOI] [PubMed] [Google Scholar]
- Rogulja D., Rauskolb C., Irvine K. D. (2008). Morphogen control of wing growth through the Fat signaling pathway. Dev. Cell 15, 309-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saburi S., Hester I., Fischer E., Pontoglio M., Eremina V., Gessler M., Quaggin S. E., Harrison R., Mount R., McNeill H. (2008). Loss of Fat4 disrupts PCP signaling and oriented cell division and leads to cystic kidney disease. Nat. Genet. 40, 1010-1015 [DOI] [PubMed] [Google Scholar]
- Shaw R. L., Kohlmaier A., Polesello C., Veelken C., Edgar B. A., Tapon N. (2010). The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development 137, 4147-4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E., Tsatskis Y., Gardano L., Tapon N., McNeill H. (2006). The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr. Biol. 16, 2081-2089 [DOI] [PubMed] [Google Scholar]
- Simmonds A. J., Liu X., Soanes K. H., Krause H. M., Irvine K. D., Bell J. B. (1998). Molecular interactions between Vestigial and Scalloped promote wing formation in Drosophila. Genes Dev. 12, 3815-3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M. A. (2004). Planar cell polarity in the Drosophila eye is directed by graded Four-jointed and Dachsous expression. Development 131, 6175-6184 [DOI] [PubMed] [Google Scholar]
- Simon M. A., Xu A., Ishikawa H. O., Irvine K. D. (2010). Modulation of FAt:Dachsous binding by the cadherin domain kinase four-jointed. Curr. Biol. 20, 811-817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skouloudaki K., Puetz M., Simons M., Courbard J. R., Boehlke C., Hartleben B., Engel C., Moeller M. J., Englert C., Bollig F., et al. (2009). Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc. Natl. Acad. Sci. USA 106, 8579-8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow M. H., Tam P. P. (1979). Is compensatory growth a complicating factor in mouse teratology? Nature 279, 555-557 [DOI] [PubMed] [Google Scholar]
- Song H., Mak K. K., Topol L., Yun K., Hu J., Garrett L., Chen Y., Park O., Chang J., Simpson R. M., et al. (2010). Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc. Natl. Acad. Sci. USA 107, 1431-1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., McNeill H. (2009). The skinny on Fat: an enormous cadherin that regulates cell adhesion, tissue growth, and planar cell polarity. Curr. Opin. Cell Biol. 21, 717-723 [DOI] [PubMed] [Google Scholar]
- Sopko R., Silva E., Clayton L., Gardano L., Barrios-Rodiles M., Wrana J., Varelas X., Arbouzova N. I., Shaw S., Saburi S., et al. (2009). Phosphorylation of the tumor suppressor fat is regulated by its ligand Dachsous and the kinase discs overgrown. Curr. Biol. 19, 1112-1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley B. K., Irvine K. D. (2010). Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr. Biol. 20, 1580-1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger B. Z. (2008). Organ size determination and the limits of regulation. Cell Cycle 7, 318-324 [DOI] [PubMed] [Google Scholar]
- Stavridi E. S., Harris K. G., Huyen Y., Bothos J., Verwoerd P. M., Stayrook S. E., Pavletich N. P., Jeffrey P. D., Luca F. C. (2003). Crystal structure of a human Mob1 protein: toward understanding Mob-regulated cell cycle pathways. Structure 11, 1163-1170 [DOI] [PubMed] [Google Scholar]
- Strano S., Munarriz E., Rossi M., Castagnoli L., Shaul Y., Sacchi A., Oren M., Sudol M., Cesareni G., Blandino G. (2001). Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J. Biol. Chem. 276, 15164-15173 [DOI] [PubMed] [Google Scholar]
- Strano S., Monti O., Pediconi N., Baccarini A., Fontemaggi G., Lapi E., Mantovani F., Damalas A., Citro G., Sacchi A., et al. (2005). The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol. Cell 18, 447-459 [DOI] [PubMed] [Google Scholar]
- Striedinger K., VandenBerg S. R., Baia G. S., McDermott M. W., Gutmann D. H., Lal A. (2008). The neurofibromatosis 2 tumor suppressor gene product, merlin, regulates human meningioma cell growth by signaling through YAP. Neoplasia 10, 1204-1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H., Strutt D. (2002). Nonautonomous planar polarity patterning in Drosophila: dishevelled-independent functions of frizzled. Dev. Cell 3, 851-863 [DOI] [PubMed] [Google Scholar]
- Sudol M. (1994). Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene 9, 2145-2152 [PubMed] [Google Scholar]
- Tanentzapf G., Smith C., McGlade J., Tepass U. (2000). Apical, lateral, and basal polarization cues contribute to the development of the follicular epithelium during Drosophila oogenesis. J. Cell Biol. 151, 891-904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T., Takeichi M. (2004). Mammalian Fat1 cadherin regulates actin dynamics and cell-cell contact. J. Cell Biol. 165, 517-528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue T., Takeichi M. (2005). New insights into Fat cadherins. J. Cell Sci. 118, 2347-2353 [DOI] [PubMed] [Google Scholar]
- Tao W., Zhang S., Turenchalk G. S., Stewart R. A., St John M. A., Chen W., Xu T. (1999). Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat. Genet. 21, 177-181 [DOI] [PubMed] [Google Scholar]
- Tapon N., Harvey K., Bell D., Wahrer D., Schiripo T., Haber D., Hariharan I. (2002). salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467 [DOI] [PubMed] [Google Scholar]
- Taylor L. K., Wang H. C., Erikson R. L. (1996). Newly identified stress-responsive protein kinases, Krs-1 and Krs-2. Proc. Natl. Acad. Sci. USA 93, 10099-10104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. J., Cohen S. M. (2006). The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126, 767-774 [DOI] [PubMed] [Google Scholar]
- Tyler D. M., Baker N. E. (2007). Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev. Biol. 305, 187-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan R. S., Kango-Singh M., Nolo R., Tao C., Halder G. (2003). Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5, 914-920 [DOI] [PubMed] [Google Scholar]
- Varelas X., Sakuma R., Samavarchi-Tehrani P., Peerani R., Rao B. M., Dembowy J., Yaffe M. B., Zandstra P. W., Wrana J. L. (2008). TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 10, 837-848 [DOI] [PubMed] [Google Scholar]
- Varelas X., Miller B. W., Sopko R., Song S., Gregorieff A., Fellouse F. A., Sakuma R., Pawson T., Hunziker W., McNeill H., et al. (2010). The Hippo pathway regulates Wnt/beta-catenin signaling. Dev. Cell 18, 579-591 [DOI] [PubMed] [Google Scholar]
- Vassilev A., Kaneko K. J., Shu H., Zhao Y., DePamphilis M. L. (2001). TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 15, 1229-1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villano J. L., Katz F. N. (1995). four-jointed is required for intermediate growth in the proximal-distal axis in Drosophila. Development 121, 2767-2777 [DOI] [PubMed] [Google Scholar]
- Wang K., Degerny C., Xu M., Yang X. J. (2009). YAP, TAZ, and Yorkie: a conserved family of signal-responsive transcriptional coregulators in animal development and human disease. Biochem. Cell Biol. 87, 77-91 [DOI] [PubMed] [Google Scholar]
- Wei X., Shimizu T., Lai Z. C. (2007). Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 26, 1772-1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke M., Hamaratoglu F., Kango-Singh M., Udan R., Chen C. L., Tao C., Zhang X., Halder G. (2006). The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr. Biol. 16, 2090-2100 [DOI] [PubMed] [Google Scholar]
- Willecke M., Hamaratoglu F., Sansores-Garcia L., Tao C., Halder G. (2008). Boundaries of Dachsous Cadherin activity modulate the Hippo signaling pathway to induce cell proliferation. Proc. Natl. Acad. Sci. USA 105, 14897-14902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Huang J., Dong J., Pan D. (2003). hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445-456 [DOI] [PubMed] [Google Scholar]
- Wu S., Liu Y., Zheng Y., Dong J., Pan D. (2008). The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14, 388-398 [DOI] [PubMed] [Google Scholar]
- Xu T., Wang W., Zhang S., Stewart R. A., Yu W. (1995). Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121, 1053-1063 [DOI] [PubMed] [Google Scholar]
- Xu Y., Stamenkovic I., Yu Q. (2010). CD44 attenuates activation of the hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res. 70, 2455-2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta N., Fujii T., Copeland N. G., Gilbert D. J., Jenkins N. A., Nishiguchi H., Endo Y., Toji S., Tanaka H., Nishimune Y., et al. (2000). Structure, expression, and chromosome mapping of LATS2, a mammalian homologue of the Drosophila tumor suppressor gene lats/warts. Genomics 63, 263-270 [DOI] [PubMed] [Google Scholar]
- Yang C. H., Axelrod J. D., Simon M. A. (2002). Regulation of Frizzled by fat-like cadherins during planar polarity signaling in the Drosophila compound eye. Cell 108, 675-688 [DOI] [PubMed] [Google Scholar]
- Yokoyama T., Osada H., Murakami H., Tatematsu Y., Taniguchi T., Kondo Y., Yatabe Y., Hasegawa Y., Shimokata K., Horio Y., et al. (2008). YAP1 is involved in mesothelioma development and negatively regulated by Merlin through phosphorylation. Carcinogenesis 29, 2139-2146 [DOI] [PubMed] [Google Scholar]
- Yu J., Poulton J., Huang Y. C., Deng W. M. (2008). The hippo pathway promotes Notch signaling in regulation of cell differentiation, proliferation, and oocyte polarity. PLoS ONE 3, e1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Zheng Y., Dong J., Klusza S., Deng W. M., Pan D. (2010). Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell 18, 288-299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S. K., Sullivan A. J., Medina R., Ito Y., van Wijnen A. J., Stein J. L., Lian J. B., Stein G. S. (2004). Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 23, 790-799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M., Struhl G. (2010). A feed-forward circuit linking wingless, fat-dachsous signaling, and the warts-hippo pathway to Drosophila wing growth. PLoS Biol. 8, e1000386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler M. P., Perrimon N., Strutt D. I. (1999). The four-jointed gene is required in the Drosophila eye for ommatidial polarity specification. Curr. Biol. 9, 1363-1372 [DOI] [PubMed] [Google Scholar]
- Zeidler M. P., Perrimon N., Strutt D. I. (2000). Multiple roles for four-jointed in planar polarity and limb patterning. Dev. Biol. 228, 181-196 [DOI] [PubMed] [Google Scholar]
- Zeng Q., Hong W. (2008). The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell 13, 188-192 [DOI] [PubMed] [Google Scholar]
- Zhang J., Smolen G. A., Haber D. A. (2008a). Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 68, 2789-2794 [DOI] [PubMed] [Google Scholar]
- Zhang L., Ren F., Zhang Q., Chen Y., Wang B., Jiang J. (2008b). The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 14, 377-387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Liu C. Y., Zha Z. Y., Zhao B., Yao J., Zhao S., Xiong Y., Lei Q. Y., Guan K. L. (2009). TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J. Biol. Chem. 284, 13355-13362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Bai H., David K. K., Dong J., Zheng Y., Cai J., Giovannini M., Liu P., Anders R. A., Pan D. (2010). The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell 19, 27-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747-2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Ye X., Yu J., Li L., Li W., Li S., Lin J. D., Wang C. Y., Chinnaiyan A. M., Lai Z. C., et al. (2008a). TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962-1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Szafranski P., Hall C. A., Goode S. (2008b). Basolateral junctions utilize warts signaling to control epithelial-mesenchymal transition and proliferation crucial for migration and invasion of Drosophila ovarian epithelial cells. Genetics 178, 1947-1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Lei Q., Guan K. L. (2010a). The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24, 862-874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Tumaneng K., Wang C. Y., Guan K. L. (2010b). A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 24, 72-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Conrad C., Xia F., Park J. S., Payer B., Yin Y., Lauwers G. Y., Thasler W., Lee J. T., Avruch J., et al. (2009). Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell 16, 425-438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziosi M., Baena-Lopez L. A., Grifoni D., Froldi F., Pession A., Garoia F., Trotta V., Bellosta P., Cavicchi S. (2010). dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLoS Genet 6, e1001140 [DOI] [PMC free article] [PubMed] [Google Scholar]