Abstract
Human germ cell tumors show a strong sensitivity to genetic background similar to Dnd1Ter/Ter mutant mice, where testicular teratomas arise only on the 129/SvJ genetic background. The introduction of the Bax mutation onto mixed background Dnd1Ter/Ter mutants, where teratomas do not typically develop, resulted in a high incidence of teratomas. However, when Dnd1Ter/Ter; Bax–/– double mutants were backcrossed to C57BL/6J, no tumors arose. Dnd1Ter/Ter germ cells show a strong downregulation of male differentiation genes including Nanos2. In susceptible strains, where teratomas initiate around E15.5-E17.5, many mutant germ cells fail to enter mitotic arrest in G0 and do not downregulate the pluripotency markers NANOG, SOX2 and OCT4. We show that DND1 directly binds a group of transcripts that encode negative regulators of the cell cycle, including p27Kip1 and p21Cip1. P27Kip1 and P21Cip1 protein are both significantly decreased in Dnd1Ter/Ter germ cells on all strain backgrounds tested, strongly suggesting that DND1 regulates mitotic arrest in male germ cells through translational regulation of cell cycle genes. Nonetheless, in C57BL/6J mutants, germ cells arrest prior to M-phase of the cell cycle and downregulate NANOG, SOX2 and OCT4. Consistent with their ability to rescue cell cycle arrest, C57BL/6J germ cells overexpress negative regulators of the cell cycle relative to 129/SvJ. This work suggests that reprogramming of pluripotency in germ cells and prevention of tumor formation requires cell cycle arrest, and that differences in the balance of cell cycle regulators between 129/SvJ and C57BL/6 might underlie differences in tumor susceptibility.
Keywords: Germ cell, Cell cycle, Testicular teratoma, Mouse
INTRODUCTION
Germ cell tumors (GCTs) are the most frequent cause of cancer in men between the ages of 15 and 40 (Hussain et al., 2008). Teratomas represent a class of non-seminomatous GCTs that arise spontaneously in young boys, and are characterized by the differentiation of a diverse array of cell and tissue types within the tumor including cartilage, muscle, hair and glandular tissue, as well as a cluster of stem-like cells from which the tumor can be propagated. The diversity of cell types that appear within teratomas is believed to reflect the latent pluripotency of germ cells (for a review, see Solter, 2006). Although there have been significant recent advances in understanding how pluripotency can be induced (Okita et al., 2008; Yamanaka, 2009), discerning the mechanisms that regulate endogenous populations of highly pluripotent stem cells lies at the heart of both cancer biology as well as efforts to manipulate various adult stem cell populations for regenerative repair.
Histological analyses carried out from the 1950s to the 1980s by Leroy Stevens and others established a foundational knowledge of teratoma formation during fetal development in the highly susceptible 129/SvJ strain and in the presence of a spontaneous mutation that arose in this strain called Ter (Noguchi and Noguchi, 1985; Noguchi and Stevens, 1982; Rivers and Hamilton, 1986; Stevens, 1967; Stevens, 1973; Stevens, 1984; Stevens and Bunker, 1964; Stevens and Hummel, 1957; Stevens and Little, 1954). In 2005, Ter was mapped to a point mutation in the third exon of the RNA-binding protein (RBP) dead end homolog 1 (Dnd1Ter); this nonsense mutation induces a premature stop codon that leads to nonsense-mediated decay and a significant loss of protein (Youngren et al., 2005).
Dnd1 was first characterized in zebrafish germ cells, where loss-of-function studies demonstrated that all primordial germ cells (PGCs) were lost owing to defects in migration (Weidinger et al., 2003). Dnd1 is also expressed in mouse germ cells (Cook et al., 2009; Youngren et al., 2005). In Dnd1Ter/Ter mouse embryos, mutant PGCs do not display defects in migration but do show a significant decrease in population size during migration and gonad colonization, due in part to active cell death (Cook et al., 2009; Noguchi and Noguchi, 1985; Sakurai et al., 1995). The few PGCs that reach the testis are lost by birth on most genetic backgrounds, but mutant germ cells are thought to give rise to teratomas in the susceptible 129/SvJ strain.
In mice, specification of PGCs during early embryogenesis is associated with the upregulation of markers of pluripotency such as Oct4 (Pou5f1; POU domain, class 5, transcription factor 1), Nanog (Nanog homeobox) and Sox2 (SRY-box containing gene 2), and repression of markers of differentiation such as homeobox genes (Saitou et al., 2002; Saitou et al., 2003; Saitou et al., 2005). Paracrine signals from the immediate environment, cell-autonomous epigenetic and transcriptional programming, and post-transcriptional control through the action of RBPs are all involved in regulating the underlying pluripotency of germ cells (Ohinata et al., 2009; Seydoux and Braun, 2006). During migration to the gonad, PGCs can be explanted and cultured in vitro to form pluripotent embryonic germ (EG) cells, which morphologically and functionally resemble embryonic stem (ES) cells (Labosky et al., 1994). Once PGCs colonize the gonad (at E11.5), they undergo a sex-specific reprogramming process that leads to the downregulation of pluripotent markers, and by E12.5 they can no longer be efficiently induced to form EG cells (Anderson et al., 2000; Molyneaux et al., 2001).
Between E12.5 and E15.5, germ cells in the testis begin differentiation as pro-spermatogonia, and enter mitotic arrest in G0 until near birth (Durcova-Hills and Capel, 2008; Matsui, 1998; McLaren, 1984; Western et al., 2008). Cell cycle regulators associated with the initiation of mitotic arrest include p27Kip1(Cdkn1b), p21Cip1(Cdkn1a), p16Ink4a(Cdkn2a) and p15Ink4b(Cdkn2b) (Western et al., 2008). Survival and commitment of germ cells to male fate is dependent on expression of Fgf9 (fibroblast growth factor 9) in somatic cells (DiNapoli et al., 2006), which upregulates Nanos2 (nanos homolog 2) in germ cells (Barrios et al., 2010; Bowles et al., 2010). Null mutations of Nanos2 lead to transient upregulation of meiotic markers (typical of female germ cells during this stage of fetal development) and subsequent germ-cell death, but do not lead to teratomas (Suzuki and Saga, 2008). Recent work has demonstrated that DND1 can bind uridine-rich regions in the 3′ untranslated region (UTR) of target transcripts and protect them from microRNA (miRNA)-mediated translational repression (Kedde et al., 2007). Nanos1 was identified as a target of DND in zebrafish PGCs, and two cell cycle genes, p27Kip1 and Lats2, were identified as targets of DND1 in human tumor cell lines (Kedde and Agami, 2008; Kedde et al., 2007). However, it is not known whether these targets were affected in Dnd1Ter/Ter mutants, what other relevant targets might be, and how this might contribute to teratoma formation.
We demonstrate here that loss of Dnd1 expression in germ cells results in a strong downregulation of male differentiation genes (including Nanos2), ectopic upregulation of meiotic markers, maintenance of pluripotency genes and a failure to enter mitotic arrest at G0. Mouse DND1 binds transcripts of a group of cell cycle genes, including p27Kip1 and p21Cip1. These two proteins are significantly decreased in Dnd1Ter/Ter mutants on all strain backgrounds tested, strongly suggesting that DND1 regulates mitotic arrest in male germ cells through translational regulation of cell cycle genes. Although C57BL/6J mutant germ cells do not arrest in G0, they arrest prior to M-phase, successfully downregulate NANOG and SOX2, and do not form teratomas. Consistent with their ability to arrest cell cycle, negative regulators of cell cycle are overexpressed in E14.5 XY C57BL/6J germ cells relative to those of 129/SvJ. These strain-specific differences in the regulation of cell cycle at mitotic arrest correlate strongly with the ability to downregulate pluripotent markers and with the incidence of teratomas, and might explain the differences in sensitivity to tumorigenesis.
MATERIALS AND METHODS
Mice, timed matings and genotyping
Dnd1Ter/+ and Bax+/– mice were maintained, genotyped and crossed as described previously (Cook et al., 2009). The mixed genetic background is a mixture of C57BL/6J, 129/SvJ and a small contribution from CD-1. For timed matings, females were inspected every morning for the presence of a vaginal plug, and noon on the day a vaginal plug was detected was considered embryonic day (E) 0.5.
Immunofluorescence
Fluorescent immunocytochemistry was performed as previously described in either whole-mount (E11.5-E14.5) or frozen section (E14.5 and later) (Cook et al., 2009). The antibodies and dilutions used were: rat anti-E-cadherin (1:500; Zymed Laboratories, San Francisco, CA, USA; cat. #13-1900), rabbit anti-SOX9 (1:500; Chemicon, Temecula, CA, USA), rabbit anti-MVH (1:500; Abcam, Cambridge, MA, USA; cat. #ab13840), rabbit anti-NANOG (1:300; Cosmo Bio, Tokyo, Japan; cat. #RCAB0002P-F), rabbit anti-SOX2 (1:1000; Chemicon, Temecula, CA, USA; cat. #Ab5603), rabbit anti-OCT4 (1:500; Abcam; cat. #ab19857-100), rabbit anti-P27Kip1 (1:250; Santa Cruz Biotechnology, Santa Cruz, CA, USA; cat. #sc-529), goat anti-P21Cip1 (1:50; Santa Cruz Biotechnology; cat. #sc-397-G), rabbit anti-Ki67 (1:500; Neomarkers, Thermo Scientific, Waltham, MA, USA; cat. #RM-9106-S), rabbit anti-Gm114 [1:500; generated in our lab (Tang et al., 2008)], rabbit anti-SCP3 (1:500; Novus, Littleton, CO, USA; cat. #300-231) and rabbit anti-STRA8 (1:500; kindly provided by Pierre Chambon, Institute for Genetics and Cellular and Molecular Biology, Strasbourg, France).
qRT-PCR
Quantitative RT-PCR (qRT-PCR) was used to determine relative expression levels of transcripts in XY gonads of different genotypes and strain backgrounds. Gonad dissection, RNA extraction and cDNA synthesis have all been described previously (Cook et al., 2009). For qPCR, each analysis was performed in triplicate in a total volume of 20 μl reaction mix containing 1.25 μl cDNA template, 10 μl 2× Quantace SensiMixPlus SYBR (Bioline, Boston, MA, USA; cat# QT615-02), 4.75 μl RNase-free water and 4 μl 1 μM gene-specific forward and reverse primers (200 nM final concentration each). qPCR was performed on an iCycler Thermal Cycler (Bio-Rad, Hercules, CA, USA; cat# 170-8720). Cycling conditions for all primers were as follows: 95°C for 3 minutes (one cycle); 95°C for 15 seconds, 62°C for 30 seconds, 72°C for 30 seconds (40 cycles); and 72°C for 5 minutes (one cycle). Specific sequences for all primers can be found in Table 1. Primer sets were tested for efficiency and found to work optimally with the delta-Ct method. Target gene Ct thresholds were determined and normalized to either Hprt or Oct4.
Table 1.
qPCR primers for each gene tested
Microarray analysis
Total RNA was extracted from Oct4-EGFP-positive sorted germ cell populations (C57BL/6J, n=3; 129/SvJ, n=2) using the RNeasy Micro Kit (Qiagen, Valencia, CA, USA; #74004) with on-column DNase digestion according to manufacturer's protocols. Total RNA was reverse-transcribed to cDNA and then transcribed in vitro into biotin-labeled cRNA using the Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX, USA; #AMIL1791) according to manufacturer's protocols. Biotin-labelled cRNA (750 ng) was hybridized to MouseRef-8 v2.0 Expression BeadChips (Illumina, Hayward, CA, USA; BD-202-0202) according to Illumina protocols. After washing steps, BeadChips were scanned on the iScan BeadArray Reader (Illumina). Raw probe-level data was imported into GenomeStudio (Illumina) software, the average was normalized and the background signal level was subtracted out. Genes that were not detected in at least one sample (detection P-value cutoff of 0.05) were removed from the analysis. The Illumina custom error model with false discovery rate (FDR) correction was applied to detect genes that were differentially expressed between C57BL/6J and 129/SvJ Oct4-EGFP-positive germ cells. Expression differences were considered significant if the calculated Diffscore was ± 13 (equivalent to P<0.05). Data are available in the GEO database (accession #GSE24340) at http://www.ncbi.nlm.nih.gov/geo.
Cell culture, western blot analysis and RNA immunoprecipitation
Although two isoforms of the DND1 protein are known, only DND1α is expressed in the fetal gonad (Bhattacharya et al., 2007) and is labeled as DND1 in these assays. Mouse 3T3 cells were transfected with vectors encoding GFP, GFP fused at the N-terminus of DND1, TAP, or TAP fused at the N-terminus of DND1. For GFP imaging, cells were fixed in 4% PFA, washed in PBS, stained for 2 hours with Propidium Iodide (PI), washed in PBS and mounted in DABCO for imaging on a Zeiss 510 Meta confocal microscope. For RNA immunoprecipitation (RIP) and western blot analysis, 3T3 cells were lysed 24 hours after transfection in appropriate lysis buffer. For western blot analysis, extracts were separated on a 4-20% gradient SDS-PAGE gel and transferred to nitrocellulose. Rabbit anti-TAP (WB 1:10,000; Open Biosystems, Huntsville, AL, USA) was used as a primary antibody and a horseradish peroxidase-conjugated anti-rabbit antibody was used as a secondary antibody at 1:5000. Western blots were developed with ECL (Amersham GE, Piscataway, NJ, USA) and exposed to film (Kodak). RIP was performed as published previously (Keene et al., 2006). Briefly, lysed cells were immediately frozen at –80°C to prevent adventitious binding. IgG Sepharose beads (Amersham) were washed three times in wash buffer and 100 μl lysate was added to 850 μl NT2 buffer, 2 μl RNaseOUT (Invitrogen, Carlsbad, CA, USA), 2 μl of 200 mM VRCs (Sigma, St Louis, MO, USA), 10 μl 1 M DTT, and 40 μl 0.5 M EDTA. After 2 hours of incubation at room temperature, the beads were pelleted and washed six times with wash buffer. TRIzol was added directly to the beads to perform an RNA extraction followed by cDNA synthesis and qRT-PCR as described above. Target genes were examined by qRT-PCR, normalized to Hprt1 and compared with cells transfected with the TAP tag alone (Gapdh represents a negative control that is not enriched).
RESULTS
Markers of pluripotency are maintained in nascent teratomas
Germ cell tumors in the human population show a strong sensitivity to genetic background that is mirrored by the Dnd1Ter/Ter mutants, where testicular teratomas arise only on the 129/SvJ genetic background. Little is known about the etiology of the teratomas that develop in 129/SvJ Dnd1Ter/Ter testes or about the cell biology of Dnd1Ter/Ter germ cells in strains susceptible to tumors versus strains that are not. Previously, it was shown that clusters of cells that are histologically distinct from wild-type prospermatogonia appear within E15.5-E17.5 cords and are the first signs of tumorigenesis (Rivers and Hamilton, 1986; Stevens, 1967; Stevens, 1973). As these clusters are believed to arise from germ cells (Solter, 2006; Stevens, 1967), we performed immunocytochemistry on samples during this transitional stage to compare mutant germ cells in a strain where tumors arise (129/SvJ) with those in a genetic background where they do not (C57BL/6J) (Noguchi et al., 1996; Stevens, 1981).
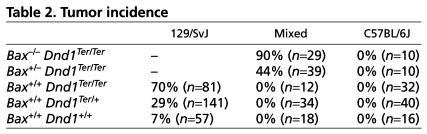
A major difficulty of this analysis is that the number of germ cells that reach the testis is significantly reduced in Dnd1Ter/Ter mutants and that the few remaining germ cells disappear around the transitional stage in C57BL/6J and other strains that are not susceptible to tumor formation. To circumvent this problem, we introduced a mutant allele of Bax onto Dnd1Ter/Ter mutants. This generated a mixed genetic background (primarily consisting of 129/SvJ and C57BL/6J), which we maintained through intercrosses at the same time we began backcrossing to transfer the mutation onto a pure C57BL/6J background. As reported previously, the introduction of the Bax mutation onto mixed-background Dnd1Ter/Ter mutants resulted in a high incidence of teratomas (90% in Dnd1Ter/Ter; Bax–/– and 44% in Dnd1Ter/Ter; Bax+/–) (Cook et al., 2009). However, when Dnd1Ter/Ter; Bax–/– double mutants were backcrossed ten generations to C57BL/6J, no tumors arose, despite the significant rescue of germ cells in the gonad (Table 2).
Table 2.
Tumor incidence
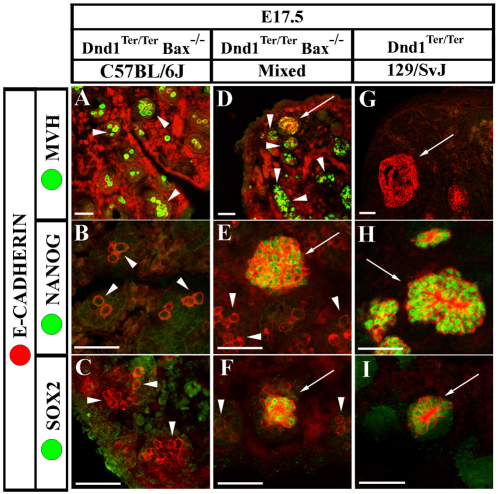
We compared a group of markers of germ cell development among the three genetic backgrounds (C57BL/6J, mixed and 129/SvJ) at E17.5. Mouse Vasa homolog (MVH) (Fujiwara et al., 1994; Toyooka et al., 2000) and E-cadherin (ECAD) (Di Carlo and De Felici, 2000; DiNapoli et al., 2006) both label male germ cells beginning at the time they enter the gonad (see Fig. 1A), whereas NANOG and SOX2 expression decline by the time germ cells enter mitotic arrest at E15.5 (Durcova-Hills and Capel, 2008). In E17.5 C57BL/6J Dnd1Ter/Ter; Bax–/– mutants, individual germ cells continued to express MVH and ECAD and downregulated NANOG and SOX2 (Fig. 1A-C, arrowheads). On a mixed genetic background, many germ cells followed this pattern at E17.5 (Fig. 1D-F, arrowheads); however, clusters of cells strongly positive for ECAD were detected and cells within these clusters strongly expressed NANOG and SOX2 (Fig. 1D-F, arrows). In 129/SvJ mutants, no individual MVH or ECAD-positive germ cells were detected. Instead, multiple clusters of strongly ECAD-positive cells were seen in each gonad and virtually all cells in each cluster were MVH-negative and NANOG- and SOX2-positive (Fig. 1G-I, arrows). Further information regarding the origin of ECAD-positive clusters within testis cords of Dnd1Ter/Ter 129/SvJ testes can be found in the supplementary material (see Figs S1, S2 in the supplementary material).
Fig. 1.
Early neoplasias express high levels of ECAD, NANOG and SOX2. Arrowheads indicate morphologically wild-type germ cells and arrows indicate neoplasias. E-cadherin (ECAD; red) labels both germ cells and neoplasias in all frames. The frequency of neoplasia formation is strain-dependent. (A-C) Neoplasias do not form in double mutants on a C57BL/6J background. Germ cells express MVH (A) and lack expression of the pluripotency genes NANOG (B) and SOX2 (C) (arrowheads). (D-F) On a mixed background, ECAD-positive, morphologically distinct neoplasias form in double mutants (arrows). These clusters do not express MVH (D, arrow), although many germ cells persist that express MVH normally (arrowheads) and lack NANOG (E, arrowheads) and SOX2 (F, arrowheads). Cells within neoplasias maintain expression of NANOG (E, arrow) and SOX2 (F, arrow). (G-I) Many neoplasias form on a 129/SvJ background that lack MVH (G) and ectopically maintain expression of NANOG (H) and SOX2 (I). No MVH-positive, morphologically normal germ cells are detected at this stage in 129/SvJ mutants. Scale bars: 50 μm.
DND1 regulates expression of NANOS2 and other germ cell differentiation genes
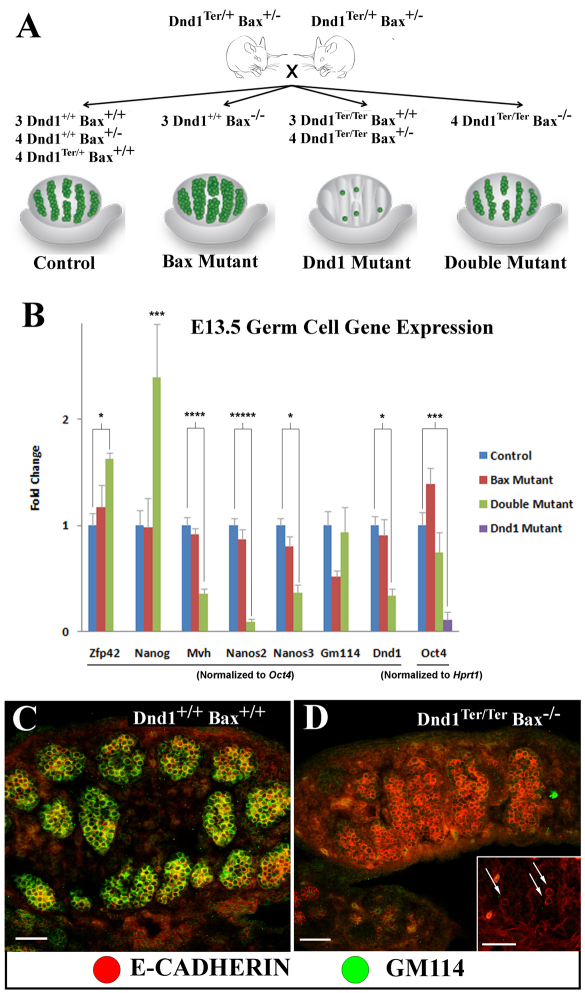
To determine how markers of fetal male germ cell differentiation were affected in Dnd1Ter/Ter mutants, we took advantage of the mixed background double mutants with rescued germ cells to quantify the expression level of eight genes specific to germ cells in the gonad, comparing single and double mutants to controls. We isolated total RNA from whole gonads dissected at E13.5 from Dnd1Ter/+; Bax+/– intercrosses and quantified the expression levels of germ cell-specific genes normalized to a germ cell-specific transcript by quantitative RT-PCR (Fig. 2A). To validate this approach we included two experimental controls. First, Oct4 (a gene specific to germ cells at this stage) was normalized to a ubiquitously expressed gene, Hprt1. Oct4 expression was reduced (P<0.005) in Dnd1Ter/Ter gonads, consistent with the presence of many fewer germ cells, whereas there was a slight increase in Oct4 expression in Dnd1+/+; Bax–/– gonads, reflecting the presence of more germ cells compared with wild-type and double-mutant testes (Fig. 2B) (see Cook et al., 2009). Second, expression of germ cell-specific Dnd1 was analyzed in wild-type, Bax mutant and double-mutant gonads. Dnd1Ter/Ter single mutants were omitted from this and following analyses because the persistence of so few germ cells led to highly variable and inconclusive results. Dnd1 expression was downregulated almost threefold (P<0.001) in germ cells from double-mutant gonads (Fig. 2B, double mutant, green bar) compared with controls (Fig. 2B, control, blue bar), as expected from nonsense-mediated decay caused by the mutation (Youngren et al., 2005). Based on these validations, we concluded that normalization of germ cell-specific genes to Oct4 expression accounted for differences in germ cell numbers in the mutants and provided a more reliable normalization than a general housekeeping gene like Hprt1.
Fig. 2.
Germ cell markers change at the transcriptional and post-transcriptional levels in E13.5 mutant germ cells. (A) Schematic representation of germ cell numbers in tissue collected for qRT-PCR analysis. The numeral indicates the number of pairs of gonads for each genotype collected. (B) Oct4 expression levels are consistent with germ cell number estimates by immunofluorescence as previously published [see cartoon in A and Cook et al. (Cook et al., 2009)]. Double mutants express lower levels of Dnd1, show transcriptional upregulation of pluripotent markers (Zfp42 and Nanog) and a decrease in differentiation markers (Mvh, Nanos3 and Nanos2) compared with controls. Although there is no significant difference in Gm114 transcript levels in double mutants (B), antibodies against GM114 (green), which label wild-type germ cells (ECAD, red) at E13.5 (C), indicate that double-mutant germ cells fail to express GM114 protein (D) on a mixed background or in Dnd1Ter/Ter mutants on a 129/SvJ background (inset, arrows). Scale bars: 50 μm. In B, each bar represents the standard error of the mean (s.e.m.) of at least three biological replicates. *P<0.05; ***P<0.005; ****P<0.001; *****P<0.0005; ******P<0.0001.
Nanog was upregulated approximately 2.5-fold (P<0.005) in double mutants (Fig. 2B). By contrast, Mvh expression was downregulated more than 2.5-fold (P<0.001) in Dnd1Ter/Ter; Bax–/– germ cells. Nanos3 expression was halved (P<0.0005), whereas Nanos2 expression was downregulated more than 10-fold (P<0.0005) in double mutants. Another germ cell-specific gene, Gm114 (Plk1s1, polo-like kinase 1 substrate 1) (Tang et al., 2008), showed no significant difference in transcript expression level in double mutants (Fig. 2B); however, GM114 protein was not detected in Dnd1Ter/Ter; Bax–/– mixed-background germ cells at this stage (Fig. 2C,D) or in Dnd1Ter/Ter 129/SvJ germ cells (Fig. 2D, inset), indicating potential translational regulation of this gene.
Mutant XY germ cells ectopically activate meiotic markers
Unlike XX germ cells, wild-type XY germ cells do not enter meiosis until after birth. This is dependent on the somatic expression of Cyp26b1 from E11.5-E13.5, which degrades the meiosis-inducing factor retinoic acid (RA) in the testis (Bowles et al., 2006) and germ cell-specific expression of Nanos2 from E13.5 until after birth (Suzuki and Saga, 2008). Loss of either Cyp26b1 or Nanos2 leads to precocious upregulation of the meiotic markers STRA8 (stimulated by retinoic acid gene 8) and SCP3 (synaptonemal complex protein 3) in mutant male germ cells. Levels of Cyp26b1 and other essential somatic genes such as Sox9, Fgf9 and Dhh were normal in mutant testes (see Fig. S3 in the supplementary material). However, the significant decrease in Nanos2 expression (Fig. 2B) was correlated with the upregulation of meiotic markers in mutant germ cells (Fig. 3).
Fig. 3.
Mutant germ cells ectopically express meiotic markers SCP3 and STRA8. (A-F) ECAD and OCT4-EGFP (red) both label germ cells. (A) E16.5 129/SvJ Dnd1Ter/Ter germ cells that are morphologically wild-type express SCP3 (green), but ECAD-positive neoplasias (arrowhead) do not (B). At E17.5, in double mutants on a mixed genetic background, many, but not all, germ cells express SCP3 (C) and STRA8 (D), although they are never expressed in neoplasias (arrowheads). E17.5 C57BL/6J double-mutant germ cells also express SCP3 (E) and STRA8 (F). Scale bars: 50 μm.
Similar to Nanos2 mouse mutants, Dnd1 mutants showed upregulation of SCP3 in male germ cells. On the 129/SvJ background, a few morphologically normal cells still present at E16.5 expressed SCP3, but SCP3 was not detected in the ECAD-positive clusters (Fig. 3A,B). A similar pattern was detected at a slightly later stage in mutants on a mixed background. At E17.5, SCP3 and STRA8 were both expressed in mutant germ cells but never in the strongly ECAD-positive clusters from which teratomas originate (Fig. 3C,D). SCP3 and STRA8 were also both detected in many individual germ cells in C57BL/6J mutants (Fig. 3E,F), indicating that loss of Dnd1 leads to the aberrant activation of meiotic markers in some XY germ cells, regardless of genetic background.
DND1 regulates germ cell mitotic arrest in G0
Previous experiments conducted in a human cell line showed that human DND1 binds transcripts for two cell cycle inhibitors, p27Kip1 and Lats2, and protects them from miRNA-mediated translational repression (Kedde et al., 2007). To determine whether mouse DND1 binds these targets or others involved in male germ cell mitotic arrest, we performed RNA immunoprecipitation (RIP) (Keene et al., 2006). We overexpressed both TAP- and GFP-tagged DND1 in mouse NIH-3T3 cells, a cell line previously used to examine the function of DND1 (Bhattacharya et al., 2008). We verified that full-length TAP-DND1 protein was produced by western analysis (Fig. 4A) and found that GFP-DND1 localized to perinuclear granules (Fig. 4B), as reported previously in both NIH-3T3 cells and in zebrafish germ cells (Bhattacharya et al., 2007; Slanchev et al., 2008).
Fig. 4.
DND1 binds a set of functionally related targets that are cell cycle regulators. (A) TAP-DND1 migrates at the predicted size of about 50 kDa (right lane) in a western blot of transfected 3T3 cells. (B) Compared with GFP alone (top panel), GFP-DND1 (green, lower panel) localizes to perinuclear granules in 3T3 cells. Scale bars: 20 μm. (C) qRT-PCR results from RNA immunoprecipitation (RIP) experiments using TAP-DND1 in 3T3 cells compared with the TAP tag alone and normalized to Hprt1. Bars represents fold-enrichment (the s.e.m. of at least three replicates) for each gene compared with the control. *P<0.05. n=the number of independent RIP replicates.
We performed RIP from 3T3 cells followed by reverse transcription (RT) and quantitative RT-PCR (qRT-PCR). TAP-DND1 was enriched for both p27Kip1 and Lats2 compared with the TAP tag alone (Fig. 4C). We also examined other candidate cell cycle regulator genes that are expressed in both 3T3 cells and E14.5 germ cells undergoing mitotic arrest (Kimura et al., 2003; Western et al., 2008). A group of cell cycle inhibitors including p21Cip1, p27Kip1, Lats2, pRB, p53 and Pten (Fig. 4C, left bracket) showed significant enrichment in this assay, suggesting that they might be targets of DND1 for translational regulation.
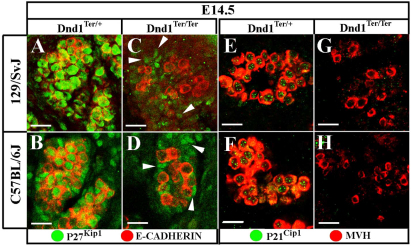
Consistent with a previous report (Western et al., 2008), P27Kip1 and P21Cip1 were detected in both 129/SvJ and C57BL/6J germ cells at E14.5 as they transitioned into a G1 or G0 arrest (Fig. 5). To determine whether these potential targets of DND1 regulation are expressed in mutant germ cells, we investigated expression of P27Kip1 and P21Cip1 protein in Dnd1Ter/Ter mutants on both 129/SvJ (tumor-susceptible) and C57BL/6J (tumor non-susceptible) genetic backgrounds. Neither P27Kip1 (Fig. 5A-D) nor P21Cip1 (Fig. 5E-H) were detected in mutant germ cells at E14.5, regardless of strain background, although P27Kip1 protein was not disrupted in somatic cells in the gonad (Fig. 5C,D, arrowheads).
Fig. 5.
At E14.5, Dnd1Ter/Ter germ cells fail to express putative targets P27Kip1 and P21Cip1. ECAD and MVH (red) both mark germ cells. (A-D) P27Kip1 (green) is robustly expressed in both 129/SvJ and C57BL/6J control germ cells (A,B), but is absent in mutant germ cells of both backgrounds (C,D). (E-H) P21Cip1 (green) is also expressed in control germ cells of both backgrounds (E,F), but not in mutants (G,H). P21Cip1 is specific to germ cells; however, note that somatic cells (C,D, arrowheads) express P27Kip1 normally in mutants. Scale bars: 20 μm.
Mutant germ cells on the teratoma-resistant C57BL/6J strain do not arrest in G0 but fail to undergo mitosis
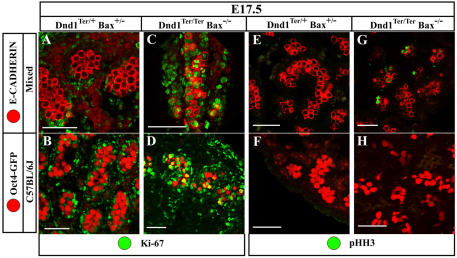
To determine whether mutant germ cells had entered mitotic arrest in G0, we investigated expression of the cell cycle marker Ki67, which labels all stages of the cell cycle except G0. As expected, Ki67 was absent from male germ cells at E17.5 in control Dnd1Ter/+ samples, regardless of the Bax genotype (Fig. 6A,B). However, consistent with the loss of cell-cycle inhibitors in Dnd1 homozygous mutants, Ki67 was still expressed in E17.5 male germ cells on both mixed and C57BL/6J genetic backgrounds (Fig. 6C,D), indicating that male germ cells had not entered G0 mitotic arrest. Surprisingly, this was true for all genetic backgrounds tested, even though C57BL/6J mutant germ cells do not develop ECAD neoplasias positive for NANOG and SOX2 (Fig. 1A-C) and teratomas do not form (Table 2).
Fig. 6.
Double-mutant C57BL/6J germ cells undergo mitotic arrest prior to M-phase. E17.5 double-mutant gonads from mixed and C57BL/6J backgrounds are compared with each other and with controls. (A,B) Control germ cells (red – note the different germ cell marker between mixed and C57BL/6J) are negative for Ki67 (green) because they have exited the cell cycle and are arrested in G0. (C,D) Double-mutant germ cells (red) on both mixed and C57BL/6J backgrounds remain in cycle and are Ki67-positive (green). (E,F) Control germ cells (red) are negative for pHH3 (green) because they have exited the cell cycle and are not undergoing active mitoses. (G,H) Double-mutant germ cells (red) on a mixed background (G) remain pHH3-positive, whereas double-mutant germ cells on a C57BL/6J background (H) are negative. This indicates that mixed-background, double-mutant germ cells continue to divide, whereas those on a C57BL/6J background successfully arrest prior to M-phase. ECAD is a membrane marker and labels germ cells in the mixed background. Oct4-EGFP is both nuclear and cytoplasmic and labels germ cells in the C57BL/6J background. Scale bars: 50 μm.
Germ cells on the C57BL/6J background might compensate for the failure to arrest at G0 and achieve mitotic arrest at another stage of the cell cycle. To test whether Dnd1Ter/Ter; Bax–/– germ cells were arrested at another point in the cell cycle in C57BL/6J mutants, testes were stained with a marker for active mitosis, phospho-histone H3 (pHH3). Strikingly, many double-mutant germ cells on a mixed background (where tumors develop) were positive for pHH3, whereas no mutant germ cells on a C57BL/6J background (which do not develop tumors) stained positive (Fig. 6E-H). This finding suggests that C57BL/6J germ cells can compensate for the loss of P27Kip1 and P21Cip1 and arrest before M-phase of the cell cycle.
We predicted that strain-specific differences in the transcriptome of C57BL/6J male germ cells might account for their ability to counteract the loss of negative regulators of the cell cycle controlled by DND1. To test this hypothesis, we crossed the Oct4-EGFP transgene separately to C57BL/6J and 129/SvJ (n=10 generations). We then isolated wild-type germ cells from each strain at E14.5 by FACS, quantified global transcript abundance in germ cell total RNA by microarray and compared strain-specific germ cell expression profiles. Genes differentially expressed between strains were organized into gene sets and functionally annotated (DAVID database, http://david.abcc.ncifcrf.gov/) (Dennis et al., 2003; Huang da et al., 2009). Genes upregulated within each strain were independently subjected to gene set enrichment analysis (GSEA) in DAVID. Interestingly, BioCarta and KEGG pathway analyses revealed a significant enrichment for genes involved in cell cycle and tumor-suppressor signaling pathways in C57BL/6J germ cells at E14.5 (Table 3). In fact, a number of well-characterized cell cycle inhibitor and death proteins were significantly upregulated (P<0.05) in C57BL/6J germ cells relative to 129/SvJ, including p15Ink4b (4.89-fold), Puma (Bbc3; 2.98-fold), Atm (1.83-fold), p53 (Trp53; 1.82-fold), Bax (1.7-fold), Bub3 (1.57-fold), Bub1b (1.44-fold) and 14-3-3 eta (YWHAH, 1.39-fold) (see Table S1 in the supplementary material). Additionally, expression of cyclin D1 (Ccnd1) was 3.43-fold downregulated in C57BL/6J germ cells. These expression differences provide further evidence that the transcription network regulating the cell cycle in tumor-resistant C57BL/6J germ cells is altered relative to tumor-susceptible 129/SvJ germ cells.
Table 3.
E14.5 XY germ cell gene expression differences
DISCUSSION
We have shown here by RNA immunoprecipitation that mouse DND1 binds a group of transcripts that are negative regulators of the cell cycle including p27Kip1, p21Cip1, p53, pRb and Pten. Both p27Kip1 and p21Cip1 are strongly implicated in male germ cell mitotic arrest (Western et al., 2008), and conditional deletion of Pten in male germ cells also leads to testicular teratoma formation (Kimura et al., 2003). Surprisingly, these cell cycle inhibitor proteins are upregulated in mitotically arresting germ cells even though the miRNAs that block their translation are also present (Hayashi et al., 2008). Our data provides evidential support for a model suggested previously (Western, 2009), where DND1 acts to promote translation of cell cycle inhibitors, most probably protecting them from miRNA-mediated translational repression. The finding that DND1 binds a group of cell cycle targets is consistent with the RNA regulon model, where a single RBP controls the translation of a functionally related set of target transcripts (Keene, 2007).
Although there is no ortholog of Dnd1 in Drosophila or C. elegans, other RBPs act as translational regulators of the cell cycle in germ cells. Nanos and pumilio function to repress translation of maternally deposited Cyclin B (CycB) in Drosophila PGCs (Kadyrova et al., 2007). In addition, GLD-1 translationally represses Cyclin E (cye-1) in C. elegans germ cells, thus preventing premature mitotic division and embryonic gene activation, ultimately leading to teratoma formation (Biedermann et al., 2009). The significant decrease in Nanos2 expression in mouse Dnd1 mutants indicates that Nanos2 is a direct or indirect target of DND1. In zebrafish, DND binds to a uridine-rich region in the 3′UTR of nanos and protects it from miR-430 inhibition during PGC development (Kedde et al., 2007). Consistent with conservation of this mechanism in mammals, the 3′UTR of Nanos2 is essential for upregulation of protein in mouse fetal male germ cells (Tsuda et al., 2006). In Dnd1 mutants, it is still not clear whether upregulation of meiotic markers, which might result from loss of Nanos2, is an antecedent to cell death or possibly to tumor formation. However, expression of STRA8 and SCP3 was never detected within cells of developing teratomas, arguing against the second possibility. Further experiments are necessary to determine whether mouse DND1 directly regulates Nanos2 transcript, or if NANOS2 protein interacts with DND1 protein in mammalian germ cells.
Gm114, a putative mammalian ortholog of another Drosophila translational regulator, bag of marbles (bam), might also be a direct or indirect target of DND1. GM114 protein was absent at all stages of fetal development in Dnd1Ter/Ter germ cells even though transcript levels were not significantly affected at E13.5 (Fig. 2B-D). Although GM114 is one of the earliest markers of pro-spermatogonial differentiation, no functional role for this protein is known (Tang et al., 2008). Recently it was reported that Drosophila Bam negatively regulates the translation of E-cadherin and is inhibited by the translation initiation factor eIF4A (Shen et al., 2009). In mice, deficiencies in the translation initiation factor eIF2S2 suppress testicular germ cell tumor development (Heaney et al., 2009). Future studies will determine whether GM114 also regulates E-cadherin and interacts with eIF2S2 in mice and whether mouse DND1 directly regulates Gm114.
The effect of Dnd1 loss on germ cell cycle is strongly influenced by genetic background. In 129/SvJ mutants, loss of Dnd1 causes all germ cells to fail to arrest active divisions. In mixed-background mutants, this occurs in some, but not all, germ cells. However, in C57BL/6J mutants, although large numbers of germ cells can be rescued by a mutation in Bax, and the markers of G1/G0 arrest, P27Kip1 and P21Cip1, are not expressed, germ cells successfully arrest their cell cycle prior to M-phase. Based on the comparison between three genetic backgrounds, we suggest that mitotic arrest is a threshold effect influenced by the levels of multiple factors that vary between strains, including positive and negative regulators of the cell cycle and possibly miRNAs. The variability that we see among genetic backgrounds reflects the etiology of the human disease, where the incidence of germ cell tumors is correlated with ethnicity and other complex genetic factors that have not been completely explained (Brown et al., 1987; Brown et al., 1986; Hussain et al., 2008; Linger et al., 2007; Looijenga, 2009; Oosterhuis and Looijenga, 2005).
Post-transcriptional regulation of the cell cycle is emerging as a central theme in the control of both germ cell and ES cell pluripotency. A rapid G1 to S transition is required to maintain pluripotent ES cells and is dependent on the suppression of p27Kip1, p21Cip1, Lats2 and other cell cycle inhibitors by miRNAs (Wang and Blelloch, 2009). In this study, cell cycle arrest prior to M-phase in C57BL/6J germ cells is associated with the downregulation of the pluripotent markers, NANOG and SOX2. We speculate that the transition from a migratory PGC program (expressing high levels of pluripotent markers) to differentiation as pro-spermatogonia is dependent on cell cycle arrest. Our data supports prior work in ES cells (Singh and Dalton, 2009; Wang and Blelloch, 2009) showing that cell-cycle arrest and reprogramming of pluripotent markers are mechanistically linked.
Our data also suggest that an enhanced checkpoint control in C57BL/6J germ cells might underlie their ability to arrest or activate cell death pathway(s) prior to division. However, strain-specific contributions to the cell cycle and reprogramming are complex. First, the genetic modifier(s) of the cell cycle in C57BL/6J that causes mutant germ cells to arrest prior to M-phase might not map to a single causative locus, but could reflect multiple differences that affect a threshold for cell cycle progression. Second, we have observed significant differences in the global transcriptional networks of C57BL/6J and 129/SvJ E11.5 gonads that could be maintained at later stages (Munger et al., 2009). These strain-specific differences might include signaling pathways that predispose the 129/SvJ background to teratoma formation. For example, the ability to derive ES cells varies between strains, with 129/SvJ being the easiest and C57BL/6J requiring a reduction in Erk activation (Batlle-Morera et al., 2008). A previous report showing that Dmrt1 mutants also develop testicular teratomas (independent of Dnd1) revealed that Eras (ES-expressing Ras) is upregulated sevenfold in 129/SvJ mutant germ cells compared with C57BL/6J mutant germ cells or wild-type cells of both strains (Krentz et al., 2009). Future studies examining both somatic and germ cell-specific transcriptional differences between mutant strains at E14.5 could reveal additional signaling components that influence tumorigenesis. We predict that identification of the global set of targets for DND1 at all stages of germ cell development will reveal novel regulatory functions for this RNA-binding protein and expose links between programming of the pluripotent genome, cell cycle and tumor biology.
Supplementary Material
Acknowledgments
We thank David Zarkower and Tony Krentz for their valuable comments and review of the manuscript, Fan Wang for providing the Bax mutant mice and a critical reading of the manuscript, Pierre Chambon for the STRA8 antibody and Heidi Munger for running the microarrays. This work was supported by grants from the Lance Armstrong Foundation, the Stewart Trust Foundation (DUMC-Cancer Center), the NIH (HD39963) to B.C., and the NCI (CA75056) to J.H.N. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.057000/-/DC1
References
- Anderson R., Copeland T. K., Scholer H., Heasman J., Wylie C. (2000). The onset of germ cell migration in the mouse embryo. Mech. Dev. 91, 61 [DOI] [PubMed] [Google Scholar]
- Barrios F., Filipponi D., Pellegrini M., Paronetto M. P., Di Siena S., Geremia R., Rossi P., De Felici M., Jannini E. A., Dolci S. (2010). Opposing effects of retinoic acid and FGF9 on Nanos2 expression and meiotic entry of mouse germ cells. J. Cell Sci. 123, 871-880 [DOI] [PubMed] [Google Scholar]
- Batlle-Morera L., Smith A., Nichols J. (2008). Parameters influencing derivation of embryonic stem cells from murine embryos. Genesis 46, 758-767 [DOI] [PubMed] [Google Scholar]
- Bhattacharya C., Aggarwal S., Zhu R., Kumar M., Zhao M., Meistrich M. L., Matin A. (2007). The mouse dead-end gene isoform alpha is necessary for germ cell and embryonic viability. Biochem. Biophys. Res. Commun. 355, 194-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya C., Aggarwal S., Kumar M., Ali A., Matin A. (2008). Mouse apolipoprotein B editing complex 3 (APOBEC3) is expressed in germ cells and interacts with dead-end (DND1). PLoS ONE 3, e2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann B., Wright J., Senften M., Kalchhauser I., Sarathy G., Lee M. H., Ciosk R. (2009). Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev. Cell 17, 355-364 [DOI] [PubMed] [Google Scholar]
- Bowles J., Knight D., Smith C., Wilhelm D., Richman J., Mamiya S., Yashiro K., Chawengsaksophak K., Wilson M. J., Rossant J., et al. (2006). Retinoid signaling determines germ cell fate in mice. Science 312, 596-600 [DOI] [PubMed] [Google Scholar]
- Bowles J., Feng C. W., Spiller C., Davidson T. L., Jackson A., Koopman P. (2010). FGF9 suppresses meiosis and promotes male germ cell fate in mice. Dev. Cell 19, 440-449 [DOI] [PubMed] [Google Scholar]
- Brown L. M., Pottern L. M., Hoover R. N., Devesa S. S., Aselton P., Flannery J. T. (1986). Testicular cancer in the united-states-trends in incidence and mortality. Int. J. Epidemiol. 15, 164-170 [DOI] [PubMed] [Google Scholar]
- Brown L. M., Pottern L. M., Hoover R. N. (1987). Testicular cancer in young men-the search for causes of the epidemic increase in the United-States. J. Epidemiol. Commun. Health 41, 349-354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook M. S., Coveney D., Batchvarov I., Nadeau J. H., Capel B. (2009). BAX-mediated cell death affects early germ cell loss and incidence of testicular teratomas in Dnd1Ter/Ter mice. Dev. Biol. 328, 377-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G., Jr, Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003). DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3 [PubMed] [Google Scholar]
- Di Carlo A., De Felici M. (2000). A role for E-cadherin in mouse primordial germ cell development. Dev. Biol. 226, 209 [DOI] [PubMed] [Google Scholar]
- DiNapoli L., Batchvarov J., Capel B. (2006). FGF9 promotes survival of germ cells in the fetal testis. Development 133, 1519-1527 [DOI] [PubMed] [Google Scholar]
- Durcova-Hills G., Capel B. (2008). Chapter 6 development of germ cells in the mouse. Curr. Top. Dev. Biol. 83, 185-212 [DOI] [PubMed] [Google Scholar]
- Fujiwara Y., Komiya T., Kawabata H., Sato M., Fujimoto H., Furusawa M., Noce T. (1994). Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proc. Natl. Acad. Sci. USA 91, 12258-12262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Chuva de Sousa Lopes S. M., Kaneda M., Tang F., Hajkova P., Lao K., O'Carroll D., Das P. P., Tarakhovsky A., Miska E. A., et al. (2008). MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE 3, e1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney J. D., Michelson M. V., Youngren K. K., Lam M. Y., Nadeau J. H. (2009). Deletion of eIF2beta suppresses testicular cancer incidence and causes recessive lethality in agouti-yellow mice. Hum. Mol. Genet. 18, 1395-1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57 [DOI] [PubMed] [Google Scholar]
- Hussain S. A., Ma Y. T., Palmer D. H., Hutton P., Cullen M. H. (2008). Biology of testicular germ cell tumors. Expert Rev. Anticancer Ther. 8, 1659-1673 [DOI] [PubMed] [Google Scholar]
- Kadyrova L. Y., Habara Y., Lee T. H., Wharton R. P. (2007). Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 134, 1519-1527 [DOI] [PubMed] [Google Scholar]
- Kedde M., Agami R. (2008). Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle 7, 899-903 [DOI] [PubMed] [Google Scholar]
- Kedde M., Strasser M. J., Boldajipour B., Oude Vrielink J. A., Slanchev K., le Sage C., Nagel R., Voorhoeve P. M., van Duijse J., Orom U. A., et al. (2007). RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131, 1273-1286 [DOI] [PubMed] [Google Scholar]
- Keene J. D. (2007). RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8, 533-543 [DOI] [PubMed] [Google Scholar]
- Keene J. D., Komisarow J. M., Friedersdorf M. B. (2006). RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 1, 302-307 [DOI] [PubMed] [Google Scholar]
- Kimura T., Suzuki A., Fujita Y., Yomogida K., Lomeli H., Asada N., Ikeuchi M., Nagy A., Mak T. W., Nakano T. (2003). Conditional loss of PTEN leads to testicular teratoma and enhances embryonic germ cell production. Development 130, 1691-1700 [DOI] [PubMed] [Google Scholar]
- Krentz A. D., Murphy M. W., Kim S., Cook M. S., Capel B., Zhu R., Matin A., Sarver A. L., Parker K. L., Griswold M. D., et al. (2009). The DM domain protein DMRT1 is a dose-sensitive regulator of fetal germ cell proliferation and pluripotency. Proc. Natl. Acad. Sci. USA 106, 22323-22328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labosky P. A., Barlow D. P., Hogan B. L. (1994). Embryonic germ cell lines and their derivation from mouse primordial germ cells. Ciba Found. Symp. 182, 157-168 [DOI] [PubMed] [Google Scholar]
- Linger R., Dudakia D., Huddart R., Easton D., Bishop D. T., Stratton M. R., Rapley E. A. (2007). A physical analysis of the Y chromosome shows no additional deletions, other than Gr/Gr, associated with testicular germ cell tumour. Br. J. Cancer 96, 357-361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looijenga L. H. (2009). Human testicular (non)seminomatous germ cell tumours: the clinical implications of recent pathobiological insights. J. Pathol. 218, 146-162 [DOI] [PubMed] [Google Scholar]
- Matsui Y. (1998). Developmental fates of the mouse germ cell line. Int. J. Dev. Biol. 42, 1037 [PubMed] [Google Scholar]
- McLaren A. (1984). Meiosis and differentiation of mouse germ cells. Symp. Soc. Exp. Biol. 38, 7-23 [PubMed] [Google Scholar]
- Molyneaux K. A., Stallock J., Schaible K., Wylie C. (2001). Time-lapse analysis of living mouse germ cell migration. Dev. Biol. 240, 488-498 [DOI] [PubMed] [Google Scholar]
- Munger S. C., Aylor D. L., Syed H. A., Magwene P. M., Threadgill D. W., Capel B. (2009). Elucidation of the transcription network governing mammalian sex determination by exploiting strain-specific susceptibility to sex reversal. Genes Dev. 23, 2521-2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T., Stevens L. C. (1982). Primordial germ cell proliferation in fetal testes in mouse strains with high and low incidences of congenital testicular teratomas. J. Natl. Cancer Inst. 69, 907-913 [PubMed] [Google Scholar]
- Noguchi T., Noguchi M. (1985). A recessive mutation (ter) causing germ cell deficiency and a high incidence of congenital testicular teratomas in 129/Sv-ter mice. J. Natl. Cancer Inst. 75, 385-392 [PubMed] [Google Scholar]
- Noguchi M., Watanabe C., Kobayashi T., Kuwashima M., Sakurai T., Katoh H., Moriwaki K. (1996). The ter mutation responsible for germ cell deficiency but not testicular nor ovarian teratocarcinogenesis in ter/ter congenic mice. Dev. Growth Differ. 38, 59-69 [DOI] [PubMed] [Google Scholar]
- Ohinata Y., Ohta H., Shigeta M., Yamanaka K., Wakayama T., Saitou M. (2009). A signaling principle for the specification of the germ cell lineage in mice. Cell 137, 571-584 [DOI] [PubMed] [Google Scholar]
- Okita K., Nakagawa M., Hyenjong H., Ichisaka T., Yamanaka S. (2008). Generation of mouse induced pluripotent stem cells without viral vectors. Science 322, 949-953 [DOI] [PubMed] [Google Scholar]
- Oosterhuis J. W., Looijenga L. H. (2005). Testicular germ-cell tumours in a broader perspective. Nat. Rev. Cancer 5, 210-222 [DOI] [PubMed] [Google Scholar]
- Rivers E. N., Hamilton D. W. (1986). Morphologic analysis of spontaneous teratocarcinogenesis in developing testes of strain 129/Sv-ter mice. Am. J. Pathol. 124, 263-280 [PMC free article] [PubMed] [Google Scholar]
- Saitou M., Barton S. C., Surani M. A. (2002). A molecular programme for the specification of germ cell fate in mice. Nature 418, 293-300 [DOI] [PubMed] [Google Scholar]
- Saitou M., Payer B., Lange U. C., Erhardt S., Barton S. C., Surani M. A. (2003). Specification of germ cell fate in mice. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 358, 1363-1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M., Payer B., O'Carroll D., Ohinata Y., Surani M. A. (2005). Blimp1 and the emergence of the germ line during development in the mouse. Cell Cycle 4, 1736-1740 [DOI] [PubMed] [Google Scholar]
- Sakurai T., Iguchi T., Moriwaki K., Noguchi M. (1995). The ter mutation first causes primordial germ cell deficiency in ter/ter mouse embryos at 8 days of gestation. Dev. Growth Differ. 37, 293-302 [DOI] [PubMed] [Google Scholar]
- Seydoux G., Braun R. E. (2006). Pathway to totipotency: lessons from germ cells. Cell 127, 891-904 [DOI] [PubMed] [Google Scholar]
- Shen R., Weng C., Yu J., Xie T. (2009). eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proc. Natl. Acad. Sci. USA 106, 11623-11628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. M., Dalton S. (2009). The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell 5, 141-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slanchev K., Stebler J., Goudarzi M., Cojocaru V., Weidinger G., Raz E. (2008). Control of Dead end localization and activity-Implications for the function of the protein in antagonizing miRNA function. Mech. Dev. 126, 270-277 [DOI] [PubMed] [Google Scholar]
- Solter D. (2006). From teratocarcinomas to embryonic stem cells and beyond: a history of embryonic stem cell research. Nat. Rev. Genet. 7, 319-327 [DOI] [PubMed] [Google Scholar]
- Stevens L. C. (1967). Origin of testicular teratomas from primordial germ cells in mice. J. Natl. Cancer Inst. 38, 549-552 [PubMed] [Google Scholar]
- Stevens L. C. (1973). A new inbred subline of mice (129-terSv) with a high incidence of spontaneous congenital testicular teratomas. J. Natl. Cancer Inst. 50, 235-242 [DOI] [PubMed] [Google Scholar]
- Stevens L. C. (1981). Genetic influences on teratocarcinogenesis and parthenogenesis. Prog. Clin. Biol. Res. 45, 93-104 [PubMed] [Google Scholar]
- Stevens L. C. (1984). Spontaneous and experimentally induced testicular teratomas in mice. Cell Differ. 15, 69-74 [DOI] [PubMed] [Google Scholar]
- Stevens L. C., Little C. C. (1954). Spontaneous testicular teratomas in an inbred strain of mice. Proc. Natl. Acad. Sci. USA 40, 1080-1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L. C., Hummel K. P. (1957). A description of spontaneous congenital testicular teratomas in strain 129 mice. J. Natl. Cancer Inst. 18, 719-747 [PubMed] [Google Scholar]
- Stevens L. C., Bunker M. C. (1964). Karyotype and sex of primary testicular teratomas in mice. J. Natl. Cancer Inst. 33, 65-78 [PubMed] [Google Scholar]
- Suzuki A., Saga Y. (2008). Nanos2 suppresses meiosis and promotes male germ cell differentiation. Genes Dev. 22, 430-435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Ross A., Capel B. (2008). Expression and functional analysis of Gm114, a putative mammalian ortholog of Drosophila bam. Dev. Biol. 318, 73-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y., Tsunekawa N., Takahashi Y., Matsui Y., Satoh M., Noce T. (2000). Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech. Dev. 93, 139 [DOI] [PubMed] [Google Scholar]
- Tsuda M., Kiso M., Saga Y. (2006). Implication of nanos2-3′UTR in the expression and function of nanos2. Mech. Dev. 123, 440-449 [DOI] [PubMed] [Google Scholar]
- Wang Y., Blelloch R. (2009). Cell cycle regulation by MicroRNAs in embryonic stem cells. Cancer Res. 69, 4093-4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger G., Stebler J., Slanchev K., Dumstrei K., Wise C., Lovell-Badge R., Thisse C., Thisse B., Raz E. (2003). dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr. Biol. 13, 1429-1434 [DOI] [PubMed] [Google Scholar]
- Western P. (2009). Foetal germ cells: striking the balance between pluripotency and differentiation. Int. J. Dev. Biol. 53, 393-409 [DOI] [PubMed] [Google Scholar]
- Western P. S., Miles D. C., van den Bergen J. A., Burton M., Sinclair A. H. (2008). Dynamic regulation of mitotic arrest in fetal male germ cells. Stem Cells 26, 339-347 [DOI] [PubMed] [Google Scholar]
- Yamanaka S. (2009). Elite and stochastic models for induced pluripotent stem cell generation. Nature 460, 49-52 [DOI] [PubMed] [Google Scholar]
- Youngren K. K., Coveney D., Peng X., Bhattacharya C., Schmidt L. S., Nickerson M. L., Lamb B. T., Deng J. M., Behringer R. R., Capel B., et al. (2005). The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 435, 360-364 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.