Abstract
BRCA2 is a key component of the homologous recombination (HR) pathway of DNA repair, acting as the loader of RAD51 recombinase at sites of double-strand breaks. Here, we demonstrate that BRCA2 associates with telomeres during S/G2 and facilitates RAD51 loading onto telomeres. Conditional Brca2 deletion and Rad51 inhibition in mouse embryonic fibroblasts (MEFs), but not Brca1 inactivation, led to telomere shortening and accumulation of fragmented telomeric signals, a hallmark of telomere fragility associated with replication defects. This suggests that BRCA2-mediated HR reactions contribute to telomere length maintenance by facilitating telomere replication and implies an essential role for BRCA2 in telomere integrity during unchallenged cell proliferation. Mouse mammary tumors lacking Brca2 accumulated telomere dysfunction-induced foci. BRCA2-mutated human breast tumors had shorter telomeres than BRCA1-mutated ones, suggesting that the genomic instability observed in BRCA2-deficient tumors is due in part to telomere dysfunction.
INTRODUCTION
Germline mutations in one allele of BRCA2 are associated with high susceptibility to breast and ovarian cancers. Female carriers of one mutant BRCA2 allele have an accumulative probability throughout their lifetime of 80% and 20% to develop breast or ovarian tumors, respectively1. At cellular level, the BRCA2 protein interacts directly with the RAD51 recombinase and regulates recombination-mediated double-strand break repair, which is thought to account for the high levels of spontaneous chromosomal aberrations seen in BRCA2-defective cells (reviewed in 2, 3, 4).
HR provides an important error-free mechanism of double-strand DNA break repair in mammalian cells and plays a major role during DNA replication, in the restart and repair of stalled or broken replication forks5,6. In addition, HR is required for telomere maintenance by providing a mechanism of telomere elongation alternative to telomerase (ALT)7,8 and a pathway for telomere capping by facilitating t-loop formation9. A central role in the process of HR is played by the RAD51 recombinase10, which assembles nucleoprotein filaments at break sites to initiate the search for a homologous recombination target sequence. RAD51 assembly at DNA breaks depends on the tumor suppressor protein BRCA2. In mammalian cells, a family of proteins known as the RAD51 paralogs consists of five proteins (RAD51B, RAD51C, RAD51D, XRCC2 and XRCC311). RAD51 paralogs also contribute to RAD51 filament assembly. In addition, a subset of the RAD51 paralogs act upstream of RAD51 and BRCA2 in DNA damage signaling through checkpoint kinases12.
Telomeres, the natural ends of linear chromosomes, consist of repetitive G-rich DNA and associated proteins. One of their prominent functions is to protect chromosome ends from degradation and fusion. Failure of telomere protection can have deleterious effects resulting in chromosomal end-to-end fusions, breakage and rearrangements13. Thus, telomere integrity is essential for genome stability.
Telomere integrity in mammalian cells encompasses two aspects: telomere length maintenance and the formation and stability of capping structures. The first aspect requires telomerase, a reverse transcriptase that uses an RNA component as a template for telomere elongation and acts after telomere replication is completed. The second aspect, formation of a protective telomere structure, is thought to involve formation of a telomeric t-loop. This protective structure is assembled when the 3′ single stranded overhang invades double-stranded telomeric DNA tracts to form a displacement loop at the invasion site. T-loop formation is essential for preventing recognition of chromosome ends as broken DNA and consequent checkpoint activation13.
Two mammalian recombination proteins, RAD51D and RAD54, were shown to act at telomeres14,15. Deletion of these HR activities leads to telomere shortening and loss of capping even in the presence of telomerase, supporting the notion of a contribution of HR to telomere protection. HR could promote telomere elongation by inter- or intra-telomere recombination or could facilitate formation of the protective t-loop structure16. Indeed, RAD51, RAD52 and XRCC3 recombination activities have been shown to associate with telomeres in S and G2 phase of the cell cycle, when capping is restored following DNA replication9. However, the mechanism by which recombination promotes telomere integrity, the contributing HR activities and the consequences of compromised telomeric recombination for tumorigenesis remain incompletely understood.
Here, we set out to investigate the contribution of the tumor suppressor BRCA2 to telomere integrity. Using ChIP assays, we show that BRCA2 associates with telomeres during the S/G2 phases of the cell cycle, similarly to RAD51, and that RAD51 telomeric association is abrogated in cells in which BRCA2 is depleted using siRNA. We demonstrate that, in addition to its role in genomic stability by promoting DNA repair, BRCA2 is crucial for telomere length maintenance by facilitating telomere replication and for chromosome end protection. This adds an unanticipated dimension to the cellular roles of this tumor suppressor. These phenotypes are recapitulated in cells defective for the HR activities of RAD51 and RAD51C, a member of RAD51 paralog family, suggesting that BRCA2 acts in telomere protection by promoting HR. Importantly, we find accumulation of dysfunctional telomeres in mouse and human tumors lacking BRCA2. This suggests that genomic instability observed in BRCA2-deficient cells and tumors is due in part to telomere dysfunction.
RESULTS
BRCA2 and RAD51 associate with telomeres during S-phase of the cell cycle
It has been shown that RAD51associates with telomeres during late S- and G2-phases of the cell cycle9, when telomere capping structures are restored after replication. Thus, RAD51 may facilitate HR-mediated capping reactions. We therefore investigated whether BRCA2, the loader of RAD51 at DNA double stranded breaks, can also be detected at telomeres during these stages of the cell cycle.
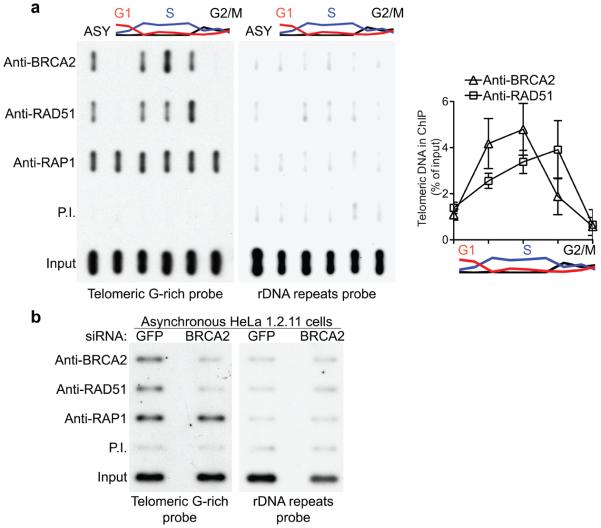
The association of RAD51 with telomeric DNA during cell cycle progression was determined using chromatin immunoprecipitation (ChIP; Fig. 1). ChIP analyses were performed using HeLa 1.2.11 cells synchronized at the G1/S transition with a double thymidine block and released in fresh medium. The DNA and associated proteins in extracts prepared at several timepoints after this release were cross-linked, fragmented by sonication, and then immunoprecipitated using various antibodies. The presence of telomeric DNA was detected using 32P-labelled probes corresponding to G-rich telomeric DNA strand (Fig. 1a). In these experiments, the telomere-associated protein RAP1 was used as a positive control. RAP1 associated with telomeres throughout the cell cycle, whilst RAD51 association occurred specifically during S and G2, as previously reported9. Importantly, when BRCA2 was similarly immunoprecipitated from synchronized HeLa extracts, it showed a telomeric association pattern similar to that of RAD51. The quantification of the ChIP signal shown in Fig. 1a indicated that BRCA2 telomeric association peaks before RAD51 during cell cycle progression. These results are consistent with the idea that BRCA2 associates with telomeres during their replication in S-phase, to load the RAD51 recombinase onto the chromosome ends. Subsequent HR reactions could facilitate both telomere replication and re-establishment of telomere protection.
Figure 1.
BRCA2 is required to recruit RAD51 to the telomeres during S/G2. (a) ChIP analyses were carried out using HeLa 1.2.11 extracts prepared at 0, 2, 4, 6, 8 hours after release from double-thymidine block or untreated cells (ASY). Error bars represent SD of three independent experiments. (b) Extracts prepared from HeLa 1.2.11 cells collected six days after the first transfection with control GFP or BRCA2 siRNAs were processed for ChIP analysis.
To test the possibility that BRCA2 functions as RAD51 loader at telomeres, we performed ChIP analyses with extracts prepared from asynchronous HeLa1.2.11 cells in which BRCA2 was depleted using siRNA (Fig. 1b). We found that the anti-BRCA2 and anti-RAD51 antibodies pulled down BRCA2 and RAD51 with associated telomeric DNA in HeLa cells treated with GFP siRNA control. When BRCA2 expression was inhibited using siRNA treatment (Supplementary Fig. 1), both BRCA2 and RAD51 binding to telomeric DNA were abrogated, as suggested by telomeric ChIP signals at or below the level of the pre-immune serum. When a non-telomeric probe such as rDNA was used in the ChIP analysis, only the input extract generated a positive signal. This shows that BRCA2 is required for telomere association of RAD51 and suggests that one of the BRCA2 roles at telomeres could be mediated by its function as RAD51 loader.
Telomere attrition due to BRCA2 deficiency in MEFs
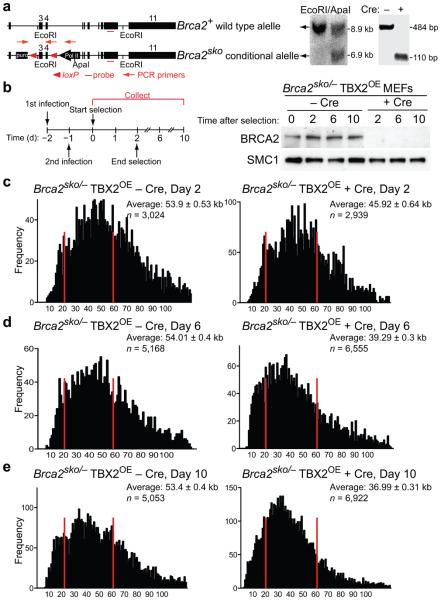
Genomic instability leading to gross chromosomal rearrangements is a hallmark of BRCA2-deficient cells and tumors2,4. Short telomeres are known to contribute to genome instability in human cancer17,18, however a possible contribution of the BRCA2 tumor suppressor to telomere maintenance has not so far been analyzed. Given the cell cycle regulated association of BRCA2 with telomeres observed in ChIP assays, we studied telomere integrity in MEFs conditionally deleted for Brca2. The Brca2sko allele contains a puromycin resistance marker and loxP sites flanking exons 3 and 4 (Fig. 2a). When treated with Cre recombinase, exons 3 and 4 of the Brca2 gene are deleted and a functional puromycin resistance gene is expressed, thus allowing selection of Brca2-deficient cells. As the Brca2 gene is essential for cell survival, we immortalized these MEFs by stable expression of TBX2, a suppressor of the p16 and p19 pathways19 to generate Brca2sko/− (TBX2OE) MEFs. Transient expression of Cre recombinase from a self-deleting (Hit&Run)20 retrovirus resulted in efficient deletion of exons 3 and 4 of the Brca2 gene, as monitored by PCR analysis (Fig. 2a) and concomitant loss of the BRCA2 protein as soon as two days after selection (Fig. 2b). BRCA2 protein remained undetectable by Western blotting up to 10 days following Cre treatment.
Figure 2.
Conditional deletion of Brca2 causes telomere shortening. (a) Diagrammatic representation of the Brca2+ and Brca2sko alleles. Wild type and sko alleles of Brca2 were visualized on a Southern blot of mouse genomic DNA. Cleavage of the Brca2sko allele following Cre treatment was detected by genomic PCR with the pair of primers indicated. (b) Cell extracts were prepared from TBX2-immortalized Brca2sko/− MEFs at the indicated times after selection and analyzed by Western blotting as indicated. SMC1 was used as a loading control. (c) − (e) Q-FISH analysis of telomere length distribution in Brca2sko/− MEFs treated with Cre (+ Cre) and control (− Cre) retroviruses and analyzed two, six and ten days after start of selection. Average telomere length values are shown with SEM. n, number of telomeres analyzed for each sample.
To study the impact of Brca2 deletion on telomere length, we performed quantitative fluorescence in situ hybridization (Q-FISH) analysis with a telomere-specific probe on metaphase chromosomes isolated from Brca2sko/− (TBX2OE) MEFs. We observed significant (P<0.0001) telomere shortening in Cre-treated compared to vector (pBabe) only-treated control cells (Fig. 2c–e). This shortening became more pronounced with increased time in culture and as cells underwent additional cell divisions. Telomere attrition progressed from 14.8% of control telomere length at day 2, to 27.2% at day 6 and 30.7% at day 10 after selection. Calculated P values (< 0.0001) for the three individual treatments shown in Fig. 2c–e indicated that Brca2 deletion resulted in a statistically significant (P<0.0001) reduction in telomere length.
To verify that the observed telomere shortening was not a consequence of the specific genetic background of these cells, we measured telomere length by Q-FISH in independently derived MEFs carrying one Brca2 null allele and one allele in which exon 11 was flanked by loxP sites (Brca2ex11/−)21. As these MEFs also fail to proliferate following Brca2 deletion, we stably infected them with a construct expressing the SV40 large T (LT) antigen to generate Brca2ex11−- LT-immortalized MEFs. Upon Cre treatment, the second copy of the Brca2 gene is inactivated. Q-FISH analysis of these MEFs showed a 20.3% reduction in the average telomere length at day 6 after puromycin addition (Supplementary Fig. 2), which was also statistically significant (P<0.0001). This indicates that BRCA2 is required for telomere length maintenance.
Telomere attrition induced by HR deficiency is telomere length-dependent
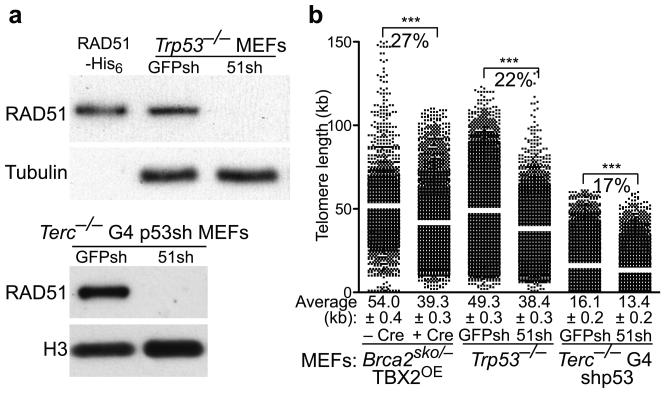
The role of BRCA2 at the telomere could be mediated by its function as RAD51 loader to initiate strand invasion and recombination reactions required for telomere capping and elongation. Thus, we next addressed whether RAD51 plays a similar role in telomere length maintenance. Like BRCA2, RAD51 is essential for cell viability. We therefore depleted this protein using shRNA in Trp53−/− MEFs, which survive the HR repair defects and the subsequent accumulation of DNA damage. MEF treatment with RAD51 shRNA led to loss of this protein as detected by Western blotting, when compared to cells treated with control shRNA (Fig. 3a).
Figure 3.
BRCA2 and RAD51 are required for telomere length maintenance in MEFs. (a) Trp53−/− MEFs or Terc−/− G4 MEFs immortalized by p53 knock-down were infected with retroviruses expressing control GFP or RAD51 shRNAs, then selected with puromycin. Cell extracts prepared six days after the first infection were analyzed by Western blotting as indicated. Recombinant human RAD51-His6 served as control. (b) Q-FISH analysis of telomere length distribution in cells treated as in (a). Average telomere length values, indicated by white bar, are shown along with SEM. Statistical analyses were performed using the Wilcoxon signed rank test. ***, P<0.0001.
We then examined telomere length in the RAD51-depleted MEFs and compared it to the Brca2-deleted MEFs. We observed a 22.3% drop in telomere length at day 6 in RAD51-depleted MEFs (Fig. 3b). Calculated P values (<0.0001) indicated that this decrease was statistically significant. A comparable decrease was observed in MEFs depleted of RAD51C (Supplementary Fig. 3a,b). This suggests that general HR activities are required for telomere length maintenance and opens the possibility that BRCA2 acts at telomeres to recruit RAD51.
We addressed whether the rapid reduction in telomere length observed as a consequence of HR deletion in MEFs was dependent on telomere length. We thus depleted RAD51 using shRNA and measured telomere length using Q-FISH in MEFs established from late generation Terc−/− G4 embryos, which show a significant (P<0.0001) decrease in telomere length due to the absence of telomerase22. shRNA-mediated depletion led to loss of detectable expression of this protein by Western blotting (Fig. 3a). Telomere length decrease was 17% relative to control cells infected with GFP shRNA, 6 days after treatment (Fig. 3b). While this shortening remained statistically significant (P<0.0001), telomere shortening due to deficient HR affected longer telomeres more strongly than shorter telomeres.
The similarity in telomere length phenotype between Brca2- and Rad51-deficient MEFs suggested that the function of BRCA2 in telomere homeostasis is mediated by recruiting RAD51 to telomeres. Consistent with RAD51 association with uncapped telomeres, we observed that approximately 40% of the wild-type MEFs carried more than 3 telomeric RAD51 foci (Supplementary Fig. 4) visualized by co-localization of RAD51 immunofluorescence-detected foci with telomeric TRF2 signals. The telomeric RAD51 staining was significantly reduced in MEFs depleted of BRCA2 after Cre-mediated excision, suggesting that BRCA2 promotes telomere association of RAD51 in this type of cells, similarly to HeLa cells.
BRCA2 and RAD51 deletions induce telomere fragility
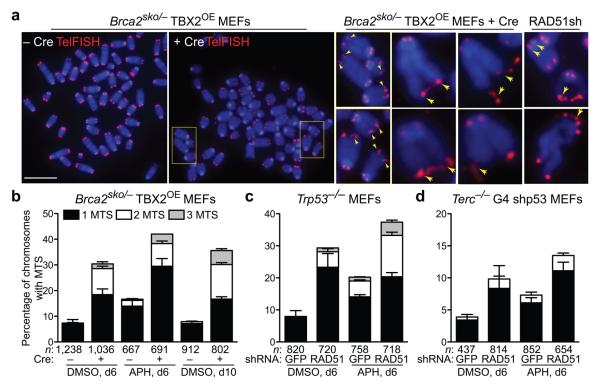
Abrogation of telomerase activity in mice results in telomere shortening at a rate of approximately 4.8 kb of telomeric repeat sequence per generation23. Our observation of the loss of up to 16 kb of telomere length within 6 days of depletion of BRCA2 or RAD51 in MEFs, corresponding to only approximately 9 cell divisions, suggested that these HR activities are required in a more profound way to sustain telomere length. A prominent telomeric phenotype of Brca2−/− and RAD51-depleted MEFs was the presence of multiple telomeric signals (MTS) at individual ends (Fig. 4a). This type of telomere aberration has been previously described in the context of TRF2 overexpression and TRF1 deficiency24-26, and more recently was associated with fragile telomeres generated by conditional deletion of TRF1 (refs. 27,28). MTS are thought to arise due to replication fork stalling and breakage within the G-rich telomeric sequence. To address whether MTS observed in BRCA2-deficient cells are due to high rate of telomere breakage, we treated wild-type and BRCA2-deficient MEFs with aphidicolin, a drug known to inhibit replication and induce fragile site instability and breakage29. MTS became detectable in wild-type cells treated with aphidicolin and further increased in cells depleted of BRCA2 (Fig. 4b). Furthermore, we observed an increase over time in MTS incidence upon Brca2-deletion (Fig. 4b).
Figure 4.
Increased telomere fragility in BRCA2- and RAD51-deficient MEFs. (a) Representative images of fragile telomeres identified by the presence of multiple telomeric signals (MTS, yellow arrowheads) in Brca2-deleted and RAD51 shRNA-depleted MEFs. Bar, 10 μm. (b) − (d) MEFs of the indicated genotypes were treated with retroviruses encoding Cre or shRNAs in the presence (APH) or absence (DMSO) of aphidicolin. The frequency of MTS was quantified in metaphase spreads following colcemid arrest. Error bars represent SD of 3 independent experiments. n, number of chromosomes scored for each sample.
A similar response to aphidicolin was observed in HeLa cells depleted of BRCA2 by siRNA (Supplementary Fig. 1) and also MEFs depleted by shRNA of RAD51 (Fig. 4c). Importantly, shRNA-mediated depletion of RAD51 in Terc−/− G4 MEFs also led to an increase in the levels of MTS (Fig. 4d), indicating that telomere fragility induced by the HR defect is also a feature of short telomeres. Consistent with the less pronounced telomere shortening in late generation Terc−/−MEFs, this increase was less pronounced than in wild-type MEFs. A comparable increase in MTS levels was observed in wild type and Terc−/− MEFs depleted of RAD51C (Supplementary Fig. 3c). These findings suggest that loss of HR activities leads to increased incidence of telomere fragility, possibly because of the HR requirement for the restart of frequently stalled replication forks at telomeres. Defective replication fork restart may in turn be the cause of the observed telomere shortening.
HR and TRF1 provide independent pathways to telomere replication
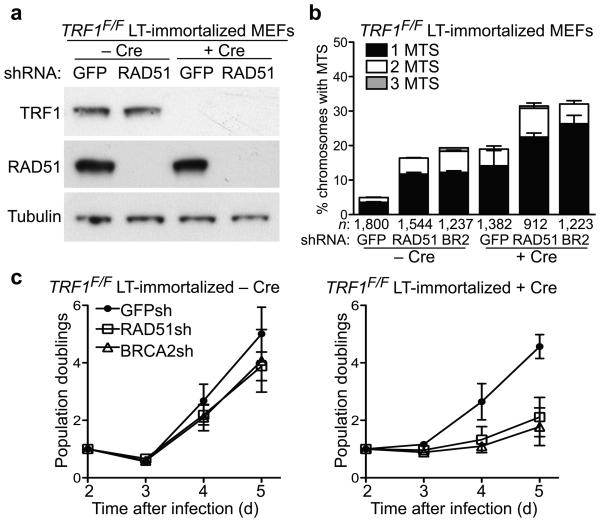
A role for TRF1 in facilitating telomere replication has been recently established27,28 and cells lacking this telomere factor showed a similar increase in MTS to that observed here in HR-defective cells. To address the relationship between these activities, we depleted BRCA2 and RAD51 using shRNA in TRF1-deleted MEFs (Fig. 5a and data not shown). Quantification of MTS showed an additive effect of loss of BRCA2 or RAD51 and TRF1 (Fig. 5b). This suggests that TRF1 and homologous recombination provide mutually independent mechanisms to promote telomere replication. Depletion of HR activities in the absence of TRF1 induced a rapid cell growth arrest (Fig. 5c). Importantly, this occurred despite immortalization of these cells by SV40 large T antigen, which is thought to suppress both p53 and pRb-dependent senescence pathways. Additional DNA damage response pathways may have been activated to cause growth arrest in the BRCA2/RAD51 and TRF1 double depleted cells.
Figure 5.
Homologous recombination activities and the telomeric factor TRF1 act independently in facilitating telomere replication. (a) Western blot detection of mouse TRF1 and RAD51 in LT-immortalized TRF1F/F MEFs treated with Cre (+ Cre) and control (− Cre) retroviruses 3 days post-selection. Tubulin was used as loading control. (b) MTS quantification in metaphase spreads following colcemid arrest cells treated as in (a). Error bars represent SD of three independent experiments. n, number of chromosomes scored for each sample. Statistical analyses were performed on total MTS using an unpaired two-tailed t-test, P<0.0001. (c) Cell proliferation assays for cells treated as in (a). Error bars represent SD of three independent experiments.
DNA damage response at uncapped telomeres in BRCA2-deficient cells
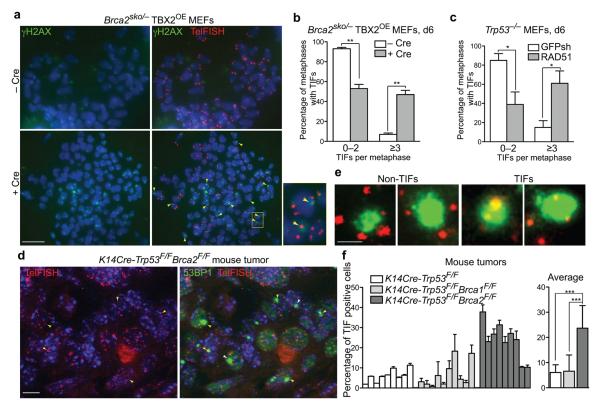
To characterize the consequences of the observed telomere dysfunction in Brca2-deficient cells, we addressed whether BRCA2 abrogation resulted in increased telomere damage. γH2AX foci are markers for double strand breaks, but also associate with short or dysfunctional telomeres, including uncapped30,31 and damaged28 telomeres. Transient expression of Cre recombinase in Brca2sko/− (TBX2OE) MEFs led to significant (P<0.001) accumulation of γH2AX signals at telomeres (telomere dysfunction-induced foci, TIFs), in addition to other sites along chromosomes (Fig. 6a). This is illustrated by γH2AX signals overlapping telomeres identified by FISH with a telomere-specific probe (Fig. 6a, inset). TIF quantification (Fig. 6b) shows that the majority of control-treated cells (95%) exhibit less than 3 TIFs per metaphase, whilst 47% of the metaphase chromosome spreads obtained from Cre-treated cells carried 3 or more TIFs. Telomere damage in Brca2-deficient cells could be mediated by a defect in RAD51 assembly. Consistent with this possibility, RAD51 depletion in MEFs using shRNA led to similar levels of TIF accumulation as Brca2 deletion (Fig. 6c). This suggests that defective RAD51 loading in the absence of BRCA2 elicits a DNA damage response at telomeres, which could be due to replication fork collapse or failed capping. Further supporting this conclusion, Replication fork progression through telomeric regions, measured using a BrdU incorporation assay9, was slower in Brca2-deficient HeLa cells compared to wild type (Supplementary Fig. 5a). The frequency of HU-induced TIF formation was correspondingly increased upon BRCA2 inactivation in human and mouse cells (Supplementary Fig. 5b,c).
Figure 6.
Telomere dysfunction-induced foci (TIFs) accumulate in BRCA2- and RAD51-deficient MEFs and in Brca2-deficient mammary tumors. (a) Immunofluorescence detection of γH2AX (green) combined with FISH staining of the telomeres (red) in Brca2sko/− MEFs immortalized by TBX2 overexpression and treated with either empty vector (− Cre) or Cre recombinase (+ Cre). Enlarged image depicts the area marked with yellow rectangle. TIFs, the sites of γH2AX co-localization with telomeres, are indicated by yellow arrowheads. Bar, 10 μm. (b) The percentage of metaphase nuclei exhibiting <3 or ≥3 TIFs was determined for at least 50 metaphases prepared as in (a) and collected 6 days post-selection. Error bars represent SD of two independent experiments. Statistical analyses were performed using an unpaired two-tailed t-test. **, P<0.001. (c) As (b), but TIFs were analyzed in Trp53−/− MEFs treated with control GFP shRNA and an shRNA against mouse RAD51. Error bars represent SD of two independent experiments. Statistical analyses were performed using an unpaired two-tailed t-test. *, P<0.05. (d) Immunofluorescence detection of 53BP1 (green) combined with telomeric FISH staining (red) in sections from a K14Cre-Trp53F/FBrca2F/F mammary tumor. Bar, 10 μm. (e) Enlarged images of TIFs, marking sites of 53BP1 co-localization with telomeres, or non-TIFs from mouse mammary tumors. Bar, 3 μm. (f) The percentage of cells containing one or more 53BP1 foci that co-localized with telomeres into TIFs was determined relative to the total number of cells analyzed (% of TIF positive cells) using paraffin-embedded sections from mouse K14Cre-Trp53F/F, K14Cre-Trp53F/FBrca1F/F or K14Cre-Trp53F/FBrca2F/F tumors. At least 100 cells were scored for each tumor. Error bars represent SD. P values were obtained using an unpaired two-tailed t-test. ***, P<0.0001.
Association of the RAD51 recombinase with mammalian telomeres occurs specifically during S and G2 (ref. 9), when telomeres are thought to become capped again following their replication. The ensuing DNA damage response facilitates assembly of telomeric capping structures mediated by HR activities. To analyze the consequence of telomere damage in the absence of HR activities, we observed chromosome morphology and telomeres in mitotically arrested cells. MEFs deficient in BRCA2, RAD51 or RAD51C showed a small, but significant (P<0.05) increase in the frequency of telomere fusions displaying telomeric DNA at the fusion site (Supplementary Fig. 6a). These fusion events are indicative of loss of telomere protection. Even more pronounced was the incidence of telomere loss, quantified as telomere-free ends. The latter observation is consistent with problems in the completion of telomere replication in the absence of HR activities. These cells also accumulated chromatid- and chromosome-type of breaks and further complex aberrations (tri-radials, quadri-radials), which could be a consequence of telomere loss-induced chromosome fusion-breakage cycles or could be due to deficient repair of DNA breaks (Supplementary Fig. 6b).
Telomere dysfunction due to loss of BRCA2 in mouse and human tumors
To study in vivo the impact of BRCA2 inactivation on telomere integrity and its possible relevance for tumorigenesis, we analyzed TIF incidence in mammary tumors from K14Cre-Trp53F/FBrca2F/Fmice21. For this, we performed a combination of antibody staining and telomeric FISH on paraffin-embedded sections of mouse mammary gland tumors. These tumors accumulate 53BP1 foci at high levels compared to K14Cre-Trp53F/Fcontrol tumors (data not shown), suggestive of persistent DNA damage. Importantly, we observed that a significant percentage (P<0.0001) of 53BP1 foci co-localized with telomeres identified by FISH (Fig. 6d) in the Brca2-deleted tumors, but much less so in tumors harboring intact BRCA2. The 53BP1 foci visualized in these tumor sections had a relatively broad appearance. Examples shown in Fig. 6e illustrate that our method could nevertheless reliably differentiate 53BP1 foci at telomeres from those localized elsewhere. Quantification of the percentage of 53BP1 foci overlapping with telomeric FISH signal relative to the total number of 53BP1 foci detected in individual tumor cells (Fig. 6f) suggested a significant (P<0.0001) increase in the frequency of uncapped telomeres in BRCA2-deficient tumors, but not in BRCA1- or p53-deficient tumors. Thus, telomere uncapping is associated with loss of BRCA2 function during tumorigenesis.
Telomeres shortening in BRCA2-deficient human mammary tumors
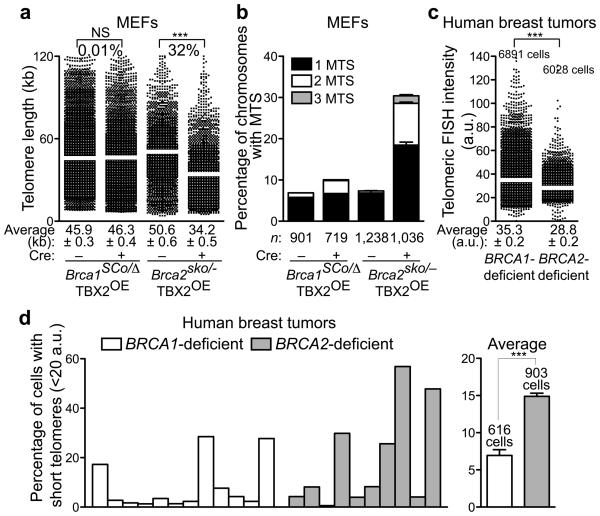
It was recently reported32 that BRCA1 and BRCA2 tumor suppressors have fundamentally distinct roles in the response to DNA damage: BRCA2 functions as RAD51 loader at the sites of damage, whilst BRCA1 is required during the very initial steps of the DNA damage signal amplification. We therefore investigated whether these two tumor suppressors also contribute differentially to telomere integrity. Brca1SCo/− MEFs contain loxP sites flanking exons 5 and 6, in addition to one null Brca1 allele. Treatment with Cre recombinase generated null Brca1 MEFs32. We measured telomere length using Q-FISH in Brca1-deleted and control MEFs, and found that the two cell populations have similar telomere length (Fig. 7a). When BRCA2 function was abrogated using Cre treatment in Brca2sko/− MEFs, telomere length was significantly shorter (P<0.0001) in the deleted cells compared to untreated control (Fig. 7a). Attrition of telomeric sequences in BRCA2-defective MEFs is linked with telomere fragility and telomere replication defects. Unlike BRCA2, loss of BRCA1 did not lead to an increase in MTS frequency (Fig. 7b), suggesting that BRCA1 is not required for telomere replication and consistent with the unaffected telomere length in Brca1-deficient cells.
Figure 7.
BRCA2 function, but not that of BRCA1, is essential for telomere maintenance in human breast tumors. (a) Conditional deletion of Brca2, but not Brca1 triggers telomere shortening in MEFs. Q-FISH analysis of telomere length distribution in Brca1SCo/− and Brca2sko/− MEFs treated with Cre (+ Cre) and control (− Cre) retroviruses and analyzed six days after start of selection. Average telomere length values, indicated by white bar, are shown along with SEM. Statistical analyses were performed using the Wilcoxon signed rank test. NS, P>0.05; ***, P<0.0001. (b) Quantification of MTS frequency in metaphase spreads from cells treated as in (a). Error bars represent SD from 2 independent experiments. n, number of chromosomes scored for each sample. (c) Q-FISH analysis of a breast tumor microarray collection consisting of 12 BRCA1-null and 10 BRCA2-null human breast tumors. The intensity of FISH telomeric signal was quantified for at least 6,000 cells for each type of tumor. a.u., arbitrary fluorescence units. Average telomere intensity values, indicated by white bar, are shown along with SEM. Statistical analyses were performed using the Wilcoxon signed rank test. ***, P<0.0001. (d) Quantification of the relative changes in the frequency of cells with short telomeres (< 20 a.u.) in BRCA1- and BRCA2-mutated human breast tumors.
The observation that loss of BRCA2, but not BRCA1, led to abrupt telomere shortening in MEFs suggested that telomere length maintenance could be relevant to the pathogenesis of breast cancer. Thus, we measured telomere length using Q-FISH on paraffin-embedded breast tumor microarrays from a collection of human breast tumors at the stage of in situ ductal carcinoma, carrying either BRCA1 or BRCA2 mutations33. Q-FISH was performed from germline mutation carriers. The collection contained 12 BRCA1-null and 10 BRCA2-null human breast tumors, from which we analyzed by Q-FISH n = 6,891 and n = 6,028 cells, respectively. Telomere length was significantly reduced (P<0.0001) in BRCA2−null tumors relative to the BRCA1-deleted (Fig. 7c and data not shown), also illustrated by the increase in the frequency of cells with short telomeres, specifically when BRCA2 function was abolished (Fig. 7d). This was consistent with the differential effects on telomere length of the conditional Brca1 or Brca2 deletions in MEFs (Fig. 7a). Thus, telomere shortening is associated specifically with loss of BRCA2 function during tumorigenesis, reflecting fundamentally different roles for the BRCA1 and BRCA2 tumor suppressors in genome integrity.
DISCUSSION
Cells harboring targeted mutations in the murine homolog of the BRCA2 gene exhibit a DNA repair defect34 and spontaneous chromosomal instability35, both stemming from a failure in recombinational DNA repair. However, the impact of BRCA2 deletion on telomere function has not been addressed so far. Here we describe for the first time that BRCA2 promotes genomic stability also by mediating HR reactions required for telomere maintenance and that this telomere function of BRCA2 may have clinical relevance.
Telomere homeostasis requires BRCA2
The data presented here indicate that BRCA2, along with the recombination activities of RAD51 and RAD51 paralogs, also act as suppressors of telomere shortening. The average telomere shortening in Brca2-deficient MEFs was approximately 14.8% at day 2, 27.2% at day 6 and 30.7% at day 10 after Brca2 inactivation by deletion of exons 3 and 4. This level of shortening was similar to that of MEFs depleted of RAD51 and RAD51C using shRNA (22% and 32%, respectively 6 days after depletion). Moreover the loss of another member of the RAD51 paralog family, RAD51D, which associates with telomeres in mammalian cells14 and RAD54 (ref. 15), which promotes Holliday junction branch migration36, lead to a comparable reduction of telomere length (21% and 32%, respectively). This suggests that HR activities play a significant role in telomere homeostasis. Most likely, this role is mediated by the ability of HR to promote efficient replication. Fork stalling and subsequent breakage has been suggested as a mechanism causing rapid loss of telomeric sequences37,38. The rapid telomere shortening in HR-deficient cells could reflect loss of large tracts of telomeric DNA due to replication fork stalling at the telomere, which are converted to double strand breaks. Supporting this hypothesis is the observation that in telomerase-negative MEFs (G4 Terc−/−) with significantly shorter telomeres than wild type (16 vs 54 kb in length; P<0.0001) the loss of telomeric sequences following depletion of HR activities of RAD51 and RAD51C is less dramatic (17% and 11%, respectively). However, even these shorter telomeres depended on BRCA2 for their maintenance. This suggests that also the maintenance of human telomeres, which are of similar length to those in G4 Terc−/− MEFs, could depend on HR reactions.
Role for BRCA2-mediated HR reactions in telomere replication and protection
Mammalian telomeres, due to their G-rich repetitive DNA sequence and protective t-loop structures, represent a natural obstacle for passing replication forks. Paradoxically, components of telomeric complexes, which bind directly to telomeric DNA, facilitate telomere replication instead of obstructing it. Fork stalling at telomeric tracts has been reported in S. pombe Taz1 mutants, lacking an essential telomere-binding factor37. More recently, the mammalian telomere-binding protein TRF1, a mammalian ortholog of Taz1, was shown to be required for efficient fork progression in mammalian cells27,28. Telomeric factors could facilitate removal of secondary structures present in the telomeric DNA, or recruit enzymatic activities (e.g. helicases, topoisomerases) that are capable of performing this function. Alternatively, a direct interaction between telomeric proteins and replication factors could be required for efficient replication of chromosome ends.
During normal progression through S-phase, stalled replication forks are processed by HR. In cell lines in which this repair pathway is compromised, single-and double-stranded DNA breaks arise spontaneously indicating that they originate during DNA replication, when stalled or broken replication forks fail to be restarted. Here, we report that in Brca2-deficient MEFs, telomeres become fragile as indicated by the high frequency of MTS, a feature associated with replicative failure27,28. Indeed, the presence of disintegrated telomeres with beads-on-a-string appearance, which often form connecting bridges between sister chromatids, is one of the most prominent features of Brca2-deficient telomeres. MEFs lacking RAD51 and RAD51C also exhibit telomere fragility phenotype. Thus, we propose that HR reactions contribute to telomere integrity by facilitating telomere replication. Consistent with this, Brca1-defective MEFs in which HR is not severely compromised32, do not show telomere replication defects and have normal telomere length.
A prediction from these results is that longer telomeres are more likely to become fragile, correlating with an increased chance of fork stalling during replication of longer G-rich regions. Consistent with this, shorter telomeres of G4 Terc−/− MEFs, although still prone to breakage in the absence of HR, accumulate MTS at lower level than wild-type counterparts. Importantly, we demonstrate an additive effect of HR abrogation and TRF1 deletion on MTS frequency. This supports the concept that both pathways act to facilitate telomere replication independently from each other. TRF1 could mediate interactions between the telomeric complex and the replication machinery important for fork progression, whilst HR could ensure restart of replication forks that break down in the G-rich telomere sequences despite the presence of TRF1.
The telomere uncapping and telomere fusion phenotype observed in HR-deficient cells suggests the involvement of HR factors in chromosome-end processing and protection. The S/G2 association of BRCA2 and RAD51 telomeres in human cells shown here by ChIP analyses, together with immunofluorescence detection of RAD51 at a subset of telomeres in wild-type MEFs, further support a role for HR reactions in telomere capping during each cell-cycle. BRCA2 stimulates formation of RAD51 helical nucleoprotein filament on single-stranded DNA substrates, which can invade homologous DNA duplexes to mediate formation of protective t-loop structures16,9. Supporting a role for BRCA2-mediated RAD51 loading in t-loop formation is the result reported here that the telomere uncapping in the RAD51-depleted MEFs (end-to-end fusions, telomere-free ends) is quantitatively similar to that of Brca2-deficient cells, suggesting that both factors have equal contributions to the capping reaction. By comparison, the telomere uncapping phenotype in RAD51C-depleted MEFs is somewhat milder, suggestive of a possible late role in t-loop HR reactions and less drastic consequences when inhibited. Thus, we favor the hypothesis that the HR-mediated mechanism of telomere capping involves conventional HR reactions, which engage the activities characteristic for this pathway of DNA repair and lead to the assembly of protective structures at telomeres.
An important consequence of telomere uncapping and replication-induced telomere breakage is activation of a DNA damage response at telomeres, as indicated by TIF formation31,27,28,30. Consistent with the uncapping and inefficient replication, we observed an increase in TIF levels in MEFs lacking BRCA2 or RAD51, similar to cells deficient in TRF1, a regulator of replication at telomere28,27. This supports the notion that BRCA2 promotes telomere replication and protection and, thus, acts as a suppressor of a DNA damage response induced by loss of telomere integrity.
Clinical implications of dysfunctional telomeres in BRCA2-deficient tumors
Accumulation of chromosome end-to-end fusions and telomere-free ends, a signature of telomere dysfunction, in addition to structural aberrations arising from stalled or broken replication forks (i.e. chromatid- and chromosome-breaks and complex aberrations), may work to initiate or promote carcinogenesis. Telomeres exposed as a consequence of uncapping or breakage can become re-ligated with each other, or with other broken DNA, to form dicentric chromosomes and initiate breakage-fusion-breakage cycles18. The high incidence of TIFs observed in Brca2-deficient mouse tumors suggests that this pathway is very likely to contribute to the radical genome instability characteristic of these tumors. The telomeric function described here adds a novel dimension to the role of BRCA2 as a genome “caretaker”. BRCA2 inactivation could promote via this new route the mutation or altered expression of “gatekeeper” genes which control cell division and death, promoting proliferation of cancer cells39. Moreover, it is known that telomerase is activated during breast cancer progression at the stage of in situ ductal carcinoma, before the invasive phenotype is acquired40. As HR reactions may contribute along with telomerase to telomere maintenance, combined therapies that target both pathways may have additive effects. Thus, drugs targeting telomerase function could be more efficient in tumors in which the HR pathway is compromised.
METHODS
Construction of the Brca2sko allele
The Brca2sko allele was generated by targeting the Brca2 locus of E14-IB10 ES cells with a targeting construct encompassing Brca2 exons 3-8 and containing a puromycin resistance marker with a floxed phosphoglycerate kinase (PGK) promoter in intron 3 and a separate RNA PolII promoter in intron 5. Following introduction of the targeting vector by electroporation and selection for puromycin resistance, ES clones were analyzed by Southern blotting using an EcoRI-ApaI digest and a Brca2 exon 10 probe. Correctly targeted ES clones were subjected to transient Cre expression to remove the floxed PGK promoter. In the resulting Brca2sko ES cells, Brca2 exons 3 and 4 are floxed and flanked by the puromycin resistance gene in intron 3 and the RNA PolII promoter in intron 4 (Fig. 2a). Switching of the conditional allele was demonstrated by PCR amplification of genomic DNA with the following primers: GTG GGC TTG TAC TCG GTC AT (forward), GTA ACC TCT GCC GTT CAG GA (reverse 1) and AGA TGT TGC TAG TCC GCC TC (reverse 2). A 110 bp fragment corresponding to the switched allele was generated upon Cre treatment. Subsequently, chimeric mice were generated by injection of mutant ES cells in C57BL6 blastocysts and germline transmission of the Brca2sko allele was obtained upon crossing with FVB/N mice. Brca2sko/+ and Brca2−/+ (ref. 21) were crossed to obtain E13.5 Brca2sko/− fetuses from which MEFs were established.
Cell culture and treatments
Primary MEFs were isolated from day 13.5 embryos as previously described23 and cultivated in DMEM medium (Invitrogen) supplemented with antibiotics and 10% foetal bovine serum (Invitrogen) in a low oxygen (3%) incubator. Brca2sko/− and Brca2ex11/− (ref. 21). MEFs were immortalized either by overexpression of TBX2 (ref. 19) or SV40 Large T antigen, then retrovirally transduced as described in Supplementary Methods. Primary MEFs isolated from p53+/+ and p53−/− littermates41 were passaged once and plated for retroviral transduction. G4 Terc−/− MEFs established from C57BL6 embryos22 were immortalized by p53 depletion (shp53)42. For metaphase chromosome preparation, cells were treated with 0.1 μg ml−1 colcemid for 4 hours or overnight, trypsinized and fixed as below. For replication arrest and fragile site analysis, MEFs and HeLa cells were treated with 0.2 μM aphidicolin (from a 10 mM stock in DMSO) overnight and released in fresh medium for 2 hours before colcemid addition. Alternatively, cells were incubated in the presence of HU (2 mM) overnight, allowed to recover in fresh media without HU for 12 hours and processed for IF-FISH analysis. The synchronization of HeLa 1.2.11 cells was performed using a double thymidine block. Exponentially growing cells were arrested by addition of thymidine (2 mM) to the growth medium for 16 hours, followed by two washes in Hank's Buffered Salt Solution HBSS and release into fresh medium for 10 hours. Cells were then arrested a second time by addition of thymidine and 16 hours incubation, then washed as above and released into fresh medium. The synchronous progression through the cell cycle was demonstrated by FACS analysis.
Preparation of metaphase spreads
Mitotic cells were collected by mitotic shake-off and swollen in hypotonic buffer (10 mM Tris-HCl, pH7.5, 10 mM NaCl, 5 mM MgCl2) at 37°C for 5 min. For combined immunofluorescence (IF)-FISH43, cells were spun onto coverslips using a cytospin centrifuge (Cytospin 4, Thermo Scientific) and dried. For Q-FISH, cells were fixed in a freshly prepared 3:1 mix of methanol:glacial acetic acid. Nuclear preparations were dropped onto slides pre-soaked in 45% acetic acid and left to dry overnight.
IF-FISH43
Metaphase spreads prepared as above were fixed in 4% paraformaldehyde with 0.1-0.5% Triton-X100 (v/v) in PBS, permeabilized with 0.5% Triton-X100 (v/v) in PBS and subjected to immunofluorescence staining as described44. After secondary antibody incubation, samples were washed and fixed again in 4% paraformaldehyde (v/v) in PBS. FISH was performed as described14 using 15 μg ml−1 Cy3-conjugated peptide nucleic acid (PNA) oligonucleotide probe with the DNA sequence [CCCTAA]3 (Applied Biosystems). Chromosomes were visualized with DAPI. Cells grown on coverslips were fixed in 4% paraformaldehyde, then permeabilized by adding 0.03% SDS (v/v) to the fixative and immunostained as described44. Telomere IF-FISH was performed on the paraffin sections as described45. Slides were antigen retrieved using sodium citrate buffer in a food steamer for 40 min. Images were acquired as z-stacks separated by 0.1–0.2 μm using a Zeiss Axiovert 200 epi-fluorescence microscope equipped with an ApoTome unit. Image processing was carried out with Axiovision Rel 4.6 software (Zeiss).
Antibodies
The following antibodies were used for immunoblotting: rabbit polyclonal antisera raised against human RAD51 (ref. 44), SMC1 (BL308, Bethyl) and human histone H3 (a gift from A. Verreault); mouse monoclonal antibodies raised against BRCA2 (OP95, Calbiochem), RAD51C (2H11)12, TRF1 (Abcam) and α-tubulin44; Cancer Research UK Monoclonal Antibody Service). Additional antibodies used for immunoflourescence detection were: mouse monoclonal antibodies raised against phosphorylated histone H2AX-Ser 139 (Upstate Biotechnology), mouse polyclonal antibody raised against full-length RAD51 (MTA2), rabbit anti-53BP1 polyclonal antibody (NB11-304, Novus) and rabbit polyclonal antisera raised against human TRF2 and RAP1 (ref. 14). For ChIP analyses, we used rabbit polyclonal antibodies against RAD51 (H92, Santa Cruz), RAP1 (ref. 14) and BRCA2 (MTA42). The rabbit polyclonal serum against human BRCA2 was generated against a polypeptide encompassing the C-terminal 322 amino acids expressed in E. coli and purified using its histidine tag.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Patrick Biggs for generating the targeting construct for the Brca2sko allele and Ian Roswell (Cancer Research UK, Clare Hall Laboratories) for help with establishing p53−/− MEFs. F. Uhlmann is thanked for critical reading and comments on the manuscript. Work in M.T. laboratory is supported by Cancer Research UK and Breast Cancer Campaign. Travel related to this project was funded by a Joint International Award to M.T. and M.A.B. from The Royal Society. Work in J.J. laboratory is supported by the Dutch Cancer Society (KWF) and the Netherlands Organisation for Scientific Research (NWO). M.A.B.'s laboratory was funded by the Spanish Ministry of Innovation and Science. U.H. was supported by the New Jersey Commission on Cancer Research (NJCCR) 09-1124-CCR-EO.
Footnotes
AUTHOR CONTRIBUTIONS
M.T., S.B and J.M.E. designed and planned the experiments. S.B and J.M.E. performed most of the experiments. P.B and J.J. generated the Brca2sko conditional mouse model, established immortalized MEFs and contributed to the results in Fig. 2a. A.R.C contributed the results in Figs. 1a,b and Sxa. M.Th. performed the IF-FISH experiments in Figs 6a,b,c, S1d and Sx b,c. M.M.G., J.B. and M.A.B. performed the experiments in Fig. 7b,c. A.S. and U.H. contributed to the results in Fig.6d,e,f. I.J. designed and validated the shRNA against mouse RAD51. M.T. made the figures and wrote the paper.
REFERENCES
- 1.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 2.Evers B, Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene. 2006;25:5885–5897. doi: 10.1038/sj.onc.1209871. [DOI] [PubMed] [Google Scholar]
- 3.Thorslund T, West SC. BRCA2: a universal recombinase regulator. Oncogene. 2007;26:7720–7730. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- 4.Venkitaraman AR. Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Annu. Rev. Pathol. 2009;4:461–487. doi: 10.1146/annurev.pathol.3.121806.151422. [DOI] [PubMed] [Google Scholar]
- 5.Nagaraju G, Scully R. Minding the gap: the underground functions of BRCA1 and BRCA2 at stalled replication forks. DNA Repair (Amst.) 2007;6:1018–1031. doi: 10.1016/j.dnarep.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsang E, Carr AM. Replication fork arrest, recombination and the maintenance of ribosomal DNA stability. DNA Repair (Amst.) 2008;7:1613–1623. doi: 10.1016/j.dnarep.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 8.Cesare AJ, Reddel RR. Telomere uncapping and alternative lengthening of telomere. Mech. Ageing Dev. 2008;129:99–108. doi: 10.1016/j.mad.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 10.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:1–11. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 11.Thacker J, Zdzienicka MZ. The mammalian XRCC genes: their roles in DNA repair and genetic stability. DNA Repair. 2003;2:655–672. doi: 10.1016/s1568-7864(03)00062-4. [DOI] [PubMed] [Google Scholar]
- 12.Badie S, et al. RAD51C facilitates checkpoint signalling by promoting CHK2 phosphorylation. J. Cell Biol. 2009;185:587–600. doi: 10.1083/jcb.200811079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:16.1–16.34. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 14.Tarsounas M, et al. Telomere maintenance requires the RAD51D recombination/repair protein. Cell. 2004;117:337–347. doi: 10.1016/s0092-8674(04)00337-x. [DOI] [PubMed] [Google Scholar]
- 15.Jaco I, et al. Role of mammalian Rad54 in telomere length maintenance. Mol. Cell. Biol. 2003;23:5572–5580. doi: 10.1128/MCB.23.16.5572-5580.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tarsounas M, West SC. Recombination at mammalian telomeres: an alternative mechanism for telomere protection and elongation. Cell Cycle. 2005;4:672–674. doi: 10.4161/cc.4.5.1689. [DOI] [PubMed] [Google Scholar]
- 17.Maser RS, DePinho RA. Telomeres and the DNA damage response: why the fox is guarding the henhouse. DNA Repair (Amst.) 2004;8-9:979–988. doi: 10.1016/j.dnarep.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Maser RS, DePinho RA. Connecting chromosomes, crisis, and cancer. Science. 2002;297:565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs JJL, et al. Senescence bypass screen identifies TBX2, which represses Cdkn2a(p19ARF) and is amplified in a subset of human breast cancers. Nat. Genet. 2000;26:291–299. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- 20.Silver DP, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 21.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 22.Herrera E, et al. Disease states associated to telomere deficiency appear earlier in mice with short telomeres. EMBO J. 1999;18:2950–2960. doi: 10.1093/emboj/18.11.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blasco MA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 24.Munoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 2005;37:1063–1071. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- 25.Blanco R, Munoz P, Flores JM, Klatt P, Blasco MA. Telomerase abrogation dramatically accelerates TRF2-induced epithelial carcinogenesis. Genes Dev. 2007;21:206–220. doi: 10.1101/gad.406207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto K, Iwano T, Tachibana M, Shinkai Y. Distinct roles of TRF1 in the regulation of telomere structure and lengthening. J. Biol. Chem. 2008;283:23981–23988. doi: 10.1074/jbc.M802395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez P, et al. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durkin SG, Glover TW. Chromosome fragile sites. Annu. Rev. Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 30.di Fagagna FD, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 31.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 32.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palacios J, et al. Immunohistochemical characteristics defined by tissue microarray of hereditary breast cancer not attributable to BRCA1 or BRCA2 mutations: differences from breast carcinomas arising in BRCA1 and BRCA2 mutation carriers. Clin. Cancer Res. 2003;9:3606–3614. [PubMed] [Google Scholar]
- 34.Connor F, et al. Tumorigenesis and a DNA-repair defect in mice with a truncating BRCA2 mutation. Nature Genet. 1997;17:423–430. doi: 10.1038/ng1297-423. [DOI] [PubMed] [Google Scholar]
- 35.Patel KJ, et al. Involvement of BRCA2 in DNA repair. Molec. Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 36.Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 37.Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 38.Karlseder J. Telomeric proteins: clearing the way for the replication fork. Nat. Struct. Mol. Biol. 2006;13:386–387. doi: 10.1038/nsmb0506-386. [DOI] [PubMed] [Google Scholar]
- 39.Kinzler KW, Vogelstein B. Cancer-susceptibility genes: gatekeepers and caretakers. Nature. 1997;386:761. doi: 10.1038/386761a0. &. [DOI] [PubMed] [Google Scholar]
- 40.Meeker AK, Argani P. Telomere shortening occurs early during breast tumorigenesis: a cause of chromosome destabilization underlying malignant transformation? J. Mammar Gland Biol. Neoplasia. 2004;9:285–296. doi: 10.1023/B:JOMG.0000048775.04140.92. [DOI] [PubMed] [Google Scholar]
- 41.Jacks T, et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 42.Dirac AM, Bernards R. Reversal of senescence in mouse fibroblasts through lentiviral suppression of p53. J. Biol. Chem. 2003;278:11731–11734. doi: 10.1074/jbc.C300023200. [DOI] [PubMed] [Google Scholar]
- 43.Thanasoula M, et al. p53 prevents entry into mitosis with uncapped telomeres. Curr. Biol. 2010;20:521–526. doi: 10.1016/j.cub.2010.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- 45.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol. Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.