Abstract
Many advances in our understanding of the molecules that regulate the development, differentiation and function of T cells have been made over the past few years. One important regulator of T-cell differentiation is the transcription factor GATA3 (GATA-binding protein 3). Although the main function of GATA3 is to act as a master transcription factor for the differentiation of T helper 2 (TH2) cells, new research has helped to uncover crucial functions of GATA3 in T cells that go beyond TH2-cell differentiation, and that are important at earlier stages of haematopoietic- and lymphoid-cell development. This Review focuses on the functions of GATA3 from early thymocyte development to effector T-cell differentiation. In addition, we discuss the interactions between GATA3 and other transcription factors and signalling pathways, and highlight the functional significance of GATA3 protein structure.
The GATA family of transcription factors are conserved proteins that contain one or two C2-C2-type zinc-finger motifs that recognize the consensus DNA sequence WGATAR (where W denotes A or T and R denotes A or G)1, 2. The six members of the mammalian GATA family (GATA1 to GATA6) contain two zinc-finger motifs, which probably arose by gene duplication (FIG. 1). The different GATA proteins have distinct and restricted patterns of tissue expression and can be divided into the haematopoietic factors (GATA1, GATA2 and GATA3) and the endodermal factors (GATA4, GATA5 and GATA6). The GATA proteins have a common structure, which comprises distinct N-terminal regions that have transactivating activity, highly conserved zinc-finger motifs in the C-terminal region, conserved basic regions located immediately after the second zinc finger and distinct C-terminal regions of varying lengths (FIG. 1). GATA3 is the main GATA-family member that is expressed by immune cells, and can be easily detected in developing and mature T cells, natural killer (NK) cells and CD1-restricted NKT cells3-5. Indeed, several recent studies have revealed an emerging role for GATA3 in invariant NKT cells (BOX 1). By contrast, mature mast cells express GATA1 and GATA2 but not GATA36, 7. Beyond the immune system, GATA3 is expressed in many embryonic and adult tissues, including the adrenal glands, kidneys, central nervous system, inner ear, hair follicles, skin and breast tissue, and important functions in several these tissues have been demonstrated in knockout and conditional knockout mouse models8-14 (for additional references, see REF. 15).
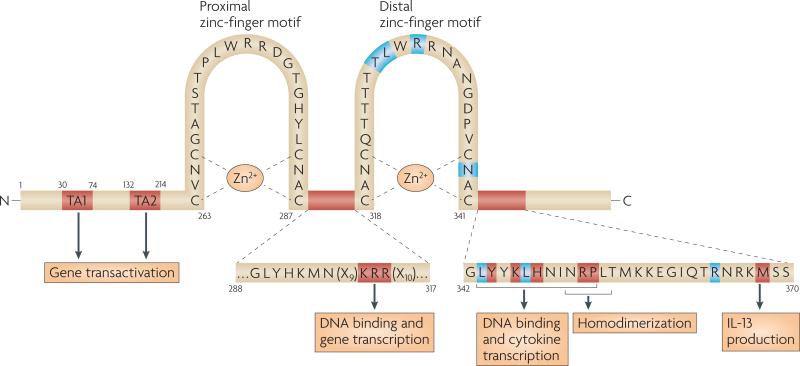
Figure 1. Functional domains and essential amino acids in mouse GATA3.
GATA-binding protein 3 (GATA3) has two amino-terminal transactivation domains TA1 and TA2, and two zinc-finger motifs, which are each followed by a conserved basic region. The C-terminal (proximal) zinc-finger motif binds the canonical GATA motif, WGATAR (where W denotes A or T, and R denotes A or G). The N-terminal (distal) zinc-finger motif seems to have broader specificity, such that the fourth position of the GATA motif can be any nucleotide111. Amino acid residues marked in blue were shown in a recent crystal structure of the distal zinc-finger motif to make direct contact with DNA111. Arginine 364 inserts deeply into the minor groove of DNA and mutation of this residue to alanine significantly disrupts binding to the WGATAR motif111. Residues marked in red have been shown to be involved in GATA3 function. For example, mutation of the amino acids KRR between the two zinc-finger motifs confers dominant negative or hypomorphic function. The YxKxHxxxRP motif (where x indicates any amino acid) is conserved between species, and mutation of any amino acid in this motif abrogates T helper 2 (TH2)-type cytokine production and DNA binding107. Methionine 368 in GATA3 (which is equivalent to proline 321 in GATA4) seems to be involved in the efficient induction of interleukin-13 (IL-13) production by GATA370. The NRPL motif that follows the distal zinc-finger motif forms the interface between two GATA3 molecules recognizing DNA sequences that contain closely arranged GATA motifs, which indicates that it may have a role in GATA3 homodimerization111.
In immune cells, GATA3 is best known to function as a master regulator of T helper 2 (TH2)-cell differentiation. However, in recent years, GATA3 has been found to have additional crucial functions in early T-cell commitment, β-selection and CD4+ T-cell development. In this Review, we highlight these new roles for GATA3 in T-cell development and differentiation, discuss the relationship of GATA3 with other transcription factors and pathways, particularly ThPOK (T-helper-inducing POZ/Kruppel-like factor; also known as cKROX) and the Notch signalling pathway (BOX 2), and describe the recent structural studies that have provided insight into GATA3 function in TH2 cells.
GATA3 before T-cell-lineage commitment
GATA3 was first discovered in a screen that sought to identify factors that bind to the human T-cell receptor-α (TCRa) enhancer3. It is among the earliest genes that are expressed by progenitor cells following commitment to the T-cell lineage. As germline deletion of Gata3 results in embryonic lethality16, analysis of the role of GATA3 in T-cell development required the generation of chimeric mice from immunodeficient (recombination-activating gene 2 (Rag2)-/-) blastocysts17. In these chimeric mice, T cells were not detected in the spleen or thymus, and there was little or negligible contribution of Gata3-/- embryonic stem cells to the rudimentary Rag2-/- thymus. B-cell and NK-cell development were unaffected. These data led to the conclusion that GATA3 expression is a hallmark of T cells, and that GATA3 is necessary for T-cell development.
But, is GATA3 expression sufficient for T-cell development? Gata3 mRNA is present in progenitors, such as fetal and adult haematopoietic stem cells (HSCs) in both mice and humans, before they commit to the T-cell lineage, which suggests that expression of GATA3 is not strictly T-cell specific18-22. Physiological expression of GATA3 is not sufficient to abrogate the development of progenitors into non-T-cell lineages, as indicated by the finding that common lymphoid progenitors (CLPs; which have a LIN-SCA1+KIT+IL-7Rα+ phenotype) express GATA3 while retaining B-cell and NK-cell developmental potential23. However, might forced overexpression of GATA3 in non-T-lineage cells be sufficient to divert their development to the T-cell lineage? This issue has been a difficult to address for two reasons. First, from a technical standpoint, overexpression of GATA3 in mouse HSCs or fetal human CD3-CD4-CD8- thymocytes in fetal thymic organ cultures (FTOCs) inhibits cell growth and/or induces apoptosis, which makes the analysis of developmental outcomes difficult24, 25. Second, forced expression of GATA3 in non-T-lineage cells may induce developmental programmes that are normally controlled by other GATA-family proteins (such as GATA1 and GATA2). For example, overexpression of GATA3 in mouse HSCs diverted these cells to develop into erythroid and megakaryocytic lineages, which are fates usually specified by GATA124. So, although GATA3 is clearly required for T-cell lineage development, its function prior to T-cell commitment remains unclear. The physiological role of GATA3 in early progenitor cell populations, such as HSCs, which co-express GATA3 and GATA219, must be defined in the future by conditional knockout approaches, to discriminate the non-redundant functions of related GATA proteins.
During early T-lymphoid development, Gata3 mRNA is detectable in HSCs and the levels increase as development proceeds through the lymphoid progenitor, double negative 1 (DN1), DN2 and DN3 stages26, 27. This expression pattern led to the suggestion that GATA3 expression occurs downstream of the Notch signalling pathway, which is known to be crucial for T-cell development. Indeed, the ligation of Notch on uncommitted precursors by culturing them with OP9 stromal cells that express the Notch ligand Delta-like ligand 1 (DLL1; the OP9-DLL1 culture system)28 forces their development into T cells and upregulates their expression of GATA329-31, an effect that is reversed when Notch ligands are removed31. As the Notch signalling pathway is both necessary and sufficient for T-cell commitment in vivo32, 33, a simple model would predict that Notch signals in the thymus promote T-cell commitment by direct or indirect upregulation of GATA3 expression and suppress B-cell commitment by inactivation of B-cell-specifying factors.
If this model were correct, forced expression of GATA3 in the absence of Notch signalling would be expected to promote T-cell development. However, a recent study from the Rothenberg laboratory34 shows that the relationship between Notch and GATA3 is more complex and that constraining GATA3 expression is important to avoid cell death and alternative lineage specification. Using the OP9-DLL1 culture system, these investigators showed that the growth of purified HSCs (LIN-SCA1+KIT+), lymphoid progenitors (LIN-SCA1+KIT+CD27+FLT3+IL-7Rα+) or DN thymocytes was inhibited by GATA3 overexpression. Moreover, the development of fetal (embryonic day 14.5) thymocytes, which comprise DN cells, that overexpress GATA3 was arrested at the DN1 and DN2 stages when cultured under T-cell-promoting conditions (that is, in the presence of Notch ligands)34, an effect that is similar to the toxic effects of GATA3 observed in human FTOCs25. Under B-cell-promoting conditions (that is, in the absence of Notch ligands), GATA3 overexpression failed to induce the differentiation of lymphoid progenitors (or fetal liver cells) into T cells34, 35. In addition, too little GATA3 expression was detrimental to progenitor survival, as GATA3-deficient fetal liver cells were profoundly depleted from OP9 co-cultures despite expression of the active, intracellular domain of Notch135. So, in the absence of Notch signalling, GATA3 overexpression is insufficient to drive T-cell differentiation, and in the presence of Notch signalling, GATA3 overexpression or lack of expression is toxic, which suggests that the levels of GATA3 must be kept ‘just right’ for thymocyte survival and development.
Unexpectedly, GATA3 overexpression by DN1 or DN2 thymocytes that were cultured in the absence of Notch ligands resulted in their differentiation into mast cells, as indicated by the upregulation of canonical mast-cell genes and a characteristic mast-cell morphology34. Overexpression of GATA3 by cells at the later, DN3 stage of development did not lead to induction of the mast-cell programme, indicating that the developmental window for diversion of progenitors to the mast-cell lineage by GATA3 expression is narrow. As GATA1 and GATA2, but not GATA3, are normally expressed by mast cells, the forced overexpression of GATA3 used in this study does not necessarily indicate a physiological role for GATA3 during the terminal stages of mast-cell development. The finding that GATA proteins induce their own expression36, 37 may be relevant in this setting, as GATA3 overexpression probably induces the mast-cell programme indirectly through auto-activation of GATA1 and/or GATA2 expression. These studies show that keeping the expression of GATA3 (or perhaps any GATA protein) at the appropriate level in DN1 and DN2 cells is essential to suppress their development into alternative cell lineages. Together, these data suggest that Notch signals regulate thymocyte development by maintaining the expression of T-cell-specific genes, preventing the induction of non-T-cell-specific transcriptional programmes and, if necessary, leading to the demise of cells that express GATA3 at high levels. Although it is clear that Notch signalling precedes the upregulation of GATA3 expression by DN thymocytes, there is no clear evidence for direct regulation of GATA3 expression by Notch. So, in the presence of Notch signals, the developmental outcome of regulated GATA3 expression seems to be influenced by other cell-type and stage-specific factors.
GATA3 after T-cell-lineage commitment
The life of a T cell can be said to begin after the so-called β-selection checkpoint, when DN3 thymocytes are tested for ‘fitness’ based on the ability to rearrange and express a functional pre-TCR. Signalling through the pre-TCR in DN3 thymocytes first induces downregulation of CD25 expression to become DN4 cells, which undergo a period of rapid proliferation, and then induces upregulation of CD4 and CD8 co-receptor expression, leading to the formation of DP thymocytes. Rearrangement at the TCRa gene locus occurs in DP thymocytes; TCRαβ-expressing thymocytes then undergo positive selection, negative selection and commitment to the CD4 or CD8 single positive (SP) lineages.
GATA3 protein expression is upregulated in thymocytes between the DN3 and DN4 stages38, although the levels of mRNA do not change markedly after β-selection and tend to decrease by the DN4 stage26, 27. Using mice in which Gata3 was specifically deleted in thymocytes at the DN3 stage of development, GATA3 was shown to be required for optimal β-selection in vivo39.[Dangling modifier?] Deletion of Gata3 led to an accumulation of DN3 cells, and a reduction of DN4 cells, DP cells and SP cells. DN3 cells that lack GATA3 expression failed to undergo the increase in cell size that accompanies pre-TCR expression, and most DN4 cells lacked TCRβ protein expression39.
It remains unclear how a lack of GATA3 impairs β-selection. Interestingly, conditional inactivation of Notch1, of the Notch signalling co-repressor RBP-J (recombination-signal-binding protein for Igκ J region; also known as CSL) or of the transcription factor MYB at the DN3 stage of development also led to a partial block in development and an accumulation of TCRβ– DN4 cells40-42. In two of these studies, the developmental block was associated with defective TCRb gene rearrangement40, 42. By contrast, GATA3-deficient DN3 thymocytes have intact rearrangement and normal levels of mRNA encoding TCRβ but no TCRβ protein, which suggests that GATA3 controls the expression of TCRβ at a post-transcriptional level. These data are consistent with the idea that during β-selection, Notch signalling and GATA3 function in series, such that Notch1 controls TCRb gene rearrangement and GATA3 controls TCRβ protein expression.
However, these studies should be interpreted with caution owing to the known limitations of models based on conditional deletion of genes using Cre-lox technology under the control of the proximal Lck promoter, which can result in ‘leaky’ expression[leaky implies inappropriate expression, whereas the problem here is incomplete or late excision, which is why I prefer variegated or incomplete]39, 43, 44. Maillard et al. used an alternative strategy to abolish all Notch signalling, in which a dominant-negative form of the Notch signalling co-activator Mastermind-like 1 (MAML1) was linked to green fluorescent protein (GFP) and expressed in a conditional manner under the control of the proximal Lck promoter, such that cells incapable of Notch signalling could be detected by GFP expression45. By excluding the ~50% of DN3 cells that did not express dominant-negative MAML1 (GFP- DN3 cells) and then injecting the GFP+ DN3 cells into the thymus of irradiated recipient mice, Maillard et al. showed that Notch-signalling-defective thymocytes had a complete block in the generation of DP thymocytes, as opposed to the partial block that was reported previously41, 42, 45. They also suggest that loss of Notch signalling causes a downregulation of CD25 expression, a potential direct target of Notch. These data imply that the ‘DN4’ cells (as defined by their CD25–CD44– phenotype) that were shown to lack TCRβ protein expression in Lck-Cre-driven Notch1-deficient (or GATA3-deficient) mice were not true DN4 cells but were instead DN3 cells that have failed to develop further and have downregulated CD25 expression. Therefore, the failure of DN3 thymocyte development may be more complete than the Lck-Cre-driven deletion models would suggest and lack of TCRβ protein expression may not be the cause of the block in development. Indeed, forced expression of TCRαβ transgenes to restore TCRβ protein levels failed to correct the block in thymocyte development in the Notch1- or GATA3-conditional-knockout mice 39, 45. These observations indicate that GATA3 is required for the effects of pre-TCR signalling or that GATA3 functions in parallel to pre-TCR signalling, perhaps by promoting cell survival or inhibiting apoptosis. A role for GATA3 in the inhibition of apoptosis is supported by our recent findings that annexin V staining is increased and proliferation is decreased (as indicated by lower rates of 7-aminoactinomycin D uptake) in GATA3-deficient ‘DN4’ cells compared with wild-type DN4 cells39 (S.-Y.P. and I.-C.H., unpublished observations). So, while GATA3 is required for optimal β-selection, the exact mechanism remains unclear.
CD4+ T-cell development: putting GATA3 into context
Rearrangement of the TCRa locus following β-selection is the first step towards positive selection, negative selection and CD4- versus CD8-lineage choice. Although these processes occur simultaneously in DP thymocytes, the regulation of each is distinct at the transcriptional level. We refer the reader to the many excellent reviews published recently46-51, and focus here on several recent advances in our understanding of CD4+ T-cell development. More specifically, we compare and contrast the roles of TOX (thymus high-mobility group box protein), ThPOK and MYB with the role of GATA3 in CD4+ T-cell development. These factors are involved during distinct stages of CD4+ T-cell development: modulation of positive selection; control of CD4 versus CD8 lineage determination; and support of CD4+ T-cell survival following lineage commitment.
Modulating positive selection
Both positive selection and lineage determination require engagement of the TCR with MHC molecules, and the duration and/or strength of the signals transduced, together with the contribution of co-receptor-mediated enhancement of TCR–MHC interactions, are thought to influence the outcome of self-antigen recognition52-54. Short-lived or weak interactions in the context of CD8 co-receptor downregulation are thought to lead to CD8+ T-cell commitment, whereas prolonged or strong interactions are thought to lead to CD4+ T-cell commitment55-57. Cells undergoing positive selection show alterations in cell-surface expression of receptors, by initially downregulating both CD4 and CD8 (resulting in cells with a CD4midCD8mid phenotype) and then by upregulating CD69 and the TCR to generate a population of transitional or post-selection thymocytes (CD4hiCD8midCD69hiTCRmid) that ultimately give rise to CD4 and CD8 SP thymocytes in an MHC-dependent manner55, 58-60.
Recently, the Kaye laboratory showed that mice lacking the transcription factor TOX undergo partial positive selection and arrest at the CD4midCD8mid stage of development, and that they lack post-selection transitional cells61. Interestingly, only mature CD8 SP thymocytes developed in these mice, a finding that might be explained by the lack of a specific CD4-lineage commitment factor, the ability of CD4midCD8mid cells to differentiate into CD8 SP thymocytes without the transitional intermediate stage and/or the absence of transitional cells that have received sufficient signals to direct thymocytes to the CD4 lineage61.
Controlling CD4-lineage commitment
A naturally occurring strain of mouse, known as the helper deficient (HD) mouse strain, lacks CD4+ T cells but maintains CD8+ T-cell development62. Positive selection is intact in these mice, but thymocytes that are destined for the CD4 lineage divert to the CD8 lineage, which suggests that the processes that regulate positive selection and lineage determination are distinct63. The phenotype of HD mice implied that there is a single gene that controls CD4-lineage determination in positively selected cells. Analysis of these mice by independent investigators using different approaches led to the identification of a transcription factor, ThPOK (which is encoded by Zbtb7b), that is both necessary and sufficient for specification of the CD4 lineage64, 65. A point mutation in Zbtb7b was shown to be responsible for the phenotype of the HD mice. CD4 SP thymocytes developed in HD mice that were transgenic for wild-type ThPOK and any of three different MHC class I-restricted TCRs, which demonstrates that ThPOK is sufficient to direct the development of positively selected cells to the CD4 lineage64, 65. Two recent publications have further shown that ThPOK prevents the downregulation of CD4 expression, stabilizes its own expression and inhibits the expression of runt-related transcription factor 3 (RUNX3), a transcription factor that promotes the development of CD8-lineage cells66, 67.
Supporting post-commitment CD4+ T-cell survival
In contrast to ThPOK deficiency, which abrogates CD4-lineage commitment, GATA3 deficiency impairs CD4+ T-cell survival or maintenance after commitment. Transgenic expression of GATA3 in adult mice suppressed CD8+ T-cell development but did not increase CD4+ T-cell development68. By contrast, retroviral expression and knockdown studies in fetal thymocytes showed that GATA3 increases CD4+ T-cell development at the expense of CD8+ T-cell development69, 70. Mice in which GATA3 expression is conditionally ablated under the control of CD4-Cre have normal maturation of CD8 SP thymocytes but a marked reduction in the number and percentage of CD4 SP thymocytes39. That transitional post-selection thymocytes and CD8+ T cells developed nearly normally in these mice indicated that positive selection did take place. Indeed, GATA3-deficient DP thymocytes in this study showed normal early TCR signalling, as measured by protein tyrosine phosphorylation and the upregulation of CD69 expression. The finding that CD8+ T-cell numbers were not increased implied that lineage determination was also not impaired. When the GATA3-deficient mice were crossed with mice that express the MHC class II-restricted TCR transgenes AND or DO11.10, neither CD4 nor CD8 SP thymocytes developed, showing that GATA3 deficiency did not divert developing CD4+ thymocytes to the CD8 lineage39 (I.-C.H. and S.-Y.P., unpublished observations). So, these results led us to propose that GATA3 is not required for lineage commitment but instead is necessary for the maintenance of CD4 SP thymocytes after lineage commitment has occurred.
The idea that factors such as ThPOK regulate lineage commitment, whereas factors such as GATA3 regulate post-commitment CD4 SP thymocyte survival or maintenance is further supported by studies of mice that are deficient in MYB. CD4-Cre-mediated T-cell-specific deletion of Myb also led to a reduction in the CD4:CD8 T-cell ratio40, mainly due to a decrease in the number of CD4 SP cells. Similar to observations in Gata3-conditional-knockout mice, introduction of a MHC class II-restricted TCR transgene (OT-II) to Myb-conditional-knockout mice did not rescue the development of CD4 SP cells or divert the development of transgene-positive cells into the CD8+ T-cell lineage. In addition, overexpression of a constitutively active form of MYB results in a block in the development of CD8 SP cells71, an effect that is similar to that occurring following forced expression of GATA3 in fetal thymocytes. Interestingly, the upregulation of GATA3 expression by positively selecting TCR signals is reduced in MYB-deficient DP thymocytes, suggesting that MYB and GATA3 work sequentially71. Consistent with this, chromatin immunoprecipitation and reporter assays showed that MYB binds directly to the promoter of Gata371.
Crosstalk between GATA3 and other factors: beyond a sequential model
On the basis of these studies defining the role of each transcription factor at different stages of CD4 SP thymocyte development a simple model could be proposed, whereby TOX drives CD4midCD8mid thymocytes to become CD4hiCD8mid cells, ThPOK expression by these CD4hiCD8mid cells then specifies CD4-lineage commitment, and committed CD4 SP cells are maintained by the expression of GATA3 and MYB (FIG. 2). However, this model has been challenged by a recent publication that supports a more complicated relationship between GATA3 and ThPOK72. In contrast to previous work, Wang et al. show that GATA3-deficient mice that are crossed with mice expressing the high-affinity MHC class II-restricted TCR transgene 5CC7 develop MHC class II-restricted CD8+ T cells, albeit 10–20-fold less efficiently than when ThPOK is absent72. They also found that GATA3 binds to the Zbtb7b promoter and that GATA3-deficient positively-selected CD69+ thymocytes had a marked decrease in ThPOK protein levels. These observations suggested that GATA3 controls ThPOK expression, possibly by direct transactivation, and raised the possibility that the failure of GATA3-deficient CD4 SP thymocytes to develop is due to the lack of ThPOK upregulation. Indeed, the lack of CD4 SP development in GATA3-deficient mice39 could be due to an inability to commit to the CD4 lineage, rather than a failure of the cells to survive after lineage commitment. However, when a Zbtb7b transgene was introduced into GATA3-deficient thymocytes, the development of mature CD4 SP thymocytes was not restored, although mature CD8 SP thymocyte development was still inhibited72. So, GATA3 seems to act both upstream and downstream of ThPOK during CD4+ T-cell development, but is dispensable for ThPOK-mediated inhibition of CD8+ T-cell development. This model is consistent with the observation that TCR stimulation upregulates GATA3 expression in DP thymocytes69, but only induces ThPOK expression in the small subset of positively selected transitional thymocytes72. So, although GATA3 is required for upregulation of ThPOK expression, the finding that ThPOK overexpression could not restore CD4 SP thymocyte development indicates that GATA3 has additional roles in supporting the survival of CD4 SP thymocytes after lineage commitment.
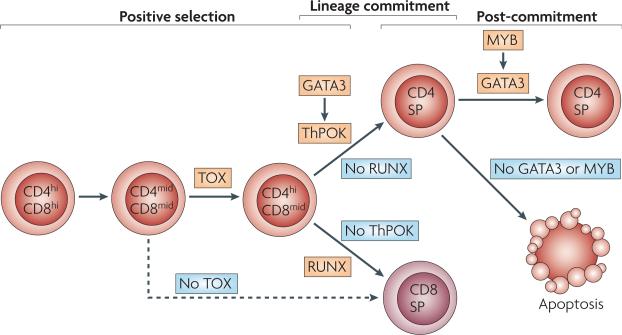
Figure 2. Transcriptional regulation of positive selection, CD4/CD8-lineage commitment and post-commitment development of thymocytes.
Positive selection and CD4- versus CD8-lineage commitment of developing thymocytes are depicted occurring sequentially. The transition from the CD4midCD8mid stage to the CD4hiCD8mid stage is controlled by the transcription factor TOX (thymus high-mobility group box protein). In the absence of TOX expression, CD4midCD8mid cells develop by ‘default’ into CD8 single positive (SP) thymocytes. The commitment of CD4hiCD8mid cells to either the CD4 SP or CD8 SP lineage requires the expression of ThPOK (T-helper-inducing POZ/Kruppel-like factor) or RUNX (runt-related transcription factor) transcription factors, respectively. Recent data show that ThPOK expression by CD4hiCD8mid cells is controlled by GATA372. The further maturation of committed CD4+ SP thymocytes is dependent on MYB and GATA3.
Recent data provide further insight into the idea of crosstalk between GATA3 and ThPOK and other factors. The low level ThPOK expression that is normally present in CD4midCD8mid thymocytes is lacking in Tox-/- mice, which suggests that TOX upregulates ThPOK in thymocytes at this stage of development61. A 500 base pair regulatory element that is located 5’ of Zbtb7b was recently found to be required for the suppression of ThPOK expression in cells other than transitional and CD4+ SP cells. This region was sufficient to drive lineage-specific ThPOK expression, and therefore, can be said to have both silencer and enhancer activity73, 74. The region contains binding sites for RUNX, GATA3, E-box-binding proteins and NF-κB (nuclear factor-κB)73, 74. Together with the observation that GATA3 is required for the expression of ThPOK, we propose that in TCR-stimulated thymocytes GATA3 cooperates with prolonged TCR signals in transitional cells to upregulate the expression of ThPOK, which, in turn, might cooperate with GATA3 in promoting post-commitment survival.
GATA3 in TH2-cell differentiation and function
Following TCR triggering by antigen, peripheral naive CD4+ T cells differentiate into effector T cells that produce high levels of cytokines. Based on the cytokines they produce, effector T cells can be divided into TH1-, TH2-, TH17- and regulatory T-cell subsets. The cytokine milieu, signalling transduction pathways and key transcription factors that are required for the differentiation of each functional CD4+ T-cell subset have been the subject of intense investigation (for reviews, see refs 75-79 and others in this issue of Nature Reviews Immunology80, 81.
STAT6-dependent pathway
A role for GATA3 in TH2-cell differentiation is well known, owing to the induction of GATA3 expression by the TH2-cell-inducing cytokine IL-4 in a signal transducer and activator of transcription 6 (STAT6)-dependent manner82. Ligation of the TCR in naive CD4+ T cells leads to the upregulation of IL4 mRNA levels within several hours83 and of Gata3 mRNA levels within 24 hours84. GATA3 facilitates the conversion of the IL4–IL5–IL13 gene locus into an “open” conformation, which allows other transcription factors that are involved in TH2-cell differentiation to access this locus85-88. It also inhibits the expression of interferon-γ (IFNγ) and directly transactivates IL5 and IL13 genes. A positive-feedback loop that further induces GATA3 expression and reinforces the differentiation of TH2 cells is established through the production of high levels of IL-4 and the auto-activation of GATA3 expression36 (FIG. 3). The source of the initial IL-4 that initiates the STAT6-dependent positive-feedback loop during in vivo TH2-cell responses is still a matter of debate. Current data suggest that memory CD44+CD4+ T cells, naive CD4+ T cells and innate immune cells, such as basophils, may all contribute to the initial source of IL-484, 89-91 (FIG. 3).
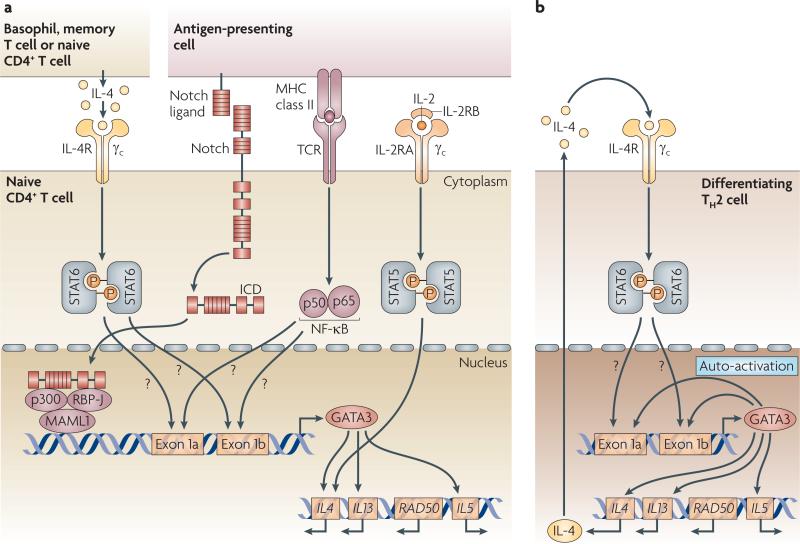
Figure 3. GATA3 is required for both STAT6-dependent and STAT6-independent pathways of TH2-cell differentiation.
a | T helper 2 (TH2)-cell differentiation can be initiated by interleukin-4 (IL-4) derived from activated TH cells through signal transducer and activator of transcription 6 (STAT6)-dependent signalling and upregulation of GATA-binding protein 3 (GATA3) expression. TH2-cell differentiation can also be initiated independently of IL-4 receptor (IL-4R) signalling. Various Notch ligands expressed by antigen-presenting cells can interact with Notch of naive CD4+ T cells, leading to the cleavage of transmembrane Notch to release the active intracellular domain of Notch (ICD). ICD then interacts with recombination-signal-binding protein for Igκ J region (RBP-J) and converts it into a transcriptional activator, which allows it to recruit Mastermind-like 1 (MAML1) and p300. The ICD–RBP-J–MAML1–p300 complex can bind to and transactivate the distal promoter of Gata3, which is located upstream of exon 1a, and subsequently leads to the production of IL-4 by the newly activated CD4+ T cells. IL-2 can activate STAT5, which binds to two DNase I hypersensitive sites in the second intron of IL4, resulting in the production of IL-4. Signals through the T-cell receptor (TCR) can also induce early IL-4 production, a process that is probably mediated by nuclear factor-κB (NF-κB). The binding sites for STAT6 or NF-κB in Gata3 remain to be determined. b | The initial burst of GATA3 and IL-4 expression induced dependently or independently of STAT6 then reinforces the expression of GATA3 through the a positive-feedback loop or GATA3 auto-activation. The cis-acting element that is responsible for GATA3 auto-activation is unknown. GATA3 modifies the IL4, IL13 and IL5 loci to create a conformation that is accessible to various other transcription factors that are involved in driving the differentiation of T cells into TH2 cells.
Although the important role of GATA3 in STAT6-dependent TH2-cell differentiation has been known for some time36, 82, 92, 93, more recent studies in GATA3-deficient mouse models have provided further confirmation that GATA3 is important for TH2-cell differentiation in vitro and in vivo94, 95. GATA3-deficient TH cells showed a profound defect in their ability to differentiate into TH2 cells and instead differentiated into IFNγ-producing cells even in conditions that were optimized for TH2-cell polarization. Interestingly, however, GATA3 was shown to be required for the continuous production of IL-5 and IL-13, but it was not required for maintaining the expression of IL-4 and IL-10 by TH2 cells95 (S.-Y.P. and I.-C.H., unpublished observations).
STAT6-independent pathway: Notch and GATA3 collaborate [I'm not sure collaborate is right, makes it sound like they physically interact? We would prefer our previous title Notch signals promote Th2 differentiation through GATA3]
The observation that TH2-cell differentiation can still occur, albeit at markedly reduced efficiency, in the absence of STAT696-98 led to the suggestion that STAT6-independent pathways of TH2-cell differentiation and GATA3 expression exist. Consistent with this idea, two recent studies indicate that Notch signals directly regulate the transcription of Gata3 and are essential for mounting TH2-cell responses99, 100. In one of the studies99, Notch signals were abrogated by T-cell-specific deletion of the co-repressor RBP-J or of both Notch1 and Notch2. In the second report100, conditional expression of dominant-negative MAML1 was used to inhibit Notch signals in naive CD4+ T cells. Both studies showed that TH2-cell responses in vivo (as indicated by basal levels of the TH2-cell-dependent antibody isotypes IgE and IgG1) and/or TH2-cell differentiation in vitro were markedly impaired in the absence of Notch signals under physiological or non-polarizing conditions. Using a reciprocal approach, forced expression of the intracellular domain of Notch increased IL-4 production and GATA3 expression, even in STAT6-deficient T cells.
Gata3 transcription can be initiated from two promoters, which are positioned ~10 kb apart, to generate two distinct non-coding first exons that become spliced to a common second exon101. Both promoters are used to transcribe Gata3 in T cells, whereas the distal promoter (exon 1a) is mainly used in brain tissue. The distal promoter upstream of exon 1a was shown to contain a consensus binding site for RBP-J, and was responsible for the induction of Gata3 transcription by Notch signalling99.
Intracellular Notch and GATA3 have been shown to synergize in the induction of GATA3 and IL-4 expression, through a STAT6-independent mechanism100. Although GATA3 can induce TH2-cell differentiation independently of Notch signalling, the TH2-cell-promoting effect of Notch signals requires GATA3, as overexpression of intracellular Notch in TH cells that are deficient in GATA3 or that express a dominant-negative form of GATA3 resulted in very little IL-4 production.
STAT6-independent pathway: IL-2R signalling boosts early IL-4 production
A second STAT6-independent pathway of TH2-cell differentiation has been described by William Paul's group, which has provided extensive evidence showing a crucial role for IL-2 receptor (IL-2R) signalling through STAT5 in TH2-cell differentiation. Exposure of naive T cells to exogenous IL-2 or forced expression of a constitutively active form of STAT5A (STAT5A1*6) in non-polarizing or TH1-cell-polarizing conditions (that is, in cells that express low but detectable levels of GATA3) led to increased TH2-cell differentiation102. Accordingly, TH2-cell differentiation in vitro and in vivo was impaired in the absence of IL-2R signalling through STAT584, 103. A role for IL-2R signalling in TH2-cell differentiation is consistent with the observation that TH2 cells express a higher level of CD25 (the α-chain of IL-2R) and are therefore more sensitive to IL-2 than TH1 cells104. The effect of STAT5A1*6 in promoting TH2-cell differentiation was still apparent, albeit to a lesser degree, in STAT6-deficient and IL-4Rα-deficient T cells103, suggesting that the mechanism is STAT6 independent. IL-2-induced STAT5 activation resulted in epigenetic modification of the IL4 locus at locations that are different to those modified by GATA3, and this led to the induction of IL-4 production by TH cells103. In addition, STAT5 and GATA3 were found to synergize in the promotion of TH2-cell differentiation103. One unexpected finding was that forced expression of STAT5A1*6, although sufficient to cause the production of TH2-type cytokines and inhibition of expression of the TH1-cell-associated transcription factor T-bet, it did not induce GATA3 expression102, which suggests that the IL-2R–STAT5 signalling pathway can drive TH2-cell differentiation in a GATA3-independent manner. However, forced expression of STAT5A1*6 alone was insufficient to induce the differentiation of GATA3-deficient T cells into TH2 cells95. These paradoxical observations fail to explain how IL-2R–STAT5 signalling promotes TH2-cell differentiation in the absence of IL-4Rα or STAT6 (therefore, in the absence of the IL-4–STAT6 positive-feedback loop) despite inducing IL-4 but not GATA3 expression (therefore, in the absence of GATA3 auto-activation).
Together, these studies indicate that TH2-cell-polarizing signals other than IL-4 can promote TH2-cell differentiation through STAT6-independent mechanisms: Notch signals, by driving the expression of GATA3 (and subsequently IL-4), and IL-2R–STAT5 signals, by directly inducing the production of IL-4 in newly activated naive T cells. These data, together with data showing that IL-4 production hours after TCR triggering is abrogated in GATA3-deficient naive T cells84, indicate that both STAT6-dependent and STAT6-independent pathways of IL-4 production require GATA3 (FIG. 3).
Structural insights into GATA3 function
The six mammalian GATA-family members are highly homologous, with a common GATA-binding motif and a common general structural organization (FIG. 1). Here, we review studies that delineate crucial structural elements of GATA3. We also discuss the idea that the structure of GATA3 has evolved features that are distinct from other GATA-family members and that are involved in controlling TH2-cell cytokine production.
Most structure–function studies of GATA3 have investigated the ability of GATA3 mutants in which large domains have been deleted to induce TH2-cell cytokine production when expressed in developing TH1 cells (Table 1). Deletion of the transactivation domains, which were defined in early reporter assay studies, abrogated GATA3 function, as did deletion of the distal zinc-finger motif87, 105, 106. The function of the proximal zinc-finger motif is less clear, as two groups found that it was dispensable87, 106 and another found that it was required105 for IL-4 production. The most C-terminal amino acids of GATA3 (371–443) were found to be dispensable for TH2-cell cytokine production 70, 107 (FIG. 1).
Table 1.
Effects of various GATA3 mutants on the function and differentiation of helper T cells36, 70, 82, 87, 99, 105-109
| GATA protein | Cytokine production | TH2 locus remodelling | DNA binding | Autoactivation | References | |||
|---|---|---|---|---|---|---|---|---|
| IL-4 | IL-5 | IL-13 | IFNγ | |||||
| Wild-type GATA3 | ↑ | ↑ | ↑ | ↓ | Yes | Yes | Yes | 36, 82, 87 |
| GATA3 ΔTA1 | ↔ | ND | ND | ↓ | ND | Yes | No | 105 |
| GATA3 ΔTA2 | ↔ | ND | ND | ↓ | ND | No | Yes | 105 |
| GATA3 ΔTA1+TA2 | ↔ | ↔ | ↔ | ↔ | Yes (IL-13) | Yes | ND | 87, 106 |
| GATA3 ΔNf | ↑, ↔ | ↔ | ↔ | ↓ | Yes | Yes/No | No | 87, 105, 106 |
| GATA3 ΔCf | ↔ | ↔ | ↔ | ↔ | No | No | No | 87, 105, 106 |
| GATA3 ΔYxKxHxxxRP | ↔ | ↔ | ↔ | ↔ | No | No | ND | 107 |
| GATA3 KRR→AAA | ↑ | ↑, ↔ | ↑ | ↓ | Yes | Yes | Yes | 82, 87, 106, 108, 109 |
| Wild-type GATA4 | ↑ | ↔ | ↔ | ↓ | Yes | Yes | Yes/No | 70, 105 |
| GATA4 Pro368→Met368 | ↑ | ↔ | ↑ | ↓ | ND | ND | ND | 70 |
↑, increased expression; ↓, decreased expression; ↔, no change; Δ, deletion; TA, transactivation domain; ND, not determined; Nf, N-terminal zinc-finger motif; Cf, C-terminal zinc-finger motif; IL, interleukin, IFNγ, interferon-γ.
Studies of GATA3 in which the three amino acids at positions 305–307 between the two zinc-finger motifs have been mutated (FIG. 1), referred to as the KRR mutant, have provided several insights into GATA3 function. The KRR mutant was shown to have dominant-negative activity in reporter assays108, impaired binding to GATA doublet elements (as found in the IL5 promoter) and reduced ability to induce IL-5 and IL-13 production in vivo when expressed as a transgene, probably by acting as a competitive inhibitor of endogenous GATA3109. In many contexts, however, the KRR mutant functions as a hypomorphic mutant of GATA3, inducing T-cell differentiation, CD4+ T-cell development and IL4 locus remodelling in early pre-committed lymphocyte precursors, thymocytes and TH1 cells, respectively, although less efficiently than native GATA334, 69, 82, 106.
More recently, key amino acids in the region distal to the second zinc finger motif (residues 342–370) have been identified (FIG. 1). In mouse TH2 cells, deletion of amino acids 349–355, or point mutations in this region, which has a conserved YxKxHxxxRP motif (where x denotes any amino acid) abrogates all GATA3 functions, including the induction of TH2-cell cytokine production and histone hyperacetylation of TH2-type cytokine loci107. Mutation of these key residues greatly reduces DNA binding in vitro107. Further downstream of the YxKxHxxxRP motif, amino acids NRKMSS (365–370) are also essential for TH2-cell function, as a deletion mutant that lacks the C-terminal tail up to and including these six residues was not able to induce the production of TH2-cell cytokines or suppress IFNγ production, whereas a mutant that includes NRKMSS has activity that is comparable to that of wild-type GATA370.
Comparison of the activity of GATA3 with the activity of related GATA-family members that are not normally expressed in T cells showed that the region following the zinc finger motifs of GATA3 specifically drives IL-13 production in CD4+ T cells70. The T-cell-specific expression of GATA3 raises the question of whether the unique functions of GATA3 are a result of its expression pattern or of biochemical features that distinguish it from other GATA-family members. To address this question directly, we tested whether another GATA-family member, GATA4, or GATA fusion proteins could substitute for the T-cell-specific functions of GATA370. We found that GATA4 supported CD4+ thymocyte development in reaggregate FTOCs, and suppressed IFNγ production as well as GATA3 in developing TH1 cells. However, GATA4 induced less IL-4 production than GATA3 and did not induce IL-5 or IL-13 production70. Interestingly, we found that replacing the entire zinc-finger and post-zinc-finger region of GATA4 with that of GATA3 restored the production of IL-13, without affecting IL-4 or IL-5 production, and that a single amino-acid substitution in GATA4, replacing proline with methionine (equivalent to methionine 368) had the same effect70. Therefore, distinct structural features of GATA3 are important for IL-13 production. A study showing that protein inhibitor of activated STAT1 (PIAS1) binds to GATA3 in TH2 cells and controls IL-13 production but not IL-5 or IL-4 production indicates that distinct co-activators may be recruited for individual cytokines110.
Conclusions and future directions
In this Review, we have discussed recent data that have expanded the role of GATA3 beyond TH2-cell differentiation, highlighting how Notch signals are incompatible with high levels of GATA3 expression in early pro-T cells, act at discrete steps in β-selection and directly induce GATA3 during TH2-cell development. The Notch signalling pathway and GATA3 intersect, but the extent of the relationship differs according to the developmental context. Although the direct effect of Notch on the Gata3 promoter during TH2-cell development has been described, the molecular mechanisms that underlie the Notch–GATA relationship at earlier stages of T-cell development remain to be elucidated.
The level of GATA3 expression must be tightly regulated, as demonstrated by the findings that GATA3 overexpression has cytotoxic effects and that inappropriate expression of GATA factors can divert the development of progenitors into alternative lineages. Although GATA3 is continuously expressed during T-cell-lineage development, it is not clear how this single transcription factor directs distinct stage-specific transcriptional programmes. Specifically, the molecular mechanism for modulation of GATA3 expression levels, which increase progressively from early thymocytes to CD4+ T cells to differentiated TH2 cells, remains elusive; the cis-acting element responsible for GATA auto-activation has not been identified. Furthermore, it is not clear how the level of GATA3 protein results in different functional outcomes. A recent report of the crystal structure of the distal zinc-finger motif of mouse GATA3 suggests that recognition of adjacent DNA-binding sites can involve either both the proximal and distal zinc-finger motifs from the same molecule or the distal zinc-finger motifs from two molecules that are self-associated111. This study also suggests that, depending on the concentration of GATA protein, one or two GATA3 molecules can bind to DNA sequences containing two motifs108. This finding provides an interesting structural explanation for the distinct functional outcomes of low versus high levels of expression of GATA proteins.
At present, the direct transcriptional targets of GATA3 in pro-T cells, CD4+ T cells and invariant NKT cells, in which the levels of expression are much lower than in TH2 cells, remain largely undefined. Studies using chromatin immunoprecipitation and genome-wide profiling are technically challenging in the setting of low protein levels and are dependent on antibodies, so new methods to delineate the transcriptional programme controlled by GATA3 will be required in the future, together with further studies using stage-specific genetic deletion. Understanding the mechanisms by which varied GATA3 expression in a developmentally regulated chromatin landscape induces distinct genetic programmes will increase our ability to dissect and modulate immune responses.
Box 1 | Emerging roles for GATA3 in invariant natural killer T cells.
Invariant natural killer T (iNKT) cells express both T helper 1 (TH1)-type and TH2-type cytokines rapidly in response to lipid antigens presented by CD1 molecules112, 113. Unlike in conventional T cells, overexpression of GATA3 in iNKT cells leads to little change in TH2-type cytokine production, implying that GATA3 may not be crucial for this function in iNKT cells114. However, recent loss-of-function studies have shown that GATA3 has important roles in the development and function of peripheral iNKT cells.
Mice in which Gata3 is deleted using CD4-cre were found to have near normal numbers of thymic iNKT cells, although they matured abnormally and, interestingly, had a selective loss of CD4+ iNKT cells despite preservation of double negative (DN) iNKT-cell numbers115, reminiscent of the loss of conventional CD4+ T cells39. In contrast to the normal number of iNKT cells in the thymus, there was a sixfold reduction in the number of iNKT cells in the spleen and almost no iNKT cells in the liver. Those peripheral iNKT cells that did develop did not mature normally, as shown by a failure to upregulate CD69 expression, and a higher rate of apoptosis, which might explain the decreased numbers. Finally, peripheral iNKT cells of GATA3-deficient mice were unresponsive to the exogenous agonist α-galactosylceramide in vivo. The refractory state could be overcome by stimulation with phorbol ester and ionomycin, which indicates that the defect was at the level of receptor-proximal signalling, but only for the production of interferon-γ115. Stimulated GATA3-deficient cells still produced very low levels of TH2-type cytokines, including interleukin-4 (IL-4) IL-5, and IL-13, which suggests that GATA3 has a similar role in iNKT cells and conventional T cells94, 95.
Box 2 | The Notch signalling pathway.
Notch is an ancient signalling pathway that has a crucial role in T-cell development and is conserved from Caenorhabditis elegans to mammals (reviewed in REFS 116, 117). Notch proteins are a family of type I transmembrane proteins. There are four Notch receptors (Notch1, Notch2, Notch3 and Notch4) in mammals that can interact with five transmembrane ligands, Delta-like ligand 1 (DLL1), DLL3, DLL4, Jagged1 and Jagged2.
In the canonical mammalian Notch signalling pathway, ligand binding triggers the cleavage of Notch by ADAM (a disintegrin and metalloproteinase; also known as TACE) family protease, thereby releasing the extracellular domain. The remaining membrane-tethered fragment of Notch undergoes two intramembranous cleavage events by the γ-secretase activity of a membrane protein complex containing presenilin, which yields the intracellular domain. The intracellular domain can translocate to the nucleus and interact with the transcription co-repressor RBP-J (recombination-signal-binding protein for Igκ J region; also known as CSL). The interaction between intracellular Notch and RBP-J converts RBP-J into an activator complex. Recruitment of co-activators, such as p300 and Mastermind-like proteins, results in the transcription of target genes.
Biography
I-Cheng Ho earned his MD from Taipei Medical University, Taiwan and PhD from University of Michigan. He received postdoctoral training at Harvard School of Public Health and rheumatology subspecialty training at Brigham and Women's Hospital. He is currently an associate professor in medicine at Harvard Medical School and an associate rheumatologist at Brigham and Women's Hospital. His research focuses on the transcriptional regulation of T-helper-cell differentiation and function.
Tzong-Shyuan Tai received his Ph.D. in immunology in 2004 from National Taiwan University, Taiwan and performed postdoctoral work at the Institute of Molecular Biology, Academia Sinica, Taiwan. He is now a postdoctoral fellow in the laboratory of I-Cheng Ho, Division of Rheumatology, Immunology and Allergy, Brigham and Women's Hospital, Boston, USA, where he is focusing on how GATA3 regulates T-cell development.
Sung-Yun Pai is a physician-scientist, who trained in paediatrics and paediatric hematology-oncology, and performed postdoctoral studies in the laboratory of I-Cheng Ho at Brigham and Women's Hospital, Boston, USA. She is currently assistant professor in paediatrics at Children's Hospital Boston and Harvard Medical School, where she studies T-cell development in mouse and human models.
Footnotes
zinc-finger motif
A DNA-binding domain in which cysteine and histidine residues are coordinated by zinc atoms and thereby form ‘fingers’ that bind to DNA.
Natural killer T cells
(NKT cells). A subpopulation of T cells that expresses both NK- and T-cell markers. In the C57BL/6 mouse strain, NKT cells express the NK1.1 (NKRP1C) molecule and the T-cell receptor (TCR). Some NKT cells recognize CD1d-associated lipid antigens and express a restricted repertoire of TCRs. After TCR stimulation of naive mice, NKT cells rapidly produce interleukin-4 and interferon-γ.
double-negative subsets
(DN subsets). The most immature thymocytes lack expression of the co-receptors CD4 and CD8, and are referred to as DN cells. This compartment can be further subdivided on the basis of CD44 and CD25 expression into four subpopulations: DN1 (CD25– CD44+), DN2 (CD25+CD44+), DN3 (CD25+CD44–) and DN4 (CD25–CD44–).
Fetal thymus organ culture
(FTOC). A system for culturing fetal thymi on a filter suspended over culture medium, which allows the growth of the organ for a longer period of time than the viability of the embryo allows and/or under various experimental conditions, for example by the addition of growth factors to the medium.
OP9-DLL1 culture system
A culture system in which stromal cells derived from osteopetrotic OP/OP mice (OP9 cells) are stably transduced to overexpress the Notch ligand Delta-like ligand 1 (DLL1) to promote T-cell lineage development of co-cultured progenitor cells. OP9 cells are useful in co-cultures initiated with myeloid progenitor cells because they do not produce macrophage colony-stimulating factor 1 that can cause excessive generation of macrophages and prevent the development of lymphoid cells.
β-selection
The controlled developmental transition beyond the double negative 3 (DN3) stage to the double positive (DP) stage that is limited to T cells that have successfully rearranged their T-cell receptor (TCR) β-chain genes to express a functional cell-surface pre-TCR. The conditional developmental arrest encountered at the DN3 stage is termed the β-selection checkpoint.
Positive selection
The process by which immature CD4+CD8+ thymocytes expressing T-cell receptors with low affinity and/or avidity for self-peptide–MHC complexes are induced to differentiate into mature CD4+ and CD8+ thymocytes.
Negative selection
The process by which CD4+CD8+ thymocytes expressing potentially autoreactive T-cell receptors are induced to undergo apoptosis in the thymus.
Cre–lox technology
A site-specific recombination system that is used to delete a gene in mouse cells using Cre recombinase. Two short DNA sequences (LoxP sites) are engineered to flank the target DNA. Expression of Cre recombinase leads to excision of the intervening sequence. Depending on the type of promoter, Cre can be expressed at specific times during development or by specific sets of cells, including embryonic stem cells.
Annexin V
A molecule that binds phosphatidylserine, which is usually located on the inner leaflet of the plasma membrane but flips to the outer leaflet during apoptosis. Positive staining with annexin V is an indicator of apoptosis.
 GATA-binding protein 3 (GATA3) is a zinc-finger transcription factor that is continuously expressed in a highly regulated manner throughout T-cell development and CD4+ T helper (TH)-cell differentiation.
GATA-binding protein 3 (GATA3) is a zinc-finger transcription factor that is continuously expressed in a highly regulated manner throughout T-cell development and CD4+ T helper (TH)-cell differentiation.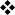 At the earliest stages of T-cell development, GATA3 is required for commitment to the T-cell lineage. However, forced expression of GATA3 in pre-committed double negative 1 (DN1) or DN2 thymocytes is toxic, and in developing B cells it diverts the development of cells to the mast-cell lineage. So, GATA3 must be expressed at levels that are ‘just right’ for successful commitment to the T-cell lineage.
At the earliest stages of T-cell development, GATA3 is required for commitment to the T-cell lineage. However, forced expression of GATA3 in pre-committed double negative 1 (DN1) or DN2 thymocytes is toxic, and in developing B cells it diverts the development of cells to the mast-cell lineage. So, GATA3 must be expressed at levels that are ‘just right’ for successful commitment to the T-cell lineage. After T-cell-lineage commitment, GATA3 has a role in β-selection, such that mice lacking GATA3 at the DN3 stage of development exhibit partial DN3 arrest, and impairment of T-cell receptor β-chain (TCRβ) protein expression. The nature and extent of this function of GATA3 are still unclear due to the current limitations of T-cell-specific conditional knockout technology.
After T-cell-lineage commitment, GATA3 has a role in β-selection, such that mice lacking GATA3 at the DN3 stage of development exhibit partial DN3 arrest, and impairment of T-cell receptor β-chain (TCRβ) protein expression. The nature and extent of this function of GATA3 are still unclear due to the current limitations of T-cell-specific conditional knockout technology.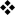 Two transcription factors, GATA3 and ThPOK (T-helper-inducing POZ/Kruppel-like factor), the gene recently found to be mutated in the helper deficient (HD) spontaneous mutant mouse, are both crucial for CD4 SP thymocyte development. The development of MHC class II-restricted thymocytes that lack or express a mutant form of ThPOK is diverted to the CD8 lineage, indicating that ThPOK functions in CD4/CD8 lineage determination. By contrast, GATA3-deficient thymocytes developing in a MHC class II-restricted environment give rise to no or few CD8-lineage cells. Delineating the distinct functions of these two factors is further complicated by evidence that GATA3 upregulates ThPOK expression but restoration of ThPOK expression in GATA3-deficient mice fails to restore CD4 SP thymocyte development.
Two transcription factors, GATA3 and ThPOK (T-helper-inducing POZ/Kruppel-like factor), the gene recently found to be mutated in the helper deficient (HD) spontaneous mutant mouse, are both crucial for CD4 SP thymocyte development. The development of MHC class II-restricted thymocytes that lack or express a mutant form of ThPOK is diverted to the CD8 lineage, indicating that ThPOK functions in CD4/CD8 lineage determination. By contrast, GATA3-deficient thymocytes developing in a MHC class II-restricted environment give rise to no or few CD8-lineage cells. Delineating the distinct functions of these two factors is further complicated by evidence that GATA3 upregulates ThPOK expression but restoration of ThPOK expression in GATA3-deficient mice fails to restore CD4 SP thymocyte development.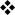 GATA3 is both necessary and sufficient for development of TH2 cells, largely because interleukin-4 receptor (IL-4R) signalling through STAT6 (signal transducer and activator of transcription 6) induces GATA3 expression in a feed-forward loop. STAT6-independent TH2-cell differentiation signals such as triggering of the TCR, activation of IL-2R–STAT5 signalling and engagement of Notch receptors also depend on GATA3 to promote IL-4 production.
GATA3 is both necessary and sufficient for development of TH2 cells, largely because interleukin-4 receptor (IL-4R) signalling through STAT6 (signal transducer and activator of transcription 6) induces GATA3 expression in a feed-forward loop. STAT6-independent TH2-cell differentiation signals such as triggering of the TCR, activation of IL-2R–STAT5 signalling and engagement of Notch receptors also depend on GATA3 to promote IL-4 production.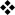 Control of production of individual TH2-type cytokines by GATA3 is accomplished by distinct mechanisms, as shown by structure-function analyses.
Control of production of individual TH2-type cytokines by GATA3 is accomplished by distinct mechanisms, as shown by structure-function analyses.
References
- 1.Ko L, Engel J. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merika M, Orkin S. Molecular and Cellular Biology. DNA-Binding Specificity of GATA Family Transcription Factors. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho I, et al. Human GATA-3: a lineage-restricted transcription factor that regulates the expression of the T cell receptor alpha gene. EMBO J. 1991;10:1187–1192. doi: 10.1002/j.1460-2075.1991.tb08059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oosterwegel M, Timmerman J, Leiden J, Clevers H. Expression of GATA-3 during lymphocyte differentiation and mouse embryogenesis. Dev Immunol. 1992;3:1–11. doi: 10.1155/1992/27903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samson SI, et al. GATA-3 promotes maturation, IFN-gamma production, and liver-specific homing of NK cells. Immunity. 2003;19:701–711. doi: 10.1016/s1074-7613(03)00294-2. [DOI] [PubMed] [Google Scholar]
- 6.Zon LI, et al. GATA-binding transcription factors in mast cells regulate the promoter of the mast cell carboxypeptidase A gene. J Biol Chem. 1991;266:22948–22953. [PubMed] [Google Scholar]
- 7.Solymar DC, Agarwal S, Bassing CH, Alt FW, Rao A. A 3' enhancer in the IL-4 gene regulates cytokine production by Th2 cells and mast cells. Immunity. 2002;17:41–50. doi: 10.1016/s1074-7613(02)00334-5. [DOI] [PubMed] [Google Scholar]
- 8.Asselin-Labat ML, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 9.de Guzman Strong C, et al. Lipid defect underlies selective skin barrier impairment of an epidermal-specific deletion of Gata-3. J Cell Biol. 2006;175:661–670. doi: 10.1083/jcb.200605057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaufman CK, et al. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev. 2003;17:2108–2122. doi: 10.1101/gad.1115203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kouros-Mehr H, et al. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell. 2008;13:141–152. doi: 10.1016/j.ccr.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim KC, et al. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat Genet. 2000;25:209–212. doi: 10.1038/76080. [DOI] [PubMed] [Google Scholar]
- 14.Tong Q, et al. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 2000;290:134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- 15.Ho IC, Pai SY. GATA-3 - not just for Th2 cells anymore. Cell Mol Immunol. 2007;4:15–29. [PubMed] [Google Scholar]
- 16.Pandolfi PP, et al. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis [see comments]. Nature Genetics. 1995;11:40–44. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- 17.Ting CN, Olson MC, Barton KP, Leiden JM. Transcription Factor Gata-3 Is Required For Development Of the T-Cell Lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [This paper established the crucial role of GATA3 in T-lineage commitment. GATA3-deficient embryonic stem cells used to complement RAG-deficient blastocysts generated B cells but not T cells.] [DOI] [PubMed] [Google Scholar]
- 18.Zhong JF, et al. Gene expression profile of murine long-term reconstituting vs. short-term reconstituting hematopoietic stem cells. Proc Natl Acad Sci USA. 2005;102:2448–2453. doi: 10.1073/pnas.0409459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertrand JY, et al. Characterization of purified intraembryonic hematopoietic stem cells as a tool to define their site of origin. Proc Natl Acad Sci USA. 2005;102:134–139. doi: 10.1073/pnas.0402270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi-Osaki M, et al. GATA motifs regulate early hematopoietic lineage-specific expression of the Gata2 gene. Mol Cell Biol. 2005;25:7005–7020. doi: 10.1128/MCB.25.16.7005-7020.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labastie MC, Cortés F, Roméo PH, Dulac C, Péault B. Molecular identity of hematopoietic precursor cells emerging in the human embryo. Blood. 1998;92:3624–3635. [PubMed] [Google Scholar]
- 22.Mouthon MA, et al. Expression of tal-1 and GATA-binding proteins during human hematopoiesis. Blood. 1993;81:647–655. [PubMed] [Google Scholar]
- 23.Dias S, Silva H, Cumano A, Vieira P. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J Exp Med. 2005;201:971–979. doi: 10.1084/jem.20042393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen D, Zhang G. Enforced expression of the GATA-3 transcription factor affects cell fate decisions in hematopoiesis. Exp Hematol. 2001;29:971–980. doi: 10.1016/s0301-472x(01)00670-1. [DOI] [PubMed] [Google Scholar]
- 25.Taghon T, et al. Enforced expression of GATA-3 severely reduces human thymic cellularity. J Immunol. 2001;167:4468–4475. doi: 10.4049/jimmunol.167.8.4468. [DOI] [PubMed] [Google Scholar]
- 26.David-Fung ES, et al. Progression of regulatory gene expression states in fetal and adult pro-T-cell development. Immunol Rev. 2006;209:212–236. doi: 10.1111/j.0105-2896.2006.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tydell CC, et al. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J Immunol. 2007;179:421–438. doi: 10.4049/jimmunol.179.1.421. [DOI] [PubMed] [Google Scholar]
- 28.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 29.Höflinger S, et al. Analysis of Notch1 function by in vitro T cell differentiation of Pax5 mutant lymphoid progenitors. J Immunol. 2004;173:3935–3944. doi: 10.4049/jimmunol.173.6.3935. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt TM, et al. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat Immunol. 2004;5:410–417. doi: 10.1038/ni1055. [DOI] [PubMed] [Google Scholar]
- 31.Taghon TN, David ES, Zúñiga-Pflücker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19:965–978. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pui JC, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 33.Radtke F, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 34.Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [By overexpressing GATA3 in DN thymocytes cultured on OP9 and OP9-DLL1 stroma, these authors show that GATA3 overexpression in the presence of Notch signals is toxic, whereas GATA3 overexpression in the absence of Notch signals diverts pre-committed thymocytes to the mast-cell lineage.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hozumi K, et al. Notch signaling is necessary for GATA3 function in the initiation of T cell development. Eur J Immunol. 2008;38:977–985. doi: 10.1002/eji.200737688. [DOI] [PubMed] [Google Scholar]
- 36.Ouyang W, et al. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12:27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- 37.Tsai S-F, Strauss E, Orkin S. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes & Development. 1991;5:919–931. doi: 10.1101/gad.5.6.919. [DOI] [PubMed] [Google Scholar]
- 38.Hendriks RW, et al. Expression of the transcription factor GATA-3 is required for the development of the earliest T cell progenitors and correlates with stages of cellular proliferation in the thymus. Eur J Immunol. 1999;29:1912–1918. doi: 10.1002/(SICI)1521-4141(199906)29:06<1912::AID-IMMU1912>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 39.Pai SY, et al. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [Using mice in which thymocytes were rendered deficient in GATA3 at the DN3 and DP stages of development, these authors showed that GATA3 is required for optimal β-selection and for CD4 SP thymocyte development.] [DOI] [PubMed] [Google Scholar]
- 40.Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol. 2004;5:721–729. doi: 10.1038/ni1085. [DOI] [PubMed] [Google Scholar]
- 41.Tanigaki K, et al. Regulation of alphabeta/gammadelta T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- 42.Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Inactivation of Notch1 impairs VDJbeta rearrangement and allows pre-TCR-independent survival of early alpha beta Lineage Thymocytes. Immunity. 2002;16:869–879. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- 43.Neilson JR, Winslow MM, Hur EM, Crabtree GR. Calcineurin B1 is essential for positive but not negative selection during thymocyte development. Immunity. 2004;20:255–266. doi: 10.1016/s1074-7613(04)00052-4. [DOI] [PubMed] [Google Scholar]
- 44.Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 45.Maillard I, et al. The requirement for Notch signaling at the beta-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J Exp Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aliahmad P, Kaye J. Commitment issues: linking positive selection signals and lineage diversification in the thymus. Immunol Rev. 2006;209:253–273. doi: 10.1111/j.0105-2896.2006.00345.x. [DOI] [PubMed] [Google Scholar]
- 47.Bosselut R. CD4/CD8-lineage differentiation in the thymus: from nuclear effectors to membrane signals. Nat Rev Immunol. 2004;4:529–540. doi: 10.1038/nri1392. [DOI] [PubMed] [Google Scholar]
- 48.He X, Kappes DJ. CD4/CD8 lineage commitment: light at the end of the tunnel? Curr Opin Immunol. 2006;18:135–142. doi: 10.1016/j.coi.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 49.Kappes DJ, He X, He X. Role of the transcription factor Th-POK in CD4:CD8 lineage commitment. Immunol Rev. 2006;209:237–252. doi: 10.1111/j.0105-2896.2006.00344.x. [DOI] [PubMed] [Google Scholar]
- 50.Laky K, Fowlkes BJ. Receptor signals and nuclear events in CD4 and CD8 T cell lineage commitment. Curr Opin Immunol. 2005;17:116–121. doi: 10.1016/j.coi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 51.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hedrick SM. T cell development: bottoms-up. Immunity. 2002;16:619–622. doi: 10.1016/s1074-7613(02)00316-3. [DOI] [PubMed] [Google Scholar]
- 53.Hogquist KA. Signal strength in thymic selection and lineage commitment. Curr Opin Immunol. 2001;13:225–231. doi: 10.1016/s0952-7915(00)00208-9. [DOI] [PubMed] [Google Scholar]
- 54.Singer A. New perspectives on a developmental dilemma: the kinetic signaling model and the importance of signal duration for the CD4/CD8 lineage decision. Curr Opin Immunol. 2002;14:207–215. doi: 10.1016/s0952-7915(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 55.Brugnera E, et al. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity. 2000;13:59–71. doi: 10.1016/s1074-7613(00)00008-x. [DOI] [PubMed] [Google Scholar]
- 56.Matechak EO, Killeen N, Hedrick SM, Fowlkes BJ. MHC class II-specific T cells can develop in the CD8 lineage when CD4 is absent. Immunity. 1996;4:337–347. doi: 10.1016/s1074-7613(00)80247-2. [DOI] [PubMed] [Google Scholar]
- 57.Sarafova SD, et al. Modulation of coreceptor transcription during positive selection dictates lineage fate independently of TCR/coreceptor specificity. Immunity. 2005;23:75–87. doi: 10.1016/j.immuni.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 58.Bosselut R, Guinter TI, Sharrow SO, Singer A. Unraveling a revealing paradox: Why major histocompatibility complex I-signaled thymocytes “paradoxically” appear as CD4+8lo transitional cells during positive selection of CD8+ T cells. J Exp Med. 2003;197:1709–1719. doi: 10.1084/jem.20030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lucas B, Germain RN. Unexpectedly complex regulation of CD4/CD8 coreceptor expression supports a revised model for CD4+CD8+ thymocyte differentiation. Immunity. 1996;5:461–477. doi: 10.1016/s1074-7613(00)80502-6. [DOI] [PubMed] [Google Scholar]
- 60.Lundberg K, Heath W, Köntgen F, Carbone FR, Shortman K. Intermediate steps in positive selection: differentiation of CD4+8int TCRint thymocytes into CD4-8+TCRhi thymocytes. J Exp Med. 1995;181:1643–1651. doi: 10.1084/jem.181.5.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med. 2008 doi: 10.1084/jem.20071944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dave VP, Allman D, Keefe R, Hardy RR, Kappes DJ. HD mice: a novel mouse mutant with a specific defect in the generation of CD4(+) T cells. Proc Natl Acad Sci USA. 1998;95:8187–8192. doi: 10.1073/pnas.95.14.8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keefe R, Dave V, Allman D, Wiest D, Kappes DJ. Regulation of lineage commitment distinct from positive selection. Science. 1999;286:1149–1153. doi: 10.1126/science.286.5442.1149. [References 62 and 63 describe the phenotype of the HD mouse strain, which lacks CD4+ T-cell development and supports the diversion of MHC class II-restricted T cells into the CD8 lineage when crossed with AND TCR-transgenic mice.] [DOI] [PubMed] [Google Scholar]
- 64.He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 65.Sun G, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [References 64 and 65 identify Zbtb7b as the gene that is mutated in HD mice. Transgenic expression of the Zbtb7b gene product (ThPOK) was sufficient to divert the development of MHC class I-restricted thymocytes to the CD4 lineage.] [DOI] [PubMed] [Google Scholar]
- 66.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muroi S, et al. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 68.Nawijn MC, et al. Enforced expression of GATA-3 during T cell development inhibits maturation of CD8 single-positive cells and induces thymic lymphoma in transgenic mice. J Immunol. 2001;167:715–723. doi: 10.4049/jimmunol.167.2.715. [DOI] [PubMed] [Google Scholar]
- 69.Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [Using overexpression and knockdown of GATA3 expression in fetal thymocytes, the authors show that GATA3 promotes CD4 SP thymocyte development.] [DOI] [PubMed] [Google Scholar]
- 70.Pai SY, et al. Distinct Structural Requirements of GATA-3 for the Regulation of Thymocyte and Th2 Cell Differentiation. J Immunol. 2008;180:1050–1059. doi: 10.4049/jimmunol.180.2.1050. [DOI] [PubMed] [Google Scholar]
- 71.Maurice D, Hooper J, Lang G, Weston K. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J. 2007;26:3629–3640. doi: 10.1038/sj.emboj.7601801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang L, et al. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat Immunol. 2008 doi: 10.1038/ni.1647. [This paper shows that GATA3 upregulates ThPOK expression in post-selection thymocytes and that GATA3 can function as a lineage-determining factor. The finding that restoration of ThPOK expression in GATA3-deficient mice failed to restore the development of CD4 SP thymocytes suggests that GATA3 has additional roles in the survival of CD4 thymocytes after lineage commitment.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He X, et al. CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 2008;28:346–358. doi: 10.1016/j.immuni.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 74.Setoguchi R, et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 75.Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- 76.Reiner SL. Development in motion: helper T cells at work. Cell. 2007;129:33–36. doi: 10.1016/j.cell.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 77.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 78.Mowen KA, Glimcher LH. Signaling pathways in Th2 development. Immunol Rev. 2004;202:203–222. doi: 10.1111/j.0105-2896.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- 79.Zhu J, Yamane H, Cote-Sierra J, Guo L, Paul WE. GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell Res. 2006;16:3–10. doi: 10.1038/sj.cr.7310002. [DOI] [PubMed] [Google Scholar]
- 80.Amsen D, Antov A, Flavell RA. The different faces of Notch in T-helper-cell differentiation. Nat Rev Immunol. 2009;9:xxx–xxx. doi: 10.1038/nri2488. [DOI] [PubMed] [Google Scholar]
- 81.Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that govern T-cell development and differentiation. Nat Rev Immunol. 2009;9:xxx–xxx. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ouyang W, et al. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 83.Grogan JL, et al. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity. 2001;14:205–215. doi: 10.1016/s1074-7613(01)00103-0. [DOI] [PubMed] [Google Scholar]
- 84.Yamane H, Zhu J, Paul WE. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J Exp Med. 2005;202:793–804. doi: 10.1084/jem.20051304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Avni O, et al. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 86.Lee GR, Fields PE, Flavell RA. Regulation of IL-4 gene expression by distal regulatory elements and GATA-3 at the chromatin level. Immunity. 2001;14:447–459. doi: 10.1016/s1074-7613(01)00125-x. [DOI] [PubMed] [Google Scholar]
- 87.Lee HJ, et al. GATA-3 Induces T Helper Cell Type 2 (Th2) Cytokine Expression and Chromatin Remondeling in Committed Th1 Cells. Journal of Experimental Medicine. 2000;192:105–115. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 89.Tanaka S, et al. The interleukin-4 enhancer CNS-2 is regulated by Notch signals and controls initial expression in NKT cells and memory-type CD4 T cells. Immunity. 2006;24:689–701. doi: 10.1016/j.immuni.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 90.Xin J, Ohmori K, Nishida J, Zhu Y, Huang H. The initial response of CD4+ IL-4-producing cells. Int Immunol. 2007;19:305–310. doi: 10.1093/intimm/dxl147. [DOI] [PubMed] [Google Scholar]
- 91.Sokol CL, Barton GM, Farr AG, Medzhitov R. A mechanism for the initiation of allergen-induced T helper type 2 responses. Nat Immunol. 2008;9:310–318. doi: 10.1038/ni1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription Factor Gata-3 Is Differentially Expressed Murine Th1 and Th2 Cells and Controls Th2-Specific Expression Of the Interleukin-5 Gene. Journal of Biological Chemistry. 1997;272:21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- 93.Zheng WP, Flavell RA. The Transcription Factor Gata-3 Is Necessary and Sufficient For Th2 Cytokine Gene Expression In Cd4 T Cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 94.Pai SY, Truitt ML, Ho IC. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc Natl Acad Sci U S A. 2004;101:1993–1998. doi: 10.1073/pnas.0308697100. [This paper and reference 95 use conditional deletion of GATA3 to confirm the requirement for GATA3 in the differentiation of TH2 cells in vitro and in vivo.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhu J, et al. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 96.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 97.Shimoda K, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 98.Takeda K, et al. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 99.Amsen D, et al. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27:89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fang TC, et al. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27:100–110. doi: 10.1016/j.immuni.2007.04.018. [References 99 and 100 show that Notch signals directly induce the transcription of GATA3 from exon 1a and that in the presence of GATA3, Notch signals can promote TH2-cell differentiation in a STAT6-independent manner.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Asnagli H, Afkarian M, Murphy KM. Cutting edge: Identification of an alternative GATA-3 promoter directing tissue-specific gene expression in mouse and human. J Immunol. 2002;168:4268–4271. doi: 10.4049/jimmunol.168.9.4268. [DOI] [PubMed] [Google Scholar]
- 102.Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 2003;19:739–748. doi: 10.1016/s1074-7613(03)00292-9. [This is the first report showing that IL-2R signalling through STAT5 can promote TH2-cell differentiation in a STAT6-independent manner.] [DOI] [PubMed] [Google Scholar]
- 103.Cote-Sierra J, et al. Interleukin 2 plays a central role in Th2 differentiation. Proc Natl Acad Sci USA. 2004;101:3880–3885. doi: 10.1073/pnas.0400339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hwang ES, White IA, Ho IC. An IL-4-independent and CD25-mediated function of c-maf in promoting the production of Th2 cytokines. Proc Natl Acad Sci USA. 2002;99:13026–13030. doi: 10.1073/pnas.202474499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ranganath S, Murphy K. Structure and specificity of GATA proteins in Th2 development. Mol Cell Biol. 2001;21:2716–2725. doi: 10.1128/MCB.21.8.2716-2725.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Takemoto N, Arai K, Miyatake S. Cutting edge: the differential involvement of the N-finger of GATA-3 in chromatin remodeling and transactivation during Th2 development. J Immunol. 2002;169:4103–4107. doi: 10.4049/jimmunol.169.8.4103. [DOI] [PubMed] [Google Scholar]
- 107.Shinnakasu R, et al. Critical YxKxHxxxRP motif in the C-terminal region of GATA3 for its DNA binding and function. J Immunol. 2006;177:5801–5810. doi: 10.4049/jimmunol.177.9.5801. [DOI] [PubMed] [Google Scholar]
- 108.Smith VM, Lee PP, Szychowski S, Winoto A. Gata-3 Dominant Negative Mutant - Functional Redundancy Of the T Cell Receptor Alpha and Beta Enhancers. Journal of Biological Chemistry. 1995;270:1515–1520. doi: 10.1074/jbc.270.4.1515. [DOI] [PubMed] [Google Scholar]
- 109.Zhang D-H, et al. Inhibition of allergic inflammation in a murine model of asthma by expression of a dominant-negative mutant of GATA-3. Immunity. 1999;11:473–482. doi: 10.1016/s1074-7613(00)80122-3. [DOI] [PubMed] [Google Scholar]
- 110.Zhao X, et al. Interaction between GATA-3 and the Transcriptional Coregulator Pias1 Is Important for the Regulation of Th2 Immune Responses. J Immunol. 2007;179:8297–8304. doi: 10.4049/jimmunol.179.12.8297. [DOI] [PubMed] [Google Scholar]
- 111.Bates DL, Chen Y, Kim G, Guo L, Chen L. Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA. J Mol Biol. 2008;381:1292–1306. doi: 10.1016/j.jmb.2008.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 113.Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. 2007;7:505–518. doi: 10.1038/nri2116. [DOI] [PubMed] [Google Scholar]
- 114.Wang ZY, et al. Regulation of Th2 cytokine expression in NKT cells: unconventional use of Stat6, GATA-3, and NFAT2. J Immunol. 2006;176:880–888. doi: 10.4049/jimmunol.176.2.880. [DOI] [PubMed] [Google Scholar]
- 115.Kim PJ, et al. GATA-3 regulates the development and function of invariant NKT cells. J Immunol. 2006;177:6650–6659. doi: 10.4049/jimmunol.177.10.6650. [DOI] [PubMed] [Google Scholar]
- 116.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 117.Osborne BA, Minter LM. Notch signalling during peripheral T-cell activation and differentiation. Nat Rev Immunol. 2007;7:64–75. doi: 10.1038/nri1998. [DOI] [PubMed] [Google Scholar]