Abstract
The study of gene expression evolution in vertebrates has hitherto focused on the analysis of transcriptomes in tissues of different species. However, because a tissue is made up of different cell types, and cell types differ with respect to their transcriptomes, the analysis of tissues offers a composite picture of transcriptome evolution. The isolation of individual cells from tissue sections opens up the opportunity to study gene expression evolution at the cell type level. We have stained neurons and endothelial cells in human brains by antibodies against cell type-specific marker proteins, isolated the cells using laser capture microdissection, and identified genes preferentially expressed in the two cell types. We analyze these two classes of genes with respect to their expression in 62 different human tissues, with respect to their expression in 44 human “postmortem” brains from different developmental stages and with respect to between-species brain expression differences. We find that genes preferentially expressed in neurons differ less across tissues and developmental stages than genes preferentially expressed in endothelial cells. We also observe less expression differences within primate species for neuronal transcriptomes. In stark contrast, we see more gene expression differences between humans, chimpanzees, and rhesus macaques relative to within-species differences in genes expressed preferentially in neurons than in genes expressed in endothelial cells. This suggests that neuronal and endothelial transcriptomes evolve at different rates within brain tissue.
Keywords: transcriptomics, neuron, evolution, brain, evo-devo, primate
DNA Sequence and Expression Evolution of Protein-Coding Genes Expressed in Brain
Generally, genes that are expressed in many tissues within an organism evolve under stronger selective constraints than genes that are specifically expressed in one tissue (Winter et al. 2004; Zhang and Li 2004; Liao and Zhang 2006). In primates, genes that are broadly expressed have lower evolutionary rates in protein-coding sequences (Khaitovich et al. 2005; Wang et al. 2007) as well as lower evolutionary rates of transcription factor binding sites (Gaffney et al. 2008) than genes that are expressed in a tissue-specific manner. These findings make intuitive sense if one imagines that a broadly expressed gene must meet the cellular needs in many different cell types—whereas a gene expressed in a single or few tissues has to meet the needs of just a single or few cell types.
In primates, gene expression in brain evolves more slowly than in other tissues (Khaitovich et al. 2005). Also, if a gene is expressed in several tissues including brain, it tends to evolve more slowly than if it is not expressed in brain (Khaitovich et al. 2005; Khaitovich, Enard, et al. 2006). This implicates that being expressed in brain represents an additional layer of constraints on transcriptome evolution (Khaitovich, Enard, et al. 2006). Recently it has been shown that a negative correlation between expression breadth and constraints exists also between brain regions (Tuller et al. 2008). Cortically expressed genes evolve under more constrains than subcortically expressed genes—both at the DNA sequence level as well as at the gene expression level in humans (Tuller et al. 2008). This is in agreement with the observation that more genes show differential expression between humans and chimpanzees in the cerebellum and the caudate nucleus than in cortical brain regions (Khaitovich et al. 2004). It has been pointed out that these findings seem to be somewhat counterintuitive because higher cognitive functions, which is a class of phenotypic traits that may set humans apart from nonhuman primates, are attributed to the cortex (Varki et al. 2008). However, it is not known whether all cell types within the primate cortex transcriptome evolve more slowly or if the observations represent an average of cell types with very different evolutionary rates. In this study, we ask if two cell types in the primate brain differ in rates of gene expression evolution.
Cell Type-Specific Expression Differences in Human Brain
We decided to study two cell types in the brain that differ drastically with regard to functions—neurons and endothelial cells. Neuronal cell bodies are found only in the nervous system, whereas endothelial cells as part of the circulatory system extend into all tissues of the body. We identified 1,000 pyramidal neurons and an equivalent area of endothelial cells by immunohistochemical staining of human “postmortem” brains (Fend et al. 1999) and isolated neurons and endothelial cells by laser capture microdissection (Emmert-Buck et al. 1996) from 6 (in the case of neurons) and 7 (in the case of endothelial cells) individuals, respectively. Examples of fluorescence immunostainings and pictures taken of brain sections during the laser capture microdissection process can be found in Harris et al. (2008).
RNA was isolated from these 13 samples, processed with protocol adjustments for low sample quantities (Harris et al. 2008), and hybridized onto microarrays (Schena et al. 1995) in order to assay transcriptome-wide RNA abundance levels with whole Human Genome U133 Plus 2.0 arrays (Affymetrix).
The transcriptomes of neurons and endothelial cells differ significantly as judged by comparisons with all possible (13!/(6! × 7!)) 1,716 sample-label permutations (Pperm = 0.0012). We identified 1,761 gene probe sets (termed “genes” below), which are more highly expressed in neurons (Pt-test ≤ 0.05; blue circles in fig. 1) and 1,315 genes, which are more highly expressed in endothelial cells (red circles in fig. 1). When we determined the cellular components associated with these genes using the gene ontology (GO) (Ashburner et al. 2000), the genes preferentially expressed in neurons (NEX) are enriched in GO groups that are “specific” for neurons, such as the groups “axons,” “synaptic vesicle membrane,” and “post synaptic membrane” (table 1). Similarly, the genes preferentially expressed in endothelial (ENDEX) cells are enriched in GO groups relevant to endothelial cell features such as “basement membrane” and “apical junction complex” (Stamatovic et al. 2008). The identified NEX and ENDEX genes and the P values from the gene by gene analysis are listed in supplementary table 1 (Supplementary Material online).
FIG. 1.—
Volcano plot of the significance level of the expression difference between neurons and endothelia cells versus the mean difference in gene expression levels. Each dot represents one gene probe set. The y axis shows negative log10 transformed P values obtained from gene by gene two-sided t-tests (e.g., 4 is equal to a P value of 10−4). Blue circles indicate the probe sets with enriched expression in neurons and red circles indicate enriched expression in endothelial cells. A gene is called enriched expressed, if it shows a statistically significant expression difference between the two cell types (P ≤ 0.05).
Table 1.
Overrepresentation of Genes That Are Enriched Expressed Genes in Neuronal (NEX) and Endothelial Cells (ENDEX) in GO Categories of the Core Taxonomy Cellular Component
| GO Identifier | GO Description | N det | N enr | Fold e |
| Enriched GO categories for NEX genes | ||||
| GO:0005834 | Trans-Golgi network | 13 | 9 | 4.2 |
| GO:0030672 | Heterotrimeric G-protein complex | 12 | 7 | 3.5 |
| GO:0033178 | Microtubule | 18 | 8 | 2.7 |
| GO:0005802 | Integral to membrane | 35 | 14 | 2.4 |
| GO:0045211 | Cell junction | 48 | 17 | 2.1 |
| GO:0030424 | Axon | 83 | 26 | 1.9 |
| GO:0034702 | Synaptic vesicle membrane | 73 | 21 | 1.7 |
| GO:0005874 | Organelle membrane | 122 | 33 | 1.6 |
| GO:0030054 | Proton-transporting two-sector ATPase complex, catalytic domain | 178 | 43 | 1.5 |
| GO:0031090 | Ion channel complex | 387 | 87 | 1.4 |
| GO:0016021 | Postsynaptic membrane | 1,401 | 271 | 1.2 |
| Enriched GO categories for ENDEX genes | ||||
| GO:0005604 | Basement membrane | 13 | 7 | 5.1 |
| GO:0042612 | Major histocompatibility complex class I protein complex | 17 | 9 | 5 |
| GO:0022626 | Cytosolic ribosome | 75 | 25 | 3.2 |
| GO:0043235 | Receptor complex | 28 | 9 | 3.1 |
| GO:0022627 | Cytosolic small ribosomal subunit | 39 | 12 | 2.9 |
| GO:0043296 | Apical junction complex | 43 | 12 | 2.7 |
| GO:0005887 | Integral to plasma membrane | 308 | 49 | 1.5 |
NOTE.—Reported are the significant outcomes from hypergeometric tests after P value correction for multiple hypothesis testing. N det, number of gene with detectable expression in the GO category; N enr, number of enriched expressed genes —either in neurons (top part of the table) or endothelial cells (bottom part); Fold e, the fold enrichment observed (relative to expected number of genes).
One key difference of the protocol described in the Materials and Methods section that we used to generate gene expression profiles from low amounts of RNA is that we performed three consecutive linear amplification steps, whereas the standard Affymetrix protocol involves only one linear amplification step. We therefore wanted to explore the biases that are introduced through the two additional linear amplification steps. More specifically, we wanted to evaluate how reproducible expression profiles generated with three rounds of linear amplification are and how they correlate with profiles generated by the standard Affymetrix protocol. In order to address these questions, we analyzed from one mouse individual liver, cortex, and cerebellum RNA with the standard Affymetrix protocol—which involves one linear amplification—and compared this to three technical replicates from each of these tissues of diluted RNA which were amplified three times, resulting in a total of 12 microarray experiments (analyzed on MG_U74Av2). For the nine experiments that involved three rounds of amplifications, input RNA amount was set to 1 ng—this is one order of magnitude lower than what we would expect to isolate from 1,000 cells given the estimates that are reported for other mammalian cells (Copois et al. 2003) and thus rather conservative. In contrast, for the three experiments that were processed with only one linear amplification step, 5 μg (5,000 times more) of input RNA were used.
Correlations of expression profiles among technical replicates of three times amplified samples range between 0.977 and 0.994 and thus is in the range of what was previously reported between technical replicates of samples processed with only one linear amplification (Zakharkin et al. 2005). Dot plots of samples that were amplified once versus samples that were amplified three times are depicted in supplementary figure 1 (Supplementary Material online). The correlation between once amplified and three times amplified samples is lower (ranging between 0.893 and 0.928) than the correlation between technical replicates. This indicates that the three rounds of amplification reveal reproducible results but introduce a systematic bias when compared with experiments that involve only one round of linear amplification. One influencing factor that we could identify is the increasing 3-prime bias of signal measurements (see supplementary fig. 1, Supplementary Material online), which was also described by others (Luzzi et al. 2003; Cope et al. 2006). This bias is thought to be the result of incomplete processivity of the polymerase during cDNA synthesis. The cDNA synthesis employed here starts at the poly-a tail of mRNA molecules by elongating an oligo-dT primer. The polymerase stochastically dissociates from the template strand before the 3-prime end of the mRNA molecule is reached. Thus, because three rounds of linear amplification involve three rounds of cDNA synthesis, we find this effect to be much more pronounced in samples that were three times amplified than in samples that went only through one cycle of amplification (supplementary fig. 2, Supplementary Material online). This bias implicates that probes that target regions that are further away from the 3-prime end of transcripts are less informative for the calculation of transcript abundance levels. Thus, less information from different probes within a probe set is useable in highly amplified samples and this most likely reduces the power to detect differences between highly amplified samples—in comparison with samples that were only amplified once.
Expression Breadth and Constraints on Expression of NEX and ENDEX Genes
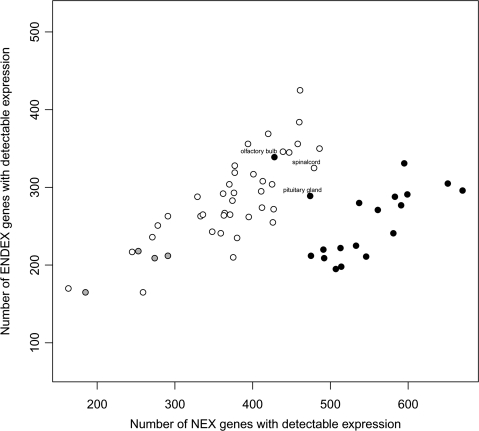
We next inferred to what extent NEX and ENDEX genes show expression in tissues other than brain. Figure 2 shows the numbers NEX and ENDEX genes with detectable expression in data collected from 62 human tissues (Su et al. 2004). Black-filled circles denote brain tissues, that is, amygdala, cerebellum peduncles, cingulate cortex, hypothalamus, medulla oblongata, occipital lobe, parietal lobe, pons, prefrontal cortex, temporal lobe, thalamus, whole brain, caudate nucleus, cerebellum, globus pallidus, subthalamic nucleus, and fetal brain. In these tissues, a higher proportion of NEX is expressed relative to ENDEX genes, when compared with all other tissues analyzed. Two brain structures, the olfactory bulb and the pituitary gland, do not group with the other brain tissues. Also the spinal cord sample, which is a part of the central nervous system, does not group with the bulk of the brain tissues. Four tissues that contained ganglia from the peripheral nervous system (the trigeminal, ciliary, superior cervical, and the dorsal root ganglion; gray filled circles in fig. 2) show no excess of NEX gene expression. This analysis shows that NEX genes tend to be specifically expressed in the brain—whereas ENDEX genes do not show brain specificity.
FIG. 2.—
Number of neuronal (x axis) and endothelial (y axis) enriched expressed genes with detectable expression in 62 tissues. A gene is called expressed in a tissue, if it showed detectable expression in at least 1 of the 2 biological replicates of a tissue. Black-filled circles indicate brain tissues—with the exception of “olfactory bulb” and “pituitary gland,” which are labeled on the plot. Gray-filled circles indicate ganglia.
In order to know whether the level of constraints on gene expression levels differ between NEX and ENDEX genes, we estimated expression diversity between two biological replicates from the 62 different human tissues (see supplementary fig. 3, Supplementary Material online). For 42 out of the 62 tissues, diversity in ENDEX genes is higher than for NEX genes (Pbinomial < 0.0072). This indicates that NEX genes are more constrained in their expression than ENDEX genes.
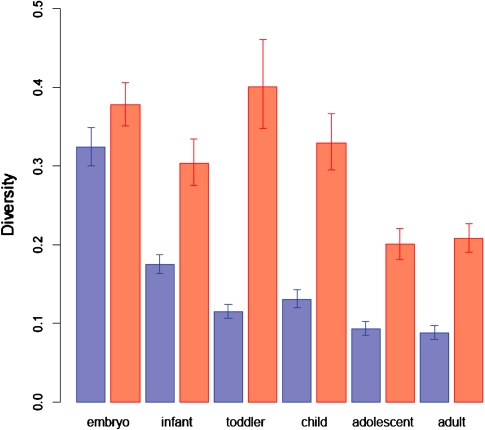
We then asked whether expression constraint, as measured by diversity, changes across lifespan, and whether constraints on expression levels of NEX genes is higher than on ENDEX genes throughout lifespan. Reanalysis of human cortex transcriptome profiles from six developmental time points—embryo, infant, toddler, child, adolescent, and adult (Johnson et al. 2009; Somel et al. 2009) suggests that expression diversity of these genes tend to increase with age and that ENDEX genes have higher levels of diversity and thus are less constrained than NEX genes across all developmental stages (fig. 3).
FIG. 3.—
Diversity estimates for NEX (blue) and ENDEX (red) genes in 4 embryos, 13 infant, 3 toddler, 5 child, 7 adolescent, and 11 adult human postmortem brains. The error bars indicate 95% CIs as determined by 10,000 bootstraps over genes. The associated bootstrap derived P values of the differences in diversity between NEX and ENDEX genes for different developmental stages are: Pembryo = 0.0012; Pinfant < 10−4; Ptoddler < 10−4; Pchild < 10−4; Padolescent < 10−4; Padult < 10−4.
Rates of Neuronal and Endothelial Transcriptome Evolution in Primate Brains
We then analyzed whether neuronal and endothelial transcriptomes might differ in their rates of evolution across species. For this, we used two data sets (Khaitovich et al. 2005; Khaitovich, Tang, et al. 2006) which consist of measurements from 6 humans and 5 chimpanzees for Brodmann Area 9 (BA9) and of 10 humans, 6 chimpanzees, and 6 rhesus macaques for Brodmann Area 46 (BA46), respectively. Both BA9 and BA46 are dorsolateral prefrontal cortex regions.
In all within-species comparisons, the transcriptome diversity in NEX genes is significantly lower than in ENDEX genes (all P values < 10−4), showing that neuronal transcriptomes are more constrained than endothelial transcriptomes also in chimpanzees and rhesus macaques.
In contrast, the divergence between species does not significantly differ between NEX and ENDEX genes in the BA46 data set, although the human–chimpanzee difference is of borderline significance (P human–chimpanzee BA46 = 0.06; P human–rhesus BA46 = 0.63; P chimpanzee–rhesus BA46 = 0.84). In the BA9 data set, transcriptome divergence for NEX genes is significantly lower (P human–chimpanzee BA9 = 0.02). The point estimates and 95% bootstrap derived confidence intervals (CIs) for divergence and diversity are reported in supplementary table 2 (Supplementary Material online).
We next compared evolutionary rates by analyzing the ratios of divergence relative with diversity (D) of NEX and ENDEX genes. Endothelial D in BA9 is 1.59, whereas D for neurons is 3.02 and thus higher than for the endothelium (Pbootstrap < 10−5; fig. 4). Endothelial D calculated for BA46 between humans and chimpanzees is 2.06, whereas neuronal D is 2.75 and thus larger than endothelial D (Pbootstrap = 0.0135; fig. 4). When neuronal D is compared with endothelial D for the human–macaque comparisons and for the chimpanzee–macaque comparisons, neuronal D is found to be 1.9 times larger (6.3 vs. 3.35) than endothelial D for the human comparison (Pbootstrap < 10−5) and 1.5 times larger (3.71 vs. 2.48) for the chimpanzee comparison (Pbootstrap = 0.0007), respectively (fig. 4). Thus, both these data sets suggest that D is higher in neuronal transcriptomes than in endothelial transcriptomes among primates.
FIG. 4.—
Pairwise comparisons of transcriptome divergence to diversity estimates (D) for different primate species in the BA9 and BA46 of the prefrontal cortex for NEX and ENDEX genes. The error bars indicate the 95% CIs of the D estimate as determined by 10,000 bootstrap replicates over genes and individuals. H: human; C: chimpanzee; R: rhesus macaque.
To analyze if D for NEX and ENDEX genes differ significantly from D estimates for other transcripts expressed in the brain, we randomly assigned genes expressed in BA9 and BA46 data sets to 10,000 groups of the same size as the NEX and ENDEX genes. The D values estimated for NEX and ENDEX genes in the two frontal cortex data sets were then compared with the distributions of D in the 10,000 random groups of genes. In all four comparisons was the observed neuronal D significantly higher than D calculated for the random gene sets (fig. 5). In fact, for BA46, none of the 10,000 random groups of genes had as high a D as the neuronal D found for the human–chimpanzee and the human–macaque comparisons. By contrast, the observed endothelial D was significantly lower than for the random genes in three out of four cases. Thus, D measured for neuronal and endothelial transcriptomes are located at two extremes of the distribution of possible D values—with the neuronal D being exceptionally high and the endothelial D exceptionally low.
FIG. 5.—
Diversity (x axis) and divergence values (y axis) for bootstrap replicates of genes expressed in BA46 and BA9. Each small blue dot indicates an outcome of bootstrapping the same number of genes as there are in the list of NEX genes; each small red dot indicates outcomes of bootstrapping the same number of genes as there are in the list of ENDEX genes. The big blue filled circles indicate the actual observation for NEX genes and the red filled circles indicate the actual observation for ENDEX genes. The P values indicate the location of the observed diversity to divergence values in the respective empirical distribution (two-sided test).
As mentioned above in the text, the differences in D values between NEX and ENDEX genes are mainly driven by differences in diversities and not by differences in divergence (fig. 5; supplementary table 2, Supplementary Material online). It remains to be elucidated why expression of NEX genes show excess divergence given the low levels of diversity. One possible explanation would be that NEX genes are more often regulated by the same transcription factors than ENDEX genes. This would allow the NEX genes to evolve in a more concerted manner (in expression) than ENDEX genes, and expression diversity would remain low—even if divergence between species increases.
Other explanations for these patterns could be that the magnitude of environmental influences on gene expression in neurons is lower than on other cells—and the magnitude of environmental influence on endothelial cells higher than on other cells in the brain. This is conceivable considering that endothelial cells are a part of the blood brain barrier and thus have a function of controlling the influx of molecules into the brain and therefore might have to react more often to environmental changes. This, however, would not explain why neuron-related genes are more constrained in their expression in nonbrain tissues or why they show more constraints in their amino acid sequences.
The study presented with this paper points at the fact that different transcriptomes of different cell types within a tissue can show different evolutionary patterns. Thus, there is a potential in better understanding transcriptome evolution if transcriptomes of more closely defined cell populations are analyzed. For primate tissue transcriptome comparisons (other than blood), laser capture microdissection provides currently the only technological solution for isolating specific cell populations from complex tissues. This is because the tissue material is collected several hours after the death of donors, which compromises tissue integrity and thus other technologies for isolating specific cell types out of a complex mixture, such as fluorescence-activated cell sorting (Arlotta et al. 2005), can therefore not be applied to such samples. Future work will aim at measuring and comparing different cell types isolated from human and chimpanzee brains and other tissues.
Materials and Methods
Samples, Laser-Assisted Microdissection and Microarrays
Postmortem human brain tissue was donated by The Stanley Medical Research Institute’s brain collection courtesy of Drs Michael B. Knable, E. Fuller Torrey, Maree J. Webster, and Robert H. Yolken. All individuals with the exception of one individual were defined as normal controls by medical examiners, with no structural brain pathology, history of focal neurological signs, or other CNS disorders, substance or alcohol abuse, or subnormal IQ. Individual one had a medical history of schizophrenia. In order to counteract a potential outlier effect that this sample could have for the identification of NEX and ENDEX genes, we collected both neuronal and endothelial cells from this individual. All individuals suffered sudden death, without brain injury. Sample properties are listed in supplementary table 3 (Supplementary Material online).
Endothelial cells were collected from seven humans and neuronal cells from six humans that partially overlap (supplementary table 3, Supplementary Material online). Cryosections (15-μm thick) (Bright Technologies) of dorsolateral prefrontal cortex were cut, mounted onto polyethylene naphthalate membrane slides (Zeiss), air dried, and fixed for 10 min in acetone. After air drying, the sections were incubated either with a polyclonal antibody to detect neurons (Neurofilament 160/200 kDa mouse; Cambridge Bioscience 13-1300) or endothelial cells (von Willebrand factor rabbit; Chemicon AB7356) for 5 min, followed by brief washing in RNase-free phosphate-buffered saline (PBS). After that, the slides were incubated with a secondary fluorescently labeled antibody for 5 min. These antibodies were Cy2-conjugated goat anti-mouse (Jackson Immunoresearch) for the neuron and Cy3-conjugated goat anti-rabbit (Jackson Immunoresearch) for the endothelial staining. All antibodies that were used in this study were polyclonal and used at 1:20 dilution with 1 unit/ml RNase inhibitor (GE Healthcare) in RNase-free PBS (Ambion). The described incubation conditions were found to give the best staining for cell identification together with optimal RNA preservation. Following antibody incubation and brief washing in PBS, sections were then dehydrated by incubation in increasing ethanol concentrations (20 s in each 30%, 50%, 70%, 90%, and absolute ethanol).
The tissue sections were subjected to laser capture microdissection immediately after staining. Laser capture microdissection was carried out using the PALM microlaser system (www.palm-microlaser.com). Pyramidal neurons were selected based on staining and morphology. Thousand neurons were captured from each subject, in two batches of 500. For the endothelial cells, we collected approximately 400,000 μm2 of vascular endothelium, which we determined as being the equivalent area of 1,000 pyramidal neurons.
Following capture, RNA was extracted from cells using the PALM RNA extraction kit (Zeiss) and amplified through two rounds using the RiboAmp HS kit (Arcturus). The resulting antisense RNA (aRNA) was assessed on an Agilent Bioanalyser Nanochip to determine length of RNA transcripts in the samples. aRNA profiles with jagged curves or pronounced skews to the left, indicating degradation of the RNA, were eliminated from the analysis. Amplified RNA was converted to cDNA using Round 2 components of the RiboAmp HS kit, labeled by in vitro transcription in the presence of biotinylated UTP (Codelink Expression Assay kit, GE Healthcare), and purified using YM-30 columns (Microcon). The quality of the amplified RNA was again assessed with a Bioanalyzer Nanochip (Agilent). Amplified cRNA samples were chemically fragmented—according to instructions in the Affymetrix sample preparation protocol —and hybridized onto whole Human Genome U133 Plus 2.0 arrays (Affymetrix). Thus, the molecules that were hybridized onto microarrays underwent three consecutive linear amplification steps. This is the number of linear amplification steps that allowed us to generate the required amount of cRNA (18 μg) for hybridization on Affymetrix microarrays.
In order to assess the impact of three rounds of linear amplification on the technical reproducibility of gene expression measurements, we performed 12 additional microarray experiments. We analyzed mouse RNA from cortex, cerebellum, and liver from one individual which served as an experimental control in a different study conducted in our laboratory (Somel et al. 2008) and thus was kept and sacrificed for other reasons than the analysis presented here. RNA was isolated using TRIzol reagent from the frozen mouse tissue according to manufacturer’s instructions. After isolation, the RNA was column purified with the Qiagen RNeasy kit.
One RNA sample of 5 μg from each of the three different tissues was processed according to the standard Affymetrix protocol and hybridized onto mouse gene expression arrays MG_U74Av2. Three additional samples from each tissue of 1 ng were processed according to the procedure that involves three linear amplifications (described above in the text) before hybridization onto the microarray chips.
All primary expression data are publicly available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE12293.
Comparisons of Expression Profiles of 1× Amplified Samples with 3× Amplified Samples
Affymetrix microarray image data were analyzed with the GeneChip operating software. Gene probe set expression values were calculated using the Robust Multichip Average for small RNA quantities (srma) algorithm (Cope et al. 2006). As probe weights for signal summary statistic calculation, we used the inverse of the probe-specific coefficient of variation calculated across chips.
Probe sets were defined as expressed if the P value determined by a signed-rank test of perfect match versus mismatch signal distributions was lower or equal 0.05 in at least three chip experiments in a tissue. This resulted in a list of 5,279 probe sets that were analyzed in liver, 5,629 in cortex, and 6,099 in cerebellum. Pairwise Pearson correlations were calculated in the statistical programming language R (R Development Core Team 2009).
Identification of NEX and ENDEX Genes
Low-level data treatment was done in the same way as described in the previous section. Probe sets were defined as expressed if the P value determined by a signed-rank test of perfect match versus mismatch signal distributions was lower or equal 0.065 in at least three chip experiments in a given cell type. Probe sets that showed statistically significant higher expression in neurons or endothelial cells in two-sided t-tests were considered as preferentially neuronal or endothelial expressed genes, respectively (see fig. 1).
For the GO analysis, we matched probe sets to GO identifier using the information available in “biomart” (www.ensemble.org) version 36 for human genes (GRCh37). We have used the information of GO term relationship as provided by the GO consortium (www.geneontology.org) on November 8, 2009. With the program “Func_hyper” as implemented in the software package Func (Prufer et al. 2007), we looked for overrepresentation of NEX and ENDEX genes in GO categories. We restricted the analysis to the core taxonomy “cellular component” (GO:0005575). P values were obtained by performing a permutation test (1,000 permutations over genes). We only considered GO groups in which more than ten genes in our analysis were annotated. In table 1, we report the GO groups that were significantly overrepresented after adjusting for multiple hypothesis testing by controlling the false discovery rate at a 5% cutoff.
Expression Analysis of 62 Human Tissues and Developmental Series
We downloaded Affymetrix microarray data for 62 human tissues (Su et al. 2004). This data set consists of two biological replicates for each one of the tissues. We considered a probe set as expressed in a tissue, if its perfect match versus mismatch signal distributions was lower or equal 0.05 in at least one of the two chip experiments. We then matched these probe sets with ENDEX and NEX probe set ids (by matching the probe set ids from both array platforms to Ensembl genes). Diversity for NEX and ENDEX genes in the respective tissues was calculated as the average squared difference in expression between the two biological replicates.
Following the same logic, we also analyzed previously published gene expression data from four prefrontal human midfetal brains (Johnson et al. 2009) and 39 postmortem cortex tissues of humans covering an age range of 0.1 to 47.4 years (Somel et al. 2009). The data were grouped according to the developmental stages embryo (midgestation, four individuals, left and right prefrontal cortex each), infant (newborns less than 1 year old; 13 individuals), toddler (between 1 and 4 years of age; three individuals), child (between 4 and 10 years old; five individuals), adolescent (between 10 and 18 years old; seven individuals), and adult (older than 18 years; 11 individuals). Ninety-five percent CIs for diversity estimates were determined by bootstrapping over genes 10,000 times, using the 2.5% and 97.5% quantiles of the bootstrap distribution as lower and upper bounds.
Expression Analysis of Cortical Regions
Two data sets containing primate expression measures of the dorsolateral prefrontal cortex (BA9 and BA46) using U133 Plus 2.0 arrays (Affymetrix) were analyzed. The BA9 data set (Khaitovich et al. 2005) consists of 6 human and 5 chimpanzee individuals, whereas the BA46 data set (Khaitovich, Tang, et al. 2006) consists of 10 human, 6 chimpanzee, and 6 rhesus macaque individuals. Five chimpanzees were analyzed in both data sets.
In order to investigate only gene probes that match the human and the chimpanzee genome equally well, oligonucleotide probes for which DNA sequence differences exist between the human genome (build 36) and the chimpanzee genome (build 2) were removed from all the analyses. Because there are also rhesus macaque individuals present in the BA46 data set, we additionally excluded in this data set all probes that did not match the rhesus genome (build 2).
Data normalization and generation of summarization signal for each probe set using the Robust Multichip Average (Irizarry et al. 2003) were done for each data set independently. Expressed probe sets were defined as those where the P values determined by a signed-rank test of perfect match versus mismatch signal distributions was ≤0.065 in at least three individuals in either species. This resulted in a list of measurements from 23,708 probe sets for the BA9 and 16,021 for the BA46 data set that were subjected for further analysis.
We calculated D, the average squared differences in expression between species divided by the average squared differences in expression within species for each data set and cell type. Ninety-five percent CIs for these estimates were determined by bootstrapping over individuals and genes (for divergence) and over genes (for diversity) 10,000 times, using the 2.5% and 97.5% quantiles of the bootstrap distribution as lower and upper bounds.
Data Deposition
The microarray data set published with this publication is deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) database with the accession number GSE12293.
Supplementary Material
Supplementary tables 1–3 and figures 1–3 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
We thank Henriette Franz, Benjamin Krinsky, Birgit Nickel, Anastasiya Masharina, and Sabrina Schaefer for assistance in the laboratory and Christine Green, Susan Ptak, and Jeffrey Good for helpful comments on the manuscript. This study was supported by grant PKB 140404 from the European Union. Research was done at Max Planck Institute for Evolutionary Anthropology in Leipzig and at the Institute of Biotechnology at the University of Cambridge.
References
- Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope L, Hartman SM, Gohlmann HW, Tiesman JP, Irizarry RA. Analysis of Affymetrix GeneChip data using amplified RNA. Biotechniques. 2006;40:165–166. doi: 10.2144/000112057. 168, 170. [DOI] [PubMed] [Google Scholar]
- Copois V, et al. Assessment of RNA quality extracted from laser-captured tissues using miniaturized capillary electrophoresis. Lab Invest. 2003;83:599–602. doi: 10.1097/01.lab.0000062853.92828.20. [DOI] [PubMed] [Google Scholar]
- Emmert-Buck MR, et al. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- Fend F, et al. Immuno-LCM: laser capture microdissection of immunostained frozen sections for mRNA analysis. Am J Pathol. 1999;154:61–66. doi: 10.1016/S0002-9440(10)65251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney DJ, Blekhman R, Majewski J. Selective constraints in experimentally defined primate regulatory regions. PLoS Genet. 2008;4:e1000157. doi: 10.1371/journal.pgen.1000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LW, Wayland M, Lan M, et al. The cerebral microvasculature in schizophrenia: a laser capture microdissection study. PLoS One. 2008;3:e3964. doi: 10.1371/journal.pone.0003964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Johnson MB, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Enard W, Lachmann M, Paabo S. Evolution of primate gene expression. Nat Rev Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, et al. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, et al. Positive selection on gene expression in the human brain. Curr Biol. 2006;16:R356–R358. doi: 10.1016/j.cub.2006.03.082. [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during mammalian evolution. Mol Biol Evol. 2006;23:1119–1128. doi: 10.1093/molbev/msj119. [DOI] [PubMed] [Google Scholar]
- Luzzi V, Mahadevappa M, Raja R, Warrington JA, Watson MA. Accurate and reproducible gene expression profiles from laser capture microdissection, transcript amplification, and high density oligonucleotide microarray analysis. J Mol Diagn. 2003;5:9–14. doi: 10.1016/S1525-1578(10)60445-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K, et al. FUNC: a package for detecting significant associations between gene sets and ontological annotations. BMC Bioinformatics. 2007;8:41. doi: 10.1186/1471-2105-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2009. [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Somel M, et al. Human and chimpanzee gene expression differences replicated in mice fed different diets. PLoS One. 2008;3:e1504. doi: 10.1371/journal.pone.0001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, et al. Transcriptional neoteny in the human brain. Proc Natl Acad Sci U S A. 2009;106:5743–5748. doi: 10.1073/pnas.0900544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatovic SM, Keep RF, Andjelkovic AV. Brain endothelial cell-cell junctions: how to “open” the blood brain barrier. Curr Neuropharmacol. 2008;6:179–192. doi: 10.2174/157015908785777210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Kupiec M, Ruppin E. Evolutionary rate and gene expression across different brain regions. Genome Biol. 2008;9:R142. doi: 10.1186/gb-2008-9-9-r142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Geschwind DH, Eichler EE. Explaining human uniqueness: genome interactions with environment, behaviour and culture. Nat Rev Genet. 2008;9:749–763. doi: 10.1038/nrg2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, et al. Rate of evolution in brain-expressed genes in humans and other primates. PLoS Biol. 2007;5:e13. doi: 10.1371/journal.pbio.0050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter EE, Goodstadt L, Ponting CP. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharkin SO, et al. Sources of variation in Affymetrix microarray experiments. BMC Bioinformatics. 2005;6:214. doi: 10.1186/1471-2105-6-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li WH. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol Biol Evol. 2004;21:236–239. doi: 10.1093/molbev/msh010. [DOI] [PubMed] [Google Scholar]