Abstract
Pheochromocytomas, catecholamine-secreting tumors of neural crest origin, are frequently hereditary1. However, the molecular basis for the majority of these tumors is unknown2. We identified the transmembrane-encoding TMEM127 gene, on chromosome 2q11, as a novel pheochromocytoma susceptibility gene. In a cohort of 103 samples, truncating germline TMEM127 mutations were detected in one-third of familial and about 3% of sporadic-appearing tumors without a known genetic cause. The wild-type allele was consistently deleted in tumor DNA, suggesting a two-hit mechanism of inactivation. Pheochromocytomas with TMEM127 mutations are transcriptionally related to NF1-mutant tumors and, similarly, show hyperphosphorylation of mTOR targets. Accordingly, in vitro gain- and loss-of-function analyses indicate that TMEM127 is a negative regulator of mTOR. TMEM127 dynamically associates with the endomembrane system and colocalizes with perinuclear (activated) mTOR, suggesting a subcompartmental-specific effect. Our studies unveil TMEM127 as a novel tumor suppressor gene and validate the power of hereditary tumors for elucidating cancer pathogenesis.
Using an integrative genomics strategy that involved linkage analysis, global expression profiling and high-density copy number mapping, we previously defined a novel pheochromocytoma susceptibility locus to chromosome 2q11, which we named FP (Familial Pheochromocytoma)3. Identification of other affected individuals in the original kindred established a dominant model of transmission (Suppl. Fig. 1) and additional linked families redefined the limits of the FP locus to a region containing 205 genes (NCBI Build 35.1, Suppl. Table 1) spanning 19.62Mb between 94,81Mbp and 114,43Mbp. Tumor DNA from one of the affected cases (index case of Family 1) was used as a template for exon-based sequencing, as described4. Using this approach, we identified a splice-site variant (IVS3-2A>C) in the provisional transmembrane-encoding gene, TMEM127, also known as FLJ20507 (Table 1, Suppl. Fig. 2A, 2B). This mutation was also present in this patient’s germline DNA as well as in each of six other affected siblings, but not in samples from two unaffected sibs.
Table 1.
Clinical and genetic features of patients and families with TMEM127 gene mutations
| ID | Family history |
Tumor location |
Age at onset (range within family) |
Number of affected individuals in family (total number of tumors) |
% Bilaterality of tumors* |
Nucleotide change (amino acid change) |
Mutation type |
Predicted protein length |
|---|---|---|---|---|---|---|---|---|
| 1 | Y | A | 34-54 | 7(10) | 42(3/7) | IVS3-2 A>C (p.Leu138fs) |
splice site |
148aa |
| 2 | U | A | 25 | 1(1) | 0 (0/1) | c.150insA (p.Pro51fs) |
insertion | 106aa |
| 3 | Y | A | 48-72 | 2(4) | 100 (2/2) | c.475C>T (p.Gln159X) |
nonsense | 158aa |
| 4 | N | A | 46 | 1(2) | 100 (1/1) | c.264- 267delCAGA (p.Thr89fs) |
deletion | 121aa |
| 5 | Y | A | 34-42 | 2 (2) | 0 (0/1) | IVS3-2 A>C (p.Leu138fs) |
splice site |
148aa |
| 6 | Y | A | 54-66 | 2(2) | 100 (2/2) | IVS2-1G>T (p.Phe83fs) |
splice site |
99aa |
| 7 | N | A | 32 | 1(1) | 0 (0/1) | c.268G>A (p.Val90Met) |
missense | 238aa |
number of cases with bilateral tumors/total number of affected cases in the family; Y= yes, N= no; U= unknown; A= adrenal
We next sequenced germline and/or tumor DNA from an additional 102 index pheochromocytomas, 19 of which had a clear familial history, and 83 were apparently sporadic cases. We detected six additional TMEM127 mutations in these samples (Table 1, Suppl. Fig. 2C). Six of the seven TMEM127 variants identified were splice-site or nonsense mutations predicted to interrupt prematurely the protein reading frame (Table 1, Suppl. Fig.2A and B). In all, four TMEM127 variants (including the index case) were detected among 12 families without mutations in other pheochromocytoma susceptibility genes (Suppl.Table 2, Suppl.Fig.2A and B). The remaining three mutations were identified among the ‘sporadic’ samples that were also wild-type for the known pheochromocytoma-related genes. One of the patients was adopted and thus heredity status was unknown (patient 2, Table1), and the other two individuals reported no family history of pheochromocytoma (patients 4 and 7, Table 1). In all cases, mutations were detected in the corresponding germline DNA and, when available, in DNA from other relatives with pheochromocytoma (Table 1). One of the identified mutations, IVS3-2A>C, was detected in two separate families (Table 1, families 1 and 5). Haplotype analysis of tumor DNA excluded a common ancestor (Suppl. Table 3). None of the sequence changes found in the pheochromocytoma samples were identified in a control group of 1064 ethnically-matched alleles, although novel and known polymorphic variants were detected (Suppl. Table 4).
Loss of heterozygosity (LOH) at the TMEM127 locus was examined in 35 pheochromocytomas: all TMEM127-mutant tumors tested (n=19, including multiple affected individuals from the same family or bilateral tumors) had LOH, in contrast with only 2 of 16 wild-type TMEM127 samples (P=5.17e-8, Suppl. Table 5). In each mutant TMEM127 pheochromocytoma, the wild-type allele was lost (Suppl.Fig.2A), in agreement with a two-hit model of tumor suppressor inactivation. The only missense mutation (V90M) in this series targeted an amino acid conserved amongst putative mammalian TMEM127 orthologs. As only germline DNA was available from this sample, LOH status could not be determined.
We next measured TMEM127 transcription in pheochromocytomas and found a 4-fold decrease in expression levels of TMEM127-mutant samples (n=7) compared to 16 non-mutated pheochromocytomas of various genetic backgrounds (P=0.002, Suppl.Fig. 2D). This degree of downregulation suggests instability of the mutant transcript, rather than low expression simply due to loss of one allele. RNA was unavailable from the two tumors that had LOH at 2q11 but no TMEM127 mutation.
Clinically, patients with TMEM127 mutations developed pheochromocytomas on average at 45.3 years of age, similar to the mean age at diagnosis of sporadic pheochromocytomas (43.6-43.9y1,5) but notably older than syndromic cases (24.9-30.2y1,5). All tumors arose from the adrenal medulla and were bilateral in approximately half of the patients (Table 1). No malignancies, defined by the presence of metastasis in nonchromaffin-derived tissues, or recurrences, were detected during follow-up of the affected cases (4 to 16 years). Four mutation carriers from Family 1 (Table 1) remain without clinical disease at 43 to 58 years of age, but a clear picture of the actual penetrance and phenotypic spectrum of TMEM127 mutations awaits larger studies. Taken together, the data above suggest that TMEM127 gene mutations are associated with pheochromocytoma predisposition and that TMEM127 has features of a classic tumor suppressor gene.
The TMEM127 gene is predicted to encode a protein with three transmembrane regions (Suppl. Fig. 2C) and no clearly recognizable functional domains. Its sequence is highly conserved throughout evolution and putative orthologs can be identified from mammals to fish (Suppl. Fig. 3A). The human gene is broadly expressed both in normal tissue (Suppl. Fig. 3B) and in a diverse panel of cancer cell lines6 (Suppl. Fig. 3C), with variable transcription levels that may reflect tissue of origin. Likewise, its putative mouse ortholog is widely expressed from an early postnatal age through adulthood (Suppl. Fig. 3D).
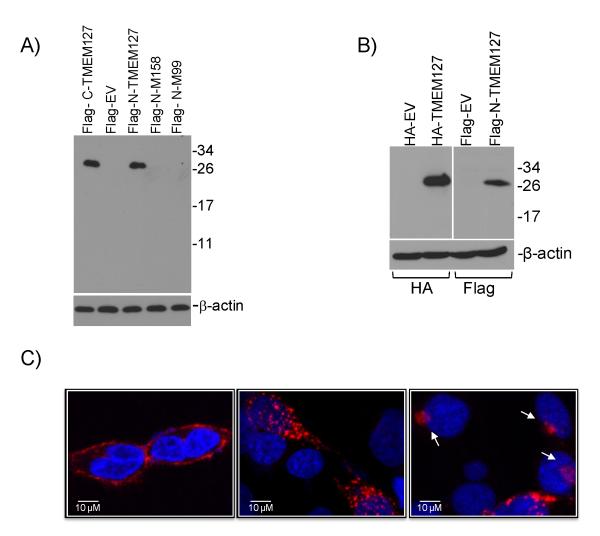
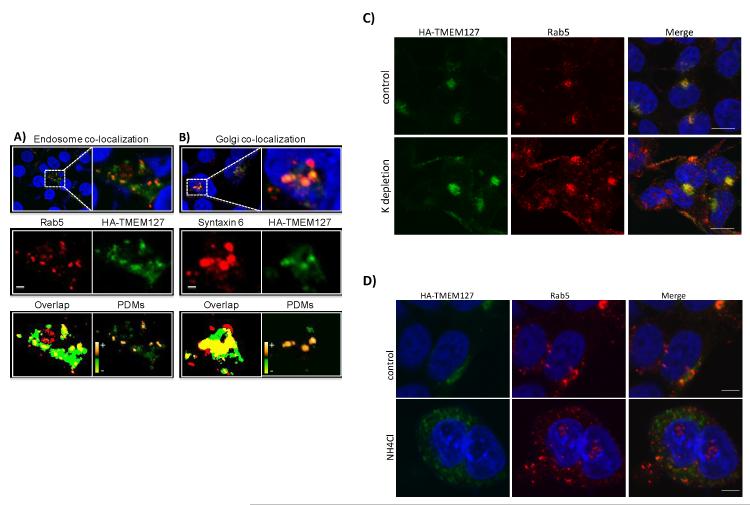
To begin to probe the functional consequences of TMEM127 mutations, we generated wild-type and mutant constructs tagged with Flag (either at the N- or C-terminus of the protein) or HA (N-terminus) epitopes. A single product of approximately 26KDa was recognized in HEK293 cells transfected with the wild-type constructs (Fig.1A, 1B). However, we were unable to detect a translation product from the mutant constructs in these cell lines (Fig.1A), despite equivalent transfection efficiencies (Suppl. Fig.4A). These data suggest that TMEM127-mutant proteins are not expressed, in support of a loss-of-function effect of these variants. Using confocal microscopy, we found that TMEM127 localizes to the plasma membrane and the cytoplasm, displaying both a punctate pattern and as perinuclear clusters (Fig.1C, Suppl. Fig. 4B). Colocalization studies showed that TMEM127 associates with a subpopulation of vesicular organelles corresponding to early endosomal structures labeled by Rab5 (Fig. 2A), the Golgi, labeled both with syntaxin 6 (Fig.2B) and N-acetylgalactosaminyltransferase-2 (Suppl. Fig.5A) and lysosomes (Suppl. Fig. 5B). We further found that association of TMEM127 with the plasma membrane is enhanced by inhibition of endocytosis7 (Fig.2C), while increasing pH in the culture media partially traps TMEM127 in endosomal structures7 (Fig.2D). These findings suggest that TMEM127 associates dynamically with endosomes and may participate in protein trafficking between the plasma membrane, the Golgi and lysosomes.
Fig.1. TMEM127 localizes to the plasma membrane and cytoplasm.
A) Western blot of HEK293 cells transfected with wild-type TMEM127 tagged with the Flag epitope at the C-terminus (C-Flag-TMEM127), N-terminus (N-Flag-TMEM127), and N-tagged TMEM127 mutants M158 (#3, Fig. 1C) and M99 (#6, Fig.1C) or empty MSCV retroviral vector (EV) and probed with a Flag antibody. A detectable product cannot be obtained from mutant constructs. β-actin was used as a loading control. B) Western blot of 293 cells transfected with TMEM127 tagged with HA at the N-terminus (HA-TMEM127) or the N-Flag-TMEM127 construct shown in (A), with respective empty vector controls, probed with an HA (left) or Flag (right) antibody. β-actin was used as a loading standard. C) Confocal microscopy of HEK293T cells expressing wild-type TMEM127 tagged with Flag on the N-terminus (left) or C-terminus (central) or with an HA N-terminal tag (right). TMEM127 immunoreactivity determined by Flag or HA (red) is present both at the plasma membrane (illustrated on the left panel) and cytoplasm, with punctate (middle and right panels) or perinuclear (right panel, arrows) signals, but is absent from the nucleus (DAPI, blue). These distinct staining patterns were observed with the various constructs in at least three independent experiments (Suppl. Fig. 4B). Plasma membrane-associated TMEM127 distribution was detected on average in 38% (±13%, n=200) of cells under regular culture conditions. A similar variance in the percentage of plasma membrane associated-signal was noted for each of the constructs.
Fig. 2. TMEM127 colocalizes with multiple components of the endomembrane system.
Confocal images of HA-TMEM127-transfected HEK293E cells fixed and immunofluorescently labeled with either the early endosome marker Rab5 antibody [red, (A)] or the Golgi marker syntaxin 6 [red, (B), HA antibody (green) and DAPI (blue)]. Regions identified by the white-dashed rectangles are presented at higher magnification in the right and lower panels. Middle panels: separate presentations of the red and green channel images. Lower panels: overlap (yellow) of red and green pixels, irrespective of their intensity. Right image panel presents intensity values of the product of the difference of the means (PDMs). Positive PDMs identify pixels where intensities in the red and green channels vary in synchrony. Perfectly synchronous signals would be indicated with a PDM value of 1. Completely asynchronous signals would have a value of −1. The PDM intensity scale ranges from the minimum to maximum values within each image panel, −0.5 to 0.5 in A and −0.6 to 0.6 in B. Optical sections are ~0.4 μm thick. Bars are 2 μm. These results were representative of multiple cells assessed in independent experiments. C) HEK293E cells stably transfected with HA-TMEM127 were left untreated (top) or were cultured in the presence of a potassium-depleting buffer (bottom), as described in Methods. Cells were fixed and stained with antibodies specific for HA (green) or, Rab5 (red) before imaging. DAPI (blue) represents nuclei. An increase in plasma-membrane associated TMEM127 and control Rab5 is seen after inhibition of endocytosis promoted by this treatment. Bars are 10μm. D) HEK293E cells stably transfected with HA-TMEM127 were left untreated (top) or were exposed to 50mM ammonium chloride (bottom) for 2 h. Cells were fixed and stained with antibodies specific for HA (green) or Rab5 (red) before imaging. DAPI (blue) represents nuclei. Expansion of endosomal structures is seen both for HA-TMEM127 and Rab5. Bars are 5μm.
We used microarray-based expression profiling to define the transcription signature of TMEM127-mutant tumors. Profiles from eight samples carrying three distinct TMEM127 mutations, as well as additional pheochromocytomas with mutations in other pheochromocytoma susceptibility genes (8 RET-, 4 NF1-, 8 VHL- and 15 SDHB- or SDHD-mutant tumors) were analyzed by unsupervised and supervised methods (Suppl. Table 6), as before8. In this sample set we confirmed the cluster association between the TMEM127-mutant tumors and pheochromocytomas with NF1 and RET mutations, suggested by our earlier study of FP-linked tumors 3. The gene set enrichment analysis (GSEA) algorithm9, used to identify pathways associated with the TMEM127-mutant tumor signature, revealed high statistical association with kinase receptor signals (Table 2, Suppl. Fig. 6A), which had been previously linked to NF1- and RET-mutants3,8. This pattern differed from the expression profiles of pheochromocytomas with mutations in VHL, SDHB or SDHD genes, which were uniquely enriched for transcripts involved in the response to hypoxia (Table 2, Suppl. Fig.6B)8.
Table 2.
TMEM127 mutant tumors are transcriptionally enriched for kinase signaling pathway.
| Gene Set (Pathway) | NOM p-val | FDR q-val |
|---|---|---|
| Tyrosine Kinase | ||
| GHa | 0 | 0.032 |
| PDGFb | 0.002 | 0.052 |
| Insulin | 0.006 | 0.068 |
| EGFc | 0.006 | 0.074 |
| IGF1d | 0.004 | 0.075 |
| Hypoxia | 0.96 | 0.97 |
growth hormone
platelet-derived growth factor
epidermal growth factor
insulin-derived growth factor
Summary of gene set enrichment analysis (GSEA) analysis performed on ranked genes according to the ratios of transcripts from TMEM127-mutant and TMEM127-wild-type pheochromocytomas. Gene sets (pathways) with a nominal P-value (NOM) <0.01 and a false discovery rate (FDR) of <0.1 were considered significant. Results of multiple kinase pathways are displayed and contrasted with a nonsignificant hypoxia gene set.
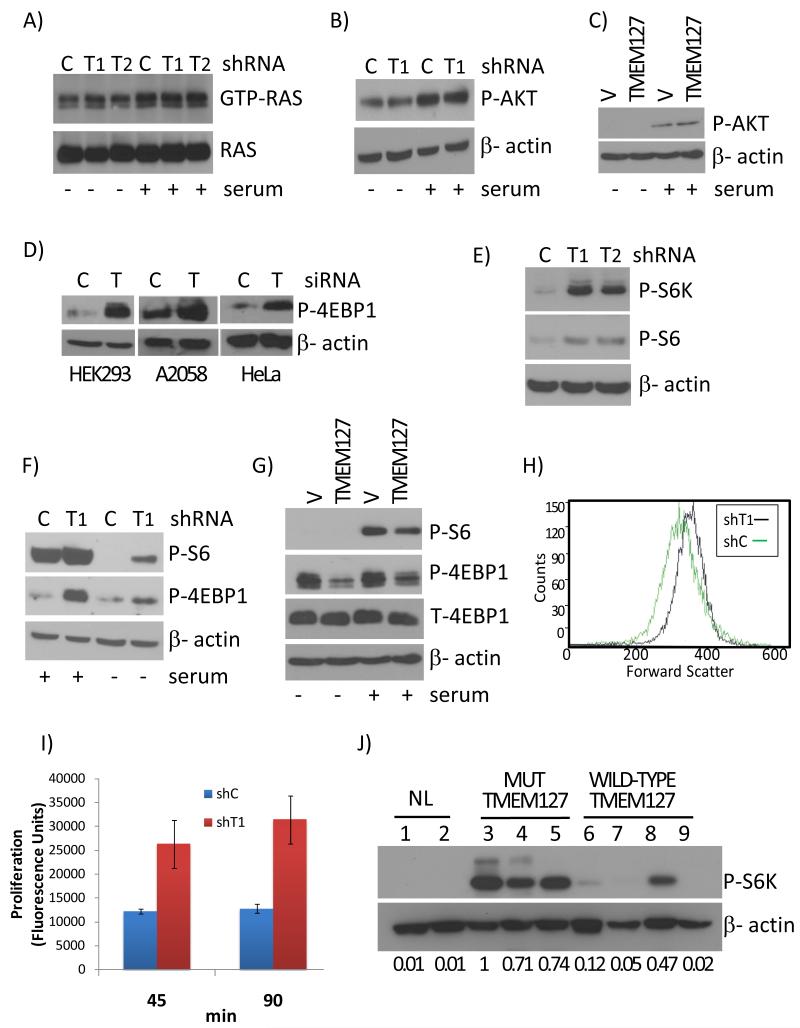
To further evaluate these findings, we knocked down TMEM127 expression by short interfering (si) or short hairpin (sh) RNA (Suppl. Fig. 6C). In agreement with the GSEA analysis, HIF1α and its targets were not induced by TMEM127 downregulation in HEK293 or Hela cells (Suppl. Fig. 6D). To explore the putative functional relationship with NF1-mutant tumors, we began by examining RAS activation, the hallmark of NF1 loss10. TMEM127 depletion by two distinct shRNA sequences did not increase RAS activity (Fig. 3A). Likewise, AKT phosphorylation, a surrogate for PI3K activity which is enhanced by both NF111 and RET12 mutations, was not increased after TMEM127 knockdown (Fig. 3B, Suppl. Fig. 7A). Deregulation of mTOR, a target of several kinase receptor pathways13, is also a feature of NF1 loss14. This fact, combined with the kinase receptor signature displayed by TMEM127-mutant tumors3, prompted us to investigate the effects of TMEM127 on mTOR signaling.
Fig.3. TMEM127 modulates mTORC1 signaling in vitro and in vivo.
A) RAS activation measured by a RAS pull down assay (RAS-GTP expression) of 293E cells with TMEM127 knockdown by two independent shRNA sequences( T1 and T2), or a control knockdown (C) in the absence or after 10 minutes of serum exposure. Total RAS is shown as sample loading control. Effectiveness of the serum treatment is shown in Suppl. Fig. 7A; B) AKT phosphorylation in TMEM127 (T1) or GFP (C) knockdown 293 cells in the presence or absence of serum; β-actin is a loading control; C) Phosphorylation of mTORC2 target AKT in 293E cells overexpressing HA-tagged TMEM127 (TMEM127) or an empty vector (V) in the presence or absence of serum. β-actin is a loading control; D) Effects of transient TMEM127 knockdown (T) on 4EBP1 phosphorylation in HEK293, A2058 and HeLa cells. C=GFP control knockdown. β-actin is a loading control; E) Phosphorylation of mTORC1 targets S6K and S6 in 293E cells depleted for TMEM127 by two independent shRNA sequences (T1 and T2); C=GFP control knockdown; F) Effects of TMEM127 knockdown (T1) on 4EBP1 and S6 phosphorylation in HEK293 in the presence or absence of 10% serum. C=GFP control knockdown; G) Phosphorylation of mTORC1 targets, as above, in 293E cells expressing HA-tagged TMEM127 (TMEM127) or a control HA-empty vector (V); T-4EBP1 = total 4EBP1. H) Forward scatter FACS analysis of TMEM127 (T1) or control (C) knockdown in HEK293 cells. Ungated results are displayed and are similar to profiles obtained at the G1 and G2 phases of the cell cycle. I) Proliferation of TMEM127 or control knockdown 293E cells measured at the indicated time points after serum addition in cells serum-starved overnight using a DNA-bound fluorescence assay(P<0.002, t-test). Error bars are ±SE of triplicate experiments J) Phosphorylation levels of S6K in pheochromocytoma lysates. Two normal adrenal medulla samples (lanes 1 and 2) were compared with tumor lysates from TMEM127 mutant (Mut TMEM127) samples (lanes 3, 4 and 5, corresponding to samples from family 5, 6 and 1, respectively) and four tumors with wild-type TMEM127 sequence (WT TMEM127), including two sporadic samples (lanes 6 and 7), one tumor from neurofibromatosis type 1 (lane 8) and one VHL-mutant tumor (lane 9). Densitometric measurements of the bands in relation to β-actin are displayed below each lane (lane 3 ratio was set to 1).
mTOR operates as two functionally distinct complexes, mTORC1 and mTORC213,15. mTORC1 promotes cell growth and protein translation, and phosphorylates 4EBP1 and S6K. The cellular role of mTORC2 is less clear and its best known substrate is AKT15. As shown above, TMEM127 knockdown did not impact on AKT activation (Fig. 3B). Likewise, enforced expression of TMEM127 did not change AKT phosphorylation (Fig. 3C), suggesting that mTORC2 is not involved in this signal. In contrast, TMEM127 knockdown led to increased phosphorylation of 4EBP1 in various cell lines (Fig.3D). These effects were observed by distinct TMEM127 shRNA target sequences (Fig. 3E) and were independent of serum in HEK293 (Fig. 3F) and A2058 cells (Suppl. Fig. 7B). Conversely, enforced expression of TMEM127 using independent constructs led to reduced mTORC1 signaling (Fig. 3G, Suppl. Fig. 7C). Consistent with the observation that mTORC1 is a critical regulator of cell growth and size in many species16, we found that TMEM127 knockdown cells were larger (Fig. 3H, Suppl. Fig.7D) and proliferated at increased rates compared to control cell lines (Fig. 3I, Suppl. Fig.7E), while cell proliferation was reduced in cells overexpressing TMEM127 (Suppl. Fig 7F). To determine the relevance of these findings in primary tumors, we examined lysates from TMEM127-mutant pheochromocytomas and found increased phosphorylation of S6K compared to normal adrenal medulla or tumors without a TMEM127 mutation (Fig.3H). In agreement with the reported effects of NF1 towards mTOR14, one NF1-mutant pheochromocytoma showed elevated S6K phosphorylation, comparable to tumors with a TMEM127 mutation. Collectively, these results suggest that TMEM127 contributes to control mTORC1 signals and support a role for TMEM127 as a tumor suppressor.
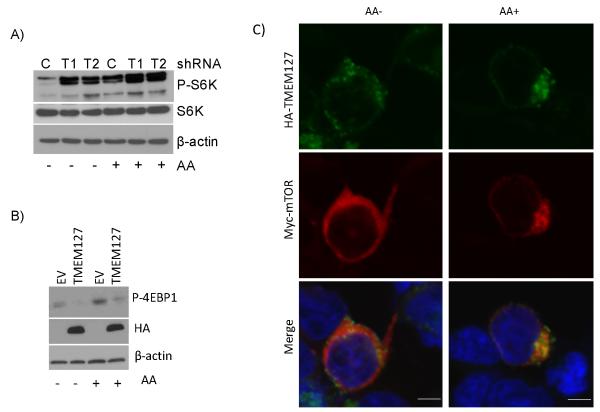
To further determine the extent to which TMEM127 influences mTOR regulation, we tested the effect of amino acids, powerful modulators of mTOR activity that supersede the actions of other regulators of this pathway17. Two independent TMEM127-specific shRNA sequences increased phosphorylation of the mTORC1 target S6K in 293E cells, both in amino acid-starved and rich conditions (Fig. 4A). In agreement with these findings, overexpression of TMEM127 decreased phosphorylation of mTORC1 targets after amino-acid repletion (Fig. 4B, Suppl. Fig. 7G). To further characterize the interplay between mTOR and TMEM127, we co-expressed myc-mTOR and HA-TMEM127 in HEK293T cells and examined their localization by confocal microscopy. In the absence of amino acid, myc-mTOR, like endogenous mTOR, localized diffusely within the cytoplasm but upon amino acid exposure, it became clustered to the perinuclear region of the cell(Fig 4C), a required step for its activation by Rheb117. We found that TMEM127 localized to the same cellular compartment as that of amino acid-activated mTOR (Fig 4C). Interestingly, the extent of myc-mTOR perinuclear localization after amino acid stimulation was significantly more pronounced in cells co-expressing TMEM127 than in control cells not overexpressing this gene (93.4±12%, vs. 64.3±12% respectively, p<0.001, Suppl. Fig. 8). This finding suggests that the effects of TMEM127 toward mTOR may be compartment-specific and influence mTOR subcellular localization.
Fig.4. TMEM127 colocalizes with amino acid-activated mTORC1.
A) Effect of 1-hour amino acid (AA) starvation, followed or not by replenishment (15 min), on S6K phosphorylation of 293E cells with TMEM127 knockdown by two independent shRNA sequences, T1 and T2. A control knockdown (C) is shown for comparison. B) Effect of amino acid (AA) treatment as in "A" on 4EBP1 phosphorylation of 293E cells overexpressing HA-TMEM127 or empty vector (EV). HA indicates TMEM127 expression. β-actin is the loading control. P-S6K, phosphorylated S6 kinase at residue T389; P-4EBP1, phosphorylated 4EBP1 at residues T37-46; S6K, total S6 kinase. C) HEK293T cells co-transfected with HA-TMEM127 and myc-mTOR were serum-starved overnight, depleted of amino acids for 1h followed by reexposure for 15 minutes (AA+). Cells were fixed and stained with antibodies specific for HA (green), myc (red) or DAPI (blue) before imaging. myc-mTOR is diffusely present in the cytoplasm in the absence of amino acid and becomes localized to a perinuclear region where TMEM127 is detected upon amino acid exposure. Similar results were obtained with two independent TMEM127 constructs. Bars, 5μm.
In summary, we identified a novel tumor suppressor gene that is associated with pheochromocytoma development. Similar to other genetic mutations reported in these tumors, all TMEM127 gene variants were present in the germline1. Our results suggest that TMEM127 limits mTORC1 activation and that it occupies the same intracellular domain as active mTOR. Interestingly, a component of the transGolgi network that shuttles between the plasma membrane and the endomembrane system has recently been implicated in cancer by modulating mTOR signals18. Taken together with other recent evidence19-20, our data give further support to an emerging role of the endomembrane and vacuolar network in mTOR regulation and in cancer. Future studies using in vivo models should determine whether mTOR activation is sufficient to explain the tumor phenotype resulting from TMEM127 mutation. Finally, our results support the notion that disruptions of TMEM127 could be the underlying basis for tumors with an aberrant mTOR pathway and validate the power of hereditary tumor models to shed light on cell growth-related signals.
Methods
Reagents
Reagents were obtained from the following sources: antibodies to phospho-T37-46-4EBP1, phospho-T389-S6K, phospho-S235/236-S6, phospho-S473-AKT, total 4EBP1, total S6K, syntaxin6, Rab5, from Cell Signaling; HRP-labeled anti-mouse and anti-rabbit secondary antibodies, and SybrGreen Supermix from BioRad; HA antibody (Covance); myc antibody (Santa Cruz Technology), RAS (Oncogene Bioscience), FLAGM2 and β-actin antibody, Dulbecco-PBS, DMEM, 50XMEM amino acid mixture, sodium orthovanadate, phenylmethylsulfonyl fluoride (PMSF), and oligonucleotides from Sigma-Aldrich; fetal calf serum, penicillin, streptomycin, trypsin, HEPES from Cell-Gro, Complete-miniEDTA Protease Cocktail from Roche, immobilized glutathione beads from Amersham Pharmacia; deoxynucleotides and Taq polymerase from Five Prime; RNAsin, random hexamers and reverse transcriptase from Promega; Trizol, Lipofectamine 2000, Alexa 488 antibody and DAPI from Invitrogen; Cy 3 and Cy5 conjugated secondary antibodies from Jackson ImmunoResearch Laboratories; Immobilon PVDF membrane and ECL kit from Millipore.
DNA and RNA isolation
Blood and tumor DNA were isolated using the Qiagen Genomic tip or the Qiagen Gentra Puregene Cell (for paraffin-embedded samples) following the manufacturer’s protocols. Total RNA was prepared from frozen tumor specimens with Trizol according to the manufacturer’s instructions.
Sequencing
FP locus sequencing strategy
The FP locus contained 205 genes (NCBI Build 35.1), of which all but 13 genes had RefSeq annotation (Suppl. Table 1). Exon-spanning primers for 85 of these genes were designed for PCR followed by sequencing using tumor DNA from one of the affected cases (index case of Family 1) as a template. Sequence variants that were present in the single nucleotide polymorphism (SNP) database or those that did not segregate with the pheochromocytoma phenotype in this family were not pursued further.
TMEM127 gene sequencing
Primers spanning the four exons of TMEM127 gene were used to amplify germline or tumor DNA from 103 pheochromocytoma patients.. Formalin-fixed paraffin embedded tumor samples were analyzed with an independent set of primers. PCR products were purified and directly sequenced by Beckman Coulter Genomics (formerly Agencourt Bioscience) as previously described3. Primer sequences and PCR conditions are available upon request.
Loss of heterozygosity (LOH) analysis
LOH analysis was performed on 35 pheochromocytomas for which blood and tumor DNA were available: 19 samples were from six index cases and eight of their affected relatives with TMEM127 mutations (5 tumors were bilateral). The remaining 16 samples were from patients without mutations in pheochromocytoma susceptibility genes. Informative chromosome 2 microsatellite markers, as reported previously8 and/or SNPs surrounding and within the TMEM127 locus (rs2276650, rs3852673, rs3739160) were used for this analysis.
Microarray-based expression profiling and gene-set enrichment analysis
Transcription profiles of 50 hereditary and sporadic pheochromocytomas were generated using Affymetrix U133 microarrays and combined with data from 76 pheochromocytomas (GEO accession GSE2841)8. In total, seven ‘FP-type’ tumors were available on the expanded dataset. Combined transcription data were normalized and used for unsupervised and supervised analysis as previously reported8. Gene set enrichment analysis (GSEA) was performed on the supervised dataset using GSEA v2.0 default parameters23. Paired analyses were done between the FP-mutants and: 1) each known hereditary pheochromocytoma class; 2) all of the hereditary tumors above combined; or 3) tumors without an identifiable TMEM127 mutation.
Real-time PCR
Quantitative real-time PCR was performed in cDNA isolated from 23 pheochromocytomas or cell lines, as indicated Quantitative real time-quantitative PCR was performed in triplicate using the iCycler iQ Real-Time PCR Detection System (BioRad). The TATA-box binding protein (TBP) gene was used as reference for calculation of 2−ΔΔ Ct as previously reported24. Primer sequences are available upon request.
Northern Blot
Northern blot was performed using the multitissue blot set (ClontechMTN Blot) containing 2μg polyA RNA from various human tissues and probed with a full-length TMEM127 insert as described previously25.
Clones and constructs
cDNA from TMEM127-mutant tumors or a full-length TMEM127 clone (pCMV6-XL5-TMEM127),obtained from Origene, were used as templates to generate constructs tagged at the N-terminus and C-terminus with a Flag epitope sequence present in one of the primers. The mutant constructs M158 and M99 (Table 1) were generated by mutagenizing the wild-type sequence using the QuickChange mutagenesis kit (Stratagene) or from the patient’s tumor cDNA, respectively. Wild-type and/or mutant constructs were cloned into the pMSCV retroviral-based vector26; pHM6 plasmid (HA tag at the N-terminus) and pEGFP-C2 vector. The corresponding empty vector was used as control for transfections. Stable clones were selected by G418 (pHM6) or GFP-sorting (MSCV and pEGFP-C2). The myc-mTOR construct was obtained from Addgene (plasmid 1861), as published27.
siRNA and shRNA
siRNA oligonucleotides directed to TMEM127 sequence was obtained from Dharmacon. GFP siRNA was used as control as previously described8. One-hundred nanomoles of each siRNA duplex were transfected with Lipofectamine 2000 and knockdown efficiency was verified by real time PCR from RNA harvested 48h after transfections. Protein lysates of efficient knockdown samples were obtained 72h post-transfection. Two of the four siRNA, siT1 and siT2 oligonucleotides(Suppl. Table 1) downregulated TMEM127 expression by over 70% of baseline levels and were used for further experiments.
Four retrovirus-based shRNA sequences targeting the TMEM127 gene in pRS-HuSH vector were obtained from Origene. Two sequences, shT1 and shT2 (Suppl. Table 1), independent of siT1 and siT2, effectively (>60%) downregulated TMEM127 expression by real time PCR.. Control sh constructs targeting GFP in pRS-HuSH vector were also obtained from Origene. These constructs were used for transfection of HEK293T, HEK293, 293E, HeLa and A2058 cell lines. Stable cell pools were selected with puromycin for 4-10 days and used within 3 weeks of selection. Efficiency of TMEM127 knockdown was determined as above.
Cell Culture and Transfections
Cell lines: 293E cells were a kind gift of Dr. John Blenis (Harvard Medical School). 293E, HEK293, HEK293T, HeLa and A2058 melanoma cells were cultured in DMEM and 10% fetal bovine serum supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin. Cells were serum starved overnight and exposed to regular serum (10%) after 10 to 30 min, as described in the text. Amino acid-response was measured by exposing overnight serum-starved cells to Dulbecco-PBS supplemented with 4.5g/l glucose for 1 hour (amino acid starvation period) followed by re-exposure to a full amino acid mixture (1X MEM amino acids solution) for 15 minutes, after which cells were harvested.
Retroviral transduction was carried out by an initial co-transfection of the relevant constructs along with pKAT and vSVG plasmids into HEK293T cells using Lipofectamine 2000. Viral supernatants were collected at 48 and 72h and used to transduce target cells in the presence of polybrene. Transduced cells were harvested at 48h for RNA and at 72h for protein analysis and selected as above. All other transfections were performed with Lipofectamine.
Western Blots
Whole cell lysates from tumors and normal adrenal medullas were obtained as previously described28. Cell lines were lysed in ice-cold buffer containing 20 mM HEPES (pH 7.4), 10% glycerol, 1 % NP-40, and protease inhibitors. . The soluble fractions were isolated by centrifugation. Fifty-ug whole cell lysates were run on 12% SDS-polyacrylamide gels, transferred to PVDF membranes and hybridized with the antibodies listed above according to the manufacturer’s instructions. Filters were developed with a chemiluminescence assay (West Pico, Pierce) and exposed to X-Ray film (Kodak).
RAS Activation Assays
HEK293, 293E or HeLa cells expressing either shTMEM127 or shGFP control were cultured as indicated above. 400μg of cell lysates were incubated with RAF1-GST fusion protein for 3h at 4C. Immunoprecipitates were washed in GST buffer and RAS-GTP was detected using a RAS antibody as previously described26. Total RAS protein and β-actin present in the whole cell lysates were determined in parallel to ensure equal loading. These results were performed five times.
Proliferation assays
Stable HEK293 or A2058 TMEM127 knockdown or control (shGFP) cells were seeded at 5 ×104 cells per well of a 6-well plate in normal (10%) growth medium or 1% serum. Daily, for 3 days, cells were trypsinized and live cells were counted on triplicate plates by trypan blue exclusion. Proliferation rates of 293E and HEK293T cells with TMEM127 knockdown or overexpression and respective control cells were also determined in the presence or absence of serum using a method based on DNA binding to a fluorescent reagent (Cyquant-NF, Invitrogen). Fluorescence was measured with a microplate reader (Optima) following the manufacturer’s guidelines. These experiments were performed three times in triplicate.
Flow Cytometry
Stable TMEM127 knockdown on HEK293 and A2058 and their respective control cell lines were fixed in ethanol, stained with propidium iodide and analyzed by forward-scattering FACS to determine cell size29. Experiments were repeated three times.
Immunofluorescence Microscopy
HEK293T cells overexpressing empty MSCV-TMEM127-Nflag or MSCV alone, or 293E cells expressing HA-TMEM127 or HA-empty vector were plated on poly-D-lysine-coated glass coverslips in 24-well tissue culture plates. After 24h,, cells were serum- or amino acid-starved and stimulated as described above, rinsed with PBS and fixed in 4% paraformaldehyde in PBS. The coverslips were permeabilized and blocked with 5%horse serum, 0.2% Triton X-100 in PBS,and incubated with primary antibody: Flag M2, HA.11, Syntaxin 6, Rab or myc, at the recommended dilutions in blocking buffer overnight at 4°C, rinsed and incubated with secondary antibody (Alexa 488, Cy3 or Cy5). .Slides were briefly exposed to DAPI and the coverslips were mounted on glass slides using Vectashield (Vector Laboratories) for imaging. Specificity of the signal was determined by omission of the primary antibody in control coverslips. Golgi staining was additionally performed using the Organelle Lights-Golgi-RFP (Invitrogen) in which the Golgi component N-acetylgalactosaminyltransferase-2 is fused to red fluorescence protein (RFP) in a baculovirus-based construct and transduced into target cells (293E stably expressing HA-TMEM127 or an empty control vector), following the manufacturer’s instructions. Images were acquired by laser scanning confocal microscopy (LSCM) with an Olympus FV1000 imaging system and IX-81 microscope. The UPLANAPO 60x oil objective (1.42 NA) was used for all datasets, and an additional electronic zoom of 3 was applied. Excitation and emission signals were respectively 405 and 425±50nm for DAPI, 488 and 500±35nm for Alexa 488 or GFP, and 543 and 560-660nm for Cy3. Quantitative co-localization was calculated using Intensity Correlation Analysis (ICA), as described30. The difference from the mean for each pixel intensity is first calculated for each channel (Ri-Rmean and Gi-Gmean). The product of the differences from the mean [PDM = (Ri-Rmean) × (Gi-Gmean]) is positive when both red and green pixel intensities are either above or below their respective means (i.e. when pixel intensities vary synchronously). ICA was performed on single optical sections ~0.4 μm thick) for at least 7 cells from multiple independent fields and three independent experiments.
Colocalization with lysosomes was performed in live HEK293T cells transfected with GFP-TMEM127. Cells were plated on poly D-lysine-coated glass plates (TissueTek) in medium without phenol red. Fifteen minutes prior to imaging, fresh media containing 25nM Lysotracker-Red-DND99 (Invitrogen) was added. Live imaging was performed using a confocal microscope (LSM 510; Carl Zeiss MicroImaging, Inc.) with a 63X/1.4 NA plan-apochromat objective. GFP and Red were excited as above. Image acquisition of the fluorescence intensity was performed with the Zeiss LSM510 Software 3.2 SP2.
To estimate the fraction of TMEM127 associated with the plasma membrane (PM), we defined fluorescence signal that was within 5 μm of the membrane and was 5% higher than background fluorescence as PM-associated TMEM127. Pixel intensities were measured in single optical sections acquired on a confocal microscope, as above. The fraction of cells with PM-associated signal was determined in relation to TMEM127-expressing cells that did not show this pattern. Data was obtained from HEK293T cells transfected with our four TMEM-tagged constructs (N- and C-flag, N-HA and GFP).
For endocytosis inhibition, 293E cells stably expressing HA-TMEM17 were processed as previously described7. Briefly, cells were rinsed in buffer A (20 mM HEPES, pH 7.4, 140 mM NaCl, 1 mM CaCl2, 1 mM MgCl2) and then incubated in hypotonic media (buffer A diluted 1:1 in water) followed by incubation in buffer A for 2 hours, fixed and stained as above. Control cells were maintained in regular media. Endosome ‘trapping’ is based on protein retention in endosomal structures by inhibition of acidification in the culture media, and was performed by incubating HA-TMEM17-expressing 293E cells in media supplemented with 50mM ammonium chloride or equal volume of vehicle for 2h7 fixed and stained as above.
mTOR and TMEM127 colocalization was performed by cotransfection of a myc-tagged mTOR construct (see above, “Constructs”) and HA-TMEM127, Flag-TMEM127 or GFP-TMEM127 in HEK293T cells. After 24h, cells were serum- and amino acid starved and reexposed to amino acids, fixed and stained as above. Comparative experiments were performed by transfection of myc-mTOR in HEK293T cells not overexpressing TMEM127 that were treated in an identical manner. Distribution of myc-mTOR localization in transfected cells was analyzed by scoring three distribution patterns: diffuse cytoplasmic, perinuclear only or vacuolar clusters (Suppl. Fig. 8) both in myc-mTOR/TMEM127 cotransfection and myc-mTOR control cells. At least 200 cells from 3 independent experiments were scored by two independent observers (Y.Q. and P.L.M.D.).
Supplementary Material
Acknowledgments
We thank Shoulei Jiang for technical assistance, Vickie Frohlich for help with optical imaging, John Blenis for kindly providing the 293E cell line, William Kaelin Jr. for his comments and suggestions, and Marion Jech, Christian Colin, Nguyen V. Nguyen, Miguel Pujana, Marc Vidal, David Hill and the Familial Pheochromocytoma Consortium members for their contribution to earlier phases of this project. We also thank the patients and families that participated in the study for their cooperation. PLMD is a Kimmel Foundation Scholar, a recipient of a Voelcker Foundation Young Investigator Award and is supported by the Cancer Therapy and Research Center (CTRC) at the University of Texas Health Science Center at San Antonio (NIH-P30 CA54174). RCTA is supported by the Voelcker Fund. Immunofluorescence images were generated in the Core Optical Imaging Facility which is supported by UTHSCSA, NIH-NCI P30 CA54174 (CTRC), NIH-NIA P30 AG013319 (Nathan Shock Center) and (NIH-NIA P01AG19316).
Footnotes
The authors declare that they have no competing interests.
References
- 1.Amar L, et al. Genetic testing in pheochromocytoma or functional paraganglioma. J Clin Oncol. 2005;23:8812–8. doi: 10.1200/JCO.2005.03.1484. [DOI] [PubMed] [Google Scholar]
- 2.Dahia PL. Evolving concepts in pheochromocytoma and paraganglioma. Curr Opin Oncol. 2006;18:1–8. doi: 10.1097/01.cco.0000198017.45982.06. [DOI] [PubMed] [Google Scholar]
- 3.Dahia PLM, et al. Novel Pheochromocytoma Susceptibility Loci Identified by Integrative Genomics. Cancer Res. 2005;65:9651–9658. doi: 10.1158/0008-5472.CAN-05-1427. [DOI] [PubMed] [Google Scholar]
- 4.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 5.Neumann HP, et al. Germ-line mutations in nonsyndromic pheochromocytoma. N Engl J Med. 2002;346:1459–66. doi: 10.1056/NEJMoa020152. [DOI] [PubMed] [Google Scholar]
- 6.Jonsson G, et al. High-resolution genomic profiles of breast cancer cell lines assessed by tiling BAC array comparative genomic hybridization. Genes Chromosomes Cancer. 2007;46:543–58. doi: 10.1002/gcc.20438. [DOI] [PubMed] [Google Scholar]
- 7.Snyder CM, Mardones GA, Ladinsky MS, Howell KE. GMx33 associates with the trans-Golgi matrix in a dynamic manner and sorts within tubules exiting the Golgi. Mol Biol Cell. 2006;17:511–24. doi: 10.1091/mbc.E05-07-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahia PLM, et al. A HIF1a regulatory loop links hypoxia and mitochondrial signals in pheochromocytomas. PLoS Genet. 2005;1:e8. doi: 10.1371/journal.pgen.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin GA, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63:843–9. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- 11.Johannessen CM, et al. The NF1 tumor suppressor critically regulates TSC2 and mTOR. PNAS. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segouffin-Cariou C, Billaud M. Transforming ability of MEN2A-RET requires activation of the phosphatidylinositol 3-Kinase/AKT signaling pathway. J. Biol. Chem. 2000;275:3568–3576. doi: 10.1074/jbc.275.5.3568. [DOI] [PubMed] [Google Scholar]
- 13.Kwiatkowski DJ, Manning BD. Tuberous sclerosis: a GAP at the crossroads of multiple signaling pathways. Hum Mol Genet. 2005;14 Spec No. 2:R251–8. doi: 10.1093/hmg/ddi260. [DOI] [PubMed] [Google Scholar]
- 14.Johannessen CM, et al. TORC1 is essential for NF1-associated malignancies. Curr Biol. 2008;18:56–62. doi: 10.1016/j.cub.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 15.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 16.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–75. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 17.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott KL, et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459:1085–90. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Binda M, et al. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35:563–73. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 20.Dippold HC, et al. GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–51. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferner RE, et al. Guidelines for the diagnosis and management of individuals with neurofibromatosis 1. J Med Genet. 2007;44:81–8. doi: 10.1136/jmg.2006.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlisio S, et al. The kinesin KIF1Bbeta acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev. 2008;22:884–93. doi: 10.1101/gad.1648608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, et al. From the Cover: Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Aguiar RC, et al. PTPROt: an alternatively spliced and developmentally regulated B-lymphoid phosphatase that promotes G0/G1 arrest. Blood. 1999;94:2403–13. [PubMed] [Google Scholar]
- 26.Smith PG, et al. The phosphodiesterase PDE4B limits cAMP-associated PI3K/AKT-dependent apoptosis in diffuse large B-cell lymphoma. Blood. 2005;105:308–16. doi: 10.1182/blood-2004-01-0240. [DOI] [PubMed] [Google Scholar]
- 27.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 28.Dahia PLM, et al. PTEN is inversely correlated with the cell survival factor Akt/PKB and is inactivated via multiple mechanismsin haematological malignancies. Hum Mol Genet. 1999;8:185–193. doi: 10.1093/hmg/8.2.185. [DOI] [PubMed] [Google Scholar]
- 29.Inoki K, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–68. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 30.Li Q, et al. A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J Neurosci. 2004;24:4070–81. doi: 10.1523/JNEUROSCI.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.