Abstract
Localized heating of droplets on an electrowetting-on-dielectric (EWOD) chip has been implemented and shown to accelerate trypsin digestion reaction rates, sample drying, and matrix crystallization for matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS). Achieving this involved extending the functionality of previous EWOD droplet-based techniques by developing a multifunctional electrode with closed-loop temperature control, while minimizing overall system complexity, and addressing challenges associated with rapid evaporation. For the EWOD chip design, we discuss the performance of multifunctional surface electrodes for actuation, localized Joule heating, and thermistic temperature sensing. Furthermore, a hydrophilic pattern is formed in the multifunctional electrode to control the location of an evaporating droplet on the electrode. To demonstrate the capabilities and limitations of this technique, we performed three experiments and measured the results using MALDI-MS: (i) insulin disulfide reductions in DTT over a range of heater temperatures (22 to 70 °C) to show how reaction rates can be affected by thermal control, (ii) insulin disulfide reductions at 130 °C in DMSO to demonstrate a reaction in a high boiling point solvent, and (iii) tryptic digestions of cytochrome c at 22 and 40 °C to show that heated droplets can yield reasonably higher peptide sequence coverage than unheated droplets. Although they do not decouple the effects of changing temperatures and concentrations, these experiments verified that thermal cycling by EWOD electrodes accelerates reaction rates in liquid droplets in air.
INTRODUCTION
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) is an important technique in proteomics research for peptide and protein detection. Sample preparation, however, can be quite tedious because of the multistep chemical processing that is required to prepare arrays of dried samples (spots) on the sample holder, or target. To streamline this process, Wheeler et al.1 and Moon et al.2 first developed the application of automated EWOD droplet microfluidics to proteomics sample preparation with direct MALDI-MS characterization. Similar platforms have been used for multi-step MALDI-MS sample processing,3,4,5 protein extraction by precipitation,6 and enzyme kinetics studies.7 With the droplet-based microfluidic platforms used in these studies, characterization follows directly after preparation because the chip is unplugged from the EWOD setup after crystal growth and placed into the MS chamber on a normal sample holder. This microfluidic approach thus removes the usual MALDI preparation steps in which liquid samples are transferred by pipette to target sites prior to matrix/sample crystallization. Microfluidic systems with continuous flows have also been used to prepare samples for mass spectrometry, including MALDI.8–11 Channel-based chips can achieve very effective coupling with other MS techniques as well, such as electrospray ionization MS (ESI-MS), by combining on-chip separation columns and electrospray tips,12 e.g., Agilent’s HPLC-Chip/MS.13 However, the digital platform seems more convenient for this MALDI-MS based application not only because of the simplicity of EWOD actuation, i.e., one can electrically dispense, transport, mix, split, and arrange sub-microliter volumes in regular arrays of isolated samples,25, 26 but also because the planar EWOD chip can be attached onto the standard target prior to MALDI-MS measurements. A chip having the multifunctional actuation elements presented here establishes the technology to design future devices for large arrays of nano- and picoliter volumes in an automated fashion, which removes the need for manual operations that are relatively slow, such as pipetting, mixing, and heating.
Various ways to heat samples have been successfully demonstrated in biochemical MEMS devices designed for miniaturized assays that require thermal cycling, such as the polymerase chain reaction (PCR).14 Common contact methods are Joule heating2–8,15 and off-chip Peltier stacks.16 Remote methods include dielectric microwave heating via a coplanar waveguide,17 induction,18 and infrared (IR) excitation.19 Sensing on-chip temperatures has been accomplished by contact methods such as resistance temperature detectors (RTD),2,8,13,14 temperature-responsive fluorescence dyes,18, 19 quantum dots,20 thermocouples,21, 22 and thermochromatic crystals,23 or non-contact (i.e., remote) methods such as IR thermometry.24 We have combined Joule heating and thermistor elements in this work because they are simple to integrate into our electrically controlled system. Specifically, to achieve Joule heating, current is passed through the EWOD actuation electrode, and to achieve thermistor sensing, the voltage drop across the EWOD actuation electrode is measured simultaneously. The method requires only electronic (software) control over current flow and no added on-chip devices (hardware modifications).
Thermal cycling on an EWOD chip has been demonstrated in various ways, e.g., hot air convection of the entire chip,27 commerical heaters and thermocouples placed against the underside of the chip,28, 29 and an on-chip incubation chamber with a thin film heater.30 Wei at al.15 reported a “versatile microelectrode,” which refers to an EWOD electrode that doubles as a resistive heater similar to the design used in this work, and demonstrated droplet heating in silicone oil. Building on preliminary studies,15, 31 we have extended this concept by: (i) adding thermistic temperature sensing to the heater and feedback control and (ii) addressing challenges of working in air. (Please refer to Supporting Information (S-1) for an illustration of the actuation and heating principles.) For the preparations in this application, where liquids should be evaporated prior to the MS measurement, it is particularly advantageous to manipulate aqueous droplets on a dry surface in air, so that samples are allowed to dry without the problem of removing a second liquid phase, such as oil.32 The effectiveness of the developed system is evaluated by performing heated protein disulfide reductions and trypsin digestions followed by MALDI-MS. Our purpose is to use relatively simple chemistries to demonstrate the built-in heating function of the EWOD chip. In the future we will explore the ability to handle more complex samples. Results obtained by others using very similar platforms lead us to believe that accommodating such samples is possible.4, 5
EXPERIMENTAL METHODS
Chemicals
All reagents used in this study were purchased from Sigma-Aldrich Corp. (St. Louis, MO, USA). All protein solutions for EWOD experiments were made within minutes of testing from stock solutions, stored at −80 °C. The stock solutions (in a 1:1 volume ratio of acetonitrile and water, or DMSO and water) used for insulin disulfide reduction experiments were diluted using the same solvent to obtain 0.5 μM insulin and 25 mM dithiothreitol (DTT). For tryptic digestions, stock solutions (in 25 mM ammonium bicarbonate in a volume ratio 1:1 acetonitrile:water) were diluted using the same solvent to obtain a 1:50 mass ratio of 10 μM cytochrome c to trypsin. 0.05 % wt. Pluronics F-68 (PL68), was added to minimize protein adsorption to the chip surface and thereby prevent contact line pinning.33 MALDI matrix solutions (3 mg/mL 2,5-dihydroxybenzoic acid (DHB)) were made fresh within minutes of testing with the same solvents used for proteins and reducing agents (excluding PL68), with the addition of 0.1% trifluoroacetic acid (TFA).
EWOD Microfluidic Platform Fabrication
The incorporation of a local temperature control function into the typical EWOD device is achieved by substituting some of the EWOD pad electrodes with the multifunctional electrodes, using essentially the same process flow as the previous EWOD devices.2 A freshly fabricated chip was used for each experiment, to eliminate uncertainties that might arise from surface degradation after repeated use and cleaning. Because we expect the eventual use of disposable chips in practical settings, we have not evaluated the performance of cleaned devices versus fresh devices. Please refer to Supporting Information (S-2) for a detailed process flow. Importantly, to heat only at desired spots and measure temperature accurately, heater leads were designed to be much more conductive than the heater; specifically, the indium-tin-oxide (ITO) heaters are about 100 times less conductive than their gold leads and chip bus contacts, and we can therefore neglect delocalized power loss.
EWOD Microfluidic System
A computer running LabView controls the off-chip driving electronics of our system. Please refer to Supporting Information (S-3) for a control system diagram. The home-built multiplexer and commercial USB digital I/O device (National Instruments DAQPad 6507) routes EWOD driving voltages (40 to 60 Vrms at 1 kHz) from an amplifier to the chip bus, and the DC power source (Keithley 2425 SourceMeter) under closed-loop proportional-integral-derivative (PID) control maintains temperatures through Joule heating and thermistor sensing. Electrical signals direct chemical processes via the EWOD chip, which is connected to the driving electronics through the chip bus like a plug in a socket. After chemical processing, the chip is unplugged, the top plate is removed, and the bottom sample plate is attached using double-sided tape (3M™) to the MALDI plate and is loaded directly into the MALDI-MS source for analysis. MALDI time-of-flight (TOF) spectrometry was performed on an Applied Biosystems Voyager-DE STR instrument and operated in linear mode with an external calibration.
Droplet Heating
A custom LabView proportional, integral, and derivative (PID) controller connected via RS-232 to a Keithley SourceMeter was used to achieve programmed set point temperatures with a sampling rate of 10 Hz. See Supporting Information (S-4) for a plot of representative heater time histories for a 1 μL water droplet over a 3 × 3 mm heater. For this scenario, we observed approximately 2 s rise time, less than 10% overshoot, and 8 s to stabilize to ± 1 °C. The PID gain parameters were tuned manually in order to achieve acceptable performance for the current device, which has a sluggish thermal response by microheater standards because most of the heat is lost to the glass substrates. To significantly lower the rise time, it will be necessary to lower the thermal capacitance of the heating element. This can be done via microchining by thinning the substrate beneath the heater so that it rests on a membrane over an air cavity that blocks power loss to the substrate. However, we opted to accept the heat loss so as to avoid complications in fabricating the EWOD chip. Additionally, three-dimensional numerical simulations were performed to generate a temperature map of the chip during heating of a single water droplet at constant temperature for one minute. The results, presented in Supporting Information (S-5), indicate that the chip substrate is predicted to be at room temperature six heater lengths away from a heated droplet. These results represent baseline performance of a chip not optimized for thermal management.
Device Layout and Operation
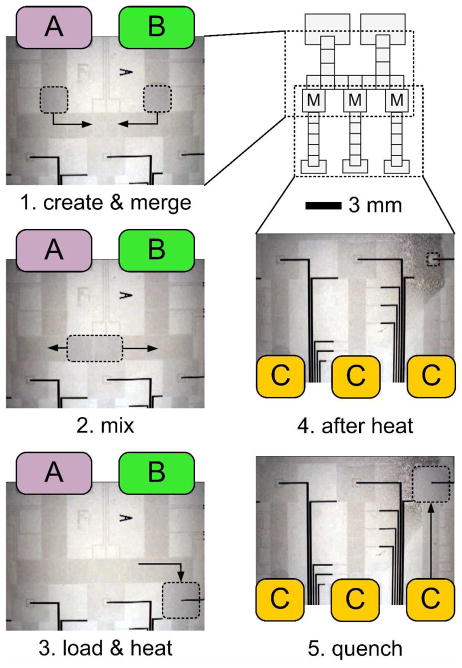
A two-dimensional array of EWOD electrodes that incorporates the multifunctional electrodes is a new device platform for reconfigurable sub-microliter droplet transport, splitting, mixing, and heating. Figure 1 shows the chip layout of a fabricated device and video frames taken during a sample preparation process. (Note: the image contrast was slightly increased using Microsoft PowerPoint 2008.) In the schematic layout (upper right), three multifunctional electrodes are the boxes labeled “M”; all other boxes are blank EWOD driving pads. The first three frames (left column) show the upper half of the layout. The heater electrode patterns are not visible at this scale. The first frame shows two droplets (dotted lines) surrounded by air, created from reservoirs A and B, on merging pathways (arrows). The second frame shows the combined droplet and its mixing pathway. The third frame shows sample loading onto a multifunctional electrode. The last two frames (right column below the layout) show the lower half of the layout. Frame 4 was taken after sample heating; the loaded sample has become smaller after significant evaporation. The final frame shows a new droplet created from reservoir C merging with the heated sample, restoring its initial volume. We call this the quench step because the third droplet can be used to quickly reduce the temperature and/or change the pH of the warmed droplet, thus controlling the reaction time for heated samples. The exact process depicted in Figure 1 was used to perform a series of protein sample preparations that is discussed later.
Figure 1.
A sample preparation process is illustrated in which: (1) 500 nL droplets are created from reservoirs A and B and merged; (2) the combine sample is mixed; (3) the mixed sample is loaded onto a heating site; (4) after heating the sample volume is decreased and (5) a quenching droplet is created from reservoir C and combined with the heated sample.
Hydrophilic Patterns for Centering Evaporating Droplets
The ability to manipulate droplets with gas as the surrounding medium is, in many cases, an advantage of EWOD droplet microfluidic systems over those that require a second liquid phase, which can introduce problems due to diffusion across the liquid-liquid interface. Also, a second liquid phase complicates the process of rendering dry samples. When volume loss due to evaporation is a problem, it can by either slowed or prevented by using a liquid surrounding medium, humidification, solvent replacement,34 or through proper packaging.35 Alternatively, when volume loss is desired, it is possible to use evaporation, a highly temperature-dependent process, for drying. In our preliminary experiments, a complicating factor of this technique was observed: with droplets on hydrophobic-coated heaters, non-uniform evaporation rates along the liquid meniscus resulted in migration of the center of mass (CM) of a heated droplet toward its coldest region. This migration occurs because the liquid nearest to the center of the heater (the hottest point) evaporates fastest, and the liquid furthest from the center of the heater evaporates slowest. Because all points on the meniscus will rarely be equidistant from the center of the heater, the droplet CM location is not stable. This problem can be complicated for samples in which solutes adsorb to the surface, as is commoon with protein solutions, because the contact line may pin to the fouled surface. We removed the uncertainty of sample location during evaporation by incorporating hydrophilic rings that hold shrinking droplets over the center of the heater.
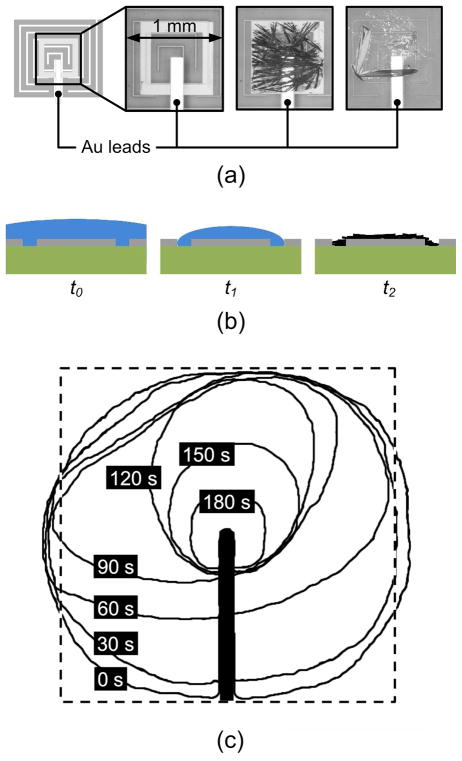
In the following section, we discuss the use of our system for on-chip preparation of peptide samples for direct characterization by MALDI-MS, an application in which hydrophilic rings perform two vital functions: (i) pinning of evaporating samples over the center of the heater, and (ii) providing proper surface conditions for growth of evenly distributed matrix crystals. On the former point, in our experiments, regardless of temperature gradients or fouling, the final resting place of a heated droplet was always over the hydrophilic ring. On the latter point, we observed that crystals from the DHB MALDI sample matrix tend to grow from many nucleation sites that form at the solid-liquid-air contact line (or dirt particles in the fluid) on the ring pattern after the matrix solution has almost completely evaporated. Without the rings, crystals tend to grow from one or a few nucleation sites on the heater. The schematic in Figure 2(a) shows a multifunctional electrode site, which corresponds to “M” in Figure 1, showing the hydrophilic square ring in the middle. The three optical pictures of Figure 2(a) magnify the crystal region before a sample is placed (left), after DHB crystals are grown inside the ring (middle), and after DHB crystals are grown by the same process on an unpatterned heater (right). We observed that crystals on rings were more slender and evenly distributed, while those on unpatterned heaters were more likely to become thick and form clumps. For MALDI-MS characterization, it is desirable to grow evenly distributed crystals because the laser spot can be positioned almost anywhere in the ring to yield good spectra. If the crystals are thick and clumped, only a few selected positions yield analyte signal. Figure 2(b) depicts a cross section of a hydrophilic ring just after top plate removal, t0, just before crystal growth, t1, and after complete evaporation of the solvent, t2. Crystals grow long and thin from many nucleation sites because a liquid film forms as the contact line sticks to the hydrophilic surface.
Figure 2.
(a) From left to right, a schematic of a multi-functioning electrode, optical picture magnifying a hydrophilic square ring, MALDI-MS matrix crystals grown on a ring, and crystals grown on a heater with no ring. (b) A cross section at times corresponding to after top plate removal, t0, just before crystal growth, t1, and after complete evaporation of the solvent, t2. (c) Meniscus profiles taken at 30 s intervals for a 1 μL aqueous droplet (initial concentrations: 0.25 μM insulin, 12 mM DTT in 1:1 volume ratio acetonitrile to water) on a 70 °C heater with hydrophilic ring for centering. After 180 s, the profile approximately matches the outline of the hydrophilic ring. The thick black line is a gold lead.
To illustrate the centering function of the rings, Figure 2(c) shows meniscus profiles of a 1 μL protein solution at 30 s intervals as it evaporated on a heater at 70 °C with an etched ring. The sequence of profiles was rendered from images by a software edge detection process (ImageJ). After 180 s, the droplet meniscus matches the hydrophilic ring outline. This exact centering makes the subsequent crystal growth and MALDI-MS measurement much more effective than with non-centering heaters.
RESULTS AND DISCUSSION
Insulin Disulfide Reductions
Insulin is composed of two polypeptides, the A- and B-chains, held together by multiple disulfide bonds that can be cleaved by a reducing agent. DTT has been used to reduce disulfide bonds in neutral-pH solution, with increased rate at elevated temperatures.36 On-chip reductions at several temperatures were performed using the process shown in Figure 1. Reservoirs A, B, and C contained insulin, DTT, and DHB solutions, respectively. Samples created in step 1 were 500 nL, and quenching droplets were 700 nL. Mixed 1μL samples were loaded onto heaters and evaporated at temperature-dependent rates. While on a heater at 70 °C for 180 s, for instance, the acetonitrile sample volume reduced by about 95%, from 1 μL to 60 nL. For the DMSO sample, a similar evaporation rate was observed at a heater temperature of 130 °C. Heating times were limited such that droplets did not completely evaporate before quenching. After matrix solution was added to the heated sample, the top plate was removed from the device prior to crystallization. Reaction time, from merging to quenching, was 5 minutes.
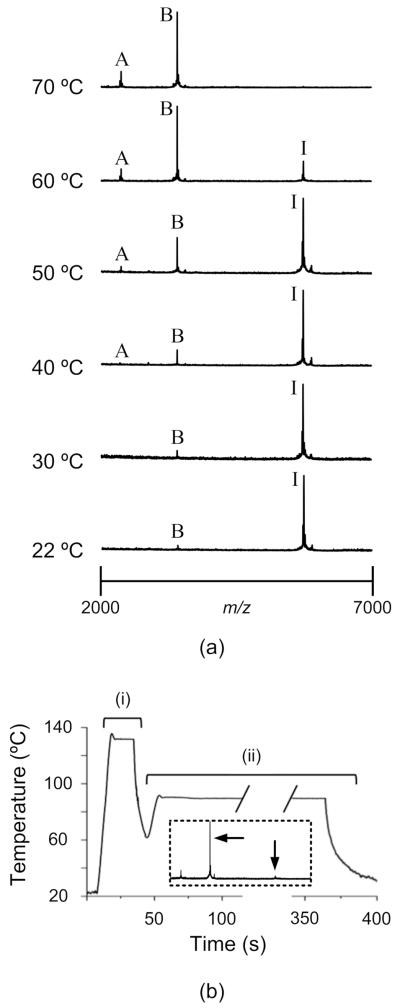
A MALDI-MS spectrum of unreduced insulin only shows a peak for the intact protein at m/z ≈ 5720. A completely reduced insulin sample has peaks only for the A-chain at m/z ≈ 2460 and B-chain at m/z ≈ 3420. Partially complete disulfide reductions give MALDI spectra showing all three peaks. By examining the spectra from a series of insulin disulfide reductions at various heater temperatures, as shown in Fig. 3a, we can clearly see that the reaction rate increases with heater temperature. That is, the spectra show that the intensity of the A- and B-chain signals increases with temperature while the intact insulin signal decreases, indicating that the rate of reduction was faster at higher heater temperatures. Each mass spectrum represents the average of 100 to 200 laser shots, taken from three different target spots. Figure 3(b) shows a heater time history and corresponding MALDI-MS spectrum for a high-temperature disulfide reduction in DMSO, which has a boiling point of 189 °C. The heater maintained a 130 °C set point for 10 s during reduction and 90 °C for 250 s during hot crystallization. The spectrum indicates the sample was almost completely reduced.
Figure 3.
(a) MALDI-MS spectra for insulin samples on heaters for 180 s at temperatures from 22 °C to 70 °C. The intact insulin signal is labeled “I” (m/z ≈ 5720), the A-chain is labeled “A” (m/z ≈ 2460), and B-chain is labeled “B” (m/z ≈ 3420). Each spectrum represents the average of 100 to 200 laser shots, taken from three different target spots. At 22 °C and 30 °C, A-chain signals could not be distinguished from noise. (b) A typical heater time history for high-temperature insulin disulfide reduction (i) and matrix crystallization (ii) in DMSO. The inset spectrum, in which arrows denote B-chain and intact insulin signals, indicates that the reduction is nearly complete after heating for 30 s.
Tryptic Digestion of Cytochrome c
Trypsin proteolytic digestion selectively cleaves peptide bonds at the carboxy-terminus of the amino acids lysine and arginine. This process is used commonly in proteomics strategies to identify unknown proteins by comparing mass spectra of cleaved proteins with previously collected or simulated peptide mass fingerprint spectra. Cytochrome c is a well-characterized 12 kDa protein used often for proteomics methods development. We performed on-chip tryptic cytochrome c digestions by the process described in Figure 1, with heaters at room temperature and 40 °C for 5 minutes. For both cases, the total reaction time, from merging until addition of matrix, was 10 minutes. The reservoirs A, B, and C contained cytochrome c, trypsin, and DHB solutions, respectively.
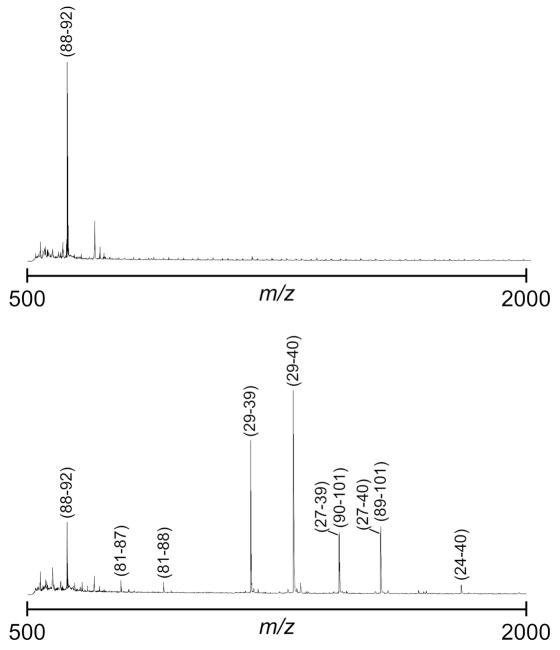
Figure 4 presents MALDI spectra from on-chip trypsin digested samples on heaters at 22 °C (upper spectrum) and 40 °C (lower spectrum). Each represents the average of 300 to 400 laser shots. The peaks that represent trypsin digestion products were identified by comparison to their theoretical m/z values and labeled by residue numbers. From three digestions at each heater temperature, we calculate average peptide sequence coverages of 15 ± 11% for room temperature and 74 ± 8% for 40 °C. As expected, the enzyme trypsin is more active near body temperature.37 While these data provide a clear comparison of heated vs. unheated droplet reactions, they do not represent optimized conditions. For example, a significantly higher room temperature coverage (~ 40%) of cytochrome c on a similar EWOD platform has been reported.5 Other, albeit more complicated, channel-based microfluidic systems can routinely yeild 60 to 100% peptide coverage,38–40 and demonstration of complete coverage, for any given protein, on the reported EWOD platform would require considerable tuning of reagent concentrations and reaction conditions. However, we have shown by our experiments that achieving high sequence coverage from trypsin digestions performed at elevated temperatures in 5-min is reasonable, and multifunctional EWOD electrodes offer a simple and effective method for tuning reaction temperature.
Figure 4.
Figure 9: MALDI-MS data for tryptic digestion of cytochrome c on heaters at 22 °C (top spectrum) and 40 °C (bottom spectrum). Residue numbers for the trypic peptides are labeled.
CONCLUSIONS
We have shown that localized droplet heating on an EWOD digital microfluidic chip can be used to accelerate reaction rates and drying times in automated proteomics sample processing. Multifunctional electrodes under feedback control have been incorporated into the existing EWOD chip design without complicating the fabrication process. A hydrophilic pattern in the multifunctional electrode effectively addresses the problem of droplet migration during rapid evaporation, enabling control of MALDI-MS target spot location.
Supplementary Material
Acknowledgments
The authors thank Dr. Prosenjit Sen, Dr. A. Jimmy Ytterberg, and all members of the Kim, Loo, and Garrell Labs. This work was supported by the NIH (R01 RR020070) and an NSF Integrative Graduate Education and Research Traineeship (DGE-0654431 to WN) through the UCLA Materials Creation Training Program (MCTP).
Footnotes
SUPPORTING INFORMATION AVAILABLE
Operating principles schematic; fabrication process flow; system diagram; device characteristics and numerical modeling. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Wheeler AR, Moon H, Bird CA, Loo RRO, Kim CJ, Loo JA, Garrell RL. Anal Chem. 2005;77:534–540. doi: 10.1021/ac048754+. [DOI] [PubMed] [Google Scholar]
- 2.Moon H, Wheeler AR, Garrell RL, Loo JA, Kim CJ. Lab Chip. 2006;6:1213–1219. doi: 10.1039/b601954d. [DOI] [PubMed] [Google Scholar]
- 3.Chatterjee D, Ytterberg AJ, Son S-U, Loo JA, Garrell RL. Proc Int Conf MicroTAS. 2008:1072–1074. [Google Scholar]
- 4.Luk VN, Wheeler AR. Anal Chem. 2009;81:4524–4530. doi: 10.1021/ac900522a. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee D, Ytterberg AJ, Son SU, Loo JA, Garrell RL. Anal Chem. 2010;82:2095–2101. doi: 10.1021/ac9029373. [DOI] [PubMed] [Google Scholar]
- 6.Jebrail MJ, Wheeler AR. Anal Chem. 2009;81:330–335. doi: 10.1021/ac8021554. [DOI] [PubMed] [Google Scholar]
- 7.Nichols KP, Gardeniers HJGE. Anal Chem. 2007;79:8699–8704. doi: 10.1021/ac071235x. [DOI] [PubMed] [Google Scholar]
- 8.Brivio M, Fokkens RH, Verboom W, Reinhoudt DN. Anal Chem. 2002;74:3972–3976. doi: 10.1021/ac020185n. [DOI] [PubMed] [Google Scholar]
- 9.Gustafsson M, Hirschberg D, Palmberg C, Jornvall H, Bergman T. Anal Chem. 2004;76:345–350. doi: 10.1021/ac030194b. [DOI] [PubMed] [Google Scholar]
- 10.Nelson RW, Nedelkov D, Tubbs KA. Electrophoresis. 2000;21:1155–1163. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1155::AID-ELPS1155>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 11.DeVoe DL, Lee CS. Electrophoresis. 2006;27:3559–3568. doi: 10.1002/elps.200600224. [DOI] [PubMed] [Google Scholar]
- 12.Jordan B, Perez-Perez MM, Ossig R, Kaforou S, Mery E, Brunet-Errard L, Paulus C, Kritsotakis M, Gerfault L, Reina C, Kafetzopoulos D, Potamias G, Ricoul F, Tsiknakis M, Schnekenburger J, Grangeat P. Proc of pHealth. 2008 [Google Scholar]
- 13.http://www.chem.agilent.com/en-US/products/instruments/lc/1200serieshplc-chipmssystem/pages/gp15400.aspx.
- 14.Zhang C, Xing D. Nucleic Acids Res. 2007;35:4223–4237. doi: 10.1093/nar/gkm389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei JH, Hsu W-S, Fan S-K. Proc IEEE Int Conf Micro/Nano Eng Mol Sys. 2007:981–984. [Google Scholar]
- 16.Fouillet Y, Jary De, Chabrol C, Claustre P, Peponnet C. Microfluid Nanofluid. 2008;4:159–165. [Google Scholar]
- 17.Shah JJ, Sundaresan SG, Geist J, Reyes DR, Booth JC, Rao MV, Gaitan M. J Micromech Microeng. 2007;17:2224–2230. [Google Scholar]
- 18.Mondal S, Venkataraman V. J Biochem Bioph Meth. 2007;70:773–777. doi: 10.1016/j.jbbm.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Roper MG, Easley CJ, Legendre LA, Humphrey JAC, Landers JP. Anal Chem. 2007;79:1294–1300. doi: 10.1021/ac0613277. [DOI] [PubMed] [Google Scholar]
- 20.Li S, Zhang K, Yang J-M, Lin L, Yang H. Proc Int Conf Transducers. 2007:1369–1372. [Google Scholar]
- 21.Wang J, Chen Z, Corstjens PLAM, Mauk MG, Bau HH. Lab Chip. 2006;6:46–53. doi: 10.1039/b511494b. [DOI] [PubMed] [Google Scholar]
- 22.Gillot F, Arata HF, Morin FO, Collard D, Fujita H. Proc Int Conf Transducers. 2007:21–23. [Google Scholar]
- 23.Hoang VN, Kaigala GV, Backhouse CJ. Lab Chip. 2008;8:484–487. doi: 10.1039/b713764h. [DOI] [PubMed] [Google Scholar]
- 24.Cheng JY, Hsieh CJ, Chuang YC, Hsieh JR. Analyst. 2005;130:931–940. doi: 10.1039/b501061f. [DOI] [PubMed] [Google Scholar]
- 25.Cho SK, Moon H, Kim CJ. J Microelectromech Syst. 2003;12:70–80. [Google Scholar]
- 26.Gong J, Kim CJ. Lab Chip. 2008;8:898–906. doi: 10.1039/b717417a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollack MG, Paik PY, Shenderov AD, Pamula VK, Dietrich FS, Fair RB. Proc Int Conf MicroTAS. 2003 [Google Scholar]
- 28.Sista R, Hua Z, Thwar P, Sudarsan A, Srinivasan V, Eckhardt A, Pollack MG, Pamula VK. Lab Chip. 2008;8:2091–2104. doi: 10.1039/b814922d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua Z, Rouse JL, Eckhardt AE, Srinivasan V, Pamula VK, Schell WA, Benton JL, Mitchell TG, Pollack MG. Anal Chem. 2010;82:2310–2316. doi: 10.1021/ac902510u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang YH, Lee GB, Huang FC, Chen YY, Lin JL. Biomed Microdevices. 2006;8:215–225. doi: 10.1007/s10544-006-8171-y. [DOI] [PubMed] [Google Scholar]
- 31.Nelson W, Peng I, Loo JA, Garrell RL, Kim C-J. Proc IEEE Conf, MEMS. :280–283. [Google Scholar]
- 32.Srinivasan V, Pamula VK, Paik PY, Fair RB. Proc SPIE Int Soc Opt Eng. 2004:26–32. [Google Scholar]
- 33.Luk VN, Mo G, Wheeler AR. Langmuir. 2008;24:6382–6389. doi: 10.1021/la7039509. [DOI] [PubMed] [Google Scholar]
- 34.Berthier E, Warrick J, Yub H, Beebe DJ. Lab Chip. 2008;8:852–859. doi: 10.1039/b717422e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong J. PhD Thesis. University of California; Los Angeles: 2007. [Google Scholar]
- 36.Iyer KS, Klee WA. J Biol Chem. 1973;248:707–710. [PubMed] [Google Scholar]
- 37.Turapov OA, Mukamolova GV, Bottrill AR, Pangburn MK. Anal Chem. 2008;80:6093–6099. doi: 10.1021/ac702527b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooper JW, Chen J, Li Y, Lee CS. Anal Chem. 2003;75:1067–1074. doi: 10.1021/ac025768b. [DOI] [PubMed] [Google Scholar]
- 39.Peterson DS, Rohr T, Svec F, Fréchet MJ. J Proteome Res. 2002;1:563–568. doi: 10.1021/pr0255452. [DOI] [PubMed] [Google Scholar]
- 40.Lee J, Musyimi HK, Soper SA, Murray KK. J Am Soc Mass Spectrom. 2008;19:964–972. doi: 10.1016/j.jasms.2008.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.