Abstract
Purpose: Tamoxifen was approved for breast cancer risk reduction in high-risk women based on the National Surgical Adjuvant Breast and Bowel Project's Breast Cancer Prevention Trial (P-1:BCPT), which showed 50% fewer breast cancers with tamoxifen versus placebo, supporting tamoxifen's efficacy in preventing breast cancer. Poor metabolizing CYP2D6 variants are currently the subject of intensive scrutiny regarding their impact on clinical outcomes in the adjuvant setting. Our study extends to variants in a wider spectrum of tamoxifen-metabolizing genes and applies to the prevention setting. Methods: Our case-only study, nested within P-1:BCPT, explored associations of polymorphisms in estrogen/tamoxifen-metabolizing genes with responsiveness to preventive tamoxifen. Thirty-nine candidate polymorphisms in 17 candidate genes were genotyped in 249 P-1:BCPT cases. Results: CVP2D6_C1111T, individually and within a CYP2D6 haplotype, showed borderline significant association with treatment arm. Path analysis of the entire tamoxifen pathway gene network showed that the tamoxifen pathway model was consistent with the pattern of observed genotype variability within the placebo-arm dataset. However, correlation of variations in genes in the tamoxifen arm differed significantly from the predictions of the tamoxifen pathway model. Strong correlations between allelic variation in the tamoxifen pathway at CYP1A1-CYP3A4, CYP3A4-CYP2C9, and CYP2C9-SULT1A2, in addition to CYP2D6 and its adjacent genes, were seen in the placebo-arm but not the tamoxifen-arm. In conclusion, beyond reinforcing a role for CYP2D6 in tamoxifen response, our pathway analysis strongly suggests that specific combinations of allelic variants in other genes make major contributions to the tamoxifen-resistance phenotype.
Keywords: Breast cancer, tamoxifen resistance, chemoprevention, pathway analysis, breast cancer risk, genomic
Introduction
Tamoxifen was the first drug to be approved by the FDA to reduce breast cancer incidence in women at increased risk of breast cancer according to Gail Model criteria [1] based on data generated from the National Surgical Adjuvant Breast and Bowel Project (NSABP)'s Breast Cancer Prevention Trial (P-1:BCPT) [2]. Among the 13,388 eligible increased-risk women in P-l, the incidence of invasive breast cancer in those randomized to tamoxifen was significantly reduced compared with those randomized to placebo, with a risk ratio [RR] of 0.51 (95% CI 0.39 -0.66) at a mean follow-up of 47.7 months.
In a subsequent study nested within P-1:BCPT, BRCA1 and BRCA2 were resequenced in constitutional DNA from 288 breast cancer cases that developed while on study [3]. Only 19 participants with breast cancer carried deleterious (cancer-associated) mutations in BRCA1, BRCA2, or both. Despite this small sample size, a tendency for BRCA2-mutation but not BRCA1-mutation carriers to benefit from tamoxifen was suggested in this BRCAl/2-carner case population.
The toxicities of tamoxifen, especially endometrial cancer and thromboembolism [2], have limited its acceptance as a breast cancer risk-reducing agent. One strategy for addressing this resistance might be to develop tools which identify women who will experience the best balance between tamoxifen's benefits and risks. If we could identify genetic modifiers of tamoxifen-related benefits and risks, stratification of potential candidates for preventive tamoxifen into those more or less likely to achieve a net benefit from treatment becomes a real possibility.
Developing a pharmacogenetic strategy for targeting women with the optimal benefitrisk ratio was the goal of our study, NSABP P-1G3. The primary P-1 endpoint of invasive breast cancer [2] allows assessment of associations between this clinical outcome and pre-selected genetic variants, in an approach analogous to that used in the case-only study of BRCA1/2 mutations [3]. Candidate genes for P-1G3 were chosen based on published relationships to breast carcinogenesis or tamoxifen response: genes involved in estrogen synthesis and metabolism (estrogen/E pathway) or tamoxifen metabolism (tamoxifen/TAM pathway); genes encoding the estrogen and progesterone receptors; and genes regulating the generation of nitrogen oxide free radicals. Selection of polymorphisms relied on prior epidemiologic or laboratory associations with breast cancer risk and/or estrogen/tamoxifen metabolism or action. In this manner, we used the available literature to identify 39 loci in 17 candidate genes for analysis in DNA from the breast cancer cases that developed while on study.
We hypothesized that one or more of the genetic polymorphisms selected for examination in this study, alone or in combination with others, would alter estrogen and tamoxifen metabolism and/or action in a manner expected to decrease (or increase) the clinical response to tamoxifen. CYP2D6 variants with decreased metabolic activity have been reported as conferring resistance to tamoxifen [4-7]. Yet, CYP2D6 is only one of the genes that we were interested in studying, given that many other genes have potential to impact response to tamoxifen. Accordingly, the goal of our research has been to expand beyond the exclusive focus on this one gene and to carry out an analysis of multiple genes taken together as a network. The pursuit of such an alternative, systems approach to pharmacogenomic analysis of tamoxifen response assumes increased importance in view of the inconsistency of recent findings regarding the impact of polymorphisms in CYP2D6 analyzed as a single gene on clinical outcomes in breast cancer patients [8-12]. Our pathway analytic approach allowed us to investigate whether polymorphic alleles of additional enzymes within either the tamoxifen or estrogen metabolic pathways were associated with lack of response to this agent. Even if the expected small effect of polymorphisms at the individual level does not translate into observable pharmacogenetic interactions, pathway analysis incorporating all genes in the E and TAM pathways might reveal genotypic differences between cases occurring in the presence versus absence of tamoxifen. To address the usefulness of this pathway approach using the limited number of case samples (n=249) available to us from P-1:BCPT, we implemented the current pilot study. Our results suggest that elucidation of pharmacogenomic associations at the level of an entire metabolic network should contribute to refinement of criteria for selecting women at increased risk who will experience the best benefitrisk balance from preventive tamoxifen use.
Materials and methods
Segment of study population examined
This genomic investigation is an ancillary study to NSABP's P-1:BCPT trial; the details of the methodology and the demographic characteristics of the study cohort in P-1 have been previously described [2]. The overall study was approved by the institutional review boards (IRBs) of the participating institutions. Women enrolled in P-1 provided written informed consent. With one exception, the IRBs of these institutions allowed informed consent forms to include the collection of blood samples that could be used in future studies for genomic analyses.
Case definition and anonymization
For purposes of this genomic analysis, cases were defined as all incident invasive breast cancers occurring before the P-1 participants were informed of their assigned treatment on April 1, 1998, as identified from the NSABP summary file with a data lock of September 30, 1999. Non-invasive breast cancers were not included. The current study utilized lymphocyte-derived DNA samples isolated from blood collected from P-1 breast cancer cases only. The actual samples consisted of DNA aliquots remaining from the DNA utilized in P-1G, a prior genetic study of BRCA1 and BRCA2 mutations in P-1 cases [3]. As described in that study, the DNA samples from the breast cancer cases were anonymized prior to delivery to study investigators.
Selection of polymorphism markers
We employed a candidate gene strategy in selecting genes for this study, reasoningthat genes encoding proteins involved in estrogen and tamoxifen synthesis, metabolism, and function would be biologically plausible candidates as genetic modifiers of breast cancer risk or response to preventive tamoxifen (Figure 1). Table 1 lists the selected genes and markers. An extensive epidemiologic literature suggesting associations between breast cancer risk and specific polymorphisms in estrogen-synthesizing/metabolizing genes and estrogen receptor genes encouraged this approach [13-26].
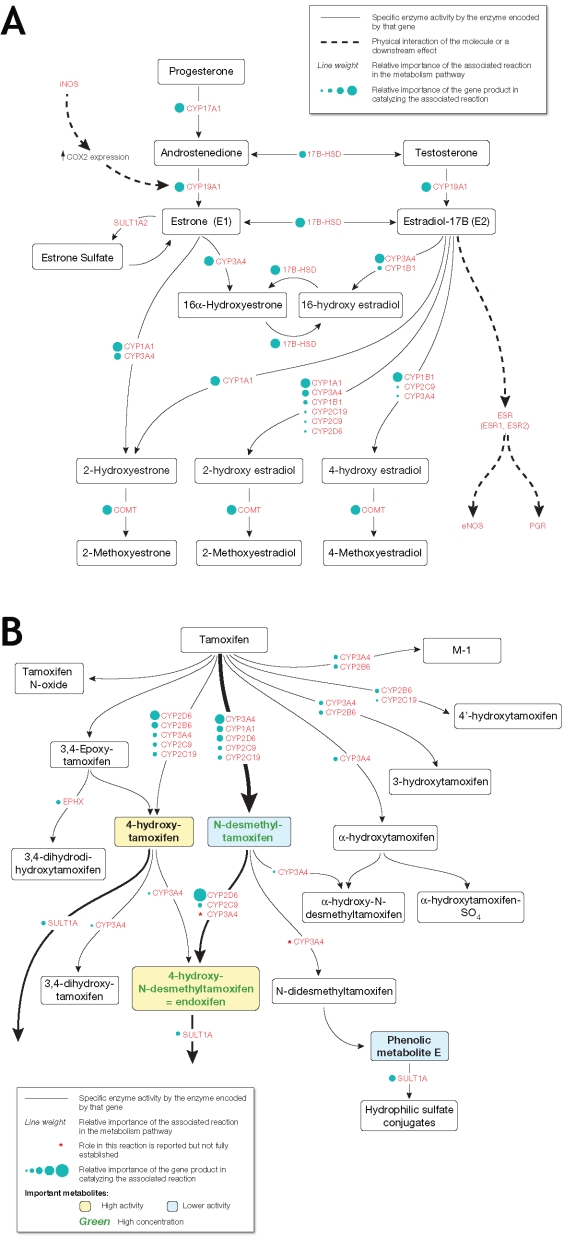
Figure 1.
Estrogen and Tamoxifen Metabolic Pathways depicted at the genomic level. The component reactions in the Estrogen (E) and Tamoxifen (TAM) metabolic pathways are depicted in terms of the genes encoding the responsible enzymes. (A) Estrogen Metabolic Pathway; (B) Tamoxifen Metabolic Pathway
Table 1.
Genes and polymorphisms selected for NSABP P-1G3 study
| Gene | Marker | dbSNP_rs | Polymorphism | cds_change | Rfreq. | Pfreq. | P(n) | Tfreq. | T(n) | P value | OR | OR CI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COMT | COMT821_V158M | 4680 | G/A | Val158Met | 0.45 | 0.542 | 167 | 0.557 | 88 | 0.769 | 1.018 | 0.694-1.495 |
| CYP17A1 | CYP17_UT34C | 743572 | T/C | 5′utr_34 | 0.40 | 0.516 | 126 | 0.508 | 62 | 0.769 | 0.984 | 0.635-1.525 |
| CYP19A1 | CYP19_C1672T | 10046 | C/T | exon 10/3′ UTR | 0.47 | 0.488 | 160 | 0.488 | 82 | 1.000 | 1.001 | 0.689-1.454 |
| CYP19A1 | CYP19_Gin6T | 4646 | G/T | intron 6 | 0.45 | 0.503 | 166 | 0.512 | 86 | 0.925 | 1.011 | 0.698-1.466 |
| CYP19A1 | CYP19_In4(TTTA)n | na | (TTTA)n | intron4 | 0.13 | 0.350 | 113 | 0.360 | 59 | 0.841 | 1.025 | 0.691-1.515 |
| CYP1A1 | CYP1A1_I462V | 1048943 | A/G | Ile462Val | 0.08 | 0.046 | 15 | 0.042 | 7 | 1.000 | 0.927 | 0.343-2.265 |
| CYP1A1 | CYP1A1_T461N | 1799814 | C/A | Thr461Asn | 0.96 | 0.039 | 13 | 0.065 | 11 | 0.268 | 1.668 | 0.711-3.844 |
| CYP1A1 | CYP1A1_T5639C | 4986884 | T/C | 3′utr_5639msp1 | 0.12 | 0.000 | 0 | 0.000 | 0 | na | na | na |
| CYP1A1 | CYP1A1_T6235C | 4646903 | T/C | 3′utr_6235msp1 | 0.12 | 0.124 | 41 | 0.143 | 24 | 0.575 | 1.157 | 0.664-1.979 |
| CYP1B1 | CYP1B1_A119S | 1056827 | G/T | Ala119Ser | 0.12 | 0.264 | 87 | 0.298 | 50 | 0.458 | 1.127 | 0.745-1.695 |
| CYP1B1 | CYP1B1_N453S | 1800440 | A/G | Asn453Ser | 0.17 | 0.200 | 66 | 0.137 | 23 | 0.085 | 0.685 | 0.400-1.136 |
| CYP1B1 | CYP1B1_V432L | 1056836 | G/C | Val432Leu | 0.41 | 0.561 | 184 | 0.500 | 84 | 0.216 | 0.889 | 0.607-1.303 |
| CYP2B6 | CYP2B6_K262R | 2279343 | A/G | Lys262Arg | 0.33 | 0.373 | 123 | 0.387 | 65 | 0.770 | 1.032 | 0.703-1.510 |
| CYP2C19 | CYP2C19_G636A** | 4986893 | G/A | TrpxStop | 0.02 | 0.000 | 0 | 0.000 | 0 | na | na | na |
| CYP2C19 | CYP2C19_G681A | 4244285 | G/A | Pro227Pro | 0.18 | 0.148 | 49 | 0.202 | 34 | 0.129 | 1.364 | 0.837-2.204 |
| CYP2C9 | CYP2C9_I359L | 1057910 | A/C | Ile359Leu | 0.06 | 0.064 | 21 | 0.060 | 10 | 1.000 | 0.948 | 0.415-2.026 |
| CYP2C9 | CYP2C9_R144C | 1799853 | C/T | Arg144Cys | 0.08 | 0.109 | 36 | 0.131 | 22 | 0.464 | 1.204 | 0.674-2.110 |
| CYP2D6 | CYP2D6_A2638del | 4986774 | A/_ | 2638A>Del(*3) Frameshift | 0.02 | 0.009 | 3 | 0.024 | 4 | 0.228 | 2.72 | 0.45-18.78 |
| CYP2D6 | CYP2D6_C1111T* | 28371706 | C/T | ThrlO7lle(*17) | a | 0.000 | 0 | 0.019 | 3 | 0.038 | na | na |
| CYP2D6 | CYP2D6_delT | 5030655 | T_ | Trp152G/R/R [DelT(*6)] | 0.01 | 0/009 | 3 | 0.006 | 1 | 1.000 | 0.65 | 0.01-8.21 |
| CYP2D6 | CYP2D6_EX4** | 3892097 | G/A | splice_defect (*4) | 0.15 | 0.000 | 0 | 0.000 | 0 | na | na | na |
| CYP2D6 | CYP2D6_P34S | 1065852 | C/T | Pro34Ser (*4) | 0.02 | 0.170 | 56 | 0.214 | 36 | 0.225 | 1.33 | 0.81-2.18 |
| CYP3A4 | CYP3A4_A290G | 2740574 | A/G | “Prom_A290G(““V””)” | 0.09 | 0.064 | 21 | 0.054 | 9 | 0.696 | 0.855 | 0.361-1.867 |
| EPHX1 | EPHX1_Y113H | 1051740 | T/C | Tyr113His | 0.64 | 0.303 | 100 | 0.319 | 53 | 0.758 | 1.052 | 0.701-1.567 |
| ESR1 | ESRl_(TA)n | na | (TA)n | Upstream from exon1 | 0.52 | 0.559 | 179 | 0.602 | 100 | 0.385 | 1.079 | 0.742-1.571 |
| ESR1 | ESR1_K303R** | na | A/G | Lys303Arg | na | 0.000 | 0 | 0.000 | 0 | na | na | na |
| ESR1 | ESR1_P | 2234693 | C/T | intron1 | 0.46 | 0.553 | 177 | 0.524 | 87 | 0.565 | 0.95 | 0.649-1.392 |
| ESR1 | ESR1_P325P | 1801132 | C/G | Pro325Pro | 0.17 | 0.227 | 75 | 0.232 | 39 | 0.911 | 1.025 | 0.655-1.588 |
| ESR1 | ESR1_X | 9340799 | A/G | intron1 | 0.32 | 0.423 | 107 | 0.450 | 59 | 0.616 | 1.059 | 0.693-1.616 |
| ESR2 | ESR2_(CA)n | na | (CA)n | intronic | 0.93 | 0.870 | 60 | 0.870 | 32 | 1.000 | 0.949 | 0.296-3.403 |
| HSD17B1 | HSD17B1_S312G | 605059 | A/G | Ser312Gly | 0.47 | 0.457 | 150 | 0.452 | 75 | 0.924 | 0.989 | 0.678-1.440 |
| NOS3 | NOS3_E298D | 1799983 | G/T | Glu298Asp | 0.24 | 0.303 | 100 | 0.339 | 57 | 0.416 | 1.112 | 0.747-1.646 |
| NOS3 | NOS3_In4VNTR | na | AGGGGTGAGG AAGTCTAGACC TGCTGC | Intron4 | 0.16 | 0.170 | 55 | 0.160 | 26 | 0.798 | 0.945 | 0.559-1.562 |
| NOS3 | NOS3_T-786C | 2070744 | T/C | 5′UTR | 0.30 | 0.367 | 116 | 0.431 | 69 | 0.196 | 1.166 | 0.795-1.708 |
| PGR | PGR_331G/A | 10895068 | G/A | promoter2 | 0.03 | 0.055 | 18 | 0.042 | 7 | 0.666 | 0.774 | 0.292-1.831 |
| PGR | PGR_44C/T | 0 | C/T | promoter1 | a | 0.003 | 1 | 0.000 | 0 | na | na | na |
| PGR | PGR_S344T | 3740753 | G/C | Ser344Thr | 0.09 | 0.152 | 50 | 0.179 | 30 | 0.519 | 1.172 | 0.707-1.917 |
| PGR | PGR_V660L | 1042838 | G/T | Val660Leu | 0.24 | 0.155 | 51 | 0.179 | 30 | 0.522 | 1.153 | 0.696-1.883 |
| SULT1A2 | SULT1A2_N235T | 1059491 | A/C | Asn235Thr | 0.38 | 0.293 | 96 | 0.357 | 60 | 0.153 | 1.219 | 0.822-1.800 |
Table 1 lists the genes and loci (markers) selected for this study with their NCBI dbSNP Identifier (dbSNP rs), the polymorphism (nucleotide change), the coding change (cds_change), reported minor allele frequency (Rfreq), the placebo arm minor allele frequency (Pfreq), the placebo arm minor allele number (Pn),the tamoxifen arm minor allele frequency (Tfreq), the tamoxifen arm minor allele number (Tn), the Fisher p-value (P value) for the difference of allele frequencies between P and T populations, and the Odds Ratio (OR) with 95% Confidence Interval (OR CI) for a given marker in the Tamoxifen versus Placebo population.
frequency not known
marker was uninformative in this sample set
Genomic assays for 39 polymorphisms in 17 genes
Buffy coat DNA samples isolated from whole blood collected from participants on entry to the P-1 trial were obtained from NSABP and stored at −20 °C until used for quality assessment and genotyping. Once a sample was thawed for use, it was stored at 4 ° C until the completion of the project. After quality control procedures were carried out (details are available in http://DunnBK_ DataSupplementAndLegends.pdf at http://sftp://caftps.nci.nih.gov), a total of 249 DNA samples were available to be genotyped for the project.
SNP genotypes were determined using homogeneous MassEXTEND™ assays with primer extension (hME™; Sequenom, San Diego, CA; http://www.sequenom.com) [27]. The hME™ is a high-throughput, multiplex SNP genotyping chemistry, which employs matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) to distinguish between the al-lele-specific extension products [28, 29]. The MALDI-TOF-based technology was chosen because, at the time of study inception, it was the best genotyping technology to enable optimization of the number of loci interrogated while keeping sample use at a minimum; using this technology, each SNP locus was assayed with 2-4 ng of DNA, enabling genotyping for all the polymorphisms listed in Table 1 with the sample quantity we obtained.
Of note, five CYP2D6 polymorphisms were selected for analysis from the literature that existed at the project inception (Table 1). Although the subsequent literature contains numerous reports of additional genomic variants that modify CYP2D6 function, we did not have access to this information at the time the study was planned. Furthermore, the technology available at that time posed challenges to the assay design due to the complexity inherent in the CYP2D6 gene [30-33]. Newer technology subsequently opened the possibility of assaying for additional polymorphisms. Although we considered using the newer methods as a follow-up to our original MALDI-TOF analysis, DNA from the tested P-1 case subjects was no longer available to us. In part this was due to the limited amount of DNA remaining after MALDI-TOF testing from the aliquots originally given to us. Importantly, however, the DNA that we received for analysis had been anonymized before transfer to our laboratory so that even if sufficient sample DNA were remaining for a subset of P-1 cases, these samples could not be linked to the samples used in our earlier analyses. Hence, the statistical analyses described below were applied to MALDI-TOF results for the SNP variants listed in Table 1. Details of the 35 SNP assay designs can be found in http://DunnBK_ TableS1_SNPAssaylnfo35_pub.xls at http://sftp://caftps.nci.nih.gov. Of note, recently Schroth, et al reported using the MALDI-TOF assay system for accurate genotyping of CYP2D6 when there was concern for limited sample quantity or quality [34].
PCR amplification of short tandem repeat polymorphic fragments (STRPs) and variable number tandem repeat (VNTR) markers were performed using custom primers (Applied Biosystems, Foster City, CA) (details in Table S2 in http://DunnBK_DataSupplementAndLegends.pdf at http://sftp://caftps.nci.nih.gov). The PCR products were detected and separated with an ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, CA) and allele sizing and calling was carried out using GeneMapper 3.0 software (Applied Biosystems, Foster City, CA).
Statistical methods
The primary efficacy endpoint in this P-1G3 sub-study of P-1:BCPT is the relative risk (RR) of having a given genotypic state while being a case, i.e. while having developed breast cancer, in the presence versus absence of tamoxifen. Genotypic states are measured in a series of progressively more complex levels, as described below.
Primary analysis - treatment relative risks (RRs) for individual polymorphic markers
All of our P-1G3 analyses included data from 249 of the individuals enrolled in P-1 who developed breast cancer during the course of follow-up in the study. Genetic assessment was performed for 39 variants of 17 genes (Table 1). The variants comprised 35 SNPs and 4 STRP/VNTRs. Each SNP was coded into three possible genetic states representing no variant, one variant copy, and two variant copies. The STRP/VNTRs were coded into all possible states depending on the length in base pairs of each of the alleles; treatment RRs were also measured for individual STRP/VNTRs collapsed into a bial-lelic format that is consistent with the literature (Table 2).
Table 2.
Conversion of STRP and VNTR marker results to biallelic form
| Loci | Rationale | Allele 1 | Allele 2 | References |
|---|---|---|---|---|
| NOS3_VNTR | The allele 417 is defined as the wild type vs all other marker alleles as variants | 417 bp | 363, 390, and/or 444 | (25, 60-61) |
| CYP19_TTTAn | The shorter repeats (<10) vs longer (>=10) | 181 bp or less | 185 bp or greater | (62-67) |
| ESR1_TAn | Shorter repeats (14 repeats or <) vs longer (>14 repeats") based on bimodal distribution with dip at 14 repeats | 170 bp or less | 172 bp or more | (68-69) |
| ESR2_CAn | Shorter repeats (<26 repeats) vs longer (= or > 26 repeats | 168 bp or less | 170 bp or greater | (70-75) |
Conversion of STRP marker results to biallelic form indicates how the results from the STRP/VNTR markers were collapsed to biallelic format for analysis. The STRP marker results were collapsed into biallelic form using the indicated cut-off points so results from this study could be compared to those from other studies in the literature. References used for each locus are given. (Abbreviations: bp - base pairs.)
The P-1G3 study design was that of a case-only analysis, in which each incident invasive breast cancer patient was studied by assessing her genotype in relation to whether or not she had been exposed to tamoxifen. This assessment of tamoxifen effect among those within a particular genetic state of SNP or STRP/VNTR was based on the testing of the treatment relative risk comparing the cases randomized to the tamoxifen group to those randomized to the placebo group. Based on Bayes' theorem, and the fact that treatment was randomly assigned in the P-1 study, the expected treatment relative risk for cases in any genetic state would be the same as the overall effect observed in the P-1 trial, specifically 0.5. The confidence intervals for these relative risks and the corresponding tests against the expected population average were calculated exactly under the corresponding binomial distributions. In addition to the assessment of tamoxifen effect within a specific genetic state, testing was performed to determine if there were tamoxifen interactions with any of the SNPs. The Mantel-Armitage test [35] was used for this analysis.
To control for multiple testing and to reduce false discovery, the polymorphisms were grouped into six families according to the functions of the genes to which they were related (Table 3). Two sets of false discovery rate (FDR) analyses were performed on each family, one for the test of the treatment relative risk within each separate genetic state of the polymorphisms, and another for the tests of tamoxifen interaction. Thus, a total of 12 FDR procedures were conducted. The FDR procedure of Benjamini and Hochberg [36] was applied to adjust for the multiple hypotheses with a threshold q=0.2. In addition, FDR-adjusted p-values were determined based on the re-sampling method of Yekutieli and Benjamini [37]. Additional analyses of individual markers (primary end-point) and pairs of markers (secondary end-point) were carried out using the chi-square test and Fisher's exact test, performed using R-project software, version 2.5.0.
Table 3.
Gene family groups
| Family | Genes involved in | Gene |
|---|---|---|
| F1 | the synthesis of estrogen | CYP17, CYP19, HSD17B1 |
| F2 | the metabolism of estrogen | CYP1A1, CYP1B1, COMT, SULT1A1, SULT1A2 |
| F3 | the metabolism of tamoxifen | CYP1A1, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP3A4, SULT1A1,SULT1A2, EPHX1 |
| F4 | regulating the structure of the estrogen receptors | ESR1, ESR2 |
| F5 | regulating the structure of the progesterone receptor | PGR |
| F6 | regulating the generation of nitrogen oxide free radicals | NOS3 |
Gene Family Groups assigns genes in the study to six families based on gene function. For each gene family, two sets of FDR (false discovery rate) analyses were conducted, one set for the tests of SNP and STRP departures from the overall tamoxifen effect, and one set for the tests of SNP-tamoxifen and STRP-tamoxifen interactions.
Secondary analysis - haplotype analysis
As a secondary analysis, we created haplotypes for the eight genes in which multiple markers were tested, using the same individual variants used in the Primary Analysis with the STRP/VNTR results converted to biallelic format according to Table 2. Following the construction of haplotypes, the 25 individual variants within the genes characterized were reduced to eight multiallelic loci, shown in Table 4 [38].
Table 4.
Haplotype analysis results: 2xk test
| Gene | Variants | chi square | df | p-value |
|---|---|---|---|---|
| CYP19A1 | In4(TTTA)n_Gin6T_C1672T | 2.4956 | 4 | 0.6454 |
| CYP1A1 | T461N_I462V_T6235C | 2.5255 | 3 | 0.4707 |
| CYP1B1 | A119S_V432L_N453S | 8.3246 | 5 | 0.1392 |
| CYP2C9 | R144C_I359L | 0.5298 | 2 | 0.7673 |
| ESR1 | (TA)n_X_P_P325P | 5.9179 | 8 | 0.6564 |
| CYP2D6 | P34S_C1111T_delT_A2638 | 9.7182 | 4 | 0.0445 |
| NOS3 | T-786C_In4VNTR_E298D | 4.3272 | 6 | 0.6325 |
| PGR | 31G/A_S344T_V66OL | 1.9026 | 4 | 0.7537 |
Table 4 lists the eight genes with multiple loci that were used in the haplotype analysis. A chi-sq test used a 2Xk approach in which for each arm (T, P) one haplotype was compared to each other haplotype in both arms (see details in the statistical methods section). This test indicates if a gene haplotype is significantly associated with either arm but does not tell which haplotype of the gene is significant. However, there were only marginally significant results for one gene, CYP2D6.
Abbreviations: df, degrees of freedom
For the work described here, haplotypes were constructed using the PHASE software, version 2.1.1 [38]. PHASE uses an expectation maximization (EM) algorithm to calculate maximum likelihood estimates of haplotype frequencies given genotype measurements which do not specify phase. Unconditional logistic regression analysis was used to calculate a chi-square statistic and the corresponding p-value for the individual locus haplotypes as a measure of association for primary and secondary endpoints described in the Primary Analysis. Logistic regression analyses were conducted using SAS version 8.2 (SAS Institute, Cary, North Carolina). Statistical tests based on haplotype counts, including the chi-square test and Fisher's exact test, were carried out using R-project software, version 2.5.0.
Tertiary analysis - pathway analysis
Gene architecture, i.e. the physical linkage of variants within a haplotype, represents only one source of lack of independence of genetic information. Lack of independence can also derive from the interactions of genes within biologic pathways, such as the tamoxifen metabolic pathway. Thus, the complex networks of biologic pathways represent an alternative to physical linkage as a framework by which the interactions of genes can be modeled. In our application of network analysis, we constructed a pathway model based on genes involved in tamoxifen metabolism and used it to test: 1) whether variations in pairs of genes are non-randomly associated in a manner dependent on their relative positions in the pathway; and 2) whether such associations differ in the two treatment arms. To accomplish this, we employed pathway analysis, a type of multiple regression analysis in which the user proposes a set of structured “causal” correlations to measure the strength of relationships between variables of interest, adjusted for other factors in the model [39-42]. In our case, the “variables of interest” were allelic variations in genes in the tamoxifen pathway. We chose to implement pathway analysis by means of Structured Equation Models (SEMs). In developing a model from biological pathways, it is essential to incorporate the adjacent nature of each set of two sequential genes, the strength of correlation of variations in these two genes, and the directionality of this sequential relationship. Influences of a single gene on multiple dependent variables can be measured simultaneously. Given a structure of correlations and genotype data, pathway analysis returns measures of the overall fit of the model, estimates of the magnitude of correlations, the proportion of variance (R2) accounted for by the model for each gene that is a dependent variable, and the significance of hypothesis tests to determine whether the pathway coefficients are statistically different from a given value (i.e. zero, or the value measured in another data classification). SEMs were used to calculate the above statistics for the primary (individual markers) and secondary (pairs of markers) endpoints described in the Primary Analysis. The fit of the pathway model was assessed for each arm, tamoxifen (T-arm) or placebo (P-arm). The parameter estimates for models derived for alternative endpoint states (in our case, the two alternative treatment arms) were tested for significant differences. SEM analyses were conducted using SAS version 8.2 (SAS Institute, Cary, North Carolina). Our application of pathway analysis is described below.
Methods used to construct pathway models
Two alternative approaches were utilized to construct the pathway models used in this study. In the first approach, pathways were constructed based on information compiled from the physiological/pharmacological literature relatingto the genes that encode enzymes involved in tamoxifen metabolism (Figure 1). In the second approach, network information derived from the Pathways Interactions Database (http://pid.nci.nih.gov/) compiled by the NCI Center for Biomedical informatics and Information Technology (NCI CBIIT) and the cancer Biomedical Informatics Grid (caBIG®) was also incorporated into the pathway models. The pathway structures derived from each of these approaches were combined to develop a pathway structure for the tamoxifen pathway. The resulting tamoxifen pathway structure (a priori tamoxifen pathway model), was then used to explicitly model interactions of the individual genotypes observed within the case population in each of the two arms (Figure 2).
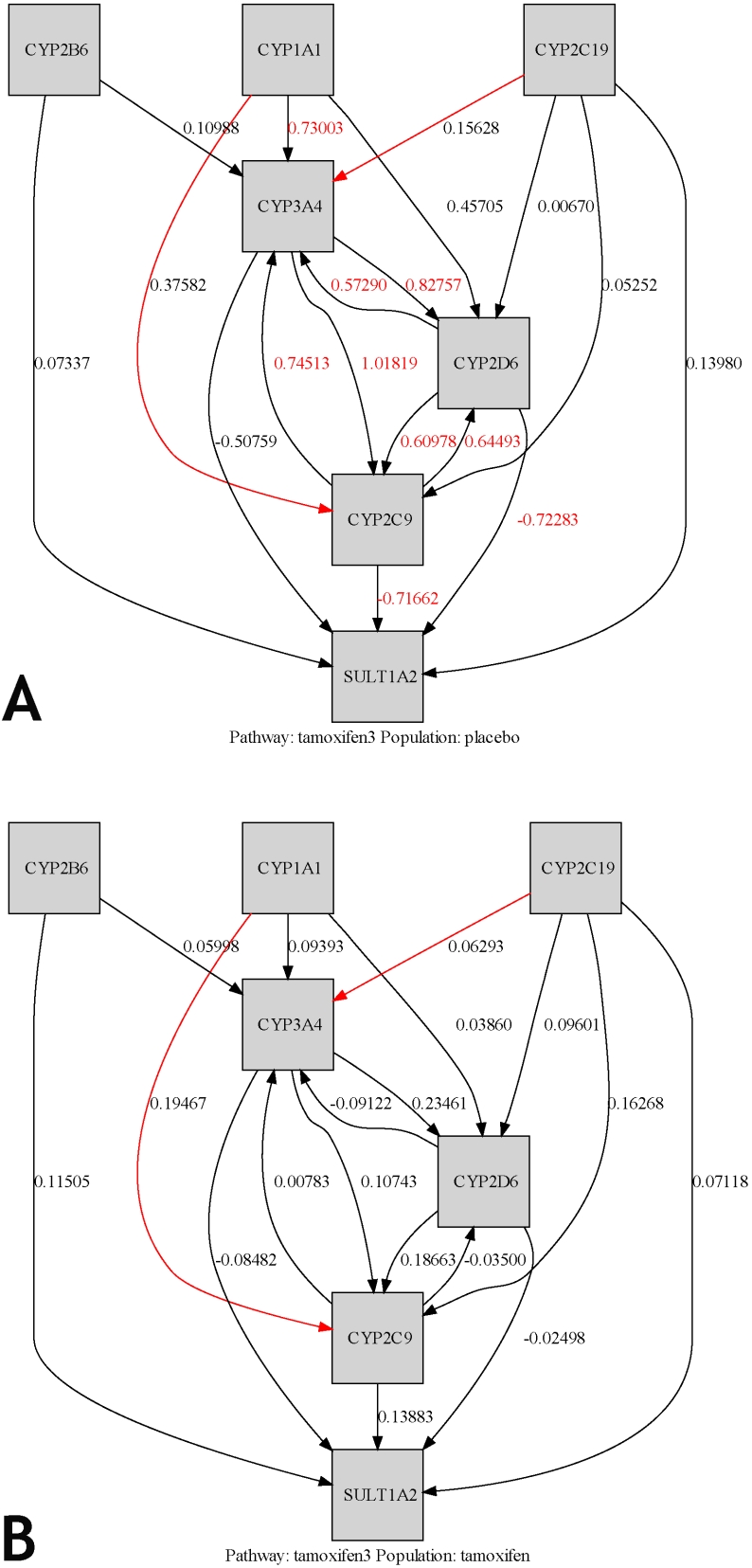
Figure 2.
Pathway Analysis Models for the Tamoxifen (TAM) Pathway tested with data from the Placebo (P) and Tamoxifen (T) arms. The pathway analysis models were generated by applying genotype data for the indicated treatment arm to the TAM pathway model. The number next to each line is a “path coefficient” which indicates how strongly the variations in the two adjacent genes are associated with each other. A zero would indicate no association between the allelic variation patterns in two adjacent genes (a red number indicates that the association is significantly stronger than a predetermined threshold level which is 0 for this figure). (A). Tamoxifen (TAM) Pathway/Placebo (P) arm. A red line indicates that there is a significant difference between the two arms with respect to the association, i.e. path coefficient, between the two linked genes. (B). Tamoxifen (TAM) Pathway/Tamoxifen (T) arm.
Analysis of pathway signatures
The individual or haplotype-based genotypic state of each locus was projected onto the pathway structure (Figure 2) as a basis for implementing our pathway analysis. For this pathway model based on the tamoxifen pathway, the interactions of the observed genotypes were modeled separately for each drug exposure arm (P or T). If the observed correlations between variation in genes in an arm are consistent with the predictions of the pathway model, the statistical analysis will reveal a “good fit”. The actual measurement that reflects how accurately the pathway model predicts the gene variants is the chi-square value for goodness of fit. A “good fit” will be associated with a non-significant chi-square p-value. Conversely, if the pathway model does not predict the correlations between variation in genes in a given drug arm, the analysis should reveal a “poor fit”. The chi-square p-value in this latter situation should be low; if the chi-square p-value <0.01, this p-value indicates a statistically significant difference between the model's prediction and the actual observations.
Results
Laboratory analysis
Genotyping assays were successfully carried out for 39 candidate loci (Table 1). Three loci, CYP2D6_EX4, CYP2C19_G636A and ESR1_ K303R, did not have a minor allele represented in this P-1 case population and thus were not included in the subsequent statistical analyses. The full set of genotype data can be obtained at http://sftp://caftps.nci.nih.gov - http://DunnBK_TableSR1_SNPFinalDataAIISNP35_pub.xls and http://DunnBK_TableSR2_STRPdata_pub.xls.
Statistical analysis
When individual polymorphisms were analyzed for association with the P- and T-arms, none of the observed associations reached statistical significance after adjustment for multiple comparisons (Table 1 p-value and OR (Cl); additional data available in Tables SR3-SR6 at http://sftp://caftps.nci.nih.gov). CYP2D6_C111T showed a borderline significant association with the T-arm (unadjusted P-value=0.038; Table 1). Similarly, no pairwise combination of polymorphic markers showed statistically significant association with either arm (Table SR6 at http://sftp://caftps.nci.nih.gov).
For each of the eight genes that contain more than one polymorphism, we constructed haplotypes. The only gene with a haplotype showing significant association with either treatment arm was CYP2D6 (p-value=0.045 in the 2Xk chi-square analysis) (Table 4). This borderline 2Xk result shows that at least one of the CYP2D6 haplotypes is associated with a drug arm but does not reveal which of the five observed haplotypes is implicated. The 2X2 chi-square analyses of CYP2D6 haplotypes showed that only the haplotype containing the variant CYP2D6_C1111T was associated with the drug arm, and this association was only of borderline significance (unadjusted p-value=0.068; Table 5).
Table 5.
CYP2D6 2×2 haplotype analysis results
| Gene | Markers | Hap ID | Hap allele | Hap P# | Other P# | HapT# | Other T# | chisq | df | p-value | Fisher p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP2D6 | P34S_C1111T_delT_A2638del | 1_1_1_1 | C_C_T_A | 268 | 62 | 124 | 44 | 3.2126 | 1 | 0.0731 | 0.0641 |
| CYP2D6 | P34S_C1111T_delT_A2638del | 1_1_1_2 | C_C_T__ | 3 | 327 | 4 | 164 | 0.8402 | 1 | 0.3593 | 0.2329 |
| CYP2D6 | P34S_C1111T_delT_A2638del | 1_1_2_1 | C_C___A | 3 | 327 | 1 | 167 | 0.0256 | 1 | 0.8730 | 1.0000 |
| CYP2D6 | P34S_C1111T_delT_A2638del | 1_2_1_1 | C_T_T_A | 0 | 330 | 3 | 165 | 3.3214 | 1 | 0.0684 | 0.0379 |
| CYP2D6 | P34S_C1111T_delT_A2638del | 2_1_1_1 | T_C_T_A | 56 | 274 | 36 | 132 | 1.1884 | 1 | 0.2756 | 0.2250 |
Table 5 includes all of the observed haplotypes for CYP2D6. In the 2×2 test, each CYP2D6 haplotype was individually tested for association with drug arm. The 2X2 chi-square analyses of CYP2D6 haplotypes show that only the haplotype containing the variant CyP2D6_C1111T was associated with arm, and this association was only of borderline significance (chi-square p-value=0.0684; Fisher p-value = 0.0379). Hap ID is the code for the haplotype (1 is the common allele and 2 is the alternate allele at a given site); Hap allele indicates the nucleotide at the 4 positions for that haplotype; HapP# is the count of the listed haplotype in the Placebo population; HapT# is the count of the listed haplotype in the Tamoxifen population; OtherP# is the count of all other haplotypes in the Placebo population; OtherT# is the count of all other haplotypes in the Tamoxifen population; df is degrees of freedom; p-value is the chi-square p-value.
Pathway analysis was performed by applying polymorphism data from each arm to the model for the TAM pathway (Figure 1), as described in Materials and Methods. The outcomes of the pathway analyses for the TAM pathway in relation to the P- and T-arms appear in the pathway projections in Figure 2. Lines connect genes that encode enzymes that catalyze sequential reactions in the TAM pathway. The number next to each line is a “path coefficient”, similar to a correlation coefficient [40-41], which indicates how strongly the variations in the two adjacent genes correlate with each other according to the pathway model (Figure 2; Table 6; Table SR7 available at http://sftp://caftps.nci.nih.gov). A zero would indicate no (i.e., random) association between the variation patterns in two adjacent genes; thus, the number next to the line reflects the degree of deviation from zero, either positive or negative, according to the model (a red number indicates that the deviation from 0 is statistically significant). Of note, the CYP2D6 gene is the hub of several strong (red numbers) correlations in the P-arm. In the P-arm population a strong correlation is observed between CYP2D6 and CYP3A4, CYP2C9, and SULT1A2. Importantly, additional strong P-arm correlations were observed between adjacent genes that do not involve CYP2D6: between CYP1A1 and CYP3A4, CYP3A4 and CYP2C9, as well as CYP2C9 and SULT1A2.
Table 6.
Path analysis data for adjacent pairs of genes in the tamoxifen pathway
| Gene_from | Gene_to | P-Arm_ path_coefficient | T-ARM_path_coefficient |
|---|---|---|---|
| CYP1A1 | CYP2C9 | 0.37582 | 0.19467 |
| CYP1A1 | CYP2D6 | 0.45705 | 0.03860 |
| CYP1A1 | CYP3A4 | 0.73003 | 0.09393 |
| CYP2B6 | CYP3A4 | 0.10988 | 0.05998 |
| CYP2B6 | SULT1A2 | 0.07337 | 0.11505 |
| CYP2C19 | CYP2C9 | 0.05252 | 0.16268 |
| CYP2C19 | CYP2D6 | 0.00670 | 0.09601 |
| CYP2C19 | CYP3A4 | 0.15628 | 0.06293 |
| CYP2C19 | SULT1A2 | 0.13980 | 0.07118 |
| CYP2C9 | CYP2D6 | 0.64493 | −0.03500 |
| CYP2C9 | CYP3A4 | 0.74513 | 0.00783 |
| CYP2C9 | SULT1A2 | −0.71662 | 0.13883 |
| CYP2D6 | CYP2C9 | 0.60978 | 0.18663 |
| CYP2D6 | CYP3A4 | 0.57290 | −0.09122 |
| CYP2D6 | SULT1A2 | −0.72283 | −0.02498 |
| CYP3A4 | CYP2C9 | 1.01819 | 0.10743 |
| CYP3A4 | CYP2D6 | 0.82757 | 0.23461 |
| CYP3A4 | SULT1A2 | −0.50759 | −0.08482 |
Table 6: Path Analysis Data for Adjacent Pairs of Genes in the Tamoxifen pathway lists the results from the path analysis generated with the Scientific Applications Software (SAS) program according to the statistical methods described in the paper. The column headers, which are standard output from the SAS program, include: gene from and gene to - indicating the direction; the path-coefficient, which is used to determine significance at the <0.05 level, listed for each arm. All analyses were run at least 100 times to ensure consistent results. These data were used to generate the pathway diagrams shown in Figure 2.
Finally, pathway analysis integrates these associations between pairs of adjacent genes (represented by the path coefficients in Figure 2) in order to elucidate the patterns of variation at the level of the entire network. The observed correlation values for an arm are then compared to the correlation coefficients predicted by the model. When the observations, taken together as a pathway, are successfully predicted by the model, then these observations are said to show a “good fit” to the model, i.e. the data fit the predictions of the model. In our dataset, pathway analysis demonstrated that our TAM pathway model is consistent with the observed genotype dataset for the P-arm cases. This consistency is reflected in a “good fit”, i.e. there is not a significant difference between the pathway model predictions and the P-arm data-set (chi-square p-value=0.4279; Table 7, Figure 2A). In essence, the TAM pathway model that we constructed successfully predicted the composite set of genetic variations in cases that had taken placebo. This contrasts with results for pathway analysis of the data from the T-arm in relation to the TAM pathway model (Figure 2B). Here, the pathway model predictions do not fit the observed patterns for allelic variations in the T-arm dataset, i.e. the goodness-of-fit is poor, and there is a significant difference between the model and the observations (chi-square p-value=0.0090; Table 7).
Table 7.
Path analysis data for the entire network of genes in the tamoxifen pathway
| Intervention Arm | Chi-square | Degrees of Freedom | Chi-square P-value |
|---|---|---|---|
| placebo | 7.0099 | 7 | 0.4279 |
| tamoxifen | 18.7549 | 7 | 0.0090 |
The path coefficients listed in Table 6 are used to model the entire network of genomic polymorphisms in the tamoxifen pathway for women in each arm. The predictions are based on prior knowledge of the pathway structure; this is the pathway model. The network derived from the polymorphisms observed in each arm is compared by a chi-square test to the network of polymorphisms predicted by the pathway model. A high p-value means the fit of the observed to the predicted polymorphism network is good and there is no statistically significant difference between them. A low p-value (p<0.01) means that the observed polymorphism network does not fit the predicted network such that a statistically significant difference exists between the observed and expected networks.
Discussion
In our pharmacogenomic analysis, we investigated in a step-wise, increasingly complex genetic fashion, the interactions between candidate polymorphisms and tamoxifen exposure among P-1 breast cancer cases. This case-only analysis revealed that only one of our genomic variants (CYP2D6_C1111T) was associated (marginal uncorrected statistical significance) with treatment arm at either the individual or the haplotype level. Thus, none of the polymorphisms or haplotypes, when tested individually or in pairs, showed a significant association with treatment arm. This limited association can be explained by: 1) In most cases, our candidate polymorphisms were selected from epidemiologic studies which involved estimates of population-based breast cancer risk, as opposed to the impact of pharmacogenomic interactions on risk. Our assumption was that genes shown to affect cancer risk would also influence the effect of gene-environment (e.g. drug) interactions on breast cancer risk; 2) The small P-1 case sample size (288), of which only 249 were available for analysis, also contributed to the lack of detection of statistically significant interaction at these levels of analysis; 3) Individually the tested polymorphisms were expected to have minor impact on phenotypic outcomes (risk or drug interaction), which likely contributed to inconsistencies in detecting risk associations in prior studies.
In contrast to the primary (SNP) and secondary (haplotype) analyses, pathway analysis is an additional approach which could potentially amplify the small risk associations occurring at the level of individual genetic variants and haplotypes by elevating the analysis to the level of a network. Multiple polymorphisms are expected to influence each other's effects and/or to show interaction with environmental factors (i.e., drug exposure) in relation to a specific shared outcome (breast cancer occurrence). The genetic polymorphisms are related to each other through a global “causal” network, or pathway. Since all the genes occupy specific places in a pathway, the biological effect of each genetic factor is not independent of the others; the genes are epistatically related to each other. An advantage of the pathway method is that it reduces the statistical complexity by limiting statistical measurements to gene-gene interactions specified by the network. Our application of pathway analysis, using SEMs [39-42], incorporated constitutional genomic variations. These candidate genomic variants had previously been shown to have a strong positive, i.e. “causal”, relationship with increased breast cancer risk, presumably via differences in the activity of their encoded enzyme products. One way to view these genotypic variants is as “perturbers” of the functioning of a metabolic pathway network [43]. We investigated how these genomic variations perturbed responsiveness to tamoxifen at both the individual gene level and the pathway network, or systems, level [44]. The pathway approach enabled us to analyze the simultaneous perturbation of tamoxifen response by all the co-existing genetic variants in the study. For example, tamoxifen was shown to reduce the risk of breast cancer in P-1:BCPT high-risk women exposed to this drug. However, it is possible for some individuals to carry minor alleles that perturb responsiveness to tamoxifen in a manner that prevents an effective response. These women could develop breast cancer despite receiving tamoxifen.
A priori, given that there was a preventive effect of tamoxifen in the overall P-1 trial, we would expect the cases in the P-arm to more closely resemble the overall P-1 population than the T-arm cases with regard to the allelic state of genes encoding tamoxifen-metabolizing enzymes. According to the results of our pathway analysis, the composite genotype dataset in the P-arm is consistent with the TAM pathway model, i.e. the observed data are not significantly different from the predictions of the model (chi-square p-value=0.4279). In contrast, the pathway is disrupted in women with breast cancer in the presence of tamoxifen such that the T-arm cases have different patterns of genotypes than the P-arm cases. Because P-1:BCPT was a randomized trial, we expect the P-arm and T-arm populations in the overall trial (i.e. cases and controls) to have the same distribution of genotypes associated with tamoxifen responsiveness and resistance. Thus, some of the alleles seen in P-arm cases would have conferred responsiveness to tamoxifen, had these women received the drug. Tamoxifen recipients who have such tamoxifen-responsive alleles, i.e. the common alleles that encode the normal metabolizing enzymes that successfully activate this pro-drug, were more likely to benefit from tamoxifen exposure, i.e., to have experienced prevention of breast cancer; they would have been selected out of the case population, leaving them under-represented among all P-1:BCPT participants who developed breast cancer, and thus, eliminated from the T-arm in this case-only study. The allelic combinations that appear in the T-arm cases would be enriched for genomic variants that encode enzymes that conjointly confer a decreased ability to metabolize tamoxifen; the T-arm cases represent a tamoxifen-resistant population. This could explain why the TAM pathway allelic observations in this ta-moxifen-resistant population do not fit the model described by the TAM pathway; in fact, there is a statistically significant difference at this network level between the observations and the predictions of the TAM pathway model for the tamoxifen-treated arm (chi-square p-value=0.0090).
The TAM pathway gene that has emerged as encoding the most important single enzyme involved in tamoxifen activation is CYP2D6, a highly polymorphic gene. An extensive pharmacogenetic literature documents the many CYP2D6 genotypes encoding isozymes with reduced activity [30-33]. Decreased activity results in poor conversion of tamoxifen to its most active metabolite, endoxifen (4-hydroxy-N-desmethyl tamoxifen) [45-47]. Women with low or absent CYP2D6 activity (due to either perturbation by an inherited CYP2D6 variant or concomitant treatment with a drug that suppresses CYP2D6 activity [32-33, 48] exhibit lower levels of endoxifen which, in turn, might be expected to impact clinical outcomes. Such an effect of CYP2D6 has been observed in several studies [4, 7, 9, 49-51]. For example, in these studies homozygosity for poor metabolizing CYP2D6 genotypes was shown to correlate with worse outcomes such as higher risk of relapse, shorter time to recurrence, and worse relapse-free survival in women with breast cancer treated with adjuvant tamoxifen [4, 7, 9, 50, 52]. As a result of such accumulating data supporting associations between CYP2D6 activity and clinical outcome, CYP2D6 genotype is beginning to be considered by some clinicians in making therapeutic decisions regarding tamoxifen use in the clinical treatment setting. However, in contrast to the findings of positive associations, a number of other studies have shown no correlation or even lower recurrence risk in tamoxifen-treated patients with poor metabolizing CYP2D6 variant status [53-57]. Among these, the updated results of an ongoing multi-center study conducted by the International Tamoxifen Pharmacogenomics Consortium, presented at the 2009 San Antonio Breast Cancer Symposium, showed no difference in clinical outcome in relation to CYP2D6 metabolizer phenotype [58, 59]. The clinical significance of CYP2D6 variants is therefore still a topic of debate among oncologists who prescribe tamoxifen in the treatment setting [8-12].
Given these contradictory findings about the impact, if any, of CYP2D6 allele status on clinical outcome, the role for factoring CYP2D6 genotype into therapeutic decision-making regarding tamoxifen requires further clarification in large prospective randomized clinical trials [9]. The NSABP P-1:BCPT offers a cohort of cases nested within such a large prospective randomized trial (13,388 participants), in this case among high-risk women. Furthermore, the inconsistency of the data regarding the impact of polymorphisms in CYP2D6 analyzed as a single gene on clinical outcomes in breast cancer patients [8-12] provides a rationale for evaluating alternative analytic strategies. Our systems approach, by incorporating multiple relevant genes taken together as a network, offers a meaningful alternative to the pharmacogenomic analysis of tamoxifen in relation to breast cancer at the single gene level. The inconsistency among study outcomes is undoubtedly due to multiple factors, both exogenous and endogenous, particularly genetic factors, that obscure the effect of variation in the single gene, CYP2D6. The systems approach to genomic analysis that we present in this paper, in providing an alternative to single gene pharmacogenetic analysis, has the potential to override the limitations inherent in focusing on one gene, even when that gene, like CYP2D6, encodes the dominant enzyme in the tamoxifen pathway. Pathway analysis offers a global analytic approach that recruits the small but additive effects of variants in multiple contributory genes that metabolize tamoxifen.
Our search for relevant CYP2D6 variants predated the current pharmacogenetic literature and was based on the then-available reports that focused on association of polymorphisms with breast cancer risk. In our candidate gene approach, five such CYP2D6 polymorphisms (Table 1) emerged from this literature search, and we tested all of them in our MALDI-TOF assay. One of the tested variants, CYP2D6_EX4, was not observed in our P-1G3 case population and was excluded from our statistical analyses. Of the four CYP2D6 variants that were subjected to statistical analyses, CYP2D6_C1111T was the only one found to exhibit significant associations with study arm. These associations were apparent when CYP2D6_C1111T was tested as a single variant or within a CYP2D6 haplotype defined for all four tested polymorphic sites (Table 4). The C1111T variant is found in haplotypes with decreased enzymatic activity and thus may be associated with resistance to tamoxifen through its failure to convert tamoxifen efficiently to endoxifen (http://www.cypalleles.ki.se/cyp2d6.htm, last accessed June 22, 2010). This mechanism could explain our statistical observations regarding CYP2D6_C1111T. Thus, the three observed C1111T alleles all occurred in the T-arm subjects, where their negative impact on endoxifen production would be expected to allow breast carcinogenesis despite the presence of the pro-drug tamoxifen.
The recent literature on CYP2D6 genotyping in relation to enzymatic function and clinical outcome reports numerous additional genomic variants that were not in the epidemiologic literature used to plan this study. Based on these updated reports we attempted to obtain additional P-1:BCPT case DNA for more complete CYP2D6 genotyping. This proved impossible because the quantity of DNA remaining in the original aliquots after our genotyping was insufficient for additional testing, and the sample anonymization required for human research subject protection prevented our matching new samples to the residual DNA.
In our analysis of the P-arm in relation to the TAM pathway, we observed that allelic variation in CYP2D6 had nonrandom associations with alleles in multiple TAM pathway genes. The CYP2D6 gene was a hub at the center of strong associations between genotypic variations only in this arm (Figure 2A). The tight correlations between allelic variation in CYP2D6 and allelic variation in adjacent pathway genes (CYP3A4, CYP2C9, and SULT1A2) are depicted as red numbers in Figure 2A. However, this cluster of nonrandom associations centered on CYP2D6 is only seen in TAM pathway analysis of the P-arm. This hub of associations disappears in the T-arm, implying a random association of CYP2D6 with specific variants in its neighboring genes (Figure 2B). This difference between the two arms with respect to the central “hub” nature of CYP2D6 suggests that the tight allelic correlations seen only in the P-arm are a major contributor to the goodness-of-fit, i.e. agreement, that the P-arm shows with the TAM pathway model. The emergence of CYP2D6 as a hub of activity in the P-arm but not the T-arm in our TAM pathway model is consistent with the key role played by this gene in tamoxifen metabolism. In this respect, our findings at the pathway, but not the individual gene, level concur with data from studies supporting an impact of CYP2D6 genotype on clinical outcomes [4-5, 7, 9, 50, 52]. The ability of pathway analysis to override the null associations seen at the individual gene level offers a possible reconciliation of the contradictory findings from other studies, all of which incorporated CYP2D6 variation in a single-gene approach.
Beyond the CYP2D6 hub, path analysis has elucidated other key foci of genetic variation. The correlations between variation in three additional pairs of adjacent genes, CYP1A1-CYP3A4, CYP3A4-CYP2C9, and CYP2C9-SULT1A2, were also significant. None of these associations involves CYP2D6, suggesting that these other pathway enzymes are meaningful contributors to tamoxifen activation. Again, these non-CYP2D6 correlations may partially explain the negative results in some of the studies that focused solely on the effect of slow-metabolizing variants of CYP2D6 on clinical outcome.
In conclusion, we have evaluated correlations between variants in multiple genes simultaneously, viewing them as a composite system. We have shown that by treating the entire system as a marker of drug response, the limitations inherent in evaluating individual genetic variants, each conferring a minor effect on tamoxifen response, may be overcome. Our underlying candidate gene approach was critical to this process, with variants identified from preexisting biological literature offering a qualitative framework on which to quantitatively model actual data from the P-1 cases. Our application of pathway analysis to the genetic network underlying tamoxifen metabolism (TAM pathway) has shown the ability to discern differences between the cases exposed to the two drugs (tamoxifen versus placebo), despite the evidence for non-significant or only marginally significant distinctions based on less complex statistical genetic comparisons. We have also demonstrated that CYP2D6 plays a key role in tamoxifen activation and clinical response at the level of pathway analysis, observations that fit with the extensive recent literature documenting its importance in metabolizing tamoxifen to its active form. Beyond reinforcing the centra I ity of CYP2D6 to tamoxifen response, our pathway analysis strongly suggests that specific combinations of allelic variants in other genes, including CYP1A1 with CYP3A4, CYP3A4 with CYP2C9, and CYP2C9 with SULT1A2, also make major contributions to the tamoxifen-resistance phe-notype.
Our observations suggest directions for pursuing more detailed analysis of pharmacogenomic interactions involving the TAM pathway. Thus, four critical foci in the TAM pathway (at CYP2D6, CYP1A1-CYP3A4, CYP3A4-CYP2C9, and CYP2C9-SULT1A2) offer a starting point for future investigations to dissect apart the specific allelic interactions that underlie these correlations. This should allow us to build more reliable genetic classifiers of tamoxifen response which could be used to offer an individualized approach to tamoxifen chemoprevention by identifying and treating only those women who are most likely to benefit from its administration.
Acknowledgments
The authors would like to thank Leslie Ford for facilitating the collaboration with NSABP; Soon-myung Paik (NSABP) for providing the samples; Alan Hoofring for graphical contributions; Carl Schaefer, Jinghui Zhang, and Sol Efroni for excellent discussions and critical reading of the manuscript; and David Flockhart for input regarding CYP2D6.
This study was supported in part by Public Health Service Grants No. U10-CA-37377, U10-CA-69974, U10-CA-12027, U10-CA-69651, and U24-CA-114732 from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services.
References
- 1.Gail MH, Brinton LA, Byar DP, Corle DK, Green SB, Schairer C, Mulvihill JJ. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 2.Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, Vogel V, Robidoux A, Dimitrov N, Atkins J, Daly M, Wieand S, Tan-Chiu E, Ford L, Wolmark N. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90:1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 3.King MC, Wieand S, Hale K, Lee M, Walsh T, Owens K, Tait J, Ford L, Dunn BK, Costantino J, Wickerham L, Wolmark N, Fisher B, National Surgical Adjuvant Breast Bowel Project Tamoxifen and breast cancer incidence among women with inherited mutations in BRCA1 and BRCA2: National Surgical Adjuvant Breast and Bowel Project (NSABP-P1) Breast Cancer Prevention Trial. Jama. 2001;286:2251–2256. doi: 10.1001/jama.286.18.2251. [DOI] [PubMed] [Google Scholar]
- 4.Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Flockhart DA, Desta Z, Perez EA, Ingle JN. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 5.Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Weinshilboum RM, Fritcher EG, Nibbe AM, Desta Z, Nguyen A, Flockhart DA, Perez EA, Ingle JN. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 6.Goetz MP, Suman VJ, Couch FJ, Ames MM, Rae JM, Erlander MG, Erlander MG, Ma XJ, Sgroi DC, Reynolds CA, Lingle WL, Weinshilboum RM, Flockhart DA, Desta Z, Perez EA, Ingle JN. Cytochrome P450 2D6 and homeobox 13/ interleukin-17B receptor: combining inherited and tumor gene markers for prediction of tamoxifen resistance. Clin Cancer Res. 2008;14:5864–5868. doi: 10.1158/1078-0432.CCR-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schroth W, Goetz MP, Hamann U, Fasching PA, Schmidt M, Winter S, Fritz P, Simon W, Suman VJ, Ames MM, Safgren SL, Kuffel MJ, Ulmer HU, Bolander J, Strick R, Beckmann MW, Koelbl H, Weinshilboum RM, Ingle JN, Eichelbaum M, Schwab M, Brauch H. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. Jama. 2009;302:1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dezentje VO, Guchelaar HJ, Nortier JW, van de Velde CJ, Gelderblom H. Clinical implications of CYP2D6 genotyping in tamoxifen treatment for breast cancer. Clin Cancer Res. 2009;15:15–21. doi: 10.1158/1078-0432.CCR-08-2006. [DOI] [PubMed] [Google Scholar]
- 9.Kuderer NM, Peppercorn J. CYP2D6 testing in breast cancer: ready for prime time? Oncology (Williston Park) 2009;23:1223–1232. [PMC free article] [PubMed] [Google Scholar]
- 10.Lash TL, Lien EA, Sorensen HT, Hamilton-Dutoit S. Genotype-guided tamoxifen therapy: time to pause for reflection? Lancet Oncol. 2009;10:825–833. doi: 10.1016/S1470-2045(09)70030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lash TL, Rosenberg CL. Evidence and practice regarding the role for CYP2D6 inhibition in decisions about tamoxifen therapy. J Clin Oncol. 2010;28:1273–1275. doi: 10.1200/JCO.2009.26.7906. [DOI] [PubMed] [Google Scholar]
- 12.Sideras K, Ingle JN, Ames MM, Loprinzi CL, Mrazek DP, Black JL, Weinshilboum RM, Hawse JR, Spelsberg TC, Goetz MP. Coprescription of tamoxifen and medications that inhibit CYP2D6. J Clin Oncol. 2010;28:2768–2776. doi: 10.1200/JCO.2009.23.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchert ET, Woosley RL, Swain SM, Oliver SJ, Coughlin SS, Pickle L, Trock B, Riegel AT. Relationship of CYP2D6 (debrisoquine hydroxylase) genotype to breast cancer susceptibility. Pharmacogenetics. 1993;3:322–327. doi: 10.1097/00008571-199312000-00006. [DOI] [PubMed] [Google Scholar]
- 14.Andersen Tl, Heimdal KR, Skrede M, Tveit K, Berg K, Borresen AL. Oestrogen receptor (ESR) polymorphisms and breast cancer susceptibility. Hum Genet. 1994;94:665–670. doi: 10.1007/BF00206961. [DOI] [PubMed] [Google Scholar]
- 15.Ambrosone CB, Freudenheim JL, Graham S, Marshall JR, Vena JE, Brasure JR, Laughlin R, Nemoto T, Michalek AM, Harrington A, Ford TD, Shields PG. Cytochrome P4501A1 and glutathione S-transferase (M1) genetic polymorphisms and postmenopausal breast cancer risk. Cancer Res. 1995;55:3483–3485. [PubMed] [Google Scholar]
- 16.Ladona MG, Abildua RE, Ladero JM, Roman JM, Plaza MA, Agundez JA, Mufioz JJ, Benftez J. CYP2D6 genotypes in Spanish women with breast cancer. Cancer Lett. 1996;99:23–28. doi: 10.1016/0304-3835(95)04033-1. [DOI] [PubMed] [Google Scholar]
- 17.Bailey LR, Roodi N, Verrier CS, Yee CJ, Dupont WD, Parl FF. Breast cancer and CYPIA1, GSTM1, and GSTT1 polymorphisms: evidence of a lack of association in Caucasians and African Americans. Cancer Res. 1998;58:65–70. [PubMed] [Google Scholar]
- 18.Thompson PA, Shields PG, Freudenheim JL, Stone A, Vena JE, Marshall JR, Graham S, Laughlin R, Nemoto T, Kadlubar FF, Ambrosone CB. Genetic polymorphisms in catechol-O-methyltransferase, menopausal status, and breast cancer risk. Cancer Res. 1998;58:2107–2110. [PubMed] [Google Scholar]
- 19.Taioli E, Bradlow HL, Garbers SV, Sepkovic DW, Osborne MP, Trachman J, Ganguly S, Garte SJ. Role of estradiol metabolism and CYP1A1 polymorphisms in breast cancer risk. Cancer Detect Prev. 1999;23:232–237. doi: 10.1046/j.1525-1500.1999.09912.x. [DOI] [PubMed] [Google Scholar]
- 20.Siegelmann-Danieli N, Buetow KH. Constitutional genetic variation at the human aromatase gene (Cyp19) and breast cancer risk. Br J Cancer. 1999;79:456–463. doi: 10.1038/sj.bjc.6690071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haiman CA, Hankinson SE, Spiegelman D, Colditz GA, Willett WC, Speizer FE, Kelsey KT, Hunter DJ. The relationship between a polymorphism in CYP17 with plasma hormone levels and breast cancer. Cancer Res. 1999;59:1015–1020. [PubMed] [Google Scholar]
- 22.Coughlin SS, Piper M. Genetic polymorphisms and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 1999;8:1023–1032. [PubMed] [Google Scholar]
- 23.Feigelson HS, McKean-Cowdin R, Coetzee GA, Stram DO, Kolonel LN, Henderson BE. Building a multigenic model of breast cancer susceptibility: CYP17 and HSD17B1 are two important candidates. Cancer Res. 2001;61:785–789. [PubMed] [Google Scholar]
- 24.Zheng W, Xie D, Cerhan JR, Sellers TA, Wen W, Folsom AR. Sulfotransferase 1A1 polymorphism, endogenous estrogen exposure, well-done meat intake, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10:89–94. [PubMed] [Google Scholar]
- 25.Tanus-Santos JE, Desai M, Flockhart DA. Effects of ethnicity on the distribution of clinically relevant endothelial nitric oxide variants. Pharmacogenetics. 2001;11:719–725. doi: 10.1097/00008571-200111000-00011. [DOI] [PubMed] [Google Scholar]
- 26.Borlak J, Thum T. Identification of major CYP2C9 and CYP2C19 polymorphisms by fluorescence resonance energy transfer analysis. Clin Chem. 2002;48:1592–1594. [PubMed] [Google Scholar]
- 27.Gut IG. DNA analysis by MALDI-TOF mass spec-trometry. Hum Mutat. 2004;23:437–441. doi: 10.1002/humu.20023. [DOI] [PubMed] [Google Scholar]
- 28.Buetow KH, Edmonson M, MacDonald R, Clifford R, Yip P, Kelley J, Little DP, Strausberg R, Koester H, Cantor CR, Braun A. High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc Natl Acad Sci U S A. 2001;98:581–584. doi: 10.1073/pnas.021506298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurinke C, van den Boom D, Cantor CR, Koster H. The use of MassARRAY technology for high throughput genotyping. Adv Biochem Eng Biotechnol. 2002;77:57–74. doi: 10.1007/3-540-45713-5_4. [DOI] [PubMed] [Google Scholar]
- 30.Zanger UM, Fischer J, Raimundo S, Stuven T, Evert BO, Schwab M, Eichelbaum M. Comprehensive analysis of the genetic factors determining expression and function of hepatic CYP2D6. Pharmacogenetics. 2001;11:573–585. doi: 10.1097/00008571-200110000-00004. [DOI] [PubMed] [Google Scholar]
- 31.de Leon J, Susce MT, Murray-Carmichael E. The AmpliChipTM CYP450 Genotyping Test. Mol Drug Ther. 2006;10:135–151. doi: 10.1007/BF03256453. [DOI] [PubMed] [Google Scholar]
- 32.Zhou SF. Polymorphism of human cytochrome P456 2D6 and its clinical significance. Part I. Clin Pharmacokinet. 2009;48:689–723. doi: 10.2165/11318030-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 33.Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance. Part II. Clin Pharmacokinet. 2009;48:761–804. doi: 10.2165/11318070-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 34.Schroth W, Hamann U, Fasching PA, Dauser S, Winter S, Eichelbaum M, Schwab M, Brauch H. CYP2D6 polymorphisms as predictors of outcome in breast cancer patients treated with tamoxifen: expanded polymorphism coverage improves risk stratification. Clin Cancer Res. 2010 doi: 10.1158/1078-0432.CCR-10-0478. Published OnlineFirst June 1, 2010; doi: 10.1158/1078-0432.CCR-10-0478. [DOI] [PubMed] [Google Scholar]
- 35.Mantel N. Chi-square tests with one degree of freedom: extensions of the Mantel-Haenszel procedure. Journal of the American Statistical Association. 1963;58:690–700. [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statistical Society. Series B. 1995;57:289–800. [Google Scholar]
- 37.Yekutieli D, Benjamini Y. Resampling-based false discovery rate controlling multiple test procedures for the correlated test statistics. J Statistical Planning Inference. 1999;82:171–196. [Google Scholar]
- 38.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wrights S. The relative importance of heredity and environment in determining the piebald pattern of guinea-pigs. Proc Natl Acad Sci U S A. 1920;6:320–332. doi: 10.1073/pnas.6.6.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li CC. Path Analysis: A primer. Pacific Grove, CA: Boxwood Press; 1975. [Google Scholar]
- 41.Rao DC, Province MA. The future of path analysis, segregation analysis, and combined models for genetic dissection of complex traits. Hum Hered. 2000;50:34–42. doi: 10.1159/000022889. [DOI] [PubMed] [Google Scholar]
- 42.Denis DJ, Legerski J. Causal Modeling and the origins of Path Analysis. Theory and Science. 2006:7. [Google Scholar]
- 43.Schadt EE. Exploiting naturally occurring DNA variation and molecular profiling data to dissect disease and drug response traits. Curr Opin Biotechnol. 2005;16:647–654. doi: 10.1016/j.copbio.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 44.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, Leonardson A, Castellini LW, Wang S, Champy MF, Zhang B, Emilsson V, Doss S, Ghazalpour A, Horvath S, Drake TA, Lusis AJ, Schadt EE. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lien EA, Solheim E, Lea OA, Lundgren S, Kvinnsland S, Ueland PM. Distribution of 4-hydroxy-N-desmethyltamoxifen and other tamoxifen metabolites in human biological fluids during tamoxifen treatment. Cancer Res. 1989;49:2175–2183. [PubMed] [Google Scholar]
- 46.Lien EA, Anker G, Lonning PE, Solheim E, Ueland PM. Decreased serum concentrations of tamoxifen and its metabolites induced by aminoglutethimide. Cancer Res. 1990;50:5851–5857. [PubMed] [Google Scholar]
- 47.Lien EA, Solheim E, Ueland PM. Distribution of tamoxifen and its metabolites in rat and human tissues during steady-state treatment. Cancer Res. 1991;51:4837–4844. [PubMed] [Google Scholar]
- 48.Stearns V, Johnson MD, Rae JM, Morocho A, Novielli A, Bhargava P, Hayes DF, Desta Z, Flockhart DA. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95:1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 49.Newman WG, Hadfield KD, Latif A, Roberts SA, Shenton A, McHague C, Lalloo F, Howell S, Evans DG. Impaired tamoxifen metabolism reduces survival in familial breast cancer patients. Clin Cancer Res. 2008;14:5913–5918. doi: 10.1158/1078-0432.CCR-07-5235. [DOI] [PubMed] [Google Scholar]
- 50.Kiyotani K, Mushiroda T, Imamura CK, Hosono N, Tsunoda T, Kubo M, Tanigawara Y, Flockhart DA, Desta Z, Skaar TC, Aki F, Hirata K, Takatsuka Y, Okazaki M, Ohsumi S, Yamakawa T, Sasa M, Nakamura Y, Zembutsu H. Significant Effect of Polymorphisms in CYP2D6 and ABCC2 on Clinical Outcomes of Adjuvant Tamoxifen Therapy for Breast Cancer Patients. J Clin Oncol. 2010;28:1287–1293. doi: 10.1200/JCO.2009.25.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramon y Cajal T, Altes A, Pare L, del Rio E, Alonso C, Barnadas A, Baiget M. Impact of CYP2D6 polymorphisms in tamoxifen adjuvant breast cancer treatment. Breast Cancer Res Treat. 2010;119:33–38. doi: 10.1007/s10549-009-0328-y. [DOI] [PubMed] [Google Scholar]
- 52.Xu Y, Sun Y, Yao L, Shi L, Wu Y, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, He L, Li P, Xie Y. Association between CYP2D6 *10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann Oncol. 2008;19:1423–1439. doi: 10.1093/annonc/mdn155. [DOI] [PubMed] [Google Scholar]
- 53.Wegman P, Vainikka L, Stal O, Nordenskjold B, Skoog L, Rutqvist LE, Wingren S. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7:R284–R290. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wegman P, Elingarami S, Carstensen J, Stal O, Nordenskjold B, Wingren S. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9:R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nowell SA, Ahn J, Rae JM, Scheys JO, Trovato A, Sweeney C, MacLeod SL, Kadlubar FF, Ambrosone CB. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91:249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 56.Okishiro M, Taguchi T, Jin Kim S, Shimazu K, Tamaki Y, Noguchi S. Genetic polymorphisms of CYP2D6 10 and CYP2C19 2, 3 are not associated with prognosis, endometrial thickness, or bone mineral density in Japanese breast cancer patients treated with adjuvant tamoxifen. Cancer. 2009;115:952–961. doi: 10.1002/cncr.24111. [DOI] [PubMed] [Google Scholar]
- 57.Dezentje VO, van Blijderveen NJ, Gelderblom H, Putter H, van Herk-Sukel MP, Casparie MK, Egberts AC, Nortier JW, Guchelaar HJ. Effect of Concomitant CYP2D6 Inhibitor Use and Tamoxifen Adherence on Breast Cancer Recurrence in Early-Stage Breast Cancer. J Clin Oncol. 2010;28:1–8. doi: 10.1200/JCO.2009.25.0894. [DOI] [PubMed] [Google Scholar]
- 58.Goetz MP, Berry DA, Klein TE. 2009. p. 492s. Adjuvant Tamoxifen Treatment Outcome According to Cytochrome P450 2D6 (CYP2D6) Phenotype in Early Stage Breast Cancer: Findings from the International Tamoxifen Pharmacogenomics Consortium, Abstract #33. Cancer Research 69: 32nd Annual San Antonio Breast Cancer Symposium; CTRC-AACR.
- 59.ITPC results incomplete; no link between CYP2D6 and tamoxifen outcome, editor. San Antonio Breast Cancer Symposium coverage. Goetz MP. Abstract #33. HemOnc Today February 25, 2010.
- 60.Hefler LA, Ludwig E, Lampe D, Zeillinger R, Leodolter S, Gitsch G, Koelbl H, Tempfer CB. Polymorphisms of the endothelial nitric oxide synthase gene in ovarian cancer. Gynecol Oncol. 2002;86:134–137. doi: 10.1006/gyno.2002.6749. [DOI] [PubMed] [Google Scholar]
- 61.Hefler LA, Grimm C, Lantzsch T, Lampe D, Koelbl H, Lebrecht A, Heinze G, Tempfer C, Reinthaller A, Zeillinger R. Polymorphisms of the endothelial nitric oxide synthase gene in breast cancer. Breast Cancer Res Treat. 2006;98:151–155. doi: 10.1007/s10549-005-9143-2. [DOI] [PubMed] [Google Scholar]
- 62.Kristensen VN, Andersen TI, Lindblom A, Erikstein B, Magnus P, Borresen-Dale AL. A rare CYP19 (aromatase) variant may increase the risk of breast cancer. Pharmacogenetics. 1998;8:43–48. doi: 10.1097/00008571-199802000-00006. [DOI] [PubMed] [Google Scholar]
- 63.Haiman CA, Hankinson SE, Spiegelman D, De Vivo I, Colditz GA, Willett WC, Speizer FE, Hunter DJ. A tetranucleotide repeat polymorphism in CYP19 and breast cancer risk. Int J Cancer. 2000;87:204–210. [PubMed] [Google Scholar]
- 64.Healey CS, Dunning AM, Durocher F, Teare D, Pharoah PD, Luben RN, Easton DF, Ponder BA. Polymorphisms in the human aromatase cytochrome P450 gene (CYP19) and breast cancer risk. Carcinogenesis. 2000;21:189–193. doi: 10.1093/carcin/21.2.189. [DOI] [PubMed] [Google Scholar]
- 65.Baxter SW, Choong DY, Eccles DM, Campbell IG. Polymorphic variation in CYP19 and the risk of breast cancer. Carcinogenesis. 2001;22:347–349. doi: 10.1093/carcin/22.2.347. [DOI] [PubMed] [Google Scholar]
- 66.Ahsan H, Whittemore AS, Chen Y, Senie RT, Hamilton SP, Wang Q, Gurvich I, Santella RM. Variants in estrogen-biosynthesis genes CYP17 and CYP19 and breast cancer risk: a family-based genetic association study. Breast Cancer Res. 2005;7:R71–R81. doi: 10.1186/bcr951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okobia MN, Bunker CH, Zmuda JM, Ezeome ER, Anyanwu SN, Uche EE, Ojukwu J, Kuller LH, Ferrell RE. Simple tandem repeat (TTTA)n polymorphism in CYP19 (aromatase) gene and breast cancer risk in Nigerian women. J Carcinog. 2006;5:12. doi: 10.1186/1477-3163-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sano M, Inoue S, Hosoi T, Ouchi Y, Emi M, Shiraki M, Orimo H. Association of estrogen receptor dinucleotide repeat polymorphism with osteoporosis. Biochem Biophys Res Commun. 1995;217:378–383. doi: 10.1006/bbrc.1995.2787. [DOI] [PubMed] [Google Scholar]
- 69.Wedren S, Lovmar L, Humphreys K, Magnusson C, Melhus H, Syvanen AC, Kindmark A, Landegren U, Fermer ML, Stiger F, Persson I, Baron J, Weiderpass E. Oestrogen receptor alpha gene haplotype and postmenopausal breast cancer risk: a case control study. Breast Cancer Res. 2004;6:R437–R449. doi: 10.1186/bcr811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsukamoto K, Inoue S, Hosoi T, Orimo H, Emi M. Isolation and radiation hybrid mapping of dinucleotide repeat polymorphism at the human estrogen receptor beta locus. J Hum Genet. 1998;43:73–74. doi: 10.1007/s100380050043. [DOI] [PubMed] [Google Scholar]
- 71.Ogawa S, Emi M, Shiraki M, Hosoi T, Ouchi Y, Inoue S. Association of estrogen receptor beta (ESR2) gene polymorphism with blood pressure. J Hum Genet. 2000;45:327–330. doi: 10.1007/s100380070002. [DOI] [PubMed] [Google Scholar]
- 72.Scariano JK, Simplicio SG, Montoya GD, Garry PJ, Baumgartner RN. Estrogen receptor beta dinucleotide (CA) repeat polymorphism is significantly associated with bone mineral density in postmenopausal women. Calcif Tissue Int. 2004;74:501–508. doi: 10.1007/s00223-003-0170-x. [DOI] [PubMed] [Google Scholar]
- 73.Gold B, Kalush F, Bergeron J, Scott K, Mitra N, Wilson K, Ellis N, Huang H, Chen M, Lippert R, Halldorsson BV, Woodworth B, White T, Clark AG, Parl FF, Broder S, Dean M, Offit K. Estrogen receptor genotypes and haplotypes associated with breast cancer risk. Cancer Res. 2004;64:8891–8900. doi: 10.1158/0008-5472.CAN-04-1256. [DOI] [PubMed] [Google Scholar]
- 74.Takeo C, Negishi E, Nakajima A, Ueno K, Tatsuno I, Saito Y, Amano K, Hirai A. Association of cyto-sine-adenine repeat polymorphism of the estrogen receptor-beta gene with menopausal symptoms. Gend Med. 2005;2:96–105. doi: 10.1016/s1550-8579(05)80016-6. [DOI] [PubMed] [Google Scholar]
- 75.Slattery ML, Sweeney C, Murtaugh M, Ma KN, Wolff RK, Potter JD, Caan BJ, Samowitz W. Associations between ERalpha, ERbeta, and AR genotypes and colon and rectal cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:2936–2942. doi: 10.1158/1055-9965.EPI-05-0514. [DOI] [PubMed] [Google Scholar]