Abstract
G protein-coupled receptors (GPCRs) represent the largest family of transmembrane receptors and are responsible for transducing extracellular signals into intracellular responses that involve complex intracellular-signaling networks. This review highlights recent research advances in fungal GPCRs, including classification, extracellular sensing, and G protein-signaling regulation. The involvement of GPCRs in pheromone and nutrient sensing has been studied extensively over the past decade. Following recent advances in fungal genome sequencing projects, a panoply of GPCR candidates has been revealed and some have been documented to play key roles sensing diverse extracellular signals, such as pheromones, sugars, amino acids, nitrogen sources, and even photons. Identification and deorphanization of additional putative GPCRs may require the development of new research tools. Here, we compare research on GPCRs in fungi with information derived from mammalian systems to provide a useful road map on how to better understand ligand–GPCR–G protein interactions in general. We also emphasize the utility of yeast as a discovery tool for systemic studies of GPCRs from other organisms.
Keywords: G protein-coupled receptor, G protein, extracellular sensing, fungus
Introduction
All living organisms are exposed to the environment and sensing environmental signals and ensuring appropriate cellular response are crucial for survival. Microorganisms have evolved elaborate mechanisms to sense and adapt to the environment in response to a variety of environmental signals. G protein-coupled receptors (GPCRs) represent the largest family of transmembrane receptors and are responsible for transmitting extracellular signals to intracellular responses by stimuli as diverse as light, protons, Ca2+, odorants, amino acids, nucleotides, proteins, peptides, steroids, and fatty acids (Maller, 2003). Despite exhibiting striking diversity in primary sequence and biological function, all GPCRs possess the same fundamental architecture consisting of seven transmembrane domains (TMs) and share common mechanisms of signal transduction. Activated GPCRs stimulate exchange of GTP for GDP on Gα proteins, dissociating Gα and Gβγ subunits that in turn trigger biological responses by binding effector proteins that regulate downstream signaling. In this review, we discuss recent advances in our understanding of how GPCRs transduce extracellular signals in fungi, the identification of novel GPCRs in fungi, and the future of GPCRs as potential targets for antifungal drug development.
Structural classification of GPCRs in fungi
The GPCR receptor family refers to proteins that contain seven TMs, localize on the plasma membrane, and sense signals outside the cell and activate intracellular G protein-mediated signal transduction pathways and cellular responses. The extracellular regions of the receptor can be glycosylated. In most studied mammalian GPCRs, these extracellular loops contain two highly conserved cysteine residues that form a disulfide bond to stabilize the receptor structure. Bovine rhodopsin is the first mammalian GPCR whose crystal structure was solved (Palczewski et al., 2000), and more recently, the first structure of a human GPCR, the β2-adrenergic receptor, was solved (Rasmussen et al., 2007). While the main feature, the seven transmembrane helices, is conserved, the relative orientation of the helices differs significantly from that of bacteriorhodopsin, which had been used as an earlier GPCR structural model (Palczewski et al., 2000; Rasmussen et al., 2007). These high-resolution structures provide a framework for understanding the wide range of biochemical and mutational data that have been amassed for GPCRs, and unveil a common activation mechanism, at least for related receptor classes.
GPCRs can be classified into six classes based on sequence homology and functional similarity, including Class A (Rhodopsin-like superfamily), Class B (Secretin receptor family), Class C (Metabotropic glutamate/pheromone receptors), Class D (Fungal mating pheromone receptors), Class E (Cyclic AMP receptors), and Class F (Frizzled/Smoothened receptors) (Attwood & Findlay, 1994; Kolakowski, 1994). This classification scheme does not include the unusual glucose/sugar sensor in yeast, Gpr1, and homologs in other fungi. Because of the unique sequence and function of these GPCRs, these can be separated as a new class. Following advances in genome sequencing projects, more GPCRs have been identified in fungi, and several GPCR classification systems have been proposed for fungal GPCRs. In one report fungal GPCRs were divided into five classes based on sequence homology and ligand sensing: classes I and II include GPCRs similar to the pheromone receptors Ste2 and Ste3, class III includes homologs of the glucose sensor Gpr1 receptor, class IV includes the nutrient sensor Stm1-like proteins, and class V includes homologs of the cAMP receptors in Dictyostelium discoideum (Han et al., 2004a). Later, this was extended to nine classes for the 16 total potential GPCRs (GprA-P and NopA) in Aspergillus nidulans (Lafon et al., 2006; Yu, 2006). Recently, a total of 10 GPCRs in Neurospora crassa were divided into five classes: pheromone receptors (Pre-1 and Pre-2), cAMP receptor-like proteins (Gpr-1, Gpr-2, and Gpr-3), carbon sensors (Gpr-4), putative nitrogen sensors (Gpr-5 and Gpr-6), and microbial opsins (Nop-1 and Orp-1) (Borkovich et al., 2004; Li et al., 2007b). In the basidiomycete Cryptococcus neoformans, we identified a large gene family of 7-TM proteins (Xue et al., 2006). Combining the classification schemes from these two fungi, we summarize the GPCRs in fungi into six classes (Table 1) (Fig. 1). Additionally, a large group of unique Pth11-like 7-TM proteins has been reported in the plant fungal pathogen Magnaporthe grisea and conserved only in the Pezizomycotina subphylum, but not in Basidiomycota or other Ascomycota subphyla (Kulkarni et al., 2005). More putative proteins with 7-TM domains exist in many fungi that require further study.
Table 1. Six classes of GPCRs in fungi.
| Species | Ste2-like pheromone receptor | Ste3-like pheromone receptor | Carbon/amino acid receptor | Putative nutrient receptor | cAMP receptor-like | Microbial Opsin |
|---|---|---|---|---|---|---|
| Ascomycetes | ||||||
| Saccharomyces cerevisiae | Ste2 | Ste3 | Gpr1 | SCRG_01312 | – | – |
| SCRG_02823 | ||||||
| SCRG_00179 | ||||||
| Schizosaccharomyces pombe | Mam2 | Map3 | Git3 | Stm1 | – | – |
| Candida albicans | Ste2 | Ste3 | Gpr1 | CAWG_02899 | – | – |
| CAWG_06059 | ||||||
| CAWG_02686 | ||||||
| Aspergillus nidulans | GprA | GprB | GprC | GprF | GprH | AN3361 |
| GprD | GprG | GprI | ||||
| GprE | AN5720 | AN8262 | ||||
| Aspergillus fumigatus | Afu3g14330 | Afu5g07880 | Afu7g04800 | Afu5g04100 | Afu3g01750 | Afu7g01430 |
| Afu1g06840 | Afu5g04140 | |||||
| Afu1g11900 | Afu3g00780 | |||||
| Neurospora crassa | Pre-2 | Pre-1 | Gpr-4 | Gpr-5 | Gpr-1 | Nop-1 |
| Gpr-6 | Gpr-2 | ORP-1 | ||||
| Gpr-3 | ||||||
| Magnaporthe grisea | MGG_04711 | MGG_06452 | MGG_08803 | MGG_04698 | MGG_06738 | MGG_09015 |
| MGG_02855 | ||||||
| Basidiomycetes | ||||||
| Cryptococcus neoformans | – | Ste3α/Ste3a | Gpr4 | Gpr2 | Gpr4 | CNAG_03572 |
| Cpr2 | Gpr3 | Gpr5 | (Ops1) | |||
| Ustilago maydis | – | Pra1 | – | UM06006 | UM03423 | UM02629 |
| Pra2 | UM01546 | UM04125 | ||||
| Coprinopsis cinerea | – | Rcb1 | – | CC1G_07132 | CC1G_02288 | – |
| Rcb2 | CC1G_04180 | CC1G_02310 | ||||
| Rcb3 | ||||||
| CC1G_02129 | ||||||
| Zygomycetes | ||||||
| Rhizopus oryzae | – | – | – | RO3G_03874 | – | – |
| RO3G_15181 | ||||||
| RO3G_13115 | ||||||
| RO3G_13187 | ||||||
| RO3G_10064 |
–, no homolog has been identified.
Fig. 1.
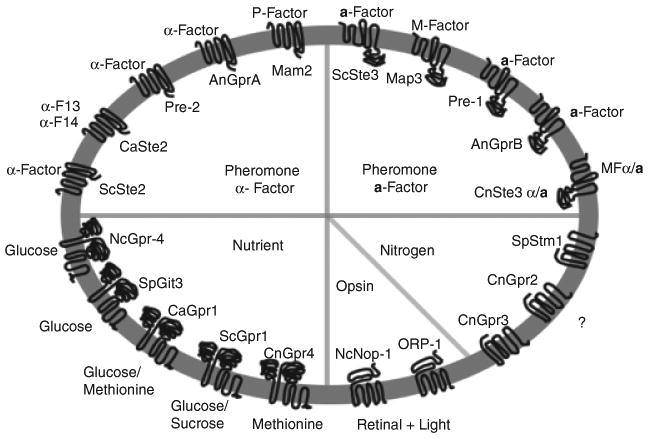
Reported GPCR classification in fungi. Fungal GPCRs can be divided into six different classes. Some representative receptors that have been studied are listed, including pheromone receptors sensing peptide pheromones (ScSte2, CaSte2, Pre-2, GprA, and Mam2), pheromone receptors sensing lipid-modified peptide pheromones (ScSte3, Map3, Pre-1, GprB, and CnSte3α/a), nutrient sensors (ScGpr1, CaGpr1, NcGpr-4, SpGit3, and CnGpr4), Stm1-like nitrogen sensors (SpStm1, CnGpr2, and CnGpr3), and microbial opsins (Nop-1 and Orp-1). This figure does not include cAMP receptor-like receptors because there are no functional studies on this group to date. Different colors represent different groups of receptors. The identified ligands are also listed corresponding to their receptors.
GPCRs in pheromone sensing
Pheromones are defined as substances that mediate communication between individuals of the same species (Karlson & Luscher, 1959). Physiological responses or behaviors are often changed when an individual senses a pheromone. Different types of pheromones such as sex pheromones, food trail pheromones, and alarm pheromones are commonly expressed in many organisms. Fungi, like many vertebrates and insects, use pheromones to attract their mate. Mating-type-specific pheromones are secreted from cells and are sensed by cells of the opposite mating-type. In ascomycetes and basidiomycetes, the pheromones are detected by pheromone receptors, which are cell surface 7-TM GPCRs; these receptors are activated upon pheromone binding and trigger the downstream-signaling pathways that lead to successful mating.
The genetic control of mating has been studied for more than three decades in the budding yeast Saccharomyces cerevisiae, which considerably advanced our understanding of how mating is controlled at a molecular level. A set of ‘sterile’ genes, which when mutated result in a sterile phenotype were discovered via genetic screens conducted in the 1970s–1980s (Mackay & Manney, 1974; Hartwell, 1980). Before cloning and sequencing of the STE genes, genes such as STE2 were already known to mediate pheromone responses in a cell type-specific manner (Hartwell, 1980). A few years later, genes such as STE2, STE3, STE4, STE6, STE7, STE11, STE12, and STE18 were all subsequently cloned and characterized (Burkholder & Hartwell, 1985; Chaleff & Tatchell, 1985; Hagen et al., 1986; Kuchler et al., 1989; Whiteway et al., 1989). As a result, components constituting the pheromone-signaling pathway were revealed. Among these STE genes, STE2 and STE3 are receptors responsible for pheromone sensing. The two genes are unrelated in sequence, but both have the signature 7-TM domain that typifies a GPCR. Ste2 is expressed in a cells and senses the mating pheromone α factor; Ste3 is expressed in α cells and senses the mating pheromone a factor. The a and α factors are both peptide pheromones. The a factor is subject to N-terminal proteolysis and C-terminal cleavage and farnesylation at the conserved CAAX motif at the C-terminus, and is secreted from a nonclassical secretion pathway that requires the pheromone transporter Ste6 (Kuchler et al., 1989; Chen et al., 1997).
Ste2 and Ste3 switch conformation from an inactive R state to an active R* state upon pheromone binding. Although unrelated in sequence, when switched to the R* state, Ste2 and Ste3 both activate the same Gα subunit Gpa1 to facilitate GDP to GTP exchange and the dissociation of Gα from the Gβγ (Ste4/Ste18) complex. In many systems both the liberated Gα and Gβγ subunits then interact with downstream effectors to engage signaling cascades. However, in the S. cerevisiae pheromone-signaling pathway, it is the Gβγ complex that functions as the main driving force to induce the downstream pheromone-signaling responses, and cells lacking either subunit of the Gβγ complex are blocked for all mating responses (Whiteway et al., 1989). The scaffold protein Ste5, the PAK kinase Ste20, and the Cdc24/Far1 complex are three main downstream targets of the Gβγ complex. When bound to Ste5, the Ste4/Ste18 complex facilitates its membrane recruitment and places the scaffold protein, the entire mitogen-activated protein kinase (MAPK) module, and Ste20 into close proximity to enable signaling circuit activation (Leeuw et al., 1998; Pryciak & Huntress, 1998). The MAPK module is a three-tiered phosphorelay system composed of Ste11 (MAPKKK), Ste7 (MAPKK) and Fus3 (MAPK). Upon signal activation, the phosphoactivated Fus3 releases the downstream transcription factor Ste12 from inhibition by Dig1/Dig2, which induces the expression of several mating-specific genes (Bardwell et al., 1994). When cells sense pheromone and activate the mating pathway, their transcriptional profile changes, cell cycle arrest occurs, and they exhibit a chemotactic response (shmooing) toward the mating partner. Components of the pheromone-signaling pathway, from the upstream receptor–G protein complex to the downstream transcription factor, are all required for these mating responses (Herskowitz, 1995).
The involvement of GPCRs in pheromone sensing is well studied in fungi, and many pheromone receptors have been identified in ascomycetes and basidiomycetes as their sequences and structure are conserved (Table 2). In ascomycetes, both Ste2- and Ste3-like pheromone receptors have been identified in the genome and many of these receptors have been demonstrated experimentally to function in mating or pheromone sensing (Kitamura & Shimoda, 1991; Tanaka et al., 1993; Kim & Borkovich, 2004, 2006; Yi et al., 2008). Interestingly, the pheromone receptors are not expressed in a cell-type-dependent manner in many ascomycetes, which is not unforeseeable for homothallic (self-fertile) species. For example, in A. nidulans, GprA and GprB were identified as pheromone receptors based on sequence homology to Ste2 and Ste3. Deletion of GPRA or GPRB impairs sexual reproduction, resulting in the production of a few small cleistothecia carrying a reduced number of ascospores, whereas gprA gprB double mutants eliminated fruiting body formation under homothallic conditions (Seo et al., 2004).
Table 2. Fungal pheromone receptor homologs.
| Species/references | Ste2-like pheromone receptor | Ste3-like pheromone receptor | Cell-type specific | Required for pheromone response/sexual development |
|---|---|---|---|---|
| Ascomycetes | ||||
| Saccharomyces cerevisiae (Burkholder & Hartwell, 1985; Hagen et al., 1986) | Ste2 | Ste3 | Y | Y |
| Schizosaccharomyces pombe (Kitamura & Shimoda, 1991; Tanaka et al., 1993) | Mam2 | Map3 | Y | Y |
| Candida albicans (Yi et al., 2008) | Ste2 | Ste3 | Y | Y |
| Candida glabrata (Muller et al., 2008) | Ste2 | Ste3 | N* | Unknown |
| Aspergillus nidulans (Seo et al., 2004) | GprA | GprB | N† | Y |
| Aspergillus fumigatus (Paoletti et al., 2005) | PreB | PreA | N† | Unknown |
| Neurospora crassa (Kim & Borkovich, 2004, 2006) | Pre2 | Pre1 | Y | Y |
| Magnaporthe grisea (Kulkarni et al., 2005) | Ste2 | Ste3 | Unknown | Unknown |
| Sordaria macrospora (Poggeler & Kuck, 2001; Mayrhofer et al., 2006) | Pre2 | Pre1 | N* | Y |
| Penicillium chrysogenum (Hoff et al., 2008) | Pcpre2 | Pcpre1 | N† | Unknown |
| Basidiomycetes | ||||
| Cryptococcus neoformans (Chung et al., 2002; Chang et al., 2003) | – | Ste3α, Ste3a | Y | Y |
| Cpr2 | N | N | ||
| Ustilago maydis (Bolker et al., 1992) | – | Pra1, Pra2 | Y | Y |
| Ustilago hordei (Anderson et al., 1999) | – | Uhpra1 Uhpra2 | Y | Y |
| Coprinellus disseminatus (James et al., 2006) | – | Cdste3.1 | N | Unknown |
| Cdste3.2 | ||||
| Cdste3.3 | ||||
| Coprinopsis cinerea (Riquelme et al., 2005) | – | Rcb1 | Y | Y |
| Rcb2 | ||||
| Rcb3 | ||||
| CC1G07395 | N | Unknown |
Homothallic species.
No sexual cycle has been described; Y, yes.
On the other hand, it is surprising that even in species in which no sexual cycle has been documented, the two pheromone receptors were found to be constitutively expressed (Table 2). For example, in the opportunistic human fungal pathogen Candida glabrata and Aspergillus fumigatus, both currently thought to be asexual species, the MAT locus has been identified with two idiomorphs, a typical organization for heterothallic fungi (Srikantha et al., 2003; Paoletti et al., 2005; Butler, 2007). The pheromone receptors were shown to be expressed constitutively in both cell types, suggesting a role independent of mating-type-specific pheromone sensing (Paoletti et al., 2005; Muller et al., 2008). Indeed, it was recently reported that instead of responding to both a and α pheromones as one would expect from a cell that express both pheromone receptors, C. glabrata cells are insensitive to either pheromone (Muller et al., 2008). Future investigations are required to examine whether the pheromone receptors have evolved new functions independent of mating, or function in novel ways during cryptic sexual cycles.
In basidiomycetes, the majority of the components in the pheromone response pathway are still conserved, but the pheromone/pheromone receptor recognition system is modified compared with the S. cerevisiae paradigm. Unlike S. cerevisiae, in which the two pheromone receptors are unrelated in sequence (Ste2 and Ste3) and recognize the peptide pheromone α factor and the lipid-modified peptide pheromone a factor, the pheromones in basidiomycetes are all lipid-modified and sensed by only Ste3-related pheromone receptors (Kronstad & Staben, 1997; Casselton, 2002). Whether Ste2-like receptors were lost from a common ancestor, or evolved only within the ascomycetes is not known, but the later seems likely given that pheromones that are known in more basal fungal lineages (Zygomycetes, Chytridiomycetes) are not peptides. Although sequence similarities between different receptors and pheromones are often observed, the receptors are still highly specific and typically do not recognize pheromones from the same-mating-type to avoid self activation.
Moreover, the pheromone/pheromone receptor genes are differentially regulated in ascomycetes and basidiomycetes. In ascomycetes, the expression of these genes are under the control of the MAT locus, while in many basidiomycetes, the pheromone/pheromone receptor genes have been incorporated as components of MAT, either in tetrapolar mating systems where they are a second unlinked sex determinant or bipolar species in which they are often encoded by MAT. The corn smut fungus Ustilago maydis and the mushroom Coprinopsis cinerea are two typical examples. In both species, two unlinked MAT loci are present in the genome: one encodes homeodomain transcription factors and the other encodes pheromone/pheromone receptor (Fraser et al., 2007). Ustilago maydis has a biallelic pheromone/pheromone receptor locus, while C. cinerea and other homobasi-diomycetes (mushroom fungi) such as Schizophyllum commune, have multiallelic pheromone/pheromone receptor loci (O'Shea et al., 1998; Kahmann & Schirrawski, 2007). The multiallelic nature of the two unlinked MAT loci creates hundreds to thousands of mating types, which promotes an outcrossing life style for these mushrooms in which inbreeding is restricted (Raper, 1966; Casselton & Olesnicky, 1998). A segmental duplication event has evidently occurred during evolution as multiple repeats of the pheromone/pheromone receptor genes forming three subloci were observed in C. cinerea (Riquelme et al., 2005). In U. maydis, pheromone/receptor recognition is essential for the cell–cell fusion event early in mating; therefore, mutants lacking receptors are sterile. However, in homobasidiomycetes that grow strictly in a filamentous form, such as C. cinerea and S. commune, hyphal–hyphal fusion during mating occurs spontaneously without a requirement for pheromone/receptor recognition. Instead, during mating, pheromone signaling regulates nuclear migration and clamp cell formation, which are required for dikaryon maintenance and completion of the sexual cycle (Casselton, 2002).
The pathogenic basidiomycetous yeast C. neoformans has a simpler bipolar mating system with two mating types, a and α (Hull & Heitman, 2002). In contrast to the small size of MAT loci in most fungi, which range from c. 700 to a few thousand base pairs, the C. neoformans MAT locus is extremely large, spanning over c. 120 kb (Lengeler et al., 2002). More than 20 genes are encoded by this locus, including the homeodomain sex regulators, pheromones, and the pheromone receptors (Lengeler et al., 2002). The fact that both key sex determinants found in tetrapolar MAT loci are now included in one large nonrecombining allele suggests that the unusual structure of the C. neoformans MAT locus likely evolved from an ancestral tetrapolar mating system (Fraser et al., 2004).
In C. neoformans, two pheromone receptors (Ste3α and Ste3a) have been identified in the MATα and MATa alleles, and studies have shown that both are responsible for pheromone sensing and critical for mating (Chung et al., 2002; Chang et al., 2003). The pheromone-signaling cascade in C. neoformans shares conserved features with that in S. cerevisiae (Lengeler et al., 2000; Hull & Heitman, 2002). When the pheromone receptor is activated by pheromone binding, a three-tiered phosphorylation cascade, composed of Ste11a/α (MAPKKK), Ste7 (MAPKK), and Cpk1 (MAPK), is sequentially activated, leading to pheromone responses (Davidson et al., 2003). One difference, however, is that in C. neoformans several components in the MAPK pathway (Ste20a/α Ste11a/α, and Ste12a/α) are encoded by MAT. Therefore, cells of different cell type express distinct alleles of these two kinases (Ste11a/α and Ste20a/α) and the transcription factor (Ste12a/α), with c. 50–70% protein sequence identity. Based on phylogenetic analysis, these three genes are thought to have been acquired into MAT early in the evolutionary history because they exhibit a lower level of amino acid sequence identity between the a and α alleles and cluster into distinct a and α clades (Fraser et al., 2004). Owing to the presence of both mating-type-specific and nonspecific elements in the MAPK cascade, Ste7 and Cpk1 must have the ability to interact with different partners in a cell-type-specific fashion. Genetic analysis has linked these three genes in a linear pathway that controls mating; cells lacking any of the three kinases exhibit in a severe unilateral mating defect and are unable to respond to pheromone in a confrontation assay (Davidson et al., 2003).
Another interesting discovery is that additional non-MAT-encoded pheromone receptors have been identified in the genomes of C. neoformans and C. cinerea. These include Cpr2 from C. neoformans and several Ste3-like genes in C. cinerea. It is unclear what roles these additional receptors play in the mushrooms, but in C. neoformans, our studies show that Cpr2 is a constitutively active GPCR (Hsueh et al., unpublished). In another bipolar mushroom, Coprinellus disseminatus, several pheromone receptor homologs have also been identified in the genome. However, genetic analysis showed that these receptors segregate independently of mating type, demonstrating that they are not part of MAT (James et al., 2006). In contrast, the MAT locus of C. disseminatus encodes only homeodomain transcription factors, which play important roles in defining sexual identity in fungi. How pheromone compatibility is achieved remains unclear; future functional studies on these receptors will provide insights into how pheromone signaling is regulated during sexual development of C. disseminatus.
Pheromone-signaling pathways are conserved among many fungal species (Lengeler et al., 2000). Based on the model established in S. cerevisiae, homologs of pheromones and pheromone receptors, G proteins, RGS proteins, and components in the conserved MAPK cascade have been discovered in many ascomycetes including Schizosaccharomyces pombe, N. crassa, M. grisea, Aspergillus spp., and Candida albicans (Nielsen & Davey, 1995; Lengeler et al., 2000; Borkovich et al., 2004; Bennett & Johnson, 2005; Paoletti et al., 2007). It is interesting that components in this pathway have even been identified in species that lack an apparent sexual cycle (Calcagno et al., 2003; Paoletti et al., 2005), suggesting that cryptic extant sexual cycles remain to be discovered in many fungi. Furthermore, this pathway has also been linked to morphogenesis and virulence in many fungal pathogens such as the rice blast fungus M. grisea, and the human pathogens C. albicans and C. glabrata (Xu & Hamer, 1996; Lo et al., 1997; Calcagno et al., 2005).
GPCRs in nutrient sensing
Nutrient sensing is central for all cells. Microorganisms developed multiple sensing systems to sense extracellular and intracellular nutrient signals to adapt to the environment and their own metabolic state. The GPCR gene family represents one important sensor system that has been found to play important roles in nutrient sensing in many fungal species (Xue et al., 1998, 2006; Lorenz et al., 2000; Bardwell, 2004; Han et al., 2004a; Lemaire et al., 2004; Miwa et al., 2004; Maidan et al., 2005a, b). Permeases and their homologs can also function as sensors for nutrients, including homologs of permeases for sugars, amino acids, ammonia, and phosphate, but will not be included in this review (Bahn et al., 2007).
GPCRs as sugar receptors
Glucose is a primary carbon and energy source for most cells, and organisms have evolved sophisticated mechanisms to sense glucose. In mammalian cells, a group of GPCRs mediate the sensation of sweet during taste (Nelson et al., 2001). Fungi, especially yeasts, developed multiple ways to sense and transport fermentable sugars like glucose, and GPCRs play important roles in sugar sensing. In the Baker's yeast S. cerevisiae, a sugar-sensing receptor (Gpr1) has been identified (Xue et al., 1998). Gpr1 and its homologs in other fungi can be grouped as a novel evolutionarily distant GPCR family, which contain a long third cytoplasmic loop, and long C-terminal tails with limited sequence similarity to other GPCR classes. Gpr1 senses glucose and sucrose to activate Gpa2, which in turn activates adenylyl cyclase to convert ATP into cAMP and thereby activate protein kinase A (PKA) (Xue et al., 1998; Kraakman et al., 1999; Lorenz et al., 2000; Lemaire et al., 2004). Interestingly, in contrast to the formation of heterotrimeric G protein complexes that function in nutrient sensing in S. pombe, two kelch-repeat proteins Gpb1/Krh2 and Gpb2/Krh1 have been identified in S. cerevisiae that associate with Gpa2 and negatively regulate Gpa2 and Gpr1 coupling, negatively regulating cAMP signaling (Harashima & Heitman, 2002, 2005; Batlle et al., 2003). A recent report has also suggested an association of these ketch-repeat proteins with Gpa2 could bypass adenylyl cyclase to regulate PKA signaling directly via interaction with the catalytic subunit of PKA (Peeters et al., 2006), but the mechanistic basis is as yet unclear. Additional targets of these ketch-repeat proteins are the neurofibromin homologs Ira1 and Ira2, which function as Ras GTPase-activating proteins (Harashima et al., 2006).
The affinity of Gpr1 for glucose is low, requiring 20–30 mM glucose for half-maximal activation (EC50) of Gpr1-dependent cAMP signaling in vivo (Versele et al., 2001). Studies have shown that the affinity of Gpr1 for sucrose is higher, with an EC50 of c. 0.5 mM. Other sugars with similar structures (such as galactose, mannose, and fructose) do not act as Gpr1 agonists, but mannose acts as an antagonist for both sucrose and glucose (Lemaire et al., 2004). Glucose phosphorylation has been found to be required for rapid activation of cAMP synthesis by glucose. The glucose phosphorylation product glucose-6-phosphate can trigger a small cAMP increase in the absence of the Gpr1 receptor, while the glucose–Gpr1 interaction alone cannot cause the rapid activation of adenylyl cyclase, suggesting that glucose-6-phosphate may function as a second messenger for cAMP activation. Some results suggest that a second G protein system may be involved in the activation of adenylyl cyclase, such as the Ras proteins (Colombo et al., 2004).
The Gpr1 protein sequence is conserved in other ascomycetes, including yeasts and filamentous fungi, but functional studies on some of these homologs revealed that not all function as carbon sensors. The human yeast pathogen C. albicans Gpr1 receptor is a homolog of Gpr1 from S. cerevisiae. Results from epistasis analysis indicate that Gpr1 functions upstream of Gα Gpa2 and directly interacts with Gpa2 in C. albicans, as in S. cerevisiae (Miwa et al., 2004; Maidan et al., 2005a, b). However, the role of Gpr1 in sugar sensing in C. albicans, if any, remains unclear. It is also unclear whether Gpa2 is part of a heterotrimeric G protein complex or associates with unique binding partners similar to Gpa2 in S. cerevisiae. Some phenotypic analysis and biochemical studies suggested that Gpr1 and Gpa2 were involved in the glucose-sensing machinery that regulates morphogenesis and hypha formation in solid media via a cAMP-dependent mechanism, but they are not required for hypha formation in liquid medium or during invasive candidiasis (Miwa et al., 2004). Independent studies also showed that Gpa2 acts downstream of Gpr1 as an activator of the cAMP-PKA pathway and that Gpr1 has only a limited role in virulence, but this study showed that deletion of either Gpr1 or Gpa2 had no effect on glucose-induced cAMP signaling (Maidan et al., 2005a, b). In contrast, glucose-induced cAMP production was abolished in strains lacking Cdc25 or Ras1, suggesting that the Cdc25-Ras1 rather than the Gpr1–Gpa2 module mediates glucose-induced cAMP signaling in C. albicans (Maidan et al., 2005a). They further demonstrated that instead of sensing glucose, Gpr1 might sense the amino acid methionine. Methionine triggers Gpr1 internalization and promotes hypha formation on solid media in a Gpr1-dependent fashion (Maidan et al., 2005a). However, the effect of methionine on hypha formation varies depending on the availability of different carbon sources in the medium, including glucose, and methionine did not induce cAMP production in C. albicans. Hence, it remains unclear whether Gpr1 senses sugars, as in S. cerevisiae, or specific amino acids like methionine, or both (Maidan et al., 2005b).
Recently, a trehalose-6-phosphate phosphatase gene (TPS2) has been shown to have a synergistic effect on virulence with Gpr1. While tps2 mutants result in reduced virulence of C. albicans due to accumulation of trehalose-6-phosphate (T6P), the gpr1 tps2 double mutants are completely avirulent in a murine model of systemic infection (Maidan et al., 2008). This synergistic effect on virulence suggests that Gpr1 and Tps2 may function independently. Thus, combination therapy targeting both proteins could prove efficacious against pathogenic fungi with increased resistance to currently used antifungal drugs (Maidan et al., 2008).
In the fission yeast S. pombe, the GPCR Git3 functions as a sensor for glucose and activates a heterotrimeric G-protein composed of the Gα Gpa2, the Gβ Git5 and the Gγ Git11, which differs from the Gpa2 protein complex composition in S. cerevisiae, where the Gpr1–Gpa2 complex controls glucose sensing but Gpa2 is not part of a heterotrimeric G protein complex, and instead associates with ketch-repeat proteins. Cells expressing Git3 fused to the Gpa1 Gα of the pheromone pathway respond to glucose with a transient activation of the pheromone pathway. Thus, Git3 is an authentic GPCR whose ligand is glucose and plays important roles in nutrient sensing (Welton & Hoffman, 2000; Hoffman, 2005).
Gpr1 homologs have also been reported in several filamentous fungi, such as N. crassa (Li & Borkovich, 2006) and M. grisea (Kulkarni et al., 2005). Gpr-4 is a Gpr1 homolog in N. crassa. Epistasis analysis indicates that Gpr-4 functions upstream of the Gα subunit Gna-1. Also, the C terminus of Gpr-4 was found to interact with Gna-1 in the yeast two-hybrid assay. Similar to S. cerevisiae, glucose induces cAMP production in N. crassa and gpr-4 mutations block this stimulation, indicating that Gpr-4 plays a role in glucose sensing and cAMP-signaling activation. Exogenous cAMP partially rescues the growth/dry mass defects of gpr-4 mutants on glycerol medium but steady-state cAMP levels are similar in wild-type strains and gpr-4 strains, suggesting that cAMP induction by glucose via Gpr-4 could be transient or localized. These results suggest that Gpr-4 is coupled to Gna-1 in a cAMP signaling pathway that regulates the response to carbon source in N. crassa (Li & Borkovich, 2006). A specific role for Gpr-4 in glucose sensing and how it regulates cAMP signaling remain to be elucidated in this case. The Gpr1 homolog in M. grisea has also been identified, but its functions remain to be analyzed (Kulkarni et al., 2005).
In basidiomycetes, no Gpr1 glucose receptor homolog has been reported. How these fungi sense sugars remains to be elucidated. One Gα protein, Gpa1 has been identified to be important for glucose-dependent cAMP-signaling activation, and a G protein complex contains Gpa1, a Gβ-like RACK homolog, and two Gγ subunits (Palmer et al., 2006). Glucose is a preferred carbon source for C. neoformans, and it activates the Gpa1-cAMP signal pathway, which is conserved with the cAMP-signaling cascade of S. cerevisiae. However, in contrast to the high affinity of Gpr1 for sucrose in S. cerevisiae, sucrose does not stimulate cAMP production in C. neoformans (C. Xue, Y.-P. Hsueh & J. Heitman, unpublished data). There is no Gpr1 sequence homolog in C. neoformans, and one GPCR, Gpr4, shares structural similarity rather than sequence identity with Gpr1 for S. cerevisiae and C. albicans. Similar to Gpr1, Gpr4 contains a long third intracellular loop and a long C-terminal tail, but our study revealed that Gpr4 is not directly implicated in glucose sensing (Xue et al., 2006).
Amino acid sensing in fungi
GPCRs are common sensors for amino acids in humans, such as the class 3 GPCRs that sense L-amino acids, and the extracellular calcium-sensing receptor (CaR), which responds to acute fluctuations in extracellular ionized Ca2+ concentration and also responds to aromatic, aliphatic, and polar chain amino acids. Heterodimeric taste receptors from mammals respond to aliphatic, polar, charged, and branched chain amino acids but not to aromatic amino acids (Conigrave & Hampson, 2006).
Amino acids are also important nutrients for fungi, and are detected by specialized sensor systems, which include the general amino acid permease Gap1 and the Ssy1–Ptr3–Ssy5 (SPS) system (Bahn et al., 2007). GPCRs have also been reported to sense amino acids in fungi. In C. albicans, methionine is important for the yeast to hypha transition on solid medium, and Gpr1 has been found to sense methionine to control filamentation in the presence of carbon sources such as glucose (Maidan et al., 2005a, b). It is presently unclear whether Gpr1 directly senses extracellular methionine or internal amino acids.
The GPCR Gpr4 identified in C. neoformans was also found to sense amino acids and activate cAMP signaling (Xue et al., 2006). The Gα protein Gpa1 in C. neoformans controls cAMP signaling and plays important roles in mating and production of virulence factors, such as capsule formation and melanin production. Gpr4 is a 7-TM protein that shares structural similarity with the Gpr1 glucose sensor in S. cerevisiae. Both are > 800 amino acids with long third cytoplasmic loops and long C-terminal tails. Mutagenesis studies revealed that Gpr4 is important for capsule production and mating, two features controlled by Gpa1-cAMP-signaling pathway. Gpr4 directly interacts with Gpa1 in a split-ubiquitin yeast two-hybrid system. Genetic and biochemical studies indicate that Gpr4 activates Gpa1 and is involved in cAMP-signaling regulation. Because Gpr4 is not important for melanin production and virulence, additional upstream receptors besides Gpr4 may govern Gpa1 function. It is also possible that other mechanisms may contribute to Gpa1 activation, such as glucose-6-phosphate has been identified as an important compound that activates cAMP signaling through Gpa2 and adenylyl cyclase in S. cerevisiae.
Interestingly, although glucose is the preferred carbon source for Cryptococcus and can induce transient cAMP production via Gpa1 G protein, Gpr4 is not important for glucose sensing based on direct cAMP assays. Similar to Gpr1 in C. albicans, Gpr4 has been found to sense amino acids such as methionine, and methionine plays a role in mating filament production (Xue et al., 2006). Methionine induces the internalization of a Gpr4–DsRED fusion protein and also induces transient cAMP production in C. neoformans, which is blocked by gpr4 mutants. But the role of methionine remains to be elucidated at a molecular level in this case. Activation of cAMP signaling by glucose and amino acids represents a nutrient coincidence detection system also shared in other pathogenic fungi such as C. albicans. There are Gpr1 and Gpr4 sequence homologs in other fungi that remain to be understood, and it would be interesting to investigate the functions of these membrane proteins.
Nitrogen sensor Stm1
A putative 7-TM protein, Stm1, which is required for proper recognition of nitrogen starvation signals, was first isolated as a multicopy suppressor of a ras1 synthetic lethal mutant under nitrogen-deficient conditions in S. pombe (Chung et al., 2001). Under nutrient replete conditions, overexpression of Stm1 inhibited vegetative cell growth, resulted in decreased intracellular cAMP levels, increased expression of the meiosis-specific proteins Ste11, Mei2, and Mam2, and facilitated sexual development in homothallic cells. The Stm1 protein was shown to interact with Gpa2 through its C-terminal transmembrane domains, and can function through Gpa2-dependent and/or -independent pathways. Stm1 could function as a sentinel molecule sensing the nutritional state of the cells, stopping the proliferative cell cycle, and preparing the cell to enter meiosis under nutritionally deficient conditions (Chung et al., 2003). However, its ligands and detailed function remain to be elucidated.
Stm1 sequences are conserved in other Ascomycetes, such as N. crassa and A. nidulans; and Basidiomycetes, such as C. neoformans and U. maydis, but their functions are as yet unclear. In N. crassa, Gpr-5 and Gpr-6 are homologous to Stm1 (Borkovich et al., 2004). Three Stm1 homologs in A. nidulans and two homologs in C. neoformans and U. maydis have been identified, suggesting this multiple member GPCR group may be of importance in these fungi (Table 1).
Microbial opsins and photochemical reactions
Opsins are a class of retinal binding, seven transmembrane helix proteins that function as light-responsive ion pumps or sensory receptors. Previously, genes encoding opsins have been identified in animals and the Archaea, and more recently in fungi and other eukaryotic microorganisms.
Light sensing is important for fungal development, especially sporulation and mating (Idnurm & Heitman, 2005). Nop-1, an archaeal opsin homolog in N. crassa, was the first opsin protein reported in fungi and has been demonstrated to bind all-trans retinal in vitro (Li et al., 2007b). The Nop-1 protein shares up to 81.8% amino acid identity with archaeal opsins in the 22 retinal-binding pocket residues, including the conserved lysine residue that forms an Schiff base linkage with retinal. Detection of gene expression and heterologous expression of NOP-1 in Pichia pastoris confirmed that Nop-1 functions as a rhodopsin in N. crassa photobiology (Bieszke et al., 1999a, b). The analysis of spectroscopic properties of Nop-1 revealed six distinct states in the Nop-1 photocycle, which associated with low efficient proton transport, similar to that of haloarchaeal sensory rhodopsin II (Brown et al., 2001). The NOP-1 gene is highly expressed in cultures that support asexual sporulation (conidiation) in N. crassa. Through analysis of NOP-1 transcript levels in wild-type strains and mutants blocked at various stages of conidiation, Nop-1 was found to be a late-stage conidiation protein that regulates the expression of conidiation-related genes Al-2, CON-10, and CON-13. The results suggest that Nop-1 directly or indirectly modulates carotenogenesis (a light-regulated carotenoid biosynthesis process) and repression of conidiation-specific gene expression in N. crassa (Bieszke et al., 2007).
Based on the sequence homology to Nop-1, more opsin-like proteins with 7-TM have been found in other fungi. An opsin gene (ops) has been characterized from Leptosphaeria maculans, the ascomycete that causes black-leg disease of Brassica species. The L. maculans opsin is transcribed at high levels in mycelia independent of light, compared with that of Nop-1 that is transcribed only in the light (Idnurm & Howlett, 2001). Recent studies revealed that this opsin protein can form a proton pump to build an electrochemical transmembrane gradient of proton (Waschuk et al., 2005). These new developments suggest that opsins in lower eukaryotes like fungi may retain the original role similar to archae as a proton pump. Detailed studies in other fungi are necessary to reveal whether opsins in fungi have conserved roles in proton gradient generation. An opsin homolog has also been identified in C. neoformans, but its function remains unclear.
Potential receptors for free fatty acids
Oxylipins comprise a family of oxygenated fatty acid-derived signaling molecules that exhibit potent biological activities in animals, plants, and fungi. Recently, free fatty acids (FFAs) have been demonstrated to serve as ligands for orphan GPCRs in mammals and have been proposed to play a critical role in glucose homeostasis. GPR40 and GPR120 are activated by medium and long-chain FFAs (Itoh et al., 2003; Tanaka et al., 2007), whereas GPR41 and GPR43 can be activated by short-chain FFAs (Xiong et al., 2004). GPR40 mediates the majority of the effects of FFAs on insulin secretion. Thus, these GPCRs have potential as novel targets for diabetes (Brown et al., 2005; Rayasam et al., 2007).
In fungi, there is no report of a fatty acid sensor, but FFAs have been shown to be important for fungal development. Oxylipins have been identified in A. nidulans and demonstrated to function as signaling molecules promoting fungal–host communication (Tsitsigiannis & Keller, 2006, 2007). Three fatty acid dioxygenases (PpoA, PpoB, and PpoC), which control biosynthesis of the oxylipin psi factors, have been identified in Aspergillus species and found to coordinate sexual and asexual sporulation, as well as host colonization and mycotoxin production (Tsitsigiannis et al., 2005b). These enzymes have sequence similarity with cyclooxygenases, the mammalian prostaglandin synthases, and have been also found to be responsible for prostaglandin production in the human fungal pathogen A. fumigatus. Ppo products, prostaglandins and/or other oxylipins may serve as activators of mammalian immune responses contributing to enhanced resistance to opportunistic fungi and as factors that modulate fungal development contributing to resistance to host defenses (Tsitsigiannis et al., 2005a). There is no homolog of the Ppo proteins or cyclooxygenase homolog in C. neoformans or C. albicans, but prostaglandin production has been detected in both (Noverr et al., 2001). PGs may be produced by alternative mechanisms or biosynthetic pathways. The laccase enzyme that controls melanin production in C. neoformans has recently been found to be critical for prostaglandin production in Cryptococcus, providing insight into a new and unique fungal prostaglandin pathway (Erb-Downward et al., 2008). How fungi sense FFAs still remains unknown. In both A. nidulans and C. neoformans, multiple novel GPCR candidates have been identified, and it is possible that one or more of these GPCR candidates senses fatty acids.
Orphan GPCRs in fungi
Besides the GPCRs described above, most fungi contain more 7-TM proteins with no or only limited knowledge as to their functions. In this review, these 7-TM proteins are considered as orphan receptors. In C. neoformans, we identified over 60 7-TM proteins. After excluding those false candidates, such as protein permease homologs, protein with incorrect N-terminal and C-terminal localization, and seven proteins related to reported GPCRs, there are still c. 24 putative 7-TM proteins with unknown function. Considering the complexity of the environmental signals during fungal–host interactions, some of these orphan receptors could sense additional ligands besides those that were described in these review. Recently, we identified some plant-derived signaling compounds that are important for the sexual development of Cryptococcus during fungal–plant interactions (Xue et al., 2007). It would be interesting to investigate whether one or more of these putative orphan 7-TM proteins are involved in plant signal sensing by this human fungal pathogen. In A. nidulans, 16 GPCRs are identified but their ligands remain to be explored. In total 76 GPCR-like proteins have been identified in M. grisea based on the genome sequence analysis, including 61 Pth11-like proteins containing PTH11 domain and 15 GPCRs similar to reported GPCRs in other fungi (Kulkarni et al., 2005). Studies on these additional orphan receptors could considerably advance our understanding of extracellular signal sensing in fungi, and potentially identify novel antifungal agents.
The recent study of insect olfactory receptors revealed interesting properties. The insect olfactory receptor gene family is a large conserved gene family with 7-TM, such as the fruitfly Drosophila melanogaster contains 62 members. New evidence revealed these receptors are not GPCRs; instead, they form heteromeric ligand-gated ion channels with distinct 7-TM topology with the amino terminus located intracellularly (Sato et al., 2008). There is no direct sequence homolog of these olfactory receptors in fungi, but we did identify some 7-TM proteins in C. neoformans containing a similar topology as these channel proteins. It is possible that they could also form ligand-gated channels. The rhodopsin homolog in the green algae Chlamydomonas reinhardtii also function as a light-gated channel (Nagel et al., 2002, 2005). Similarly, the rhodopsin protein in L. maculans can also function as a proton pump (Waschuk et al., 2005), suggesting the function of opsins in green algae and fungi may be conserved. These findings offer the caveat that some orphan receptors in fungi, which were identified because of their putative 7-TM topology, may not be GPCRs, and instead should possess ligand-gated channel receptors properties.
Regulation of GPCR signaling
GPCR-mediated signal transduction involves a complicated intracellular network of signaling molecules, including G proteins and their regulators such as regulator of G protein signaling (RGS) proteins. G proteins usually contain Gα, Gβ, and Gγ heterotrimeric subunits with some exceptions such as in S. cerevisiae in which the Gα Gpa2 subunit associates with ketch-repeat proteins, and play a central role in transducing extracellular signals into intrinsic signals and effecting appropriate biochemical and physiological responses. The basic principles of G protein regulation in both cAMP and pheromone-signaling pathways in fungi have been elucidated in S. cerevisiae; however, recently several unexpected new facets in the pheromone-signaling regulation were revealed (Dohlman & Slessareva, 2006). For a long time, the sole function of the Gα subunit Gpa1 was thought to be sequestration of the Gβγ complex as a negative regulator, yet new evidence supports a revised model in which pheromone signaling is also positively transmitted via Gpa1 (Guo et al., 2003). More specifically, the GTP-bound form of Gpa1 can induce mating-specific transcription and morphogenesis in the absence of pheromone, via the phosphatidylinositol-3-kinase Vps34 and its regulator Vps15, which form a complex on the endosomal membrane to regulate protein sorting (Slessareva et al., 2006). Activated Gpa1 colocalizes with the Vps34/Vps15 complex on endosomes, and binds directly to Vps34 to induce phosphatidylinositol-3-phosphate production. Furthermore, Vps15 has seven WD40 repeats and was found to preferentially interact with the GDP-bound Gpa1, mimicking hallmarks of a Gβ subunit. Traditionally, heterotrimeric G proteins are thought to transduce signals at the plasma membrane. Thus, the ability of Gpa1 to translocate into intracellular compartments to activate downstream effectors provides a new paradigm for G protein-signaling regulation (Koelle, 2006).
In general, heterotrimeric G proteins are activated by GPCRs, and GPCRs function as guanine nucleotide exchange factors (GEF) for G proteins. The conformational change of a GPCR following ligand binding enhances binding to the corresponding Gα subunit to promote exchange of GDP to GTP, leading to the dissociation of Gα from Gβγ. Freed Gα and Gβγ can each activate downstream-signaling pathways.
Besides GPCRs, activator of G protein-signaling (AGS) proteins have been identified in mammals and Caenorhabditis elegans and function as another G protein activator gene family (Blumer et al., 2007). AGS proteins are representative of a growing number of accessory proteins that influence signal propagation, facilitate cross talk between various types of signaling pathways, and provide a platform for diverse functions of both the heterotrimeric Gαβγ and individual Gα and Gβγ subunits. Most AGS proteins contain G protein regulatory (GPR) domains and their functions are independent of G protein activation via GPCRs. They compete with GPCRs for G protein activation, and have additional functions in other signaling pathways. In fungi, there is no direct report of AGS proteins. A recent study on the functions of Arr4/Get3 in S. cerevisiae suggests that this intracellular GEF may function similarly to the AGS proteins (Lee & Dohlman, 2008).
RGS proteins are GTPase-activating proteins for Gα, and they function primarily as GTPase-accelerating proteins (GAPs) to increase the hydrolysis rate of GTP bound to Gα subunits, thereby inactivating Gα. After GTP hydrolysis, the Gα subunit returns to its GDP-bound, inactive state that then sequesters the Gβγ complex into a heterotrimer, leading to down regulation of G protein signaling (Dohlman et al., 1996; Tesmer et al., 1997; Dohlman & Thorner, 2001; Siderovski & Willard, 2005). RGS proteins are, therefore, physiologically important negative regulators of GPCR signaling.
Following their discovery in the 1990s, RGS proteins have emerged as crucial regulators of GPCR signaling. In humans, over 20 RGS proteins have been identified (Jean-Baptiste et al., 2006; Wieland et al., 2007). In S. cerevisiae, the RGS protein Sst2 was first identified as a negative regulator of the pheromone response pathway, which is controlled by the pheromone receptors Ste2 and Ste3, and the coupled Gα subunit Gpa1 (Dohlman et al., 1996; Apanovitch et al., 1998). New features of this regulator have been revealed recently, indicating Sst2 can directly bind via its DEP domain to the C-terminal tail of the pheromone receptor Ste2, and thereby functions as a principal regulator of mating pheromone signaling (Ballon et al., 2006; Chasse et al., 2006). In addition, binding to the C-terminal tail of the receptor places Sst2 in close proximity to its substrate Gpa1, ensuring that regulation is both rapid and specific. This finding not only revealed a new interaction in the pheromone response pathway, but also has further implications on the roles of DEP-domain containing proteins in signal transduction (Ballon et al., 2006). Rgs2 is the second RGS domain protein in S. cerevisiae and negatively regulates glucose signaling via the GPCR Gpr1 and its coupled Gα subunit Gpa2, which control cAMP-PKA signaling (Versele et al., 1999).
The importance of RGS proteins has been studied in various other fungal species (Fig. 2). Similar to S. cerevisiae, the Sst2 homolog in the human fungal pathogen C. albicans also controls mating responses (Dignard & Whiteway, 2006). While only two RGS proteins are present in the model yeast S. cerevisiae, more RGS proteins exist in most other fungal systems that have been studied. Functional studies in filamentous fungi revealed RGS proteins regulate signals that control vegetative growth, sporulation, stress responses, and pathogenicity, in organisms as diverse as A. nidulans (Han et al., 2004b; Lafon et al., 2005, 2006; Yu, 2006), the rice blast fungus M. grisea (Liu et al., 2007), the chestnut blight fungus Cryphonectria parasitica (Segers et al., 2004), the mushroom S. commune (Fowler & Mitton, 2000), and the insect fungal pathogen Metarhizium anisopliae (Fang et al., 2007).
Fig. 2.
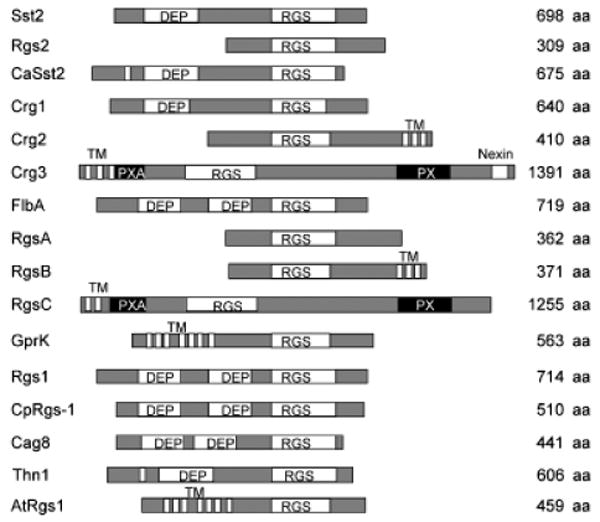
Schematic of RGS proteins in fungi. A schematic of RGS domain proteins in Saccharomyces cerevisiae (Sst2 and Rgs2), Candida albicans (CaSst2), Cryptococcus neoformans (Crg1, Crg2, and Crg3), Aspergillus nidulans (FlbA, RgsA, RgsB, RgsC, and GprK), Schizophyllum commune (Thn1), Magnaporthe grisea (Rgs1), Cryphonectria parasitica (CpRgs-1), Metarhizium anisopliae (Cag8), and the plant Arabidopsis thaliana (AtRGS1). Names of the different domains are indicated. DEP, domain found in Dishevelled, Egl-10, and Pleckstrin; RGS, regulator of G protein-signaling domain; TM, transmembrane domain; PXA, PX association domain; PX, Phox homology domain, a novel phosphoinositide (PI)-binding domain.
Five proteins containing RGS domains have been identified in A. nidulans, including GprK that contains both 7-TM and an RGS domain, similar to AtRGS1 in Arabidopsis, which plays an important role in plant cell proliferation (Chen et al., 2003). FlbA is the first RGS protein identified in A. nidulans, and contains one RGS domain and two DEP domains, similar to Sst2 in S. cerevisiae (Han et al., 2004b). FlbA regulates the Gα protein FadA to control hyphal proliferation, development, and biosynthesis of secondary metabolites (Yu et al., 1996; Hicks et al., 1997; Yu, 2006). RgsA is a homolog of Rgs2 in S. cerevisiae and negatively regulates stress responses and asexual sporulation via GanB signaling (Han et al., 2004b). Other putative RGS proteins, such as RgsB, RgsC, and GprK, remain to be studied. The Rgs1 protein in M. grisea is an analog of FlbA and has recently been found to interact with all three Gα subunits (MagA, MagB, and MagC) and regulates asexual development, thigmotropism, and pathogenicity (Liu et al., 2007).
The G protein-signaling pathways are well studied in the basidiomycete C. neoformans, which is considered a model pathogenic fungus (Fig. 3). Three G protein α subunits control two major G protein-signaling pathways that are important for cell development and virulence. In the pheromone response pathway, the pheromone receptors Ste3α/a sense pheromones from cells of the opposite mating type and activate a G protein complex that includes the G protein α subunits Gpa2 and Gpa3, the Gβ subunit Gpb1, and the Gγ subunits Gpg1 and Gpg2 (Hsueh et al., 2007; Li et al., 2007a). Following activation of Gpa2 and Gpa3, the Gβ subunit Gpb1 is released to activate the downstream MAPK cascade to trigger mating responses (Hsueh et al., 2007; Li et al., 2007a). One RGS protein, Crg1, has been identified as a homolog of Sst2 and a negative regulator of the pheromone response pathway (Nielsen et al., 2003; Wang et al., 2004). Cells lacking Crg1 are hypersensitive to mating pheromones and produce abundant conjugation tubes in confrontation assays (Wang et al., 2004). Recently, a second RGS protein, Crg2, was also shown to play an important role in modulating mating (Hsueh et al., 2007). crg2 deletion mutations promote enhanced mating filament production. Protein–protein interaction studies using a split-ubiquitin system revealed that Crg2 directly interacts with the pheromone receptor Ste3 and both Gα subunits Gpa2 and Gpa3 that control mating (Hsueh et al., 2007; Xue et al., 2008). Also expression of the MFα1 pheromone gene is significantly enhanced in a crg2 mutant background during mating (Hsueh et al., 2007).
Fig. 3.
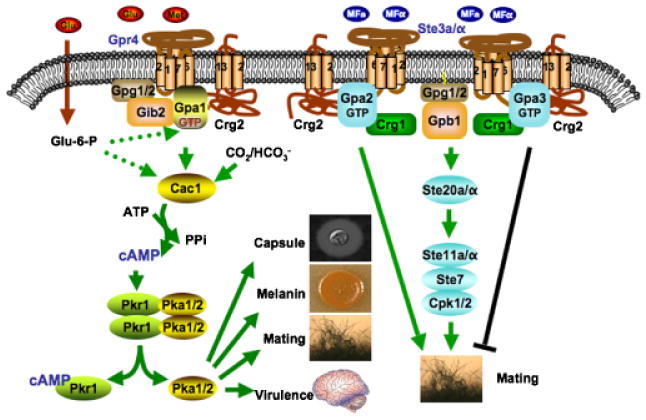
GPCR-signaling regulation in the human yeast pathogen Cryptococcus neoformans. In this model, the Gpr4 receptor activates Gpa1 and Crg2 functions as an RGS protein-regulating Gpa1 inactivation. Crg2, Gpa1, and Gpr4 form a functional protein complex. Gib2 and Gpg1 are two other proteins that function as βγ subunits with the Gpa1 Gα subunit to govern Gpa1-cAMP signaling. Upon activation, the expression of Gpa2 is induced, and Gpa2 controls signaling by binding and releasing the Gβγ subunits. The active form of Gpa2 also plays a positive role in pheromone response that leads to mating, whereas the activated form of Gpa3 inhibits mating. Both Crg1 and Crg2 interact with the pheromone receptor Ste3 and the Gα subunits Gpa2 and Gpa3, and function to constrain Gpa2 and Gpa3 signaling by stimulating GTPase activity.
In addition to the pheromone response pathway, the Gpa1-cAMP-signaling pathway has also been discovered to play a central role in virulence of C. neoformans (Fig. 3) (Wang & Heitman, 1999; Lengeler et al., 2000; Pukkila-Worley & Alspaugh, 2004). In this pathway, Gpr4 is involved in activating the downstream Gα protein Gpa1 (Xue et al., 2006; Li et al., 2007b). Because gpa1 mutants have additional phenotypes compared with gpr4, additional receptors could be involved in Gpa1 activation. Also other mechanisms might contribute to the Gpa1 function, such as involvement of AGS proteins or glucose-6-phosphate. Once activated, Gpa1 governs the production of the second messenger cAMP and activation of PKA to promote melanin and capsule production and thereby control virulence. Recently, Gib2, a novel Gβ-like/RACK1 protein homolog, was identified as a Gβ subunit that interacts with both Gpa1 and the Gγ subunits Gpg1 and Gpg2 to form a G protein complex (Palmer et al., 2006). Crg2 also functions as an RGS protein for the Gpa1-cAMP-signaling pathway controlling virulence of C. neoformans. We found that Crg2 physically interacts with Gpa1 and Gpr4 to form a protein complex and crg2 mutations cause an increase in cAMP production, providing direct evidence on the role of Crg2 in Gpa1-cAMP-signaling regulation. crg2 mutations enhance mating filament production, but reduce cell–cell fusion and sporulation efficiency during mating. crg2 mutants showed attenuated virulence in a murine model. We conclude that Crg2 participates in regulating both Gpa1-cAMP and pheromone signaling and hypothesized it may serve as a molecular interface between these two central signaling conduits (Xue et al., 2008). Our results were recently confirmed by another independent study (Shen et al., 2008). Taken together, our studies define a novel paradigm for G protein-signaling regulation, which may be conserved in other fungi as well as in multicellular eukaryotes. Two more RGS proteins (Crg3 and Crg4) have been identified recently, and their functions remain to be elucidated.
GPCR-signaling regulation is a finely tuned process that involves complex regulation at multiple levels. Besides the regulatory mechanisms that target GPCR-G protein association, several other mechanisms that directly regulate GPCR stability and activation have also been studied extensively, particularly involving regulation of the pheromone receptor Ste2 in S. cerevisiae. The C-terminus of Ste2 is important for GPCR functions, including ligand-triggered endocytosis and internalization (Zanolari et al., 1992; Schandel & Jenness, 1994). As a consequence of ligand binding, the C-terminal domain of Ste2 is ubiquitinated and triggers endocytosis and vacuolar degradation as part of the normal receptor-trafficking process (Hicke & Riezman, 1996; Hanyaloglu & von Zastrow, 2008). Phosphorylation of the C-terminus of Ste2 receptor in S. cerevisiae has been found to play an important role in GPCR-signaling regulation, both with respect to receptor desensitization (Reneke et al., 1988; Chen & Konopka, 1996) and mono-ubiquitination, which in turn triggers receptor endocytosis and degradation (Hicke et al., 1998; Terrell et al., 1998). Mutation of four phosphorylation sites in the C-terminus of Ste2 caused an increased sensitivity to α-factor and delayed recovery from a pulse of α-factor (Chen & Konopka, 1996).
A series of proteins have been identified to negatively regulate pheromone signaling in S. cerevisiae. Afr1 is a novel protein and was identified in a screen for α factor resistant (AFR) mutants (Konopka, 1993). Overexpression of AFR1 inhibits pheromone signaling, while afr1 mutants show a defect in yeast conjugation formation during mating similar to that of the ste2 mutants without the C-terminal regulatory domain (Davis et al., 1998). Further studies suggest that Afr1 inhibits the activation of G protein signaling via the Ste2 receptor, independent of receptor phosphorylation and endocytosis (Davis et al., 1998). Msg5 is a MAPK phosphatase that inhibits the MAPK Fus3 to inactivate pheromone signaling (Andersson et al., 2004). Asg7 is an a-cell-specific protein that acts in concert with the a-factor receptor Ste3 to inhibit G protein signaling (Rivers & Sprague, 2003). Detailed studies on the regulation and activation of pheromone receptor controlled pheromone response pathway have prompted the utilization of this pathway as a model system for studying other GPCRs in yeast (Minic et al., 2005b).
G protein-independent GPCR signaling
In general, GPCR signal activation requires the interaction and activation of G proteins in which GPCRs function as guanine-nucleotide exchange factors (GEFs). In mammals, the activation and desensitization of GPCRs involving GPCR kinases (GRKs) and β-arrestins have been well studied (Lefkowitz & Shenoy, 2005). GRKs phosphorylate GPCRs on serine and threonine residues and arrestins bind to the phosphorylated receptors to desensitize G protein-dependent-signaling pathways. Recent developments suggest that β-arrestins have additional functions besides the desensitization of G protein signaling, such as functioning as important adaptors that link receptors to the clathrin-dependent pathway of internalization without any direct involvement of G proteins, directing the activity of several nonreceptor tyrosine kinases in response to 7-TM receptor stimulation, and activation of the MAPK-signaling cascades (Lefkowitz & Shenoy, 2005).
So far, arrestins have only been identified in animals. No arrestin-related proteins of plant origin have been described. In fungi, even though no true arrestin protein has been found, arrestin-related proteins have been reported. One arrestin-related protein, PalF, contains N-terminal and C-terminal domains of arrestin and binds to the 7-TM pH sensor PalH to mediate pH signaling in A. nidulans (Herranz et al., 2005). Similar to mammalian β-arrestins, the phosphorylation and ubiquitination status of PalF is ligand (ambient pH) and receptor (PalH) dependent, suggesting similar regulatory mechanisms may be involved. The failure to reveal an involvement of G proteins in pH-signaling regulation suggests that the PalH-PalF pH-signaling response could be G protein-independent (Herranz et al., 2005). A similar arrestin-related protein, Rod1, has been identified in S. cerevisiae and shown to be phosphorylated by the kinase Snf1 (Shinoda & Kikuchi, 2007). In this case, it is still unknown whether any of the known yeast GPCRs binds to Rod1.
Oligomerization of GPCRs
Recent evidence reveals that many GPCRs oligomerize in living cells, and different GPCRs can form hetero-oligomers that are critical for receptor biogenesis and function (George et al., 2002; Bai, 2004; Ladds et al., 2005; Overton et al., 2005). Although it is well established that homo-dimerization is common, recent studies have sought to explore the physical basis of these interactions and the role of dimerization in signal transduction. Growing evidence supports the existence of higher-order organization of individual GPCRs and the potential for hetero-dimerization between pairs of coexpressed GPCRs. Although there may be exceptions (Meyer et al., 2006) and monomers may also function effectively (Ernst et al., 2007; Whorton et al., 2007), a great deal of recent evidence has indicated that most GPCRs do not exist as monomers but rather as dimers or, potentially, within higher-order oligomers (Milligan, 2004, 2006; Park et al., 2004).
While research advances indicate oligomerization of GPCRs is a universal phenomenon across kingdoms, there are only a few reports on dimerization of GPCRs in fungi. The functional significance of GPCR oligomerization remains poorly understood. Recent developments on the S. cerevisiae α-factor receptor Ste2 provided some insights into this complex phenomenon. Ste2 has been reported to be oligomeric in intact cells and this involves the GXXXG motif in transmembrane 1 (TM1) based on fluorescence resonance energy transfer (FRET) studies (Overton & Blumer, 2000; Overton et al., 2003). Mutation of this motif of Ste2 does not affect ligand binding but abolishes cell signaling, suggesting that oligomerization is not involved in ligand binding but important for signal transduction (Overton et al., 2003). The homo-oligomerized Ste2 complex functions as a unit to participate in ligand-dependent receptor endocytosis (Yesilaltay & Jenness, 2000). Heterodimers formed between Ste2 and its dominant negative form fail to signal, further supporting the importance of oligomerization of GPCRs (Gehret et al., 2006). These results suggest that oligomerization is likely to govern GPCR signaling and regulation.
The S. pombe pheromone receptor Mam2 was also found to form dimers during a study of a constitutively active mutant (Ladds et al., 2005). In C. neoformans, domains of Gpr4 can also interact in a yeast two-hybrid system (Xue et al., 2006). These reports suggest that, similar to GPCRs in other models, oligomerization is a common phenomenon that may mediate important physiological functions of GPCRs in fungi.
Dominant GPCR alleles
Some GPCRs can signal in the absence of any external chemical ligand and this idea was first supported by studies of the opioid and β2-adrenergic receptors (Koski et al., 1982; Cerione et al., 1984). These constitutively active receptors spontaneously adopt an active R* conformation independent of agonist binding (Seifert & Wenzel-Seifert, 2002). Many constitutively active mutant (CAM) GPCRs have been generated artificially by mutagenesis, including several examples of the pheromone receptors in budding and fission yeasts (Konopka et al., 1996; Stefan et al., 1998; Ladds et al., 2005). On the other hand, naturally occurring point mutations that result in constitutive activity have also been identified and related to human diseases (Van Sande et al., 1995). Moreover, c. 60 wild-type GPCRs from human, mouse or rat exhibit considerable constitutive activity (Seifert & Wenzel-Seifert, 2002). Recently, our studies have shown that in C. neoformans, a wild-type GPCR that shares sequence homology with pheromone receptors (Cpr2) exhibits ligand-independent activity (Hsueh et al., unpublished). Therefore, it is likely that similar examples exist in other fungal species, providing a new paradigm for fungal GPCR-mediated signaling.
Dominant negative (DN) receptor mutants represent another important class of molecules that can interfere with receptor function by out-competing wild-type receptors or hetero-oligomerization with wild-type receptors. Overexpression of dominant negative receptors often confers a loss-of-function phenotype on G protein signaling. There is no report of naturally occurring dominant negative GPCRs in fungi. The studies on the function of Ste2 revealed that loss-of-function Ste2 mutants can be isolated following mutagenesis. Two groups have reported Ste2 dominant negative mutants by screening Ste2 random point mutant libraries (Dosil et al., 1998; Leavitt et al., 1999). In one study, 16 such mutants were identified based on their failure to respond to mating pheromone even though they were all normally expressed, and detailed functional studies were conducted for two of them. Both mutants exhibited normal localization and stability similar to the wild type. Interestingly, all mutations were located at the extracellular ends of transmembrane segments and the corresponding mutants competed with wild-type receptor for G protein binding, suggest these sites may be important for ligand binding (Dosil et al., 1998). Another group identified four other mutation sites on the fourth to seventh transmembrane regions of Ste2 that converted Ste2 into a dominant negative form. Based on their study of two such mutations, a high level of the mutant receptor and Sst2 are required for the mutants to exert a dominant negative effect via outcompeting the normal receptor for G protein binding (Leavitt et al., 1999).
GPCR receptors as potential antifungal drug targets
GPCRs are key regulators of several physiological functions. Their roles in cellular signal transduction have made them the target for the majority of all currently prescribed drugs. Additionally, there are many orphan GPCRs that provide potential novel therapeutic targets. In mammalian cells, c. 720 GPCRs have been identified, and many serve as drug targets (Kostenis, 2004). GPCRs are the targets of > 40% of all drugs used clinically, as well as many drugs of abuse (Kenakin, 2005). Novel therapeutics will be developed as the functions of ‘orphan’ GPCRs and the pathways they control are elucidated. Furthermore, drugs with improved specificity and efficacy can be developed as GPCR signaling and regulatory mechanisms are understood at the molecular level.
Despite their importance in signaling regulation and drug development, only a few GPCRs other than pheromone receptors have been studied in fungi, such as Gpr1 in S. cerevisiae and C. albicans, Git3 and Stm1 in S. pombe, Gpr-4 in N. crassa, and Gpr4 and Cpr2 in C. neoformans (Xue et al., 1998, 2006; Lorenz et al., 2000; Han et al., 2004a; Lemaire et al., 2004; Miwa et al., 2004; Maidan et al., 2005a, b). Fungal GPCRs have not yet been targeted by antifungal agents. There are several major groups of antifungal drugs available for systemic fungal infections: the polyenes (such as amphotericin B) that target ergosterol on the cell membrane; the azoles (such as fluconazole, ketoconazole, itraconazole, and voriconazole) that target the enzyme lanosterol 14α-demethylase (Erg11), which converts lanosterol to ergosterol and is required in fungal cell membrane synthesis; Allylamines (such as terbinafine, amorolfine, natifine, and butenafine) that inhibit the enzyme squalene epoxidase, another enzyme required for ergosterol synthesis; and the echinocandins (such as caspofungin, micafungin, and anidulafungin) that inhibit the synthesis of glucan in the cell wall via the enzyme β-1-3-glucan synthase (Bowman & Free, 2006; Perlin, 2007). Because this antifungal drug repertoire is limited and drug resistance occurs, there is an ongoing need to identify new targets and develop novel therapeutic interventions. GPCR studies in fungi have the potential to be developed into new antifungal drug targets.
Utilization of the yeast-mating pathway for GPCR and ligand identification
Even though GPCRs in fungi have not yet been advanced in practice for disease control, GPCR-related-signaling pathways have been utilized in the deorphanization of GPCRs. The yeast pheromone response pathway, which is activated by the interaction of the pheromone receptor and Gpa1 Gα protein, has been well studied and successfully utilized to understand ligand–GPCR interactions, not only for other fungi but also animals (Fig. 4).
Fig. 4.
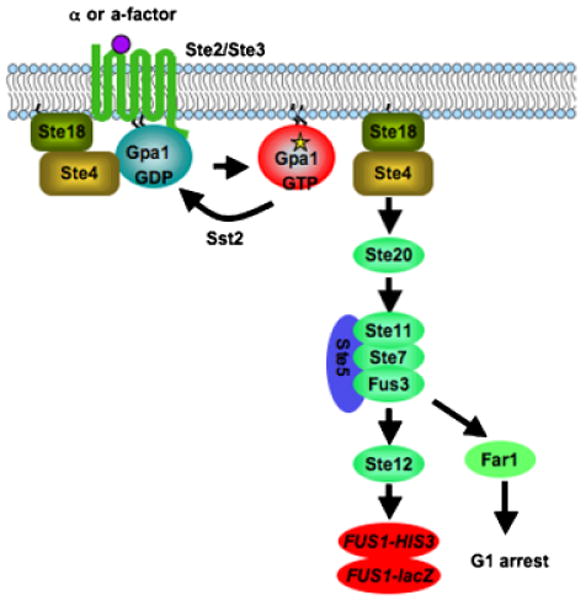
Yeast pheromone response pathway as a heterologous expression system. The pheromone receptor Ste2 activates the Gα subunit Gpa1 in yeast and promotes the release of the Gβ Ste4 and Gγ Ste18 subunits from the heterotrimeric G protein complex, which in turn activates the PAK kinase Ste20 and downstream MAPK cascade, Ste11, Ste7, and Fus3. The activation of transcription factor Ste12 promotes mating. To use this signaling pathway as a heterologous expression system, the RGS protein Sst2 and the cyclin-dependent kinase inhibitor Far1 are deleted to optimize signal output. The promoter of the pheromone inducible gene FUS1 is fused to a reporter gene, commonly HIS3 or lacZ, to quantitatively measure the pheromone response signal. Also the pheromone receptors Ste2 and Ste3 are deleted to eliminate signal interference by endogenous receptors.
The S. cerevisiae system for the expression of recombinant mammalian proteins has long been appreciated because of its low cost, simplicity, and conserved cellular pathways. A bioassay based on the S. cerevisiae pheromone response pathway has been developed to characterize heterologous GPCRs but several preconditions must be met. First, proper plasma membrane expression of the target GPCR is needed; second, the foreign receptor should properly couple to the Gα protein Gpa1 to activate the downstream signal; third, a reporter gene in this pathway is required to monitor the GPCR expression and signal activation. Finally, the pathway needs to be optimized to enhance signal sensitivity.
To engineer the yeast pheromone response pathway as a heterologous expression system, several modifications were necessary to optimize signal output. The cyclin-dependent kinase inhibitor Far1 promotes yeast cell arrest in G1 in response to pheromone, and was deleted to allow continued cell division in cells responding to pheromone or a heterologous ligand. The RGS protein Sst2 functions as a negative regulator of Gpa1 activity and deletion of Sst2 significantly increases the sensitivity of pheromone response and the yeast heterologous GPCR expression signal. To monitor the pheromone response signal for large-scale screens, the pheromone inducible gene FUS1 is commonly fused with either the lacZ gene or the HIS3 gene. Finally, to avoid interference by the endogenous receptors, the yeast pheromone receptor genes STE2 or STE3 are deleted. Some foreign GPCR genes can be directly expressed in this system and coupled to the Gpa1 G protein to activate pathway and FUS1-lacZ or FUS1-HIS3 expression. This includes some pheromone receptors from S. commune (Fowler et al., 1999), Cpr2 in C. neoformans (Hsueh et al., unpublished data), the SSTR2 receptor (Price et al., 1995), the adenosine A2 receptor (Price et al., 1996), the melatonin Mel1a receptor (Kokkola et al., 1998), and the UDP-glucose receptor (Chambers et al., 2000) (Table 3). However, in most cases, modifications of Gpa1 or the GPCR itself are necessary to promote proper GPCR-G protein interaction and pathway activation. For example, most GPCRs couple to the C terminus of the G protein, and one way to improve coupling between foreign receptors and Gpa1 without interfering with Gpa1 activity is to generate a chimeric Gα by replacing the Gpa1 C-terminal region with the corresponding protein of the foreign Gα. Some studies indicate that replacing only five amino acids is sufficient to promote specific GPCR-Gα coupling (Komatsuzaki et al., 1997; Kostenis et al., 1997; Mentesana et al., 2002). Another approach that has been successfully a pplied is to generate a fusion protein by fusing adrenergic receptors and their cognate Gα subunit (Bertin et al., 1994; Wise et al., 1997). However, this approach is not suitable for all receptors, suggesting this approach may be more complex.
Table 3. Examples of successful expression of heterologous GPCRs in yeast.
| Receptor | Species | G protein | Ligand | References |
|---|---|---|---|---|
| Cpr2 | C. neoformans | Gpa1 | N/A | Hsueh et al. (submitted) |
| Bbr1, Bbr2 | S. commune | Gpa1 | Pheromones 1 and 4 | Fowler et al. (1999) |
| Edg-2 | Human | Gpa1 | LPA | Erickson et al. (1998) |
| KIAA0001-UDP-glucose receptor | Human | Gpa1 | UDP-glucose | Chambers et al. (2000) |
| Adenosine A2a | Rat | Gpa1 | NECA | Price et al. (1996) |
| Neurotensin NT1 | Human | Gpa1 | Neurotensin | Leplatois et al. (2001) |
| Somatostatin SSTR2 | Rat | Gpa1 | Somatostatin 14 | Price et al. (1996) |
| Frizzled receptors (Fz1 and Fz2) | Human | Gpa1 | Wnt ligands | Dirnberger & Seuwen (2007) |
| Adrenergic β2 | Human | Gαs | ISO; EPI; NOR | King et al. (1990) |
| Heteromer CRLR+RAMP | Human | Gαs | ADM; CGRP | Miret et al. (2002) |
| Purinergic P2Y1 | Human | Gpa1–Gα14 | UDP | Brown et al. (2000) |
| Serotonin 5-HT1A | Human | Gpa1–Gαi0 | Serotonin | Brown et al. (2000) |
| Chemoattractant C5a | Human | Gpa1–Gαi1 | Hexapeptide C064 | Floyd et al. (2003) |
| FPRL-1 | Human | Gpa1–Gαi2 | Surrogate peptides | Klein et al. (1998) |
| Olfactory receptor I7 | Rat | Gpa1–Gαi2 | Heptanal, Octanal, Nonanal | Minic et al. (2005a) and Pajot-Augy et al. (2003) |
| Melatonin Mel1B | Human | Gpa1–Gαi16 | Melatonin | Brown et al. (2000) |
| Adenosine A2b | Human | Gpa1–Gαs | NECA | Brown et al. (2000) |
| GHRH receptor | Human | Gpa1–Gαs | GHRH | Kajkowski et al. (1997) |
| Vasopressin V2 | Human | Gpa1–Gαs | AVP | Erlenbach et al. (2001b) |
| GPR41 | Human | Gpa1–Gαq | Pentanose | Brown et al. (2003) |
| GPR43 | Human | Gpa1–Gαq | Propionate | Brown et al. (2003) |
| Muscarinic M1, M3, M5 | Human | Gpa1–Gαq | Carbachol | Erlenbach et al. (2001a) |
Modifying the receptor itself is another way to improve foreign receptor expression and signal activation in yeast. Foreign GPCRs fused with the cytoplasmic domain of the Ste2 pheromone receptor serve to enforce coupling between receptor and Gpa1 (King et al., 1990). This approach is particularly useful for GPCRs with unknown cognate Gα subunits (Yin et al., 2004).
Following the yeast pheromone response pathway-based bioassays first successful use to express the β2-adrenergic receptor by generating a chimeric receptor with Ste2 (King et al., 1990), many receptors have been expressed in this system, including the deorphanization of several receptors (Table 3). This system has also been successfully applied in functional studies of key receptor residues. We identified a second pheromone receptor Ste3 homolog in C. neoformans, which is located outside of the mating type locus. This receptor, Cpr2, was expressed in a yeast heterologous system and found to constitutively activate the FUS1-lacZ reporter gene (Hsueh et al., unpublished).
In the fission yeast S. pombe, the GPCR Stm1 has been identified as a nutrient sensor as described above. Overexpression of STM1 promotes uncontrolled cell division of yeast cells, which triggers a severe growth defect and also conversion of diploid to haploid cells (Chung et al., 2001, 2003). This phenotype has been successfully utilized for high throughput screens of Stm1 inhibitors. Chemical compounds were applied to an STM1 overexpression strain and inhibitors of Stm1 rescued the growth defect of the test strain (Chung et al., 2007). Because Stm1 interacts with the G protein Gpa2 to exert its effect on cell growth (Chung et al., 2003), this system can also be used to test other heterologous GPCRs for their interactions with Gpa2 and screen for potential modulators.
Concluding remarks
The GPCR receptor family is one of the most important sensory systems for all eukaryotic organisms to sense extracellular signals. Studies on GPCRs in animal models provide a useful road map on how to better understand ligand–GPCR–G protein interactions in microorganisms such as fungi. On the other hand, the relative simplicity of unicellular eukaryotic organisms such as yeasts provides exemplary model systems and tools for improving our knowledge on GPCRs and their signaling in multicellular eukaryotes. The utilization of the yeast pheromone response pathway as a surrogate to define receptor function and specificity is a perfect example. Also, many fundamental discoveries in modern life science are based on pioneering studies in yeast, which was the first eukaryotic genome to be sequenced. Many proteins important in human biology were first discovered by studying their homologs in yeast, such as proteins involved in the cell cycle, signal transduction, and protein-processing enzymes. The successful development of drug discovery based on GPCR studies shows a promising future of GPCR research in microbial pathogens to develop new agents to combat infectious diseases, both in agriculture and medicine. Although the pheromone receptors in yeast have been well studied, GPCR studies and related drug development in fungi are still in their infancy and many questions remain to be addressed. More GPCR candidates have been revealed recently following advances in fungal genome sequencing projects, and it is still unclear how many GPCRs exist in each of these fungi with completed genome sequences. For most putative GPCR proteins, the ligands and downstream intracellular-signaling pathways they activate remain to be defined. How these GPCRs impact cell development and pathogenicity will prove fertile ground for research in the decade ahead.
References
- Anderson CM, Willits DA, Kosted PJ, Ford EJ, Martinez-Espinoza AD, Sherwood JE. Molecular analysis of the pheromone and pheromone receptor genes of Ustilago hordei. Gene. 1999;240:89–97. doi: 10.1016/s0378-1119(99)00428-x. [DOI] [PubMed] [Google Scholar]
- Andersson J, Simpson DM, Qi M, Wang Y, Elion EA. Differential input by Ste5 scaffold and Msg5 phosphatase route a MAPK cascade to multiple outcomes. EMBO J. 2004;23:2564–2576. doi: 10.1038/sj.emboj.7600250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apanovitch DM, Slep KC, Sigler PB, Dohlman HG. Sst2 is a GTPase-activating protein for Gpa1: purification and characterization of a cognate RGS-Galpha protein pair in yeast. Biochemistry. 1998;37:4815–4822. doi: 10.1021/bi9729965. [DOI] [PubMed] [Google Scholar]
- Attwood TK, Findlay JB. Fingerprinting G-protein-coupled receptors. Protein Eng. 1994;7:195–203. doi: 10.1093/protein/7.2.195. [DOI] [PubMed] [Google Scholar]
- Bahn YS, Xue C, Idnurm A, Rutherford JC, Heitman J, Cardenas ME. Sensing the environment: lessons from fungi. Nat Rev Microbiol. 2007;5:57–69. doi: 10.1038/nrmicro1578. [DOI] [PubMed] [Google Scholar]
- Bai M. Dimerization of G-protein-coupled receptors: roles in signal transduction. Cell Signal. 2004;16:175–186. doi: 10.1016/s0898-6568(03)00128-1. [DOI] [PubMed] [Google Scholar]
- Ballon DR, Flanary PL, Gladue DP, Konopka JB, Dohlman HG, Thorner J. DEP-domain-mediated regulation of GPCR signaling responses. Cell. 2006;126:1079–1093. doi: 10.1016/j.cell.2006.07.030. [DOI] [PubMed] [Google Scholar]
- Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2004;25:1465–1476. doi: 10.1016/j.peptides.2003.10.022. [DOI] [PubMed] [Google Scholar]
- Bardwell L, Cook JG, Inouye CJ, Thorner J. Signal propagation and regulation in the mating pheromone response pathway of the yeast Saccharomyces cerevisiae. Dev Biol. 1994;166:363–379. doi: 10.1006/dbio.1994.1323. [DOI] [PubMed] [Google Scholar]
- Batlle M, Lu A, Green DA, Xue Y, Hirsch JP. Krh1p and Krh2p act downstream of the Gpa2p G(alpha) subunit to negatively regulate haploid invasive growth. J Cell Sci. 2003;116:701–710. doi: 10.1242/jcs.00266. [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Johnson AD. Mating in Candida albicans and the search for a sexual cycle. Annu Rev Microbiol. 2005;59:233–255. doi: 10.1146/annurev.micro.59.030804.121310. [DOI] [PubMed] [Google Scholar]
- Bertin B, Freissmuth M, Jockers R, Strosberg AD, Marullo S. Cellular signaling by an agonist-activated receptor/Gs alpha fusion protein. Proc Natl Acad Sci USA. 1994;91:8827–8831. doi: 10.1073/pnas.91.19.8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieszke JA, Braun EL, Bean LE, Kang S, Natvig DO, Borkovich KA. The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc Natl Acad Sci USA. 1999a;96:8034–8039. doi: 10.1073/pnas.96.14.8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieszke JA, Spudich EN, Scott KL, Borkovich KA, Spudich JL. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry. 1999b;38:14138–14145. doi: 10.1021/bi9916170. [DOI] [PubMed] [Google Scholar]
- Bieszke JA, Li L, Borkovich KA. The fungal opsin gene nop-1 is negatively-regulated by a component of the blue light sensing pathway and influences conidiation-specific gene expression in Neurospora crassa. Curr Genet. 2007;52:149–157. doi: 10.1007/s00294-007-0148-8. [DOI] [PubMed] [Google Scholar]
- Blumer JB, Smrcka AV, Lanier SM. Mechanistic pathways and biological roles for receptor-independent activators of G-protein signaling. Pharmacol Ther. 2007;113:488–506. doi: 10.1016/j.pharmthera.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolker M, Urban M, Kahmann R. The a mating type locus of U. maydis specifies cell signaling components. Cell. 1992;68:441–450. doi: 10.1016/0092-8674(92)90182-c. [DOI] [PubMed] [Google Scholar]
- Borkovich KA, Alex LA, Yarden O, et al. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SM, Free SJ. The structure and synthesis of the fungal cell wall. Bioessays. 2006;28:799–808. doi: 10.1002/bies.20441. [DOI] [PubMed] [Google Scholar]
- Brown AJ, Dyos SL, Whiteway MS, et al. Functional coupling of mammalian receptors to the yeast mating pathway using novel yeast/mammalian G protein alpha-subunit chimeras. Yeast. 2000;16:11–22. doi: 10.1002/(SICI)1097-0061(20000115)16:1<11::AID-YEA502>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Brown AJ, Goldsworthy SM, Barnes AA, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- Brown AJ, Jupe S, Briscoe CP. A family of fatty acid binding receptors. DNA Cell Biol. 2005;24:54–61. doi: 10.1089/dna.2005.24.54. [DOI] [PubMed] [Google Scholar]
- Brown LS, Dioumaev AK, Lanyi JK, Spudich EN, Spudich JL. Photochemical reaction cycle and proton transfers in Neurospora rhodopsin. J Biol Chem. 2001;276:32495–32505. doi: 10.1074/jbc.M102652200. [DOI] [PubMed] [Google Scholar]
- Burkholder AC, Hartwell LH. The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res. 1985;13:8463–8475. doi: 10.1093/nar/13.23.8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G. The evolution of MAT: the ascomycetes. In: Heitman J, Kronstad JW, Taylor JW, Casselton LA, editors. Sex in Fungi. ASM Press; Washington, DC: 2007. pp. 3–18. [Google Scholar]
- Calcagno AM, Bignell E, Warn P, Jones MD, Denning DW, Muhlschlegel FA, Rogers TR, Haynes K. Candida glabrata STE12 is required for wild-type levels of virulence and nitrogen starvation induced filamentation. Mol Microbiol. 2003;50:1309–1318. doi: 10.1046/j.1365-2958.2003.03755.x. [DOI] [PubMed] [Google Scholar]
- Calcagno AM, Bignell E, Rogers TR, Jones MD, Muhlschlegel FA, Haynes K. Candida glabrata Ste11 is involved in adaptation to hypertonic stress, maintenance of wild-type levels of filamentation and plays a role in virulence. Med Mycol. 2005;43:355–364. doi: 10.1080/13693780400006088. [DOI] [PubMed] [Google Scholar]
- Casselton LA. Mate recognition in fungi. Heredity. 2002;88:142–147. doi: 10.1038/sj.hdy.6800035. [DOI] [PubMed] [Google Scholar]
- Casselton LA, Olesnicky NS. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol Rev. 1998;62:55–70. doi: 10.1128/mmbr.62.1.55-70.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerione RA, Codina J, Benovic JL, Lefkowitz RJ, Birnbaumer L, Caron MG. The mammalian beta 2-adrenergic receptor: reconstitution of functional interactions between pure receptor and pure stimulatory nucleotide binding protein of the adenylate cyclase system. Biochemistry. 1984;23:4519–4525. doi: 10.1021/bi00315a003. [DOI] [PubMed] [Google Scholar]
- Chaleff DT, Tatchell K. Molecular cloning and characterization of the STE7 and STE11 genes of Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:1878–1886. doi: 10.1128/mcb.5.8.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JK, Macdonald LE, Sarau HM, et al. A G protein-coupled receptor for UDP-glucose. J Biol Chem. 2000;275:10767–10771. doi: 10.1074/jbc.275.15.10767. [DOI] [PubMed] [Google Scholar]
- Chang YC, Miller GF, Kwon-Chung KJ. Importance of a developmentally regulated pheromone receptor of Cryptococcus neoformans for virulence. Infect Immun. 2003;71:4953–4960. doi: 10.1128/IAI.71.9.4953-4960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasse SA, Flanary P, Parnell SC, Hao N, Cha JY, Siderovski DP, Dohlman HG. Genome-scale analysis reveals Sst2 as the principal regulator of mating pheromone signaling in the yeast Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:330–346. doi: 10.1128/EC.5.2.330-346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science. 2003;301:1728–1731. doi: 10.1126/science.1087790. [DOI] [PubMed] [Google Scholar]
- Chen P, Sapperstein SK, Choi JD, Michaelis S. Biogenesis of the Saccharomyces cerevisiae mating pheromone a-factor. J Cell Biol. 1997;136:251–269. doi: 10.1083/jcb.136.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Konopka JB. Regulation of the G-protein-coupled alpha-factor pheromone receptor by phosphorylation. Mol Cell Biol. 1996;16:247–257. doi: 10.1128/mcb.16.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KS, Won M, Lee SB, Jang YJ, Hoe KL, Kim DU, Lee JW, Kim KW, Yoo HS. Isolation of a novel gene from Schizosaccharomyces pombe: stm1+ encoding a seven-transmembrane loop protein that may couple with the heterotrimeric Galpha 2 protein, Gpa2. J Biol Chem. 2001;276:40190–40201. doi: 10.1074/jbc.M100341200. [DOI] [PubMed] [Google Scholar]
- Chung KS, Kim DU, Ryoo SW, et al. Functional overexpression of the Stm1 protein, a G-protein-coupled receptor, in Schizosaccharomyces pombe. Biotechnol Lett. 2003;25:267–272. doi: 10.1023/a:1022355102192. [DOI] [PubMed] [Google Scholar]
- Chung KS, Won M, Lee JJ, Ahn J, Hoe KL, Kim DU, Song KB, Yoo HS. Yeast-based screening to identify modulators of G-protein signaling using uncontrolled cell division cycle by overexpression of Stm1. J Biotechnol. 2007;129:547–554. doi: 10.1016/j.jbiotec.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Chung S, Karos M, Chang YC, Lukszo J, Wickes BL, Kwon-Chung KJ. Molecular analysis of CPRalpha, a MATalpha-specific pheromone receptor gene of Cryptococcus neoformans. Eukaryot Cell. 2002;1:432–439. doi: 10.1128/EC.1.3.432-439.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S, Ronchetti D, Thevelein JM, Winderickx J, Martegani E. Activation state of the Ras2 protein and glucose-induced signaling in Saccharomyces cerevisiae. J Biol Chem. 2004;279:46715–46722. doi: 10.1074/jbc.M405136200. [DOI] [PubMed] [Google Scholar]
- Conigrave AD, Hampson DR. Broad-spectrum L-amino acid sensing by class 3 G-protein-coupled receptors. Trends Endocrinol Metab. 2006;17:398–407. doi: 10.1016/j.tem.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Davidson RC, Nichols CB, Cox GM, Perfect JR, Heitman J. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2003;49:469–485. doi: 10.1046/j.1365-2958.2003.03563.x. [DOI] [PubMed] [Google Scholar]
- Davis C, Dube P, Konopka JB. Afr1p regulates the Saccharomyces cerevisiae alpha-factor receptor by a mechanism that is distinct from receptor phosphorylation and endocytosis. Genetics. 1998;148:625–635. doi: 10.1093/genetics/148.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignard D, Whiteway M. SST2, a regulator of G-protein signaling for the Candida albicans mating response pathway. Eukaryot Cell. 2006;5:192–202. doi: 10.1128/EC.5.1.192-202.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirnberger D, Seuwen K. Signaling of human frizzled receptors to the mating pathway in yeast. PLoS One. 2007;2:e954. doi: 10.1371/journal.pone.0000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- Dohlman HG, Slessareva JE. Pheromone signaling pathways in yeast. Sci STKE. 2006;364:cm6. doi: 10.1126/stke.3642006cm6. [DOI] [PubMed] [Google Scholar]
- Dohlman HG, Song J, Ma D, Courchesne WE, Thorner J. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein alpha subunit) Mol Cell Biol. 1996;16:5194–5209. doi: 10.1128/mcb.16.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosil M, Giot L, Davis C, Konopka JB. Dominant-negative mutations in the G-protein-coupled alpha-factor receptor map to the extracellular ends of the transmembrane segments. Mol Cell Biol. 1998;18:5981–5991. doi: 10.1128/mcb.18.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb-Downward JR, Noggle RM, Williamson PR, Huffnagle GB. The role of laccase in prostaglandin production by Cryptococcus neoformans. Mol Microbiol. 2008;68:1428–1437. doi: 10.1111/j.1365-2958.2008.06245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JR, Wu JJ, Goddard JG, Tigyi G, Kawanishi K, Tomei LD, Kiefer MC. Edg-2/Vzg-1 couples to the yeast pheromone response pathway selectively in response to lysophosphatidic acid. J Biol Chem. 1998;273:1506–1510. doi: 10.1074/jbc.273.3.1506. [DOI] [PubMed] [Google Scholar]
- Erlenbach I, Kostenis E, Schmidt C, Hamdan FF, Pausch MH, Wess J. Functional expression of M(1), M(3) and M(5) muscarinic acetylcholine receptors in yeast. J Neurochem. 2001a;77:1327–1337. doi: 10.1046/j.1471-4159.2001.00344.x. [DOI] [PubMed] [Google Scholar]
- Erlenbach I, Kostenis E, Schmidt C, Serradeil-Le Gal C, Raufaste D, Dumont ME, Pausch MH, Wess J. Single amino acid substitutions and deletions that alter the G protein coupling properties of the V2 vasopressin receptor identified in yeast by receptor random mutagenesis. J Biol Chem. 2001b;276:29382–29392. doi: 10.1074/jbc.M103203200. [DOI] [PubMed] [Google Scholar]
- Ernst OP, Gramse V, Kolbe M, Hofman KP, Heck M. Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc Natl Acad Sci USA. 2007;104:10859–10864. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Pei Y, Bidochka MJ. A regulator of a G protein signalling (RGS) gene, cag8, from the insect-pathogenic fungus Metarhizium anisopliae is involved in conidiation, virulence and hydrophobin synthesis. Microbiology. 2007;153:1017–1025. doi: 10.1099/mic.0.2006/002105-0. [DOI] [PubMed] [Google Scholar]
- Floyd DH, Geva A, Bruinsma SP, Overton MC, Blumer KJ, Baranski TJ. C5a receptor oligomerization. II. Fluorescence resonance energy transfer studies of a human G protein-coupled receptor expressed in yeast. J Biol Chem. 2003;278:35354–35361. doi: 10.1074/jbc.M305607200. [DOI] [PubMed] [Google Scholar]
- Fowler TJ, Mitton MF. Scooter, a new active transposon in Schizophyllum commune, has disrupted two genes regulating signal transduction. Genetics. 2000;156:1585–1594. doi: 10.1093/genetics/156.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler TJ, DeSimone SM, Mitton MF, Kurjan J, Raper CA. Multiple sex pheromones and receptors of a mushroom-producing fungus elicit mating in yeast. Mol Biol Cell. 1999;10:2559–2572. doi: 10.1091/mbc.10.8.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Diezmann S, Subaran RL, Allen A, Lengeler KB, Dietrich FS, Heitman J. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2004;2:e384. doi: 10.1371/journal.pbio.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Hsueh YP, Findley KM, Heitman J. Evolution of the mating-type locus: the Basidiomycetes. In: Heitman J, Kronstad JW, Taylor JW, Casselton LA, editors. Sex in Fungi. ASM Press; Washington, DC: 2007. pp. 19–34. [Google Scholar]
- Gehret AU, Bajaj A, Naider F, Dumont ME. Oligomerization of the yeast alpha-factor receptor: implications for dominant negative effects of mutant receptors. J Biol Chem. 2006;281:20698–20714. doi: 10.1074/jbc.M513642200. [DOI] [PubMed] [Google Scholar]
- George SR, O'Dowd BF, Lee SP. G-protein-coupled receptor oligomerization and its potential for drug discovery. Nat Rev Drug Discov. 2002;1:808–820. doi: 10.1038/nrd913. [DOI] [PubMed] [Google Scholar]
- Guo M, Aston C, Burchett SA, Dyke C, Fields S, Rajarao SJ, Uetz P, Wang Y, Young K, Dohlman HG. The yeast G protein alpha subunit Gpa1 transmits a signal through an RNA binding effector protein Scp160. Mol Cell. 2003;12:517–524. doi: 10.1016/s1097-2765(03)00307-1. [DOI] [PubMed] [Google Scholar]
- Hagen DC, McCaffrey G, Sprague GF., Jr Evidence the yeast STE3 gene encodes a receptor for the peptide pheromone a factor: gene sequence and implications for the structure of the presumed receptor. Proc Natl Acad Sci USA. 1986;83:1418–1422. doi: 10.1073/pnas.83.5.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KH, Seo JA, Yu JH. A putative G protein-coupled receptor negatively controls sexual development in Aspergillus nidulans. Mol Microbiol. 2004a;51:1333–1345. doi: 10.1111/j.1365-2958.2003.03940.x. [DOI] [PubMed] [Google Scholar]
- Han KH, Seo JA, Yu JH. Regulators of G-protein signalling in Aspergillus nidulans: RgsA downregulates stress response and stimulates asexual sporulation through attenuation of GanB (Galpha) signalling. Mol Microbiol. 2004b;53:529–540. doi: 10.1111/j.1365-2958.2004.04163.x. [DOI] [PubMed] [Google Scholar]
- Hanyaloglu AC, von Zastrow M. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu Rev Pharmacol Toxicol. 2008;48:537–568. doi: 10.1146/annurev.pharmtox.48.113006.094830. [DOI] [PubMed] [Google Scholar]
- Harashima T, Heitman J. The Galpha protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gbeta subunits. Mol Cell. 2002;10:163–173. doi: 10.1016/s1097-2765(02)00569-5. [DOI] [PubMed] [Google Scholar]
- Harashima T, Heitman J. Galpha subunit Gpa2 recruits kelch repeat subunits that inhibit receptor-G protein coupling during cAMP-induced dimorphic transitions in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:4557–4571. doi: 10.1091/mbc.E05-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima T, Anderson S, Yates JR, III, Heitman J. The kelch proteins Gpb1 and Gpb2 inhibit Ras activity via association with the yeast RasGAP neurofibromin homologs Ira1 and Ira2. Mol Cell. 2006;22:819–830. doi: 10.1016/j.molcel.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J Cell Biol. 1980;85:811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz S, Rodriguez JM, Bussink HJ, Sanchez-Ferrero JC, Arst HN, Jr, Penalva MA, Vincent O. Arrestin-related proteins mediate pH signaling in fungi. Proc Natl Acad Sci USA. 2005;102:12141–12146. doi: 10.1073/pnas.0504776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- Hicke L, Zanolari B, Riezman H. Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J Cell Biol. 1998;141:349–358. doi: 10.1083/jcb.141.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JK, Yu JH, Keller NP, Adams TH. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G alpha protein-dependent signaling pathway. EMBO J. 1997;16:4916–4923. doi: 10.1093/emboj/16.16.4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff B, Poggeler S, Kuck U. Eighty years after its discovery, Fleming's Penicillium strain discloses the secret of its sex. Eukaryot Cell. 2008;7:465–470. doi: 10.1128/EC.00430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS. Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem Soc Trans. 2005;33:257–260. doi: 10.1042/BST0330257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Xue C, Heitman J. G protein signaling governing cell fate decisions involves opposing Galpha subunits in Cryptococcus neoformans. Mol Biol Cell. 2007;18:3237–3249. doi: 10.1091/mbc.E07-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Heitman J. Genetics of Cryptococcus neoformans. Annu Rev Genet. 2002;36:557–615. doi: 10.1146/annurev.genet.36.052402.152652. [DOI] [PubMed] [Google Scholar]
- Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005;3:e95. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Howlett BJ. Characterization of an opsin gene from the ascomycete Leptosphaeria maculans. Genome. 2001;44:167–171. doi: 10.1139/g00-113. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Kawamata Y, Harada M, et al. Free fatty acids regulate insulin secretion from pancreatic beta cells through GPR40. Nature. 2003;422:173–176. doi: 10.1038/nature01478. [DOI] [PubMed] [Google Scholar]
- James TY, Srivilai P, Kues U, Vilgalys R. Evolution of the bipolar mating system of the mushroom Coprinellus disseminatus from its tetrapolar ancestors involves loss of mating-type-specific pheromone receptor function. Genetics. 2006;172:1877–1891. doi: 10.1534/genetics.105.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Baptiste G, Yang Z, Greenwood MT. Regulatory mechanisms involved in modulating RGS function. Cell Mol Life Sci. 2006;63:1969–1985. doi: 10.1007/s00018-006-6066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahmann R, Schirrawski J. Mating in the smut fungi: from a to b to the downstream cascades. In: Heitman J, Kronstad JW, Taylor JW, Casselton LA, editors. Sex in Fungi. ASM Press; Washington, DC: 2007. pp. 377–387. [Google Scholar]
- Kajkowski EM, Price LA, Pausch MH, Young KH, Ozenberger BA. Investigation of growth hormone releasing hormone receptor structure and activity using yeast expression technologies. J Recept Signal Transduct Res. 1997;17:293–303. doi: 10.3109/10799899709036610. [DOI] [PubMed] [Google Scholar]
- Karlson P, Luscher M. Pheromones: a new term for a class of biologically active substances. Nature. 1959;183:55–56. doi: 10.1038/183055a0. [DOI] [PubMed] [Google Scholar]
- Kenakin TP. New eyes to see texture in ligand efficacy. Nat Methods. 2005;2:163–164. doi: 10.1038/nmeth0305-163. [DOI] [PubMed] [Google Scholar]
- Kim H, Borkovich KA. A pheromone receptor gene, pre-1, is essential for mating type-specific directional growth and fusion of trichogynes and female fertility in Neurospora crassa. Mol Microbiol. 2004;52:1781–1798. doi: 10.1111/j.1365-2958.2004.04096.x. [DOI] [PubMed] [Google Scholar]
- Kim H, Borkovich KA. Pheromones are essential for male fertility and sufficient to direct chemotropic polarized growth of trichogynes during mating in Neurospora crassa. Eukaryot Cell. 2006;5:544–554. doi: 10.1128/EC.5.3.544-554.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K, Dohlman HG, Thorner J, Caron MG, Lefkowitz RJ. Control of yeast mating signal transduction by a mammalian beta 2-adrenergic receptor and Gs alpha subunit. Science. 1990;250:121–123. doi: 10.1126/science.2171146. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Shimoda C. The Schizosaccharomyces pombe mam2 gene encodes a putative pheromone receptor which has a significant homology with the Saccharomyces cerevisiae Ste2 protein. EMBO J. 1991;10:3743–3751. doi: 10.1002/j.1460-2075.1991.tb04943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C, Paul JI, Sauve K, Schmidt MM, Arcangeli L, Ransom J, Trueheart J, Manfredi JP, Broach JR, Murphy AJ. Identification of surrogate agonists for the human FPRL-1 receptor by autocrine selection in yeast. Nat Biotechnol. 1998;16:1334–1337. doi: 10.1038/4310. [DOI] [PubMed] [Google Scholar]
- Koelle MR. Heterotrimeric G protein signaling: getting inside the cell. Cell. 2006;126:25–27. doi: 10.1016/j.cell.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kokkola T, Watson MA, White J, Dowell S, Foord SM, Laitinen JT. Mutagenesis of human Mel1a melatonin receptor expressed in yeast reveals domains important for receptor function. Biochem Biophys Res Commun. 1998;249:531–536. doi: 10.1006/bbrc.1998.9182. [DOI] [PubMed] [Google Scholar]
- Kolakowski LF., Jr GCRDb: a G-protein-coupled receptor database. Receptors Channels. 1994;2:1–7. [PubMed] [Google Scholar]
- Komatsuzaki K, Murayama Y, Giambarella U, Ogata E, Seino S, Nishimoto I. A novel system that reports the G-proteins linked to a given receptor: a study of type 3 somatostatin receptor. FEBS Lett. 1997;406:165–170. doi: 10.1016/s0014-5793(97)00257-3. [DOI] [PubMed] [Google Scholar]
- Konopka JB. AFR1 acts in conjunction with the alpha-factor receptor to promote morphogenesis and adaptation. Mol Cell Biol. 1993;13:6876–6888. doi: 10.1128/mcb.13.11.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka JB, Margarit SM, Dube P. Mutation of Pro-258 in transmembrane domain 6 constitutively activates the G protein-coupled alpha-factor receptor. Proc Natl Acad Sci USA. 1996;93:6764–6769. doi: 10.1073/pnas.93.13.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski G, Streaty RA, Klee WA. Modulation of sodium-sensitive GTPase by partial opiate agonists. An explanation for the dual requirement for Na+and GTP in inhibitory regulation of adenylate cyclase. J Biol Chem. 1982;257:14035–14040. [PubMed] [Google Scholar]
- Kostenis E. A glance at G-protein-coupled receptors for lipid mediators: a growing receptor family with remarkably diverse ligands. Pharmacol Ther. 2004;102:243–257. doi: 10.1016/j.pharmthera.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Kostenis E, Gomeza J, Lerche C, Wess J. Genetic analysis of receptor-Galphaq coupling selectivity. J Biol Chem. 1997;272:23675–23681. doi: 10.1074/jbc.272.38.23675. [DOI] [PubMed] [Google Scholar]
- Kraakman L, Lemaire K, Ma P, Teunissen AW, Donaton MC, Van Dijck P, Winderickx J, de Winde JH, Thevelein JM. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol. 1999;32:1002–1012. doi: 10.1046/j.1365-2958.1999.01413.x. [DOI] [PubMed] [Google Scholar]
- Kronstad JW, Staben C. Mating type in filamentous fungi. Annu Rev Genet. 1997;31:245–276. doi: 10.1146/annurev.genet.31.1.245. [DOI] [PubMed] [Google Scholar]
- Kuchler K, Sterne RE, Thorner J. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 1989;8:3973–3984. doi: 10.1002/j.1460-2075.1989.tb08580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni RD, Thon MR, Pan H, Dean RA. Novel G-protein-coupled receptor-like proteins in the plant pathogenic fungus Magnaporthe grisea. Genome Biol. 2005;6:R24. doi: 10.1186/gb-2005-6-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladds G, Davis K, Das A, Davey J. A constitutively active GPCR retains its G protein specificity and the ability to form dimers. Mol Microbiol. 2005;55:482–497. doi: 10.1111/j.1365-2958.2004.04394.x. [DOI] [PubMed] [Google Scholar]
- Lafon A, Seo JA, Han KH, Yu JH, d'Enfert C. The heterotrimeric G-protein GanB(alpha)-SfaD(beta)-GpgA(gamma) is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans. Genetics. 2005;171:71–80. doi: 10.1534/genetics.105.040584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon A, Han KH, Seo JA, Yu JH, d'Enfert C. G-protein and cAMP-mediated signaling in aspergilli: a genomic perspective. Fungal Genet Biol. 2006;43:490–502. doi: 10.1016/j.fgb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Leavitt LM, Macaluso CR, Kim KS, Martin NP, Dumont ME. Dominant negative mutations in the alpha-factor receptor, a G protein-coupled receptor encoded by the STE2 gene of the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1999;261:917–932. doi: 10.1007/s004380051039. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Dohlman HG. Coactivation of G protein signaling by cell-surface receptors and an intracellular exchange factor. Curr Biol. 2008;18:211–215. doi: 10.1016/j.cub.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuw T, Wu C, Schrag JD, Whiteway M, Thomas DY, Leberer E. Interaction of a G-protein beta-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- Lemaire K, Van de Velde S, Van Dijck P, Thevelein JM. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol Cell. 2004;16:293–299. doi: 10.1016/j.molcel.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Lengeler KB, Davidson RC, D'Souza C, Harashima T, Shen WC, Wang P, Pan X, Waugh M, Heitman J. Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev. 2000;64:746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Fox DS, Fraser JA, Allen A, Forrester K, Dietrich FS, Heitman J. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot Cell. 2002;1:704–718. doi: 10.1128/EC.1.5.704-718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplatois P, Josse A, Guillemot M, Febvre M, Vita N, Ferrara P, Loison G. Neurotensin induces mating in Saccharomyces cerevisiae cells that express human neurotensin receptor type 1 in place of the endogenous pheromone receptor. Eur J Biochem. 2001;268:4860–4867. doi: 10.1046/j.0014-2956.2001.02407.x. [DOI] [PubMed] [Google Scholar]
- Li L, Borkovich KA. GPR-4 is a predicted G-protein-coupled receptor required for carbon source-dependent asexual growth and development in Neurospora crassa. Eukaryot Cell. 2006;5:1287–1300. doi: 10.1128/EC.00109-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shen G, Zhang ZG, Wang YL, Thompson JK, Wang P. Canonical heterotrimeric G proteins regulating mating and virulence of Cryptococcus neoformans. Mol Biol Cell. 2007a;18:4201–4209. doi: 10.1091/mbc.E07-02-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wright SJ, Krystofova S, Park G, Borkovich KA. Heterotrimeric G protein signaling in filamentous fungi. Annu Rev Microbiol. 2007b;61:423–452. doi: 10.1146/annurev.micro.61.080706.093432. [DOI] [PubMed] [Google Scholar]
- Liu H, Suresh A, Willard FS, Siderovski DP, Lu S, Naqvi NI. Rgs1 regulates multiple Galpha subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism. EMBO J. 2007;26:690–700. doi: 10.1038/sj.emboj.7601536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Pan X, Harashima T, Cardenas ME, Xue Y, Hirsch JP, Heitman J. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics. 2000;154:609–622. doi: 10.1093/genetics/154.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay V, Manney TR. Mutations affecting sexual conjugation and related processes in Saccharomyces cerevisiae. I. Isolation and phenotypic characterization of nonmating mutants. Genetics. 1974;76:255–271. doi: 10.1093/genetics/76.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidan MM, De Rop L, Serneels J, Exler S, Rupp S, Tournu H, Thevelein JM, Van Dijck P. The G protein-coupled receptor Gpr1 and the G{alpha} protein Gpa2 act through the cAMP-protein kinase A pathway to induce morphogenesis in Candida albicans. Mol Biol Cell. 2005a;16:1971–1986. doi: 10.1091/mbc.E04-09-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidan MM, Thevelein JM, Van Dijck P. Carbon source induced yeast-to-hypha transition in Candida albicans is dependent on the presence of amino acids and on the G-protein-coupled receptor Gpr1. Biochem Soc Trans. 2005b;33:291–293. doi: 10.1042/BST0330291. [DOI] [PubMed] [Google Scholar]
- Maidan MM, De Rop L, Relloso M, Diez-Orejas R, Thevelein JM, Van Dijck P. Combined inactivation of the Candida albicans GPR1 and TPS2 genes results in avirulence in a mouse model for systemic infection. Infect Immun. 2008;76:1686–1694. doi: 10.1128/IAI.01497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller JL. Signal transduction. Fishing at the cell surface. Science. 2003;300:594–595. doi: 10.1126/science.1083725. [DOI] [PubMed] [Google Scholar]
- Mayrhofer S, Weber JM, Poggeler S. Pheromones and pheromone receptors are required for proper sexual development in the homothallic ascomycete Sordaria macrospora. Genetics. 2006;172:1521–1533. doi: 10.1534/genetics.105.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentesana PE, Dosil M, Konopka JB. Functional assays for mammalian G-protein-coupled receptors in yeast. Methods Enzymol. 2002;344:92–111. doi: 10.1016/s0076-6879(02)44708-8. [DOI] [PubMed] [Google Scholar]
- Meyer BH, Segura JM, Martinez KL, Hovius R, George N, Johnsson K, Vogel H. FRET imaging reveals that functional neurokinin-1 receptors are monomeric and reside in membrane microdomains of live cells. Proc Natl Acad Sci USA. 2006;103:2138–2143. doi: 10.1073/pnas.0507686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan G. G protein-coupled receptor dimerization: function and ligand pharmacology. Mol Pharmacol. 2004;66:1–7. doi: 10.1124/mol.104.000497.. [DOI] [PubMed] [Google Scholar]
- Milligan G. G-protein-coupled receptor heterodimers: pharmacology, function and relevance to drug discovery. Drug Discov Today. 2006;11:541–549. doi: 10.1016/j.drudis.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Minic J, Persuy MA, Godel E, Aioun J, Connerton I, Salesse R, Pajot-Augy E. Functional expression of olfactory receptors in yeast and development of a bioassay for odorant screening. FEBS J. 2005a;272:524–537. doi: 10.1111/j.1742-4658.2004.04494.x. [DOI] [PubMed] [Google Scholar]
- Minic J, Sautel M, Salesse R, Pajot-Augy E. Yeast system as a screening tool for pharmacological assessment of G protein coupled receptors. Curr Med Chem. 2005b;12:961–969. doi: 10.2174/0929867053507261. [DOI] [PubMed] [Google Scholar]
- Miret JJ, Rakhilina L, Silverman L, Oehlen B. Functional expression of heteromeric calcitonin gene-related peptide and adrenomedullin receptors in yeast. J Biol Chem. 2002;277:6881–6887. doi: 10.1074/jbc.M107384200. [DOI] [PubMed] [Google Scholar]
- Miwa T, Takagi Y, Shinozaki M, Yun CW, Schell WA, Perfect JR, Kumagai H, Tamaki H. Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot Cell. 2004;3:919–931. doi: 10.1128/EC.3.4.919-931.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Hennequin C, Gallaud J, Dujon B, Fairhead C. The asexual yeast Candida glabrata maintains distinct a and alpha haploid mating types. Eukaryot Cell. 2008;7:848–858. doi: 10.1128/EC.00456-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Kateriya S, Adeishvili N, Hegemann P, Bamberg E. Channelrhodopsins: directly light-gated cation channels. Biochem Soc Trans. 2005;33:863–866. doi: 10.1042/BST0330863. [DOI] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect Immun. 2003;71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen O, Davey J. Pheromone communication in the fission yeast Schizosaccharomyces pombe. Semin Cell Biol. 1995;6:95–104. doi: 10.1016/1043-4682(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Noverr MC, Phare SM, Toews GB, Coffey MJ, Huffnagle GB. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect Immun. 2001;69:2957–2963. doi: 10.1128/IAI.69.5.2957-2963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea SF, Chaure PT, Halsall JR, Olesnicky NS, Leibbrandt A, Connerton IF, Casselton LA. A large pheromone and receptor gene complex determines multiple B mating type specificities in Coprinus cinereus. Genetics. 1998;148:1081–1090. doi: 10.1093/genetics/148.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton MC, Blumer KJ. G-protein-coupled receptors function as oligomers in vivo. Curr Biol. 2000;10:341–344. doi: 10.1016/s0960-9822(00)00386-9. [DOI] [PubMed] [Google Scholar]
- Overton MC, Chinault SL, Blumer KJ. Oligomerization, biogenesis, and signaling is promoted by a glycophorin A-like dimerization motif in transmembrane domain 1 of a yeast G protein-coupled receptor. J Biol Chem. 2003;278:49369–49377. doi: 10.1074/jbc.M308654200. [DOI] [PubMed] [Google Scholar]
- Overton MC, Chinault SL, Blumer KJ. Oligomerization of G-protein-coupled receptors: lessons from the yeast Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1963–1970. doi: 10.1128/EC.4.12.1963-1970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajot-Augy E, Crowe M, Levasseur G, Salesse R, Connerton I. Engineered yeasts as reporter systems for odorant detection. J Recept Signal Transduct Res. 2003;23:155–171. doi: 10.1081/rrs-120025196. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Palmer DA, Thompson JK, Li L, Prat A, Wang P. Gib2, a novel Gbeta-like/RACK1 homolog, functions as a Gbeta subunit in cAMP signaling and is essential in Cryptococcus neoformans. J Biol Chem. 2006;281:32596–32605. doi: 10.1074/jbc.M602768200. [DOI] [PubMed] [Google Scholar]
- Paoletti M, Rydholm C, Schwier EU, Anderson MJ, Szakacs G, Lutzoni F, Debeaupuis JP, Latge JP, Denning DW, Dyer PS. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr Biol. 2005;15:1242–1248. doi: 10.1016/j.cub.2005.05.045. [DOI] [PubMed] [Google Scholar]
- Paoletti M, Seymour FA, Alcocer MJ, Kaur N, Calvo AM, Archer DB, Dyer PS. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr Biol. 2007;17:1384–1389. doi: 10.1016/j.cub.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Park PS, Filipek S, Wells JW, Palczewski K. Oligomerization of G protein-coupled receptors: past, present, and future. Biochemistry. 2004;43:15643–15656. doi: 10.1021/bi047907k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters T, Louwet W, Gelade R, Nauwelaers D, Thevelein JM, Versele M. Kelch-repeat proteins interacting with the Galpha protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc Natl Acad Sci USA. 2006;103:13034–13039. doi: 10.1073/pnas.0509644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin DS. Resistance to echinocandin-class antifungal drugs. Drug Resist Updat. 2007;10:121–130. doi: 10.1016/j.drup.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggeler S, Kuck U. Identification of transcriptionally expressed pheromone receptor genes in filamentous ascomycetes. Gene. 2001;280:9–17. doi: 10.1016/s0378-1119(01)00786-7. [DOI] [PubMed] [Google Scholar]
- Price LA, Kajkowski EM, Hadcock JR, Ozenberger BA, Pausch MH. Functional coupling of a mammalian somatostatin receptor to the yeast pheromone response pathway. Mol Cell Biol. 1995;15:6188–6195. doi: 10.1128/mcb.15.11.6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price LA, Strnad J, Pausch MH, Hadcock JR. Pharmacological characterization of the rat A2a adenosine receptor functionally coupled to the yeast pheromone response pathway. Mol Pharmacol. 1996;50:829–837. [PubMed] [Google Scholar]
- Pryciak PM, Huntress FA. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R, Alspaugh JA. Cyclic AMP signaling in Cryptococcus neoformans. FEMS Yeast Res. 2004;4:361–367. doi: 10.1016/S1567-1356(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Raper J. Genetics of Sexuality in Higher Fungi. The Ronald Press; New York: 1966. [Google Scholar]
- Rasmussen SG, Choi HJ, Rosenbaum DM, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Rayasam GV, Tulasi VK, Davis JA, Bansal VS. Fatty acid receptors as new therapeutic targets for diabetes. Expert Opin Ther Targets. 2007;11:661–671. doi: 10.1517/14728222.11.5.661. [DOI] [PubMed] [Google Scholar]
- Reneke JE, Blumer KJ, Courchesne WE, Thorner J. The carboxy-terminal segment of the yeast alpha-factor receptor is a regulatory domain. Cell. 1988;55:221–234. doi: 10.1016/0092-8674(88)90045-1. [DOI] [PubMed] [Google Scholar]
- Riquelme M, Challen MP, Casselton LA, Brown AJ. The origin of multiple B mating specificities in Coprinus cinereus. Genetics. 2005;170:1105–1119. doi: 10.1534/genetics.105.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers DM, Sprague GF., Jr Autocrine activation of the pheromone response pathway in matalpha2- cells is attenuated by SST2- and ASG7-dependent mechanisms. Mol Genet Genomics. 2003;270:225–233. doi: 10.1007/s00438-003-0914-3. [DOI] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Schandel KA, Jenness DD. Direct evidence for ligand-induced internalization of the yeast alpha-factor pheromone receptor. Mol Cell Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers GC, Regier JC, Nuss DL. Evidence for a role of the regulator of G-protein signaling protein CPRGS-1 in Galpha subunit CPG-1-mediated regulation of fungal virulence, conidiation, and hydrophobin synthesis in the chestnut blight fungus Cryphonectria parasitica. Eukaryot Cell. 2004;3:1454–1463. doi: 10.1128/EC.3.6.1454-1463.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert R, Wenzel-Seifert K. Constitutive activity of G-protein-coupled receptors: cause of disease and common property of wild-type receptors. Naunyn Schmiedebergs Arch Pharmacol. 2002;366:381–416. doi: 10.1007/s00210-002-0588-0. [DOI] [PubMed] [Google Scholar]
- Seo JA, Han KH, Yu JH. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol Microbiol. 2004;53:1611–1623. doi: 10.1111/j.1365-2958.2004.04232.x. [DOI] [PubMed] [Google Scholar]
- Shen G, Wang YL, Whittington A, Li L, Wang P. The RGS protein Crg2 regulates pheromone and cAMP signaling in Cryptococcus neoformans. Eukaryot Cell. 2008 doi: 10.1128/EC.00154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda J, Kikuchi Y. Rod1, an arrestin-related protein, is phosphorylated by Snf1-kinase in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2007;364:258–263. doi: 10.1016/j.bbrc.2007.09.134. [DOI] [PubMed] [Google Scholar]
- Siderovski DP, Willard FS. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci. 2005;1:51–66. doi: 10.7150/ijbs.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slessareva JE, Routt SM, Temple B, Bankaitis VA, Dohlman HG. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell. 2006;126:191–203. doi: 10.1016/j.cell.2006.04.045. [DOI] [PubMed] [Google Scholar]
- Srikantha T, Lachke SA, Soll DR. Three mating type-like loci in Candida glabrata. Eukaryot Cell. 2003;2:328–340. doi: 10.1128/EC.2.2.328-340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Overton MC, Blumer KJ. Mechanisms governing the activation and trafficking of yeast G protein-coupled receptors. Mol Biol Cell. 1998;9:885–899. doi: 10.1091/mbc.9.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Davey J, Imai Y, Yamamoto M. Schizosaccharomyces pombe map3+ encodes the putative M-factor receptor. Mol Cell Biol. 1993;13:80–88. doi: 10.1128/mcb.13.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Katsuma S, Adachi T, Koshimizu TA, Hirasawa A, Tsujimoto G. Free fatty acids induce cholecystokinin secretion through GPR120. Naunyn Schmiedebergs Arch Pharmacol. 2007;377:523–527. doi: 10.1007/s00210-007-0200-8. [DOI] [PubMed] [Google Scholar]
- Terrell J, Shih S, Dunn R, Hicke L. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol Cell. 1998;1:193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4–activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- Tsitsigiannis DI, Keller NP. Oxylipins act as determinants of natural product biosynthesis and seed colonization in Aspergillus nidulans. Mol Microbiol. 2006;59:882–892. doi: 10.1111/j.1365-2958.2005.05000.x. [DOI] [PubMed] [Google Scholar]
- Tsitsigiannis DI, Keller NP. Oxylipins as developmental and host-fungal communication signals. Trends Microbiol. 2007;15:109–118. doi: 10.1016/j.tim.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Tsitsigiannis DI, Bok JW, Andes D, Nielsen KF, Frisvad JC, Keller NP. Aspergillus cyclooxygenase-like enzymes are associated with prostaglandin production and virulence. Infect Immun. 2005a;73:4548–4559. doi: 10.1128/IAI.73.8.4548-4559.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis DI, Kowieski TM, Zarnowski R, Keller NP. Three putative oxylipin biosynthetic genes integrate sexual and asexual development in Aspergillus nidulans. Microbiology. 2005b;151:1809–1821. doi: 10.1099/mic.0.27880-0. [DOI] [PubMed] [Google Scholar]
- Van Sande J, Parma J, Tonacchera M, Swillens S, Dumont J, Vassart G. Somatic and germline mutations of the TSH receptor gene in thyroid diseases. J Clin Endocrinol Metab. 1995;80:2577–2585. doi: 10.1210/jcem.80.9.7673398. [DOI] [PubMed] [Google Scholar]
- Versele M, de Winde JH, Thevelein JM. A novel regulator of G protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J. 1999;18:5577–5591. doi: 10.1093/emboj/18.20.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele M, Lemaire K, Thevelein JM. Sex and sugar in yeast: two distinct GPCR systems. EMBO Rep. 2001;2:574–579. doi: 10.1093/embo-reports/kve132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Heitman J. Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans. Curr Opin Microbiol. 1999;2:358–362. doi: 10.1016/S1369-5274(99)80063-0. [DOI] [PubMed] [Google Scholar]
- Wang P, Cutler J, King J, Palmer D. Mutation of the regulator of G protein signaling Crg1 increases virulence in Cryptococcus neoformans. Eukaryot Cell. 2004;3:1028–1035. doi: 10.1128/EC.3.4.1028-1035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waschuk SA, Bezerra AG, Jr, Shi L, Brown LS. Leptosphaeria rhodopsin: bacteriorhodopsin-like proton pump from a eukaryote. Proc Natl Acad Sci USA. 2005;102:6879–6883. doi: 10.1073/pnas.0409659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welton RM, Hoffman CS. Glucose monitoring in fission yeast via the Gpa2 Galpha, the Git5 Gbeta and the Git3 putative glucose receptor. Genetics. 2000;156:513–521. doi: 10.1093/genetics/156.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M, Hougan L, Dignard D, Thomas DY, Bell L, Saari GC, Grant FJ, O'Hara P, MacKay VL. The STE4 and STE18 genes of yeast encode potential beta and gamma subunits of the mating factor receptor-coupled G protein. Cell. 1989;56:467–477. doi: 10.1016/0092-8674(89)90249-3. [DOI] [PubMed] [Google Scholar]
- Whorton MR, Bokoch MP, Rasmussen SG, Huang B, Zare RN, Kobilka B, Sunahara RK. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci USA. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland T, Lutz S, Chidiac P. Regulators of G protein signalling: a spotlight on emerging functions in the cardiovascular system. Curr Opin Pharmacol. 2007;7:201–207. doi: 10.1016/j.coph.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Wise A, Carr IC, Groarke DA, Milligan G. Measurement of agonist efficacy using an alpha2A-adrenoceptor-Gi1alpha fusion protein. FEBS Lett. 1997;419:141–146. doi: 10.1016/s0014-5793(97)01431-2. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Miyamoto N, Shibata K, Valasek MA, Motoike T, Kedzierski RM, Yanagisawa M. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc Natl Acad Sci USA. 2004;101:1045–1050. doi: 10.1073/pnas.2637002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JR, Hamer JE. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- Xue C, Bahn YS, Cox GM, Heitman J. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol Biol Cell. 2006;17:667–679. doi: 10.1091/mbc.E05-07-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Tada Y, Dong X, Heitman J. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe. 2007;1:263–273. doi: 10.1016/j.chom.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Xue C, Hsueh Y, Chen L, Heitman J. The RGS protein Crg2 coordinates pheromone and cAMP signaling in Cryptococcus neoformans. Mol Microbiol. 2008 doi: 10.1111/j.1365-2958.2008.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Batlle M, Hirsch JP. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesilaltay A, Jenness DD. Homo-oligomeric complexes of the yeast alpha-factor pheromone receptor are functional units of endocytosis. Mol Biol Cell. 2000;11:2873–2884. doi: 10.1091/mbc.11.9.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Sahni N, Daniels KJ, Pujol C, Srikantha T, Soll DR. The same receptor, G Protein, and mitogen-activated protein kinase pathway activate different downstream regulators in the alternative white and opaque pheromone responses of Candida albicans. Mol Biol Cell. 2008;19:957–970. doi: 10.1091/mbc.E07-07-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D, Gavi S, Wang HY, Malbon CC. Probing receptor structure/function with chimeric G-protein-coupled receptors. Mol Pharmacol. 2004;65:1323–1332. doi: 10.1124/mol.65.6.1323. [DOI] [PubMed] [Google Scholar]
- Yu JH. Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J Microbiol. 2006;44:145–154. [PubMed] [Google Scholar]
- Yu JH, Wieser J, Adams TH. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 1996;15:5184–5190. [PMC free article] [PubMed] [Google Scholar]
- Zanolari B, Raths S, Singer-Kruger B, Riezman H. Yeast pheromone receptor endocytosis and hyperphosphorylation are independent of G protein-mediated signal transduction. Cell. 1992;71:755–763. doi: 10.1016/0092-8674(92)90552-n. [DOI] [PubMed] [Google Scholar]


