Abstract
We describe a molecularly defined duplication kit for the X chromosome of Drosophila melanogaster. A set of 408 overlapping P[acman] BAC clones was used to create small duplications (average length 88 kb) covering the 22-Mb sequenced portion of the chromosome. The BAC clones were inserted into an attP docking site on chromosome 3L using ΦC31 integrase, allowing direct comparison of different transgenes. The insertions complement 92% of the essential and viable mutations and deletions tested, demonstrating that almost all Drosophila genes are compact and that the current annotations of the genome are reasonably accurate. Moreover, almost all genes are tolerated at twice the normal dosage. Finally, we more precisely mapped two regions at which duplications cause diplo-lethality in males. This collection comprises the first molecularly defined duplication set to cover a whole chromosome in a multicellular organism. The work presented removes a long-standing barrier to genetic analysis of the Drosophila X chromosome, will greatly facilitate functional assays of X-linked genes in vivo, and provides a model for functional analyses of entire chromosomes in other species.
THE X chromosome of Drosophila melanogaster contains ∼2300 protein-coding genes or ∼15% of such genes in the genome. It contains 22 Mb of euchromatic DNA (Adams et al. 2000). About one-third of these genes are predicted to be mutable to a phenotype that can be scored, e.g., lethality, sterility, or abnormal behavior (Peter et al. 2002). However, most molecularly recognized X-linked genes have not been associated with mutations or studied in any detail (http://flybase.org) (Drysdale 2008). Indeed, one hallmark of the X chromosome in D. melanogaster and many other species is that it is haploid in males. In addition, the presence of one copy of the X in an otherwise diploid animal leads to the phenomenon of dosage compensation, a process that essentially doubles the expression of X-linked genes in Drosophila males (Gelbart and Kuroda 2009).
The presence of a single X chromosome in males facilitates screens for behavioral or visible mutant phenotypes in the hemizygous male progeny of a single-generation cross. For this reason, the X chromosome has been well saturated for viable mutations. However, many of these mutations have not been mapped since existing methods are tedious. Moreover, mutations in essential genes and genes required for male fertility cannot be propagated and genetically characterized unless they are complemented with a duplication maintained in the male. Hence, the X chromosome has been significantly less studied than the autosomes for mutations in essential and male fertility genes. For many of those mutations, the genes associated with these phenotypes have been elusive due to the lack of appropriate genetic reagents. Thus, X-linked genes in critical developmental and regulatory pathways are underrepresented in reported analyses as compared to similar classes of genes on the autosomes.
Mutations in essential and male fertility genes on the X chromosome can be mapped using a variety of techniques. One approach is to rely on recombination in females and perform meiotic mapping against visible markers (Lindsley and Zimm 1992), P-element insertions (Zhai et al. 2003), or SNPs (Berger et al. 2001; Hoskins et al. 2001; Martin et al. 2001; Nairz et al. 2002; Chen et al. 2008), all of which are labor-intensive strategies or require specialized infrastructure. An alternative is complementation mapping using deficiencies, which requires only a single cross. This approach is possible for viable mutations but not for X-linked lethal and sterile mutations since those cannot be propagated through males. Instead, complementation rescue tests need to be carried out using a segregating duplication, e.g., an X chromosome fragment on the Y chromosome [Dp(1;Y)], an autosome [Dp(1;A)], or a free duplication [Dp(1;f)] (Lindsley and Zimm 1992). Currently, duplications that encompass ∼90% of the X chromosome are available. Only three cytological regions at 13A–13F (∼1 Mb), 16D7–16F4 (∼0.3 Mb), and 18A–18F (∼0.8 Mb) are not covered. Unfortunately, these duplications are typically very large (∼1–1.5 Mb) (http://flybase.org/) (Drysdale 2008), limiting their utility for fine mapping. Moreover, most available duplications were isolated following X-ray mutagenesis, and their breakpoints are poorly defined.
Hence, a complete set of small molecularly defined duplications of the X chromosome would be extremely useful for identifying mutations in essential and male fertility genes and for fine-scale mapping of any mutation, including recessive viable mutations. In addition to promoting new genetic screens, a duplication set would allow one to map and assess the numerous, poorly characterized X-linked lethal mutants. Moreover, if molecularly defined genomic DNA clones are used to create the duplication set, then epitope tagging using recombineering would permit determination of expression patterns of genes included in the duplications (Venken et al. 2008, 2009; Ejsmont et al. 2009). Finally, such defined duplications would allow one to carry out structure–function analyses of genes through recombineering by introducing point mutations and small deletions into a gene of interest at unprecedented speed (Sharan et al. 2009).
Previously, we created the P[acman] (P/ΦC31 artificial chromosome for manipulation) transgenesis platform (Venken and Bellen 2005, 2007; Venken et al. 2006) for retrieval and manipulation of large DNA fragments in a conditionally amplifiable BAC (Wild et al. 2002). Genomic clones inserted into this vector can be subjected to recombineering (Sharan et al. 2009) and used for transformation of these fragments (up to at least 146 kb) into the genome of flies that carry a defined attP docking site using the ΦC31 integrase system (Groth et al. 2004; Venken et al. 2006; Bischof et al. 2007; Markstein et al. 2008). In a next step, we constructed two genomic BAC libraries, one with an average insert size of 21 kb (CHORI-322) and another with an average insert size of 83 kb (CHORI-321) (Venken et al. 2009). These BAC libraries were end-sequenced and mapped onto the genome sequence and are publicly available (http://pacmanfly.org) and distributed (http://bacpac.chori.org/). Here we bring these resources to a next level: BAC TransgeneOmics (Poser et al. 2008) of an entire chromosome in vivo. The 8.2-fold coverage of the X chromosome in mapped clones from the CHORI-321 library allowed us to select a tiled path of overlapping BACs containing almost all of the annotated genes on this chromosome. Here we describe the creation of the first set of molecularly defined duplications covering an entire chromosome of a multicellular organism, and we illustrate its utility for X-chromosome genetics in several experimental paradigms.
MATERIALS AND METHODS
Clone verification:
Selected BACs were streaked on LB plates (12.5 μg/ml chloramphenicol). Single colonies were used to produce primary working glycerol stocks. An aliquot of primary culture was used to inoculate a secondary culture, induce high plasmid copy number with CopyControl solution (Epicentre), perform paired end sequencing, and analyze sequences to determine BAC end coordinates in the genome, as previously described (Venken et al. 2009). The sequence data were curated to verify the identity of each BAC clone and ensure precise mapping of BAC end coordinates in the genome sequence.
BAC DNA preparation:
Working glycerol stocks were restreaked on LB plates (12.5 μg/ml chloramphenicol). A single colony was grown in 1 ml LB (12.5 μg/ml chloramphenicol) for 17 hr at 37°. The plasmid copy number was induced for five additional hours at 37° by adding 9 ml LB (12.5 μg/ml chloramphenicol) containing 2 μl CopyControl solution (Epicentre). The culture was spun down and the bacterial pellet frozen at −20°. BAC DNA was isolated with the PureLink HiPure plasmid kit (Invitrogen) according to the manufacturer's instructions with the following modifications. The bacterial pellet was resuspended in 0.4 ml R3 buffer and transferred to a microcentrifuge tube. Lysis was performed with 0.4 ml L7 buffer, gentle inversions (10 times), and incubation at room temperature for no longer than 4 min. Neutralization was performed with 0.4 ml N3 buffer, gentle inversions (10 times), and incubation on ice for 4 min. Precipitation was performed by centrifugation for 10 min at 4° at full speed. EQ1 buffer (2 ml) was added to the gravity purification column for equilibration. After centrifugation, the supernatant was loaded onto the column. The column was washed twice with 2.5 ml W8 buffer. The DNA sample was eluted with 850 μl E4 buffer, prewarmed to 50°, into a microcentrifuge tube. DNA was precipitated with 595 μl isopropanol and centrifugation for 20 min at 4° at full speed. The DNA pellet was washed with 800 μl 70% ethanol and centrifugation for 2 min at 4° at full speed. The DNA pellet was air-dried for 4 min at room temperature and rehydrated in 20 μl EB Buffer (Qiagen: 10 mm Tris–Cl, pH 8.5). The DNA sample was allowed to dissolve overnight at 4°. The sample was then centrifuged for 2 min at 4° at full speed and transferred to a new microcentrifuge tube, avoiding the remaining pellet. Two microliters of the sample was used for an OD measurement, and 1 μl of the sample was used to assess the yield and supercoiled quality of the DNA preparation using a 0.7% agarose gel. The remaining 17 μl of the DNA sample was adjusted to a concentration of 15 ng/μl for each 10 kb of plasmid length, a concentration that was decided upon after extensive testing. The diluted DNA was stored at −20°.
Transformation:
DNA was injected into y1 M{vas-int.Dm}ZH-2A w*;PBac{y+-attP-3B}VK00033 embryos. Adult flies were crossed to five w1118;TM2/TM6C,Sb. Initially, we transferred the adults to fresh vials three times and screened the G1 progeny for mini-white expressing transformants once a week for 3 weeks. Analysis of 150 transformation experiments showed that 89% of those producing transformants did so in the first week. Therefore, to save time, we transferred adults to fresh vials once and screened all flies simultaneously when both vials were producing adult progeny. We ceased screening once two independent lines, one transgenic from two independent vials, were identified. If two independent lines were not identified, two lines were maintained from sibling transformants where possible. Individual balanced G1 transformed flies were backcrossed to w1118;TM2/TM6C,Sb. A single G2 male was backcrossed to w1118;TM2/TM6C,Sb, and a sibling was used for PCR confirmation of proper integration. Sometimes, transgenic progeny were obtained from a female injected animal, and the integrase containing the X chromosome may still have been present. Hence, these flies were screened for absence of dsRed fluorescence in the eye at this stage. Virgin G3 females and males were crossed to establish the balanced line (TM6C,Sb). Homozygous viability and fertility were assessed in the G4 and homozygous lines established when possible. The six male lethal or subvital lines were propagated through virgin females and lines established with dsxD,e1,Sb1 and TM2.
PCR confirmation of integration:
PCR confirmation of insertion into the docking site was performed on DNA isolated from single flies using the “squish” method (Engels et al. 1990). PCR primers and conditions are described (Venken et al. 2009). When possible, we tested at least two lines for each clone injected. Of the 408 transformed clones tested, 382 gave the appropriate PCR pattern in at least one line. Forty-four of these 382 also produced an incorrect PCR pattern in another line, indicating a low percentage of defective integration.
Complementation testing:
Rescue experiments were performed with standard Drosophila crossing protocols using the alleles described in supporting information, Table S2.
Accession numbers:
BAC end sequences that improve on previously published data (Venken et al. 2009) have been submitted as updates to GenBank. New BAC end sequences have been submitted as new entries under accession nos. HN280414–HN280449 to GenBank.
RESULTS
Selection of a tiling path of P[acman] clones spanning the X chromosome:
Using BAC end sequence coordinates (Venken et al. 2009) and FlyBase gene annotations (Misra et al. 2002; Drysdale 2008), we selected a tiling path of P[acman] BAC clones from the CHORI-321 library spanning the sequenced portion of the D. melanogaster X chromosome: Release 5 armX (22,423 kb) (http://www.fruitfly.org) and XHet (153 kb) (Hoskins et al. 2007) sequences. The clones are contained within a vector backbone harboring an attB site for ΦC31-mediated transgenesis (Groth et al. 2004; Bischof et al. 2007) and the dominant mini-white eye marker for the identification of transgenic animals (Venken et al. 2006, 2009). Our aim was to minimize the number of clones in the tiling path while maximizing coverage of complete gene annotations and unannotated 5′ control regions. As some portions of the X chromosome are not represented by mapped 80-kb CHORI-321 clones, we selected six clones from the 21-kb insert CHORI-322 library to cover some of these regions. We selected 582 clones that were streaked from 384-well plates (Venken et al. 2009) for single colonies, and the DNA sequence was verified for each. This resulted in 566 verified clones with an average insert length of 87,710 bp and an average overlap of 47,774 bp. The resulting tiling path covers the X chromosome from the telomeric to pericentric heterochromatin. Of the 2210 annotated protein-coding genes present on the X chromosome (http://flybase.org/; FlyBase release 5.12) (Adams et al. 2000; Drysdale 2008), all but a small number are contained within at least one clone in the tiling path. We were not able to find appropriately mapped clones for 18 genes (Table S1). Some of these genes may be better represented by clones in the unmapped fraction of the CHORI-321 library. Other genes are very large and will not be covered by any clone in this library. In addition, 12 regions encompass a minimal overlap between clones (7) or a gap in mapped clone coverage (5) (Table S1). Four of these gaps are not represented in any other mapped BAC library (Celniker et al. 2002; Hoskins et al. 2007), suggesting that these regions of the Drosophila genome cannot be stably cloned in Escherichia coli.
Generation of transgenic Drosophila lines:
The P[acman] clones in the tiling path were injected into embryos that carried the VK33 attP docking site at polytene location 65B2 on chromosome arm 3L (Venken et al. 2006) and a Drosophila codon optimized ΦC31 integrase driven in the germline by the vasa promoter (Bischof et al. 2007). The VK33 integration site was chosen because it is homozygous viable, is isogenic since it was obtained as a single balanced transgenic animal (Venken et al. 2006), and had been shown to be a reliable site for the recovery of transgenic inserts (Venken et al. 2009). When the VK33 insert was originally recovered and mapped, there were no annotated genes in the genomic interval. Subsequently, it was shown that the interval does contain a gene, CG42747 (http://flybase.org/; FlyBase release 5.30) (Adams et al. 2000; Drysdale 2008), and that the VK33 insertion lies in a 5′ intron of this gene. The gene is transcribed at relatively low levels, and its function is unknown. The fact that the VK33 insert is homozygous viable and fertile indicates that, if CG42747 has an essential role, then the insert does not compromise that function. Transgenic progeny expressing mini-white were identified as described (Venken et al. 2006) and balanced and tested for proper integration into the docking site using a multiplex PCR procedure as described (Venken et al. 2009). Transgenic lines were tested for homozygous viability and fertility. Homozygous transgenic lines were established whenever possible.
We injected 461 P[acman] clones and obtained transgenics for 408 of them (88%). Multiplex PCR revealed that 382 (94%) of the recovered lines integrated into the proper docking site. The improper events resulted in three different types of PCR patterns: (1) an empty docking site pattern or (2) no pattern at all—both results suggesting an insertion at one of several pseudo-attP integration sites located within the fly genome (Groth et al. 2004)—or (3) differently sized bands, which suggest an imprecise integration event at the docking site. All duplications are stably maintained since loss of the mini-white marker has not been observed over many generations. The 382 correctly targeted duplications cover ∼96% of the euchromatic portion of the X chromosome and extend into the pericentric heterochromatin (Figure 1, Figure S1, and Table S2), with the largest contig being 6,121,885 bp in length and the second largest being 3,269,101 bp. The transgenic flies are currently available as the “Duplication Consortium X Chromosome” Duplications from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu/Browse/dp/DC-Dps.php).
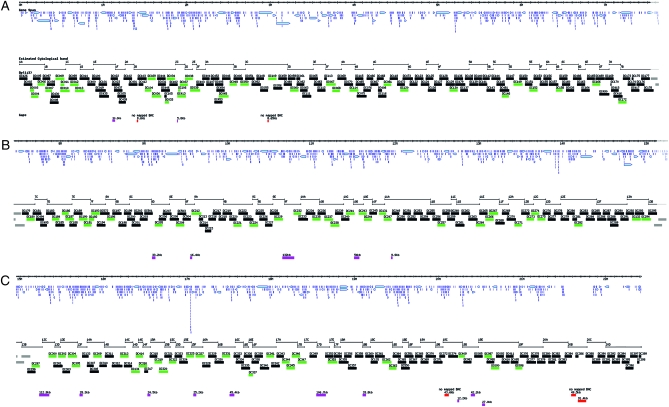
Figure 1.—
Overview of the molecularly defined X-chromosome duplication kit. The hash marked line of the X chromosome at the top of A, B, and C represents the coordinates (Mb) from the telomeric (top left) to the pericentromeric heterochromatin (bottom right). Each panel overlaps the next by 10 kb; the region of overlap is indicated in gray at the right end of each panel. The distribution of annotated gene spans (light blue arrows) is shown. Below are the estimated extents of the polytene chromosome bands from 1A to 20D. The extent of molecularly mapped duplications generated in this work is shown: duplications that complement molecularly defined mutations are indicated in green; others are indicated in black. The current gaps in coverage are indicated below by pink and red bars with the size of the gaps indicated above each bar. The pink gaps are represented by identified P[acman] BACs while the five red gaps are not represented by mapped clones in the CH321 and CH322 libraries. The map was derived from the GBrowse representation of the X chromosome in FlyBase.
There are 22 small gaps in our current coverage (Figure 1). Five of these gaps are not covered by mapped clones in the CH321 and CH322 BAC libraries (Figure 1, red bars, and Table S2). The other 17 gaps for which CH321 clones do exist (Figure 1, pink bars) will be retested by additional injections. The gaps in the current path are likely covered by larger, Y-chromosome-linked Bloomington Stock Center (BSC) duplications (Cook et al. 2010). This is illustrated for a 3-Mb region extending from 14B to 18A (Figure 2).
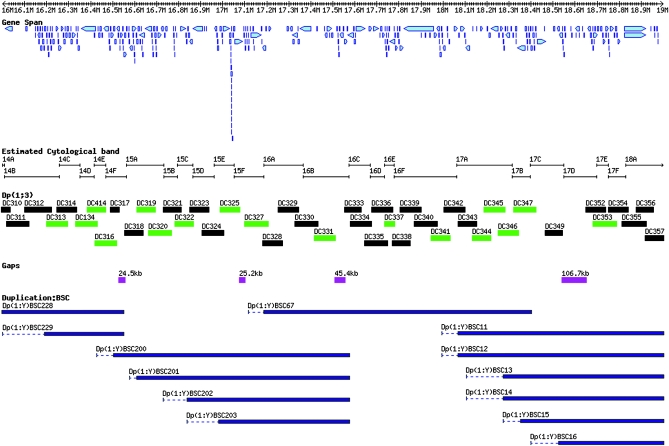
Figure 2.—
Overview of the polytene chromosome bands 14B to 18A, illustrating the complementary nature of the two new sets of X-chromosome duplications. The region illustrates the density of the DC duplications (this work) and compares them to the larger BSC duplications (Cook et al. 2010) in the same interval. This region contains four gaps in DC duplication coverage that are well covered by the BSC duplications. The tiers are the same as in Figure 1 with the BSC duplications added at the bottom in dark blue.
The average transformation efficiency was one transformant-producing fly for every 54 fertile G0 animals. For the 382 clones that produced transgenic animals, we recovered two independent lines for 214 (56%), while the remainder produced only a single transgenic line. We recovered correctly inserted clones in 66% of cases during a first injection round. Re-injections yielded 80% of appropriate transformants. Hence we conclude that our failure to recover lines for 53 clones is likely due to the small number of fertile G0 flies screened.
Interestingly, while multiple lines from an individual clone exhibit the same orange eye color from the mini-white marker gene, lines of different clones can vary in the degree of mini-white expression (from yellow to red). Since all clones are incorporated into the same docking site, the variation in eye color is most likely due to the sequences of the genomic insert, as previously reported (Venken et al. 2006).
Preliminary characterization of the duplication stocks:
Of the 367 current, characterized duplication lines, 302 (82%) are homozygous viable and fertile with no obvious phenotype. However, viability is often reduced in homozygotes. Mendelian ratios of homozygous adult progeny from crossing heterozygotes are generally decreased to 10% from the expected 33%. Nevertheless, most of the transgenic duplications are tolerated in three copies in males and four copies in females. This is somewhat surprising as dosage compensation in males does occur for most X-linked material duplicated on autosomes (Alekseyenko et al. 2008). Homozygotes of the remaining 65 transgenic lines (18%) either exhibit an obvious phenotype or have severely reduced viability (Table S3). The main phenotypes observed are Minute-like, similar to a dominant ribosomal protein deficiency phenotype; wings out, similar to the held out wings (how) phenotype; male lethality, and sex-specific sterility. Interestingly, in ∼50% of these stocks males are more severely affected than females, suggesting that dosage compensation is responsible for the severity of these phenotypes. Similarly, the eye color of males is in general darker than that of females, suggesting that dosage compensation acts on the mini-white marker of the P[acman] transgene.
We have characterized differences between isolates for 269 of the 367 transgenic strains. Fifty exhibited differences in homozygous viability or fertility between independent lines (37) containing the same P[acman] clone as well as between transgenic siblings from the same G0 parent (13). This suggests that some chromosomes may carry a second-site lethal or sterile mutation resulting either from a cryptic mutation that originated in the injection stock after isogenization several years ago (Venken et al. 2006) or from mutations caused during the trangenesis procedure. ΦC31 integrase has been shown to induce DNA damage and chromosome rearrangements (Ehrhardt et al. 2006; Liu et al. 2006, 2009) although this has been reported to be relatively rare in Drosophila (Bischof et al. 2007). We therefore outcrossed 18 of the 37 insertion stocks to a wild-type isogenized third chromosome for two to three generations. In this short period of time, 10 of 18 cases produced viable and fertile homozygotes, suggesting that distantly linked second-site mutations, and not the insertions, were the cause of the observed phenotypes.
P[acman] duplications rescue 90% of mutants tested:
To ensure that integrated P[acman] clones are functional duplications of the X chromosome, we tested 112 different transgenic lines for their ability to rescue known molecularly mapped mutations, including both lethal and viable mutant alleles. As shown in Table 1 and Table S2, the rescue experiments demonstrate that the duplications complement 92% of the tested mutations. This indicates that the majority of Drosophila genes have their required regulatory elements in the vicinity of the currently annotated transcripts (http://flybase.org/) (Drysdale 2008). Lack of rescue of lethal mutations, however, does not necessarily mean that the transgene does not contain the full-length gene with all its regulatory elements. It is possible that the chromosome bearing the mutation being tested also carries unidentified second-site lesions. This is illustrated for genes for which multiple alleles were tested, e.g., squash, TATA box binding protein-related factor 2, and cut up: some alleles are complemented whereas others are not by the same transgene, indicating that second-site mutations are present on some of these chromosomes (Table S2). However, a few examples of transgenes that fail to complement the corresponding mutations are noteworthy. On the basis of current gene annotation, mutations in roughest (rst) should have been rescued by two independent duplications, Dp(1;3)DC052 (CH321-04A01) and Dp(1;3)DC108 (CH321-65P11). However, these duplications do not rescue, suggesting that essential distant regulatory regions are lacking in the duplications. Indeed, recent RNA-Seq data (http://www.modencode.org and R. Chen, personal communication) have shown that rst has an unannotated 5′ exon 13.8 kb upstream from the currently annotated transcription start site that is absent in both clones. Similarly, mutations in cut (ct) are not rescued by duplication Dp(1;3)DC178 (CH321-62C02). This was anticipated as the known regulatory elements of ct extend over >80 kb from the annotated gene (Jack and DeLotto 1995) and thus beyond the extent of the duplication.
TABLE 1.
Complementation data
| Duplication | Clone | Predicted cytology | Rescue | No rescue |
|---|---|---|---|---|
| Dp(1;3)DC003 | CH321-12B18 | 1A1;1A1 | ewg, cin | |
| Dp(1;3)DC004 | CH321-46A16 | 1A1;1A3 | ewg | cin |
| Dp(1;3)DC006 | CH321-32O15 | 1A3;1A8 | ac, sc | |
| Dp(1;3)DC007 | CH321-34A23 | 1B1;1B5 | Exp6 | |
| Dp(1;3)DC009 | CH321-46B03 | 1B9;1B13 | RpL36 | |
| Dp(1;3)DC010 | CH321-04A18 | 1B11;1C3 | skpA, RpL36 | |
| Dp(1;3)DC011 | CH321-11D11 | 1B11;1B13 | RpL36 | |
| Dp(1;3)DC012 | CH321-32G05 | 1C3;1C5 | Rbf | |
| Dp(1;3)DC029 | CH321-35P01 | 2B4;2B8 | dor | |
| Dp(1;3)DC030 | CH321-02D20 | 2B7;2B9 | l(1)G0284 | |
| Dp(1;3)DC033 | CH321-17O06 | 2B12;2B14 | l(1)G0355 | |
| Dp(1;3)DC034 | CH321-22J19 | 2B13;2B17 | l(1)G0355, arm | |
| Dp(1;3)DC037 | CH321-24E10 | 2C4;2D2 | Unc-76, usp, csw | |
| Dp(1;3)DC038 | CH321-04O23 | 2D1;2E1 | ph-d, Pgd | wapl |
| Dp(1;3)DC039 | CH321-43N04 | 2D5;2F2 | crn, pn | |
| Dp(1;3)DC045 | CH321-25N08 | 3A4;3A6 | wds, egh | |
| Dp(1;3)DC048 | CH321-34G02 | 3A8;3B2 | sgg | |
| Dp(1;3)DC050 | CH321-23A22 | 3B4;3C1 | w | |
| Dp(1;3)DC052 | CH321-04A01 | 3C2;3C5 | rst | |
| Dp(1;3)DC060 | CH321-42E18 | 3D1;3D3 | dm | |
| Dp(1;3)DC067 | CH321-32O23 | 3F1;3F4 | ec | |
| Dp(1;3)DC068 | CH321-33A07 | 3F3;3F7 | VhaAC39 | |
| Dp(1;3)DC081 | CH321-29F10 | 4C4;4C7 | CHOp24 | rb |
| Dp(1;3)DC087 | CH321-01B20 | 19E1;19E3 | run | |
| Dp(1;3)DC090 | CH321-27E22 | 19E3;19E4 | shakB | |
| Dp(1;3)DC092 | CH321-05L19 | 19E3;19E4 | l(1)19Ec | |
| Dp(1;3)DC096 | CH321-23M15 | 19E7;19F1 | unc | |
| Dp(1;3)DC104 | CH321-90H13 | 2B3;2B6 | dor | |
| Dp(1;3)DC106 | CH322-76B11 | 2C1;2C5 | east | |
| Dp(1;3)DC108 | CH321-65P11 | 3C3;3C5 | rst | |
| Dp(1;3)DC109 | CH321-91P23 | 3C6;3C9 | N | |
| Dp(1;3)DC114 | CH321-85H19 | 4A3;4B2 | brn | |
| Dp(1;3)DC120 | CH321-61L05 | 4C11;4C16 | rap, ctp | |
| Dp(1;3)DC126 | CH321-22I01 | 4E1;4E2 | ovo | |
| Dp(1;3)DC130 | CH321-70G03 | 4F3;4F5 | Df(1)BSC823 | |
| Dp(1;3)DC131 | CH321-60D21 | 10F6;11A1 | cac | |
| Dp(1;3)DC132 | CH321-77E01 | 13A1;13A5 | eag | |
| Dp(1;3)DC134 | CH321-18K02 | 14C4;14E1 | CG4420, Rbp2, para, Df(1)FDD-0230908 | |
| Dp(1;3)DC143 | CH321-73A03 | 5A13;5C2 | l(1)G0060 | |
| Dp(1;3)DC146 | CH321-64D18 | 5C5;5C10 | Act5C | |
| Dp(1;3)DC149 | CH321-84E21 | 5D2;5D5 | rux | |
| Dp(1;3)DC152 | CH321-17L08 | 5E1;5E6 | sqh | |
| Dp(1;3)DC158 | CH321-50D09 | 6B1;6C1 | dx | |
| Dp(1;3)DC166 | CH321-65P16 | 6E3;6F1 | cm | |
| Dp(1;3)DC172 | CH321-80M12 | 7A7;7B2 | brk | |
| Dp(1;3)DC178 | CH321-62C02 | 7B4;7B6 | ct | |
| Dp(1;3)DC180 | CH321-59L01 | 7B6;7B8 | Tom40 | |
| Dp(1;3)DC184 | CH321-60P23 | 7D2;7D6 | mys | fs(1)h |
| Dp(1;3)DC185 | CH321-24M24 | 7D5;7D16 | mys, Smox, CG2263 | fs(1)h |
| Dp(1;3)DC186 | CH321-18C02 | 7D12;7D18 | Smox | |
| Dp(1;3)DC187 | CH321-74I15 | 7D17;7E1 | sdt | |
| Dp(1;3)DC190 | CH321-75D02 | 7E2;7E8 | CG10555 | |
| Dp(1;3)DC191 | CH321-09O19 | 7E6;7F1 | CG10555, Trf2 | |
| Dp(1;3)DC192 | CH321-28F18 | 7E9;7F1 | otu | |
| Dp(1;3)DC193 | CH321-80I04 | 7F1;7F6 | otu | Nrg |
| Dp(1;3)DC196 | CH321-64J01 | 8A1;8A5 | l(1)G0020 | |
| Dp(1;3)DC205 | CH321-25D20 | 8D2;8D7 | amx, lz, i | |
| Dp(1;3)DC209 | CH321-01K05 | 8E3;8E10 | l(1)G0320 | |
| Dp(1;3)DC212 | CH321-44B04 | 8F8;9A1 | CG15321, btd | |
| Dp(1;3)DC221 | CH321-69O03 | 9B2;9B5 | Hk, l(1)G0230 | |
| Dp(1;3)DC223 | CH321-67L01 | 9B6;9C1 | l(1)G0289 | |
| Dp(1;3)DC224 | CH321-64N06 | 9B12;9C6 | flw | |
| Dp(1;3)DC229 | CH321-18K03 | 9E1;9E4 | ras | |
| Dp(1;3)DC232 | CH321-47A13 | 9F8;10A1 | v | |
| Dp(1;3)DC235 | CH321-50P18 | 10A6;10B1 | ran, rtv, Dlic | |
| Dp(1;3)DC237 | CH321-64M10 | 10B2;10B5 | dsh, l(1)10Bb, Kap3 | |
| Dp(1;3)DC238 | CH321-67L16 | 10B3;10B12 | dsh, hop, dlg1 | |
| Dp(1;3)DC241 | CH321-05O16 | 10C2;10D1 | Rpll215, Kmn1 | |
| Dp(1;3)DC243 | CH321-74F04 | 10D4;10E2 | Df(1)Excel9050 | |
| Dp(1;3)DC244 | CH321-26N14 | 10E1;10E6 | m, dy | |
| Dp(1;3)DC246 | CH321-76G11 | 10F1;10F7 | wisp, pot | |
| Dp(1;3)DC247 | CH321-60H11 | 10F11;11A2 | gd, tsg, fw | Usp7 |
| Dp(1;3)DC257 | CH321-47M02 | 11B3;11B10 | CkIalpha | |
| Dp(1;3)DC264 | CH321-38B18 | 11D9;11E3 | hep, lic | |
| Dp(1;3)DC266 | CH321-03G21 | 11E3;11E8 | sno, mew | |
| Dp(1;3)DC267 | CH321-48B18 | 11E6;11E11 | comt | |
| Dp(1;3)DC271 | CH321-77D16 | 12A3;12A8 | up | |
| Dp(1;3)DC273 | CH321-23B06 | 12B2;12C1 | g, Df(1)BSC546 | CG9940, CG32627 |
| Dp(1;3)DC274 | CH321-75E20 | 12B4;12C6 | l(1)dd4, Rtc1, rdgB | |
| Dp(1;3)DC275 | CH321-70M13 | 12C5;12C7 | Clic | mamo |
| Dp(1;3)DC282 | CH321-60I22 | 12E5;12E8 | na | |
| Dp(1;3)DC294 | CH321-38B08 | 13A5;13B1 | CG5599, drd | |
| Dp(1;3)DC297 | CH321-60I24 | 13B4;13B6 | Top1, dah | |
| Dp(1;3)DC300 | CH321-59E06 | 13C3;13D1 | shtd | Gmap |
| Dp(1;3)DC302 | CH321-25M17 | 13D3;13E3 | sog | |
| Dp(1;3)DC305 | CH321-82A12 | 13E12;13F4 | l(1)G0136, Tcp-1zeta, mRpL3 | |
| Dp(1;3)DC309 | CH321-12N23 | 14A5;14A8 | exd | |
| Dp(1;3)DC313 | CH321-28P07 | 14B9;14C3 | sl | |
| Dp(1;3)DC316 | CH321-89E23 | 14E1;14F2 | Arp14D | |
| Dp(1;3)DC317 | CH321-20I24 | 14F2;14F4 | rok | |
| Dp(1;3)DC319 | CH321-08C14 | 15A3;15A9 | if | |
| Dp(1;3)DC320 | CH321-49N08 | 15A6;15B2 | CG9609 | |
| Dp(1;3)DC322 | CH321-62B18 | 15B5;15D1 | wus | |
| Dp(1;3)DC325 | CH321-64E02 | 15E3;15F1 | RpS5a, xmas-2 | |
| Dp(1;3)DC327 | CH321-56I13 | 15F3;16A1 | f | |
| Dp(1;3)DC330 | CH321-57P05 | 16A5;16B4 | CG8557 | |
| Dp(1;3)DC331 | CH321-09C05 | 16B1;16B8 | CG8557, l(1)G0222 | |
| Dp(1;3)DC337 | CH321-16E18 | 16E1;16F1 | CG32557 | |
| Dp(1;3)DC341 | CH321-90A16 | 16F5;16F7 | scu | |
| Dp(1;3)DC344 | CH321-35O24 | 17A3;17A5 | os | |
| Dp(1;3)DC345 | CH321-91O05 | 17A4;17A9 | os | |
| Dp(1;3)DC346 | CH321-32C05 | 17A7;17B1 | por | CrebB-17a |
| Dp(1;3)DC347 | CH321-90G02 | 17B1;17C1 | Aats-his | |
| Dp(1;3)DC353 | CH321-59N15 | 17D6;17F2 | Pvf | |
| Dp(1;3)DC363 | CH321-63C20 | 18C8;18D3 | car | |
| Dp(1;3)DC409 | CH321-05J07 | 19C5;19D1 | l(10G0004 | |
| Dp(1;3)DC414 | CH321-80H02 | 14D1;14E4 | Arp14D | |
| Dp(1;3)DC362 | CH321-71B04 | 18C5;18C8 | l(1)G0156 | |
| Dp(1;3)DC364 | CH321-35A10 | 18D1;18D13 | e(y)3, car | RpS10b |
| Dp(1;3)DC365 | CH321-35O18 | 18D7;18E3 | e(y)3, dome, Mer, Cdc42 | |
| Dp(1;3)DC413 | CH321-72K03 | 2C1;2D1 | east, Actn, usp, Unc-76 | |
| Dp(1;3)DC550 | CH321-51D08 | 18D13;18F1 | dome, Mer, Cdc42 |
We expected Dp(1;3)DC572 (CH321-82G19), which encompasses ocelliless (oc), to rescue mutations in that gene. We found that the duplication modifies the ocelliless phenotype of oc1 but does not restore the missing ocelli. The dorsal region that normally contains the ocelli is more like wild type but ocelli are still absent. In one instance, however, a single ocellus was restored. These results are consistent with an increase in, but not a normal level of, oc gene expression. This supports a report that increasing the levels of oc expression with a heat-shock-driven transgene in an oc mutant background improved the ocellarless phenotype (Royet and Finkelstein 1995). A second, slightly smaller duplication, Dp(1;3)DC195 (CH321-05H15), also failed to rescue oc1. Notably, larger duplications containing significantly more sequence 5′ of the gene, such as Dp(1;Y)BSC39, fully rescue the ocelliless phenotype of oc1 (Cook et al. 2010), suggesting that oc requires a very large upstream regulatory region (>41 kb) for normal transcription.
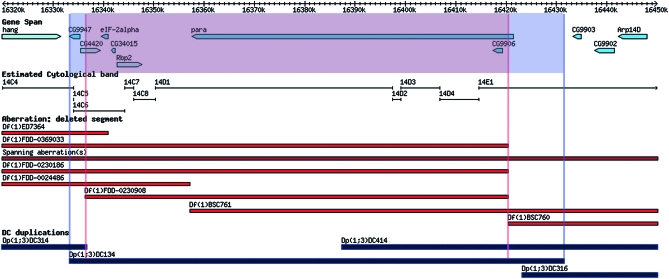
The molecularly defined duplications can also be used to rescue molecularly defined deletions previously generated by Flp/FRT-mediated recombinational excision (Parks et al. 2004; Ryder et al. 2004, 2007). For example, Dp(1;3)DC134 (CH321-18K02) rescues two paralytic (para) alleles as well as Df(1)FDD-0230908 (Ryder et al. 2004, 2007) (Figure 3). Similarly, the duplications Dp(1;3)DC130 (CH321-70G03), Dp(1;3)DC205 (CH321-25D20), Dp(1;3)DC243 (CH321-74F04), and Dp(1;3)DC273 (CH321-23B06) rescue Df(1)BSC823, Df(1)Excel9049, Df(1)Excel9050, and Df(1)BSC546, respectively (Table 1 and Table S2).
Figure 3.—
Example of gene and deficiency complementation with a molecularly defined duplication. Dp(1;3)DC134 (CH321-18K02) complements the large gene paralytic (para), demonstrating that all the regulatory sequences necessary for its function reside within the duplicated sequence and illustrating the compactness of the gene. Additionally, the duplication complements the recessive lethality associated with Df(1)FDD-0230908. The gene spans of the loci covered by both the deficiency and the duplication are indicated. Annotated gene spans are indicated in light blue, molecularly defined deficiencies are in red, and the DC duplications are in black. The map was derived from the GBrowse representation of the X chromosome in FlyBase.
Finally, 11 Minute loci are present on the X chromosome (Marygold et al. 2007). Minutes display a variety of cellular and developmental defects associated with a dominant haplo-insufficient phenotype due to a ribosomal protein deficiency. Two Minutes were tested for complementation: Dp(1;3)DC009 (CH321-46B03), Dp(1;3)DC010 (CH321-04A18), and Dp(1;3)DC011 (CH321-11D11) rescue RpL36 while Dp(1;3)DC325 (CH321-64E02) rescues RpS5a (Table 1 and Table S2).
Aneuploid-sensitive loci associated with obvious visible phenotypes:
Twelve duplication lines exhibit an obvious aneuploid-associated phenotype in animals heterozygous for the duplication (Table S3). Five of the lines have transgenes that encompass aneuploid-sensitive loci associated with obvious visible phenotypes, whereas two lines are associated with known diplo-lethal regions and five lines are not associated with known diplo-lethal regions (see below). Flies that carry one or two extra copies of Dp(1;3)DC006 (CH321-32O15), which encompasses the achaete (ac) and scute (sc) genes, display a Hairy wing (Hw) phenotype. Dp(1;3)DC097 (CH321-82N07) encompasses sc but not ac and homozygous adults exhibit a much weaker Hw phenotype. Hw mutations have been associated with overexpression of ac or sc (Balcells et al. 1988), and our data suggest that just one extra copy of ac and/or sc is sufficient to cause a Hw phenotype in females. This phenotype is also observed in other small duplications that cover this region, including Dp(1;Y)y+, which duplicates only ac and not sc (Muller 1948; Lindsley and Zimm 1992), suggesting that the Hw phenotype is not due to ectopic expression but to elevated expression levels of ac and/or sc within their normal expression domains.
Flies that carry an extra copy of Dp(1;3)DC109 (CH321-91P23) exhibit a Confluens (Co) phenotype (Lyman and Young 1993). This wing-vein phenotype is typically associated with an extra copy of Notch (N), and this clone includes four genes: Notch, Follicle cell protein 3C, CG18508, and a portion of kirre. Interestingly, the Co phenotype is also dose dependent as two extra copies of Dp(1;3)DC109 cause a more extreme phenotype than a single extra copy.
Similarly, an extra copy of Dp(1;3)DC329 (CH321-85I09), which encompasses the BarH1 gene, causes a Bar eye phenotype in males and females. This result is consistent with the fact that the dominant Bar mutations are associated with unequal crossover events and an increase in the copy number of BarH1 and/or BarH2 genes (Sturtevant and Morgan 1923; Gabay and Laughnan 1973). Interestingly, Dp(1;3)DC328 (CH321-04D11), which encompasses BarH2 but does not contain BarH1, does not exhibit a Bar phenotype, therefore supporting the idea that the Bar eye phenotype is due to extra copies of the BarH1 gene alone (Kojima et al. 1991, 1993).
Males homozygous for Dp(1;3)DC327 (CH321-56I13), which encompasses forked (f), exhibit bent macrochaete and microchaete, a phenotype similar to that observed in flies containing four copies of an f transgene in an f+ background (Petersen et al. 1994; Tilney et al. 1998, 2004). Homozygous females do not exhibit this bristle phenotype, suggesting that dosage compensation causes an increased expression of f in these Dp males. Males homozygous for this Dp could be expressing the equivalent of six doses of f resulting in the bent bristle phenotype. Notably, the original transgene causing the bent bristle phenotype (Tilney et al. 2004) encodes only four of the six f transcripts. Dp(1;3)DC327 encompasses all six transcripts. Thus the additional transcripts may contribute to the bent bristle phenotype even if the level of expression from the duplication is not quite equivalent to six doses.
Flies containing one copy of Dp(1;3)DC197 (CH321-38K07) exhibit necrotic wings and slightly misshapen eyes. The only annotated gene contained within this duplication is Lim1. Interestingly, a second independent transgenic line, Dp(1;3)DC500 (CH321-51H02) that also encompasses Lim1, exhibits the same phenotype. However, larger duplications that contain significantly more material 5′ of the Lim1 gene do not have this phenotype, e.g., Dp(1;Y)BSC41 and Dp(1;Y)BSC42 (Cook et al. 2010). Therefore, it would appear that the observed defects in animals carrying the two small duplications is not caused by aneuploidy alone. The phenotype thus may be due to abnormal expression of Lim1 either from a loss of normal regulatory sequences further upstream or from a position effect. In summary, all of these clones with obvious visible phenotypes can now be used as new dominant markers.
Aneuploid-sensitive loci associated with known diplo-lethal regions:
The set of duplications reported here has allowed a more precise localization of dosage-dependent lethal regions of the X chromosome, which are typically difficult to identify and map. The X chromosome was originally reported to contain only one hyperploid-sensitive locus (Beadex at 17A-C) and one locus associated with visible phenotypes when present in excess (triplo-abnormal) or when reduced to a single copy in a female (haplo-abnormal) (Notch at 3C7) (Lindsley et al. 1972). Subsequently, a duplication of the 11E-12B region was discovered to be lethal in males (Stewart and Merriam 1975). The cause of the lethality was hypothesized but not demonstrated to be due to mutations in upheld (up), which was proposed to be both diplo- and haplo-lethal, lethal as two copies in males or one copy in females, respectively (Homyk and Emerson 1988).
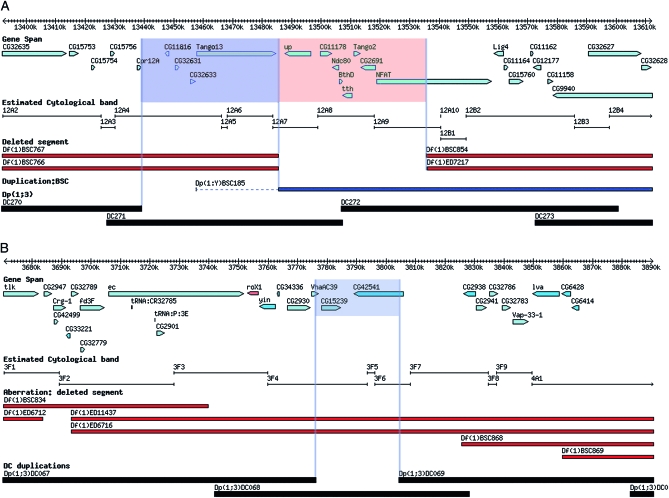
Consistent with this prior mapping, Dp(1;3)DC271 (CH321-77D16), which covers polytene region 12A4–7, is associated with diplo-lethality in males. It encompasses eight loci, including up (Figure 4A). However, partial loss-of-function mutations of up, up1, and up101 (null alleles and deficiencies do not exist) do not suppress the diplo-lethality, suggesting that up hyperploidy may not be causing the lethality or that the partial loss-of-function alleles do not affect hyperploid male lethality. Interestingly, Dp(1;Y)BSC185 also extends into this interval but is not associated with male diplo-lethality (Figure 4A) (Cook et al. 2010). Complementation analysis using this duplication with up alleles has shown that these lesions are complemented, similar to results using Dp(1;3)DC271 (Cook et al. 2010). This result appears to rule out up or any of the genes mapping to its right as the cause of the observed diplo-lethatlity (Figure 4A, pink box) and instead implicates one of the loci mapping to the left of up (Figure 4A, blue box). The fact that deficiencies including the region bounded by the pink box in Figure 4A have not been recovered implicates that region as the cause of the haplo-lethality. Thus the combined behavior of the recovered duplications and deficiencies in this region has shown that the haplo- and diplo-lethality mapped to this interval are likely caused by two different albeit tightly linked loci. We have not resolved which of the potential four diplo- and eight haplo-lethal genes is responsible but the analysis has dramatically narrowed the search and points to the resolving power afforded by these new reagents.
Figure 4.—
Aneuploid-sensitive loci associated with known diplo-lethal regions. (A) The haplo- and diplo-lethal region in cytological division 12A. The annotated genes potentially associated with diplo-lethality (blue) and the genes associated with haplo-lethality (red) are indicated. (B) The diplo-lethal interval in cytological division 3F. Two genes that are potentially associated with the lethality are indicated. The tiers and derivation of the maps are the same as in the Figure 1.
Several labs have reported that duplication of a region in 3F causes male lethality (a male-specific diplo-lethal region) (Cline 1988; Oliver et al. 1988). Here we provide molecular data that refine the mapping to a small number of loci. Dp(1;3)DC068 (CH321-33A07), which covers much of the 3F cytological region, is essentially diplo-lethal in males. Overlapping duplications that do not exhibit this phenotype allowed us to exclude some of the resident loci and suggest that any of the following three loci or a combination thereof cause male lethality when present in two copies in males: Vacuolar H+-ATPase C39 subunit (VhaAc39), CG15239, and/or CG42541 (Figure 4B). VhaAc39 has recently been shown to impinge on Notch signaling (Yan et al. 2009). Since N is one of very few loci that are haplo-insufficient and hyperploid sensitive, an additional copy of VhaAc39 could possibly lead to a gain-of-function phenotype of Notch signaling. However, complementation crosses involving a mutant allele of VhaAc39 and Dp(1;3)DC068 demonstrate that the duplication rescues the recessive lethality of this locus in females but that the VhaAc39/Y; Dp(1;3)DC068/+ genotype is male lethal. Hence, either CG15239 or CG42541, or an unannotated feature in this region, is associated with the male diplo-lethality.
Aneuploid-sensitive duplications not associated with known diplo-lethal regions:
There are five duplications that affect male viability but do not map to known dosage-sensitive regions. In all five cases, larger duplications encompassing the Duplication Consortium (DC) duplications do not exhibit an effect on male viability. The male lethal Dp(1;3)DC194 (CH321-69A10) is covered by a larger duplication that does not affect male viability (Dp(1;Y)BSC39), suggesting that the lethality may be due to truncation of a gene carried at one of the ends of the duplication, creating a dominant-negative protein, either Neuroglian (Nrg) or ocelliless (oc), or by a position effect of the DNA surrounding the docking site on chromosome arm 3L (Figure 5A).
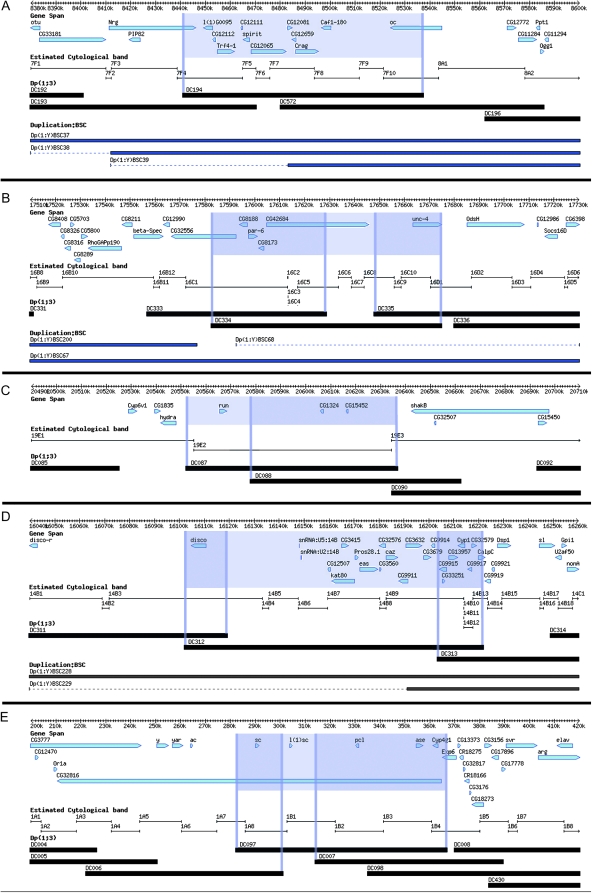
Figure 5.—
Aneuploid-sensitive duplications not associated with known diplo-lethal regions. (A) Truncation of the Nrg and/or oc genes. (B) Position effects associated with CG42684. Only Dp(1;3)DC334 (CH321-16L02) results in male lethality, indicating that CG42684 is the culprit. (C) Position effects on the run gene may cause the male viability defects seen with Dp(1;3)DC087 (CH321-01B20). (D) Position effects on any of 14 different genes may cause the male viability defects of Dp(1;3)DC312 (CH321-48H12). (E) The male lethality observed with Dp(1;3)DC097 (CH321-82N07) could be caused by truncation of CG32816 or by position effects on l(1)sc and/or sc. The tiers and derivation of the maps are the same as in Figure 1.
The male lethal Dp(1;3)DC334 (CH321-16L02) is contained within the larger duplication, Dp(1:Y)BSC67, which does not affect male viability (http://flystocks.bio.indiana.edu/Browse/dp/BDSC-Dps.php) (Cook et al. 2010). Dp(1;3)DC334 contains CG8188, par-6, CG8173, CG42684, unc-4, and part of CG32556 (Figure 5B). Overlapping duplications do not affect male viability and encompass all but the CG42684 gene. Hence, the male lethality may be due to mis-expression of CG42684 or a position effect associated with the insertion that becomes neutralized within the larger duplication Dp(1:Y)BSC67.
Males carrying a single copy of Dp(1;3)DC087 (CH321-01B20) are reduced in number (approximately one-half to two-thirds the expected number of males in the balanced stock). In addition, homozygotes are rare. There are several cytologically mapped viable duplications that cover this region (http://flystocks.bio.indiana.edu/Browse/dp/BDSC-Dps.php). This duplication contains three annotated genes, two of which are contained within Dp(1;3)DC088 (CH321-25A14), which does not affect male viability (Figure 5C). This leaves runt (run) as the candidate for the cause of the male subviability by this duplication. Further testing will be required to confirm that hyperploidy for run is indeed associated with reduced viability.
Males with one copy of Dp(1;3)DC312 (CH321-48H12) are reduced in number with very reduced fertility. Again, this region is contained within a larger duplication that does not affect male viability [Dp(1;Y)BSC228] (Cook et al. 2010). It is unclear what is causing the male viability problems associated with this duplication. The duplication contains 21 genes, 11 of which are not covered by any other DC duplication (Figure 5D). Both Dp(1;3)DC312 and Dp(1;3)DC311 (CH321-93B12) encompass disco, but male viability is unaffected in DC311. However, males with two copies of Dp(1;3)DC311 are sterile. Hence, it is possible that disco is involved in the reduced male fertility. It is unclear which genes are involved in the reduced male viability. Again, further analysis will be required to determine the cause of both effects.
Finally, flies homozygous for Dp(1;3)DC097 (CH321-82N07) are viable and both sexes are fertile, but males are extremely rare. This duplication contains sc, l(1)sc, pcl, ase, and Cyp4gi (Figure 5E). The homozygous male lethality is probably not due to pcl, ase, or Cyp4gi because these are contained within Dp(1;3)DC007 (CH321-34A23), which is homozygous viable and fertile. Both male and female homozygotes of Dp(1;3)DC006 (CH321-32O15), which contains sc, are rare. It is possible that l(1)sc or a combination of sc and l(1)sc cause the observed lethality of homozygous males in this duplication. Alternatively, the lethality could be caused by truncation or altered expression of CG32816, which extends the length of the duplication.
DISCUSSION
We describe the creation of a collection of molecularly defined duplications that will allow a much better characterization of >95% of the genes on the X chromosome of D. melanogaster. Several conclusions can be drawn from our analyses of these duplication lines. First, the efficiency of transformation with large-insert P[acman] clones is quite high: 66% upon a first injection attempt, which is better than achieved previously (Venken et al. 2006, 2009). Subsequent re-injection of clones that failed on the first attempt led to 80% transformation efficiency, suggesting that >90% of large-insert clones can be integrated into the fly genome provided that ∼100 fertile injected G0 animals are obtained. Second, second-site mutations are created during the transformation process at a frequency of 9%. Similar observations were reported previously in other experimental paradigms using ΦC31 integrase (Ehrhardt et al. 2006; Liu et al. 2006, 2009). Hence, we suggest outcrossing any transgenic chromosome that produces an unanticipated phenotype before attributing that phenotype to the inserted DNA, unless two independently generated transgenic lines produce the same phenotype. Third, our data show that most fly genes are quite compact: enhancers and other regulatory elements are generally located near the transcription units, as illustrated for several large genes such as para and shakB, which are rescued by clones containing little additional genomic DNA on either end of the current transcript annotation. Although we have identified a few exceptions, we conclude that a majority of the current FlyBase gene annotations (http://flybase.org/) are an excellent guide for selecting rescue constructs. Fourth, aneuploidy of >90% of the genes is well tolerated, even when four copies are present in females. Similarly, males with three copies of most genes, which may effectively correspond to six copies due to dosage compensation (Gelbart and Kuroda 2009), are often viable and fertile and display no obvious abnormal phenotypes, except for reduced Mendelian ratios in their progeny. It will be interesting to establish how dosage compensation is affected in these males. Fifth, consistent with previous work, very few small duplications of the X chromosome cause diplo-lethality in males, and this set of duplications has allowed the refined mapping of two diplo-lethal regions on the X chromosome. Overlapping duplications have identified potential culprits that may cause these phenotypes: two genes at cytological band 3F and four genes at cytological band 12F.
Very few genes on the X chromosome are not covered by mapped P[acman] clones. Some genes, such as Tenascin accessory (Ten-a) and dunce (dnc), are simply too large to be contained within a single P[acman] clone from the library used here. For others, we were unable to find an appropriate clone due to the finite number of clones mapped and their nonrandom distribution. We are therefore planning to upgrade existing low-copy-number, large-insert BAC clones from available mapped BAC libraries (Hoskins et al. 2000; Benos et al. 2001) by insertion of a retrofitting plasmid that contains the required elements for P[acman] transgenesis; this technique was recently used to retrofit clones from available fosmid and BAC libraries (Kondo et al. 2009). However, it remains to be determined whether retrofitted BACs from the large-insert RPCI-98 library (Hoskins et al. 2000), with an average insert size of 165 kb, can be integrated into attP docking sites in the fly genome. Alternatively, fragment size can be reduced through gap repair (Venken et al. 2006) or BAC trimming (Hill et al. 2000). In addition to the gene size problem, entire annotated genes in four regions of the X chromosome are not represented in mapped P[acman] clones due to minimal clone overlap or gaps in mapped clone coverage. Three of these regions are represented in large-insert BAC clones from other mapped libraries, and these regions should be amenable to our molecularly defined duplication strategy. The one remaining region containing annotated genes within the sequenced portion of the X chromosome, at polytene location 9A, is not represented in any available large-insert genomic library (http://www.fruitfly.org) (Hoskins et al. 2007), apparently due to problems associated with bacterial cloning of certain regions of the fly genome in an Escherichia coli bacterial host. One potential solution is to switch the cloning organism to, for example, Saccharomyces cerevisiae. Recombinogenic technologies, such as transformation-associated recombination cloning (Kouprina and Larionov 2008), have been used numerous times to retrieve large genomic regions through cotransformation-mediated gap repair directly from high-molecular-weight DNA into a linearized vector backbone (Kouprina and Larionov 2006). We conclude that very few genes are not represented in the selected tiling path of P[acman] clones. We are continuing to work on the remaining unrepresented regions to complete the duplication kit.
We note that it may be possible to create large transgenic duplications using our set of small Drosophila X chromosome duplications. Since all the clones have been integrated into the same attP docking site, a large duplication could potentially be generated through in vivo meiotic recombination between the overlapping regions of two smaller duplications. We are currently testing this possibility.
The collection of duplications that we have described will be useful for many purposes. It will be very valuable for rapidly mapping mutants, including the numerous publicly available and poorly mapped X-linked viable and lethal mutations, at high resolution. This should greatly accelerate mutation identification on the X chromosome: a first set of crosses with ∼20–30 large duplications (http://flystocks.bio.indiana.edu/Browse/dp/BDSC-Dps.php) (Cook et al. 2010) will in general allow mapping to an interval of a few 100 kb. A second set of crosses with a few P[acman] duplications will allow mapping to a 20- to 30-kb interval encompassing on average two to three genes. This can be followed by Sanger sequencing of the annotated protein-coding sequences in the region (H. J. Bellen, unpublished data). On the basis of our current experience, this strategy is much cheaper and more effective than whole-genome sequencing or gene-capture sequencing technologies (Mamanova et al. 2010; Metzker 2010). Indeed, the large number of SNPs and the bioinformatic burden associated with next-generation sequencing technologies make that approach much more expensive than setting up 30 fly crosses. Moreover, any sequencing-based strategy will still require a rescue strategy at the end of the sequencing process, as 10–100 SNPs per chromosome arm are identified, depending on the concentration of the chemical mutagens used to generate the mutations. It is therefore much more efficient to set up duplication crosses to map the mutation to a small genomic interval and then to carry out Sanger sequencing or targeted next-generation sequencing to identify the causal mutation.
The rescue of mutations will also provide an extremely useful framework for tagging and manipulating genes of interest using P[acman] clones and recombineering technology (Venken et al. 2008, 2009; Ejsmont et al. 2009). Thus, the collection of molecularly defined duplications that we have described will allow new experimental designs and strategies and will significantly expand the repertoire of manipulations of the X chromosome in Drosophila. Finally, we propose that the strategy that we have used is a model for future analysis of other chromosomes in Drosophila and other species.
Acknowledgments
We thank members of the Bloomington Drosophila Stock Center for providing flies; Johannes Bischof, Konrad Basler, and François Karch for providing germline ΦC31 sources; Roger Karess for providing flies; Kenneth Wan for technical assistance with BAC end sequencing; and Yuchun He and Hongling Pan for help in defining optimal parameters for microinjections of differently sized DNA. We are grateful to Melissa Phelps, Robert “Tank” Eisman, and David Miller for technical assistance during the screening and recovery of transgenic lines in Bloomington. We thank Rui Chen for communicating results before publication. We are grateful to Kevin Cook for sharing results before publication. We thank Shinya Yamamoto, Rick Kelley, and Herman Dierick for critical comments on the manuscript. This work was funded by National Institutes of Health grant 1R01 GM080415 (to H.J.B.), the Indiana Genomics Initiative, and the Indiana Metabolomics and Cytomics Initiative (T.C.K.). H.J.B. is an Investigator of the Howard Hughes Medical Institute.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.121285/DC1.
Available freely online through the author-supported open access option.
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287 2185–2195. [DOI] [PubMed] [Google Scholar]
- Alekseyenko, A. A., S. Peng, E. Larschan, A. A. Gorchakov, O. K. Lee et al., 2008. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcells, L., J. Modolell and M. Ruiz-Gomez, 1988. A unitary basis for different Hairy-wing mutations of Drosophila melanogaster. EMBO J. 7 3899–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benos, P. V., M. K. Gatt, L. Murphy, D. Harris, B. Barrell et al., 2001. From first base: the sequence of the tip of the X chromosome of Drosophila melanogaster, a comparison of two sequencing strategies. Genome Res. 11 710–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, J., T. Suzuki, K. A. Senti, J. Stubbs, G. Schaffner et al., 2001. Genetic mapping with SNP markers in Drosophila. Nat. Genet. 29 475–481. [DOI] [PubMed] [Google Scholar]
- Bischof, J., R. K. Maeda, M. Hediger, F. Karch and K. Basler, 2007. An optimized transgenesis system for Drosophila using germ-line-specific φC31 integrases. Proc. Natl. Acad. Sci. USA 104 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker, S. E., D. A. Wheeler, B. Kronmiller, J. W. Carlson, A. Halpern et al. 2002. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3 RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., A. Ahlford, F. Schnorrer, I. Kalchhauser, M. Fellner et al., 2008. High-resolution, high-throughput SNP mapping in Drosophila melanogaster. Nat. Methods 5 323–329. [DOI] [PubMed] [Google Scholar]
- Cline, T. W., 1988. Evidence that sisterless-a and sisterless-b are two of several discrete “numerator elements” of the X/A sex determination signal in Drosophila that switch Sxl between two alternative stable expression states. Genetics 119 829–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, R. K., M. E. Deal, J. A. Deal, R. D. Garton, C. A. Brown et al., 2010. A new resource for characterizing X-linked genes in Drosophila melanogaster: systematic coverage and subdivision of the X chromosome with nested, Y-linked duplications. Genetics 186 1095–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale, R., 2008. FlyBase: a database for the Drosophila research community. Methods Mol.Biol. 420 45–59. [DOI] [PubMed] [Google Scholar]
- Ehrhardt, A., J. A. Engler, H. Xu, A. M. Cherry and M. A. Kay, 2006. Molecular analysis of chromosomal rearrangements in mammalian cells after φC31-mediated integration. Hum. Genet. Ther. 17 1077–1094. [DOI] [PubMed] [Google Scholar]
- Ejsmont, R. K., M. Sarov, S. Winkler, K. A. Lipinsk and P. Tomancak, 2009. A toolkit for high-throughput, cross-species gene engineering in Drosophila. Nat. Methods 6 435–437. [DOI] [PubMed] [Google Scholar]
- Engels, W. R., D. M. Johnson-Schlitz, W. B. Eggleston and J. Sved, 1990. High-frequency P element loss in Drosophila is homolog dependent. Cell 62 515–525. [DOI] [PubMed] [Google Scholar]
- Gabay, S. J., and J. R. Laughnan, 1973. Recombination at the bar locus in an inverted attached-X system in Drosophila melanogaster. Genetics 75 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart, M. E., and M. I. Kuroda, 2009. Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth, A. C., M. Fish, R. Nusse and M. P. Calos, 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage φC31. Genetics 166 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, F., V. Benes, D. Thomasova, A. F. Stewart, F. C. Kafatos et al., 2000. BAC trimming: minimizing clone overlaps. Genomics 64 111–113. [DOI] [PubMed] [Google Scholar]
- Homyk, T., Jr., and C. P. Emerson, Jr., 1988. Functional interactions between unlinked muscle genes within haploinsufficient regions of the Drosophila genome. Genetics 119 105–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins, R. A., C. R. Nelson, B. P. Berman, T. R. Laverty, R. A. George et al., 2000. A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science 287 2271–2274. [DOI] [PubMed] [Google Scholar]
- Hoskins, R. A., A. C. Phan, M. Naeemuddin, F. A. Mapa, D. A. Ruddy et al., 2001. Single nucleotide polymorphism markers for genetic mapping in Drosophila melanogaster. Genome Res. 11 1100–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins, R. A., J. W. Carlson, C. Kennedy, D. Acevedo, M. Evans-Holm et al., 2007. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 316 1625–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, J., and Y. DeLotto, 1995. Structure and regulation of a complex locus: the cut gene of Drosophila. Genetics 139 1689–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, T., S. Ishimaru, S. Higashijima, E. Takayama, H. Akimaru et al., 1991. Identification of a different-type homeobox gene, BarH1, possibly causing Bar (B) and Om(1D) mutations in Drosophila. Proc. Natl. Acad. Sci. USA 88 4343–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, T., M. Sone, T. Michiue and K. Saigo, 1993. Mechanism of induction of Bar-like eye malformation by transient overexpression of Bar homeobox genes in Drosophila melanogaster. Genetica 88 85–91. [DOI] [PubMed] [Google Scholar]
- Kondo, S., M. Booker and N. Perrimon, 2009. Cross-species RNAi rescue platform in Drosophila melanogaster. Genetics 183 1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina, N., and V. Larionov, 2006. TAR cloning: insights into gene function, long-range haplotypes and genome structure and evolution. Nat. Rev. Genet. 7 805–812. [DOI] [PubMed] [Google Scholar]
- Kouprina, N., and V. Larionov, 2008. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat. Protoc. 3 371–377. [DOI] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego.
- Lindsley, D. L., L. Sandler, B. S. Baker, A. T. Carpenter, R. E. Denell et al., 1972. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics 71 157–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., I. Jeppesen, K. Nielsen and T. G. Jensen, 2006. Phic31 integrase induces chromosomal aberrations in primary human fibroblasts. Gene Ther. 13 1188–1190. [DOI] [PubMed] [Google Scholar]
- Liu, J., T. Skjorringe, T. Gjetting and T. G. Jensen, 2009. PhiC31 integrase induces a DNA damage response and chromosomal rearrangements in human adult fibroblasts. BMC Biotechnol. 9 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman, D., and M. W. Young, 1993. Further evidence for function of the Drosophila Notch protein as a transmembrane receptor. Proc. Natl. Acad. Sci. USA 90 10395–10399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamanova, L., A. J. Coffey, C. E. Scott, I. Kozarewa, E. H. Turner et al., 2010. Target-enrichment strategies for next-generation sequencing. Nat. Methods 7 111–118. [DOI] [PubMed] [Google Scholar]
- Markstein, M., C. Pitsouli, C. Villalta, S. E. Celniker and N. Perrimon, 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. G., K. C. Dobi and St. Johnston D., 2001. A rapid method to map mutations in Drosophila. Genome Biol. 2 RESEARCH0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold, S. J., J. Roote, G. Reuter, A. Lambertsson, M. Ashburner et al., 2007. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 8 R216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker, M. L., 2010. Sequencing technologies: the next generation. Nat. Rev. Genet. 11 31–46. [DOI] [PubMed] [Google Scholar]
- Misra, S., M. A. Crosby, C. J. Mungall, B. B. Matthews, K. S. Campbell et al. 2002. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 3 RESEARCH0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1948. The construction of several new types of Y chromosomes. Drosoph. Inf. Serv. 22 73–74. [Google Scholar]
- Nairz, K., H. Stocker, B. Schindelholz and E. Hafen, 2002. High-resolution SNP mapping by denaturing HPLC. Proc. Natl. Acad. Sci. USA 99 10575–10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, B., N. Perrimon and A. P. Mahowald, 1988. Genetic evidence that the sans fille locus is involved in Drosophila sex determination. Genetics 120 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, A. L., K. R. Cook, M. Belvin, N. A. Dompe, R. Fawcett et al., 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36 288–292. [DOI] [PubMed] [Google Scholar]
- Peter, A., P. Schottler, M. Werner, N. Beinert, G. Dowe et al., 2002. Mapping and identification of essential gene functions on the X chromosome of Drosophila. EMBO Rep. 3 34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, N. S., D. H. Lankenau, H. K. Mitchell, P. Young and V. G. Corces, 1994. forked proteins are components of fiber bundles present in developing bristles of Drosophila melanogaster. Genetics 136 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser, I., M. Sarov, J. R. Hutchins, J. K. Heriche, Y. Toyoda et al., 2008. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods 5 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royet, J., and R. Finkelstein, 1995. Pattern formation in Drosophila head development: the role of the orthodenticle homeobox gene. Development 121 3561–3572. [DOI] [PubMed] [Google Scholar]
- Ryder, E., F. Blows, M. Ashburner, R. Bautista-Llacer, D. Coulson et al., 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167 797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder, E., M. Ashburner, R. Bautista-Llacer, J. Drummond, J. Webster et al., 2007. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics 177 615–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan, S. K., L. C. Thomason, S. G. Kuznetsov and D. L. Court, 2009. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 4 206–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, B. R., and J. R. Merriam, 1975. Regulation of gene activity by dosage compensation at the chromosomal level in Drosophila. Genetics 79 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A. H., and T. H. Morgan, 1923. Reverse mutation of the Bar gene correlated with crossing over. Science 57 746–747. [DOI] [PubMed] [Google Scholar]
- Tilney, L. G., P. S. Connelly, K. A. Vranich, M. K. Shaw and G. M. Guild, 1998. Why are two different cross-linkers necessary for actin bundle formation in vivo and what does each cross-link contribute? J. Cell Biol. 143 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney, L. G., P. S. Connelly and G. M. Guild, 2004. Microvilli appear to represent the first step in actin bundle formation in Drosophila bristles. J. Cell Sci. 117 3531–3538. [DOI] [PubMed] [Google Scholar]
- Venken, K. J., and H. J. Bellen, 2005. Emerging technologies for gene manipulation in Drosophila melanogaster. Nat. Rev. Genet. 6 167–178. [DOI] [PubMed] [Google Scholar]
- Venken, K. J., and H. J. Bellen, 2007. Transgenesis upgrades for Drosophila melanogaster. Development 134 3571–3584. [DOI] [PubMed] [Google Scholar]
- Venken, K. J., Y. He, R. A. Hoskins and H. J. Bellen, 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314 1747–1751. [DOI] [PubMed] [Google Scholar]
- Venken, K. J., J. Kasprowicz, S. Kuenen, J. Yan, B. A. Hassan et al., 2008. Recombineering-mediated tagging of Drosophila genomic constructs for in vivo localization and acute protein inactivation. Nucleic Acids Res. 36 e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken, K. J., J. W. Carlson, K. L. Schulze, H. Pan, Y. He et al., 2009. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild, J., Z. Hradecna and W. Szybalski, 2002. Conditionally amplifiable BACs: switching from single-copy to high-copy vectors and genomic clones. Genome Res. 12 1434–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y., N. Denef and T. Schupbach, 2009. The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev. Cell 17 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, R. G., P. R. Hiesinger, T. W. Koh, P. Verstreken, K. L. Schulze et al., 2003. Mapping Drosophila mutations with molecularly defined P element insertions. Proc. Natl. Acad. Sci. USA 100 10860–10865. [DOI] [PMC free article] [PubMed] [Google Scholar]