Abstract
Many novel and important mutations arise in model organisms and human patients that can be difficult or impossible to identify using standard genetic approaches, especially for complex traits. Working with a previously uncharacterized dominant Saccharomyces cerevisiae mutant with impaired vacuole inheritance, we developed a pooled linkage strategy based on next-generation DNA sequencing to specifically identify functional mutations from among a large excess of polymorphisms, incidental mutations, and sequencing errors. The VAC6-1 mutation was verified to correspond to PHO81-R701S, the highest priority candidate reported by VAMP, the new software platform developed for these studies. Sequence data further revealed the large extent of strain background polymorphisms and structural alterations present in the host strain, which occurred by several mechanisms including a novel Ty insertion. The results provide a snapshot of the ongoing genomic changes that ultimately result in strain divergence and evolution, as well as a general model for the discovery of functional mutations in many organisms.
THE Saccharomyces cerevisiae genome sequence was completed in 1996 and represented the first complete eukaryotic genome (Cherry et al. 1997). It was a revolutionary tool for yeast researchers and provided a model for functional genome analyses in all organisms. Something it did not routinely allow, however, was the interrogation of additional strains for novel mutations. Identification of functional mutations arising spontaneously or in screens still relies primarily on classical techniques such as linkage analysis and plasmid complementation that are effective but cumbersome and can fail with dominant mutations, large genes, and when extragenic suppressors are common. The challenges of identifying target mutations are only magnified in obligatory diploid organisms with larger and more complex genomes such as mammals.
Comprehensive and unbiased discovery of new or interesting genetic differences requires the repeated application of DNA sequencing on the whole-genome scale, which for many years remained outside the reach of experimentalists. The advent of high-throughput short-read sequencing technologies has dramatically changed this status quo. The common basis of most of these new sequencing platforms is the physical separation of single DNA molecules into an array, typically with in situ amplification to increase the signal yield, followed by various chemistries to reveal the base-by-base sequence at each array position using advanced imaging techniques (Metzker 2010). Platforms now allow >100 Gb of sequence to be obtained in a single run in the form of millions of reads of <100 bp. Although generally insufficient to assemble a genome de novo, such short reads can be mapped to a reference genome, allowing differences between the study sample and the reference sequence to be identified.
Despite their raw power, there are still many obstacles to realizing the experimental utility of short-read sequencing technologies. The first is the need for efficient computational tools to deal with the large amount of generated data. Moreover, the accuracy of current short-read technologies is lower than standard sequencing so one must sort out real mutations from sequencing errors. When the experimental goal is to discover the mutation associated with a specific phenotype, one will also need to distinguish the causative allele from other mutations that are present. Finally, chromosomal alterations such as translocations operate on an inherently larger scale than <100-bp reads, and variant approaches are required to identify them.
In this study, we sought to establish approaches to genome sequencing via short-read technologies that would satisfy the above needs. We started with an uncharacterized yeast mutant with impaired vacuole inheritance, VAC6-1 (Gomes de Mesquita et al. 1996). We describe how genetic linkage in a single backcross was exploited to rapidly identify the VAC6-1 allele from among >10,000 other strain mutations and polymorphisms. To maximize information quality and yield, data were generated using mate-pair technology in which both ends of genomic DNA fragments are sequenced (Dew et al. 2005; Korbel et al. 2007), which allowed a nearly complete description of the structural alterations present. Together, the results provide broadly applicable computational tools and approaches to mutation identification whose logic is readily extendable to higher eukaryotes with appropriate modifications. In addition, the comprehensive analysis of genome alterations in our strain provides a snapshot of the striking genetic differences present in laboratory organisms.
MATERIALS AND METHODS
Yeast strains:
The yeast strains used in this study were obtained from the strain archive of the Weisman laboratory. JBY009/VAC6-1 was the kind gift of Daniel Gomes de Mesquita and Conrad Woldringh (Gomes de Mesquita et al. 1996). To perform the screen for VAC mutants, the PEP4 gene had first been knocked out of SEY6210 (MATα ura3-52 his3-Δ200 leu2-3,112 lys2-801 trp1-Δ901 suc2-Δ9) to generate RHY6210 (SEY6210 pep4-Δ1137). SEY6210 itself (Robinson et al. 1988) was derived by crossing strains from the laboratories of Gerald Fink, Ronald Davis, David Botstein, Fred Sherman, and Randy Schekman and is commonly used in laboratories that study vacuole-related processes (see http://wiki.yeastgenome.org/index.php/Commonly_used_strains). JBY009 (RHY6210 VAC6-1) was the product of a screen in which RHY6210 was mutagenized with UV irradiation (254 nm, 1 J m−2, 20 sec) to 40% viability (Gomes de Mesquita et al. 1996). For backcrossing, we introduced plasmid pGAL-HO into a PEP4 version of a strain that we believed to be otherwise isogenic with RHY6210 to generate a MATa strain that was used as the backcross parent. Eight dissected asci from a first backcross of JBY009 had been archived and were recovered from the freezer for sequencing. The yeast used to demonstrate the detection of structural variations were from the wild-type segregant pool obtained from sporulation of diploid strain LWY10741 (MATa/α ura3-52/ura3-52 his3-Δ200/his3-Δ200 leu2-3,112/leu2-3,112 lys2-801/lys2-801 trp1-Δ901/trp1-Δ901 suc2-Δ9/suc2-Δ9 VAC22/vac22-1), which also derived from SEY6210.
Pooling and sequencing:
The statistical assessments and modeling used to derive the probabilities of identifying causative mutations and excluding incidental mutations are described in supporting information, Materials and Methods, File S1. Because the pooled linkage strategy assumes equal representation of all segregants in a pool, we took special care when making genomic DNA. All spore clones obtained from the eight dissected VAC6-1 heterozygous asci were grown overnight at 30° in individual 25-ml YPAD cultures (1% yeast extract, 2% peptone, 40 μg/ml adenine, 2% dextrose). The OD600 of the cultures was determined and used to calculate the appropriate volume of each strain to mix to achieve equal numbers of cells. Pools were made for the wild-type and mutant strains and genomic DNA was prepared without further outgrowth. Wild-type and mutant mate-pair libraries were made using the Illumina Mate Pair Library Prep Kit according to the manufacturer's instructions. Briefly, the process entailed shearing genomic DNA to ∼3-kb fragments and preparing the two fragment ends for sequencing via steps including circularization, reshearing, ligation of sequencing adapters, and limited PCR (see Figure S1 in File S1). Paired-end sequencing was finally performed on the Illumina Genome Analyzer by the University of Michigan DNA Sequencing Core. Sequence image analysis and base calling were performed using the Illumina Firecrest and Bustard algorithms, respectively, according to the instructions. All called sequence reads are available in FASTQ format from the National Center for Biotechnology Information Sequence Read Archive under submission SRA023658, study SRP003355.
Mutation finding:
All subsequent sequence data analyses were performed using the informatics platform that we developed called VAMP, for Visualization and Analysis of Mate-Pairs, which is available for download at http://tewlab.path.med.umich.edu/vamp.html. The methods and logic used by VAMP are described in SI Material and Methods and Figure S1, Figure S2, Figure S3, and Figure S4 in File S1. Mapping was performed using the PASS program (Campagna et al. 2009), which supports the identification of small indels in addition to point mutations, as coordinated by the VAMP wrapper. The reference genome was the 2003 release of the S. cerevisiae genome (sacCer2). Mapping filters allowed up to five discrepancies (mismatches or indels) relative to sacCer2 and up to 10 initial genome map positions. Best mappings were chosen by giving priority to those mate-pairs with the expected separation of ∼3 kb or those consistent with the orientation artifact described in Figure S1 in File S1. Candidate VAC6-1 causative alleles were defined as sequence alterations that showed: (i) a predicted coding change in a yeast gene, (ii) at least three crossing reads in each of the wild-type and mutant pools, (iii) no more than 10% of wild-type pool reads consistent with the mutation, and (iv) at least 90% of mutant pool reads consistent with the mutation. To find Ty elements inserted relative to sacCer2, we used VAMP's modified mapping algorithm (see SI Materials and Methods, File S1) with the panel of all known yeast Ty elements as the training set. Implicated Ty reads were assembled into a contig by iterative refinement against known Ty sequences.
Validation of the VAC6-1 allele:
A 5.7-kb PHO81 DNA fragment (−1695 to 4062 bp) from wild-type or VAC6-1 genomic DNA was subcloned into the SacII and SalI sites of pRS413 (CEN, HIS3) by fusing four tandem PCR fragments. Yeast strain LWY7235 (MATa leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9) (Bonangelino et al. 1997) was transformed with these plasmids or pRS413 and cells labeled with the fluorescent dye FM4-64 for microscopic visualization of the vacuole (Vida and Emr 1995).
RESULTS
Linkage analysis by sequencing bulk segregants:
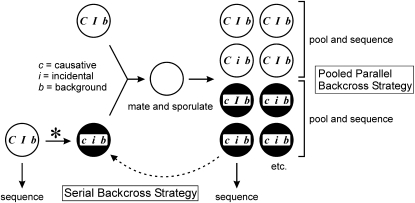
To identify the genetic alteration underlying an observable yeast phenotype by genome sequencing, the causative mutation must be both positively identified and distinguished from sequencing errors, strain polymorphisms, and, in the case of a screen isolate, noncausative mutations incidentally induced by the source mutagen. We considered two strategies for achieving these goals (Figure 1). In the first, standard serial backcrossing of the mutant strain is performed against an isogenic wild-type strain, selecting a single segregant that displays the mutant phenotype at each cross. Sequencing is ultimately applied to the last selected mutant segregant and the parent strain. The alternative approach, which we employed, entails a single backcross with many asci obtained in parallel. Instead of sequencing pure clonal isolates, all mutant segregants from a series of asci are pooled and bulk genomic DNA and a corresponding library are prepared and sequenced. The pool of all wild-type segregants from the same asci is sequenced in parallel for the same total of two required sequencing runs as the serial backcross strategy.
Figure 1.—
The pooled parallel backcross strategy. In the scenario depicted, a yeast strain with a novel phenotype (indicated by a solid circle) is derived by mutagenesis of a parental wild-type strain (indicated by an open circle). The mutant is backcrossed against the parental strain and asci are scored. In the serial backcross strategy, this process is repeated until a single mutant segregant is ultimately chosen for sequencing. In the pooled parallel backcross strategy, all wild-type and all mutant segregants from several asci from the first backcross are pooled and sequenced in two separate libraries. Letters indicate the three classes of mutation that must be tracked. C/c refers to wild-type and mutant alleles of the gene bearing the causative mutation, which by definition always cosegregates with the mutant phenotype. I/i refers to an unlinked incidental mutation induced by EMS, which will sort randomly with respect to the phenotype. B/b refers to a mutation present in the strain background prior to EMS mutagenesis.
The net result of the pooled parallel backcross strategy, also known as “bulk segregants” (Brauer et al. 2006; Ehrenreich et al. 2010; Wenger et al. 2010), is a powerful linkage experiment (Figures 1 and 2). Tight linkage and presumed possible causality are revealed by mutations that are present in 100% of mutant pool and 0% of wild-type pool sequence reads. Unlinked incidental mutations as well as sequencing errors are expected to yield a mixture of wild-type and mutant bases in both the wild-type and the mutant pools. Strain background polymorphisms will be present in 100% of reads in both pools, assuming that an isogenic wild-type strain is used in the backcross. If an isogenic strain is not used, differences between the mated strains will again sort between wild-type and mutant pools at a frequency consistent with their degree of linkage to the causative allele.
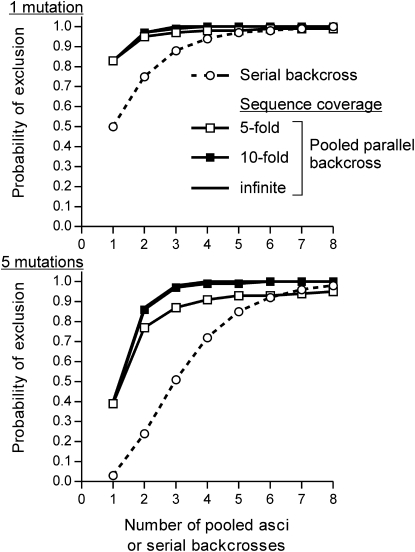
Figure 2.—
Statistical power of pooled linkage analysis by sequencing. Graphs show the probability of excluding either one (top) or five (bottom) incidental mutation(s) as the causative allele as a function of the number of pooled asci (solid lines) or serial backcrosses (dashed lines with open circles). For the pooled parallel backcross strategy, an obscured solid line indicates the theoretical maximum probability of exclusion corresponding to infinitely deep sequencing of the strain pools, while solid lines with open and solid squares show the results of a 10,000-iteration simulation conducted at 5- and 10-fold average genome coverage per pool, respectively.
Figure 2 compares the statistical power of the serial and parallel approaches. Serial backcrosses have relatively low discriminatory power for the exclusion of incidental mutations over the range of backcross iterations typically used by most yeast researchers. Pooled parallel backcrosses have much greater discriminatory power mainly because the total information content of every ascus can be brought to bear in the analysis. Surprisingly, few asci need to be sampled to exclude the large majority of incidental mutations, mainly because the probability of selecting only parental ditype asci rapidly becomes very small. The power is dependent on the sequence coverage obtained, but Figure 2 demonstrates that 10-fold genome coverage of each pool is sufficient to capture nearly all of the available information. Critically, at 10-fold coverage the probability that the causative mutation will be sequenced by at least three independent reads is 0.997 (0.875 and 1.000 for 5- and 25-fold coverage, respectively). This is severalfold less coverage than provided by a single Illumina sequencing lane (Table 1).
TABLE 1.
Sequencing pool summary statistics
|
VAC6-1 |
|||
|---|---|---|---|
| Wild type | Mutant | LWY10741 | |
| Solexa lanes | 1 | 1 | 1 |
| Read length (bp) | 36 | 36 | 39 |
| Mean fragment size (bp) | 3085 | 2610 | 2799 |
| Unique mate pairs | 1.4 × 107 | 1.3 × 107 | 4.1 × 106 |
| Allowed pair mappings | 4.5 × 106 | 5.2 × 106 | 2.0 × 106 |
| Mapped reads | 9.0 × 106 | 1.0 × 107 | 4.1 × 106 |
| Mean base coverage | 27 | 31 | 13 |
| Median base coverage | 25 | 27 | 12 |
| Base mismatch rate (%) | 0.6 | 0.5 | 0.4 |
Point mutation content of the VAC6-1 mutant strain:
To test the parallel backcross strategy, we applied it to identification of the VAC6-1 mutation, which causes a defect in vacuolar inheritance scored by microscopic examination of fluorescently labeled cells (Gomes de Mesquita et al. 1996; Wang et al. 1996). Because VAC6-1 is a dominant allele, it could not be identified by standard complementation cloning and thus the genetic basis of the defect was unknown. We pooled archived wild-type and mutant segregants from eight asci from the first backcross of JBY009, the VAC6-1 strain obtained from a UV mutagenesis screen of parental strain RHY6210 (Gomes de Mesquita et al. 1996), itself a derivative of SEY6210 (Robinson et al. 1988). A 3-kb mate-pair library was prepared for each of the wild-type and mutant spore pools, and each was sequenced in a single Illumina lane using paired reads (Table 1). Sequence reads were mapped to the yeast reference genome and analyzed using the VAMP software platform. Comparison of Figure 2 to the run data in Table 1 indicated that the coverage obtained would be sufficient to both identify the VAC6-1 mutation and to exclude all but the most closely linked incidental mutations.
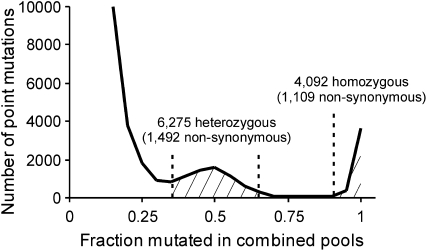
Figure 3 shows a histogram of the number of sequence changes that we observed relative to S288C, the strain represented by the reference genome, according to the fractional representation in the combined wild-type and mutant pool data. This is equivalent to what would have been obtained had the backcross diploid itself been sequenced. Three features are evident. The first is a very large number of sequence changes substantially below the 50% frequency expected for diploid heterozygous mutations. These correspond primarily to sporadic sequencing errors, which occurred in our samples at a frequency of ∼0.5% (Table 1). More important were two peaks corresponding to ∼50% and ∼100% mutation frequencies in the combined pools, sequence changes inferred to have been heterozygous and homozygous in the backcross diploid, respectively. Strikingly, >6000 heterozygous and 4000 homozygous changes were called (see Table S1). Inspection of individual calls provided clear corroboration (see Figure S5 in File S1). Further inspection confirmed that the large majority of alterations (85%) corresponded to population polymorphisms present in at least 1 of the 38 strains sequenced by the Saccharomyces Genome Resequencing Project (Liti et al. 2009), with 7403 (71%) present in five or more strains. The strongest match was to laboratory strain SK1, which shared 5339 (51%) of the changes that we observed, but similarly high rates of correspondence were seen with natural S. cerevisiae isolates, such as RM11-1a, which shared 5076 (49%).
Figure 3.—
Frequency distribution of observed point mutations and SNPs. Sequence data from the VAC6 wild-type and mutant spore pools were combined to show the point mutation content in the diploid backcross strain. True sequence changes are expected to cluster in peaks at frequencies of 50% and 100%, corresponding to heterozygous and homozygous changes in the diploid strain, respectively. Frequencies need not be precisely 50% or 100% because of stochastic effects in pool sampling. Heterozygous (35–65% frequency) and homozygous (>90% frequency) mutation counts are shown. The off-scale peak of sequence changes at <25% frequency corresponds to sequencing errors.
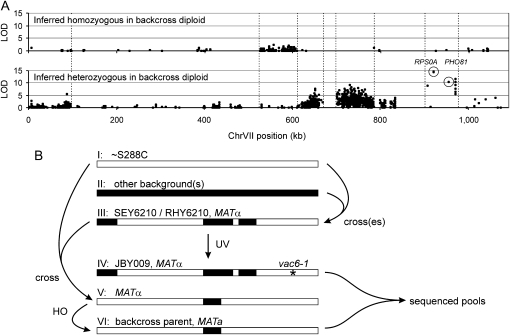
A strongly nonrandom distribution of both homozygous and heterozygous sequence changes was observed throughout the genome (Figure 4A). We interpret the obvious clustering of most polymorphisms as recombination blocks created by crosses that occurred previously in the history of our strains relative to S288C. Regions of low mutation density were inherited from a strain(s) closely related to S288C during the complex history that gave rise to RHY6210 and SEY6210 (Figure 4B and materials and methods). In contrast, homozygous high-density mutation blocks represent RHY6210 chromosome regions inherited from a background other than S288C. The observed heterozygous high-density mutation blocks were unexpected, however, since we had believed JBY009/VAC6-1 and its backcross parent to be isogenic. Examination of the sequence data revealed that the SEY6210 auxotrophic markers his3Δ-200, trp1-Δ901, and leu2-3,112 were also unexpectedly heterozygous, and indeed phenotypic testing showed the backcross parent to be His+, Trp+, and Leu+. We thus infer that this strain had in fact been crossed to a strain more closely related to S288C prior to the manipulations that we performed (Figure 4B and materials and methods). Notably, 24% and 27% of the heterozygous and homozygous mutations, respectively, changed coding of a yeast gene (Figure 3), including nonsense mutations in LYS2 (a known auxotrophic marker), YFR057W, SIM1, YKL133C, SPH1, RPS22B, UFO1, YML082W, ISW2, and GRE2, underscoring the large genetic differences that can exist between laboratory yeast strains (Liti et al. 2009).
Figure 4.—
Most VAC6-1 strain mutations occur in nonrandom blocks. (A) Chromosome distribution plots were constructed for all inferred homozygous (top) and heterozygous (bottom) mutations observed in the VAC6 wild-type and mutant pools (see Figure 3). Only chromosome VII, which contains the causative PH081 mutation, is shown (similar trends could be observed on all chromosomes). Every mutation is plotted as a single point, although there is often insufficient resolution to visualize all loci. The y-axes are the linkage LOD scores for the mutation relative to the VAC6 mutant phenotype. Mutations from the high-priority candidate list (Table 2) are circled and labeled. Vertical dashed lines indicate the edge of chromosome blocks that contain clustered mutations. (B) Drawings depict the strain history of JBY009/VAC6-1 to illustrate the inferred origin of high-density mutation blocks from prior meiotic recombination events. For each strain, chromosome VII is depicted as open when it generally matches the S288C reference genome and as solid when a high density of non-S288C values are present. Early crosses led to the mosaic strain III used for VAC6-1 mutagenesis. Inferred but undocumented crossing of strains I and III allowed further partial recombination of some high-density blocks in strain V and then strain VI, the strain ultimately used as the VAC6-1 backcross parent, leading to the zygosity pattern observed in A.
PHO81-R701S encodes VAC6-1:
Although noteworthy, no high-density mutation block in Figure 4 was likely to contain the VAC6-1 causative allele because none consistently showed the highest degree of linkage required of a causative locus. Indeed, nonsynonymous mutations that showed at least 90% segregation into the VAC6-1 mutant pool affected only two genes, RPS0A and PHO81 (Table 2). Because these genes are near each other on chromosome VII, it was very likely that one of them was the causative allele and that the other was linked to it (Figure 4A). However, even one failed correspondence between phenotype and the candidate mutation is theoretical cause for exclusion, and neither of the candidates showed perfect segregation. This prompted us to rescore the phenotype of the segregants present in the wild-type and mutant pools, which revealed that two strains had in fact been scored incorrectly and been switched, a corruption consistent with the read frequencies in Table 2. This demonstrates the power of pooled linkage analysis even when assignment errors are possible with a complex and difficult-to-score phenotype.
TABLE 2.
Nonsynonymous mutations with at least 90% segregation into the VAC6-1 mutant pool
| Sequence pool |
|||||||
|---|---|---|---|---|---|---|---|
| Chromosome | Position(s) | Mutation | Wild type | Mutant | LOD | Gene | Mutation |
| VII | 956,111 | C to A | 0/19 | 25/27 | 10.2 | PHO81 | R701S |
| VII | 921,514 and 921,515 | CC to AT | 3/34 | 45/49 | 14.5 | RPS0A | P161S |
Figure 4A revealed the presence of several other mutations in the RPS0A/PHO81 region of chromosome VII that showed LOD scores similar to these genes but that did not appear on the candidate mutation list. Closer examination of the counts of these mutations (see Table S1) revealed that all were in fact anti-correlated to the VAC6-1 mutant phenotype in being present in ∼90% of reads from the wild type, not the mutant, pool. We infer that these mutations were on the other copy of chromosome VII in contrast to the RPS0A and PHO81 mutations. This demonstrates the power of linkage for the identification of target genomic loci even when the mutations scored are not causative.
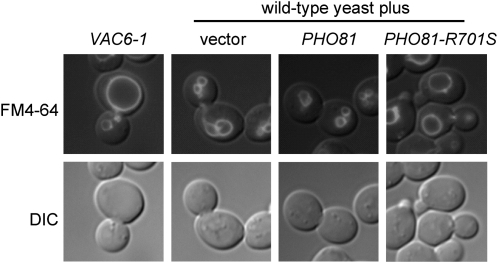
With two genes on the candidate list, final prioritization was based on function. Of the candidates, RPS0A encodes a ribosome component with no obvious connection to vacuole biology. Moreover, it is substantially redundant with RPS0B (Demianova et al. 1996). In marked contrast, candidate PHO81 encodes a cyclin-dependent kinase (CDK) inhibitor that regulates the activity of the Pho80/85 CDK complex. This made PHO81-R701S a strong candidate since we have shown that vacuolar mutant vac5 results from mutation of PHO80 (Nicolson et al. 1995). Because VAC6-1 is a dominant mutation, we tested our hypothesis that PHO81-R701S encodes VAC6-1 by recovering the PHO81 allele from wild-type and VAC6-1 mutant strains, cloning them into a plasmid vector and transforming these into wild-type yeast. Introduction of the wild-type vector had no effect on vacuole inheritance whereas the VAC6-1-mutant PHO81 vector precisely recapitulated the VAC6-1 phenotype (Figure 5). Because standard sequencing of the recovered PHO81 alleles confirmed the presence of the R701S mutation, we conclude that VAC6-1 is PHO81-R701S.
Figure 5.—
PHO81-R701S is the causative mutation in VAC6-1. Transformation of wild-type yeast with either pRS413 vector or pRS413-PHO81 had no effect on vacuole inheritance, whereas transformation with pRS413-PHO81-R701S caused an enlarged vacuole in the mother cell and a vacuole inheritance defect (right panels). Wild-type cells bearing pRS413-PHO81-R701S showed the same phenotype as VAC6-1 itself (left).
Finding yeast structural variations:
UV and EMS mutagenesis principally induce point mutations, but there are many scenarios where large-scale structural alterations of the yeast genome must be tracked. Because of technical limitations with the VAC6-1 sequence pools (see above and Figure S1 in File S1), it is more straightforward to present these approaches using sequence obtained with a different yeast strain, LWY10741 (see materials and methods).
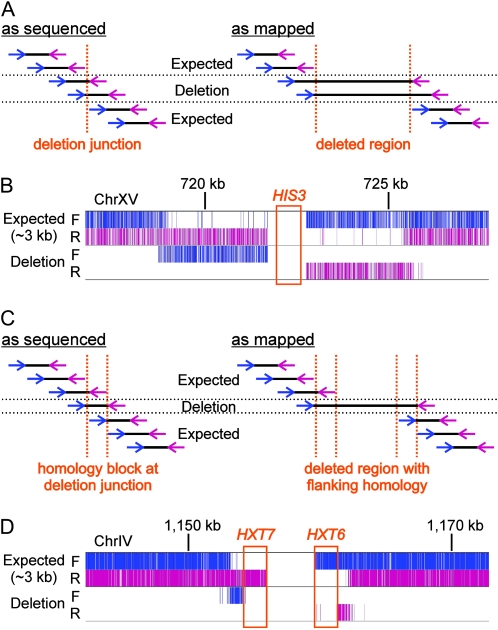
The ability to score structural variations depends on the use of paired reads during sequencing, as illustrated in Figure 6A and described in SI Materials and Methods according to published logic (Dew et al. 2005; Korbel et al. 2007). The concept is to identify discrepancies between the orientation and separation of paired reads inferred from genome mapping as compared to what is expected for the library. In this way, 14 genome deletions relative to sacCer2 were called for LWY10741 (Table 3; Figure 6; Figure S6, Figure S7, and Figure S8 in File S1). No insertions, inversions, or duplications were detected that were not within tandem LTR repeats, subtelomeric regions, or rDNA where mapping is untrustworthy. Three of the observed deletions were expected since his3-Δ200 (Figure 6B), trp1-Δ901, and suc2-Δ9 are known mutations in the sequenced strain, providing internal validation of the results. Because, to our knowledge, the origin and structure of suc2-Δ9 has never been reported (Emr et al. 1983), we reconstructed the allele and found that it was created by a microhomology mechanism corresponding to an EcoRI site (see Figure S7 in File S1). The last non-Ty deletion was unanticipated but equally clear. It removed the intergenic region between HXT6 and HXT7, and the read pattern demonstrated that this event occurred by homologous recombination between these nearly identical genes (Figure 6, C and D).
Figure 6.—
Two mechanisms of chromosome deletion. (A) It is assumed that “as sequenced” all mate-pairs corresponded to ∼3-kb physical DNA fragments present in the strain genome. However, “as mapped” to the reference genome mate-pairs flanking a deletion junction show an excessively large spacing. (B) An example deletion corresponding to his3-Δ200. All expected (∼3 kb) and deletion reads in both the forward (F) and reverse (R) orientations in the displayed region of chromosome XV are drawn as vertical lines. Forward and reverse read colors match the arrows in A. Although not illustrated, every forward deletion read was paired with a corresponding reverse deletion read on the opposite side of HIS3. The presence of a homozygous deletion is confirmed by the loss of expected reads within and surrounding the deleted segment in a pattern consistent with A. (C) Similar to A, showing the expected pattern of reads when the deletion occurs by homologous recombination via a homology block flanking the deleted locus. (D) Similar to B, showing a deletion inferred to have occurred by homologous recombination between HXT7 and HXT6 by the logic in C.
TABLE 3.
LWY10741 genome deletions
| Chromosome | Start | End | Size (bp) | Genes affected | LTR present |
|---|---|---|---|---|---|
| IV | 461,743 | 463,183 | 1441 | TRP1 | NA |
| IV | 1,155,970a | 1,159,523 | 3553 | HXT7, HXT6 | NA |
| IX | 36,451 | 40,812 | 4361 | SUC2 | NA |
| XV | 721,747 | 722,769 | 1023 | HIS3 | NA |
| II | 221,045 | 226,969 | 5925 | YBLWTy1-1 | — |
| II | 258,752 | 266,237 | 7486 | YBRWTy1-2 | Yes |
| IV | 645,271 | 651,442 | 6172 | YDRCTy1-1 | Yes |
| IV | 1,206,705 | 1,212,619 | 5915 | YDRWTy1-5 | — |
| VII | 560,879 | 567,760 | 6882 | YGRCTy1-2 | — |
| VII | 568,747 | 576,272 | 7526 | YGRCTy2-1 | — |
| VII | 535,768 | 541,681 | 5914 | YGRWTy1-1 | Yes |
| XII | 593,146 | 599,058 | 5913 | YLRWTy1-2 | Yes |
| XIII | 196,329 | 202,193 | 5865 | YMLWTy1-2 | Yes |
| XIII | 184,168 | 190,077 | 5910 | YMLWTy1-1 | — |
Coordinates for this event refer to the deleted intergenic region, exclusive of HXT7 and HXT6. One copy of this duplicate gene pair is deleted in addition to the indicated span.
The remaining 10 called “deletions” all corresponded to Ty retrotransposon elements (Voytas and Boeke 1993; Kim et al. 1998) in the sacCer2 reference sequence (Table 3). The robustness of these calls is supported by comparing the read patterns for the non-Ty deletions (Figure 6) to those seen for deleted and nondeleted Ty elements (see Figure S8 in File S1). Thus, 24% of the 50 annotated Ty elements were not present in our strain. Two equally frequent and distinct patterns were observed for the missing Ty elements (see Figure S8 in File S1). In one pattern, Ty LTRs were found to be residual at the locus, suggesting that a Ty had been present but was deleted by homologous recombination between the flanking LTRs. In the other pattern, no LTRs were apparent, which might reflect a different loss mechanism or that the Ty was never present in LWY10741.
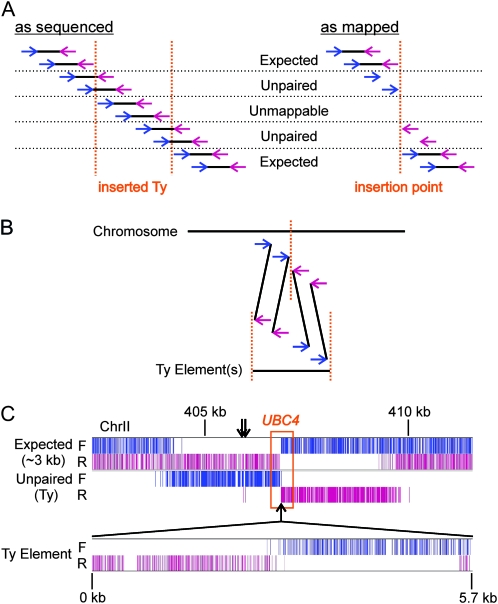
We next asked whether our strain might contain unknown Ty elements. Importantly, an intact ∼6-kb Ty is too large to be flanked by a 3-kb DNA fragment so that reads near a novel Ty will be “unpaired” (Figure 7A). We therefore wrote algorithms to establish the location of unpaired reads whose partner read could be mapped to any one of the highly related known yeast Ty elements (Figure 7B). By examining the orientation and clustering of such reads, we identified one previously unknown Ty element in our strain (Figure 7C). Assembling the sequence contig from the partner reads (see Figure S9 in File S1) showed it to be of the Ty1 family, mostly closely related to YJRWTy1-2 with 98% sequence identity >5.5 kb. There were sequence differences relative to all known Ty elements, however. Unsurprisingly, the insertion site is within 1 kb of two tRNA genes (Figure 7C), a known feature of Ty genome locations (Bolton and Boeke 2003). Strikingly, the novel Ty is within and disrupts gene UBC4, which encodes a ubiquitin-conjugating enzyme (Seufert and Jentsch 1990), again emphasizing the substantial genetic differences that can exist between laboratory yeast strains.
Figure 7.—
A novel Ty insertion. (A) Similar to Figure 6A, showing the expected orientation of reads and fragments in the vicinity of a Ty element not present in the reference genome. (B) The strategy for identifying novel Ty insertions by comparing independent mappings of mate-pairs to (i) chromosome sequences and (ii) a training set of known Ty repeat elements. (C) An identified novel Ty insertion, illustrated as in Figure 6B, now with a track corresponding to those unpaired reads whose partners independently mapped to a Ty element(s). (Bottom) The partner reads aligned to the Ty element assembled from the data. Arrows denote the location of two closely spaced tRNA genes, tR(UCU)B and tD(GUC)B.
Surprisingly, the query for new Ty elements did not return the URA3 locus. We had expected this since LWY10741 is homozygous for ura3-52, a well described 6-kb Ty insertion mutation (Rose and Winston 1984). Examination of URA3 revealed that no sequence reads had mapped across the ura3-52 insertion site (chromosome V, position 116,282), consistent with an insertion at this point, and that there were indeed Ty mate-pairs in the flanking regions (not shown). However, there were not enough such pairs to pass the threshold we used when calling Ty sets. This paucity of fragments would be explained by a small Ty insertion, which restricts the number of reads that can map within it. Accordingly, nearly all mate-pairs that spanned the Ty insertion point were individually too close to the library mean fragment size to be called as deviant, but when considered together, these 417 fragments predicted a net insertion of 278 ± 202 bp (P = 3 × 10−98 by the t-test relative to an expected insertion of 0 bp). We conclude that the LWY10741 ura3 allele did derive from ura3-52 but that the Ty element itself was subsequently deleted so that only a residual LTR remains.
DISCUSSION
Identifying yeast strain mutations by pooled linkage analysis:
Tracking linkage during meiotic recombination has been an invaluable technique in yeast genetic analysis for many decades (Mortimer and Hawthorne 1975; Mortimer and Schild 1985). Here, we demonstrate how to apply this concept to the efficient identification of uncharacterized mutations in light of an emergent ability to rapidly resequence the yeast genome with short-read technologies (Figure 1) (Metzker 2010). Tracking linkage in pools requires no more sequencing than other approaches and all but eliminates concerns over sequence errors and off-target mutations. Statistical modeling (Figure 2) and a practical example (Table 2 and Figure 5) confirmed that mutation identification requires only one backcross, surprisingly few asci, and little genome coverage. A similar approach also recently identified a novel xylose utilization gene (Wenger et al. 2010).
A main alternative to using sequencing for bulk segregant analysis is to map SNPs via microarrays to identify loci of interest (Brauer et al. 2006). When the goal is to identify an experimentally (UV or EMS) induced alteration likely to correspond to a simple point mutation, the newer sequencing approach is clearly more powerful as it can directly and positively identify the mutation. There is no need for a custom SNP array nor a backcross strain known to create extensive heterozygosity, which could have unanticipated and undesirable effects on phenotype expression. Indeed, an isogenic backcross strain is ideal since no more than a few induced mutations will likely occur within linkage distance.
The situation is different if the target mutation might be of a type difficult to discover by sequencing. Some genomic loci, such as those found in subtelomeric regions and repetitive genes, are inherently problematic (see Figure S6 in File S1). Other mutations, notably indels, are difficult because of their specific nature. For example, a 10-bp deletion would be evident only as an absence of reads crossing the variant position. Finally, target genetic differences might be associated with genes absent from the S288C reference genome, especially when the goal is to identify loci associated with phenotypes that differ between extant yeast strains (Wenger et al. 2010). Regardless of the reason that a mutation is difficult to discover, linkage information provided by SNP analysis can provide the key impetus for examining a regional sequence in more detail (Wenger et al. 2010).
SNP analysis can be comprehensively performed with sequencing (Figure 4 and Ehrenreich et al. 2010), so this method is still preferred over microarrays. The main decision point for most researchers will thus be whether to perform sequencing using (i) an isogenic backcross strain, ideal for simple point mutations in known genes; (ii) a backcross strain deliberately chosen to yield a large number of SNPs, ideal for linkage mapping of problematic loci (Wenger et al. 2010); or (iii) no backcrossing at all, ideal when the goal is to catalog all changes in a strain (Araya et al. 2010). Importantly, with the latter options it may not be possible to positively identify even a simple causative mutation within a linked locus, and especially not within an entire genome, due to the high density of nonsynonymous sequence alterations typically present (Figures 3 and 4) (Liti et al. 2009). Additional analyses can be brought to bear, including predictions of function and querying which mutations correspond to known benign polymorphisms (Ng et al. 2010). These assessments are weakly informative, however, as demonstrated by the fact that 15% of the sequence changes that we observed could not be accounted for by known yeast polymorphisms (Liti et al. 2009).
In this report, we assumed that the mutation of interest is inherited in a single-gene Mendelian fashion. However, the results give confidence that pool sequencing could also be used in the context of multigenic traits since linkage of phenotype-associated alleles will remain true as a fundamental principle. Indeed, Ehrenreich et al. (2010) recently exploited bulk segregant analysis in the study of multigenic quantitative trait loci. A difference is that these investigators used methods restricted to phenotypes that can be selected in pooled outgrowth cultures. Results presented here explored a complex phenotype that can be scored only individually by microscopy. Further investigation will be required to determine whether complex multigenic traits can be efficiently analyzed when only a relatively small collection of segregants can be pooled for sequencing.
For single-gene mutations, data here already demonstrate that a single Illumina sequencing lane is more than is required for a pool, and sequencing capacity is still rapidly increasing. Future efforts should thus implement multiplexing. The main approach is to use primers in library construction that contain fixed index sequences that identify each source sample and allow libraries to be sequenced together in a lane (Maeda et al. 2008). Assuming the need for 10-fold coverage, it should soon be routinely possible to sequence as many as 10 pools per lane. Importantly, it is not necessary to continue sequencing the wild-type pool for mutants derived from the same parent strain. VAMP includes algorithms to reconstruct the host genome to provide a reference for all mutants. In this way, a single lane can characterize numerous yeast mutants at a cost approaching $100 per mutant. It thus becomes practical to sequence arrays of mutants derived from a screen with minimal prior analysis.
Extension to other organisms:
The pooling approach described here could be immediately applied to any organism for which the phenotypes of meiotic progeny can be assessed and for which a reference genome is available. Pooling can also be applied to organisms for which diploid offspring must be obtained to allow phenotypic assessment (Schneeberger et al. 2009). As an example, one could mate mice bearing a recessive mutation to wild-type individuals to create an obligatory heterozygous F1 generation (see Figure S10 in File S1). After mating F1 mice, a causative mutation would be present in 100% of alleles of the target gene from the pool of affected mice and in only 33% of alleles from the phenotypically normal pool.
Yeast genetic variation:
To a remarkable extent, sequencing of only two related strains revealed a microcosm of the modes of genetic variation at play in yeast and other organisms (Scannell et al. 2007). Point mutations were the most numerous, with 10,367 called events affecting ∼0.1% of the yeast genome (Figure 3). The strong bias of the mutations toward transitions (71% vs. a random expected frequency of 33%) was consistent with them being derived from biological mutagenesis (Sinha and Haimes 1981; Zhang and Gerstein 2003), and indeed most corresponded to known yeast SNPs (Liti et al. 2009). Mutations were not randomly distributed but strongly reflected the reassortment of chromosome segments via meiotic recombination (Figure 4). We also called 331 indels (see Table S1). The ratio of indels to base substitutions (0.03) is very similar to other yeast strains (0.06) (Liti et al. 2009), but seemingly low in the face of studies showing that indels in homopolymer runs (HPRs) are the most frequent form of spontaneous mutation (Lynch et al. 2008). Only 55 (17%) of our called indels were in HPRs of five or more bases, which might reflect a bias against the detection of HPR indels by short-read sequencing.
Still fewer large-scale alterations of chromosome structure were found, but because of their size one or many genes were clearly disrupted (Table 3). Each of the main modes of chromosome rearrangement (Tsai and Lieber 2010) were described by different deletions: homologous recombination within a related gene cluster (Figure 6D) and nonhomologous recombination via junctional microhomology (see Figure S7, File S1, perhaps an experimentally created alteration). Finally, we observed the ongoing role of mobile genetic elements in shaping genomes (Table 3) (Garfinkel 2005; Cordaux and Batzer 2009). The Ty content of our strain and S288C differed markedly, including (i) the apparent deletion of ancient Ty elements from our strain, evidenced by the LTR that they left behind (see Figure S8C in File S1); (ii) the possible addition of Ty elements in S288C, evidenced by their complete absence from our strain (see Figure S8A in File S1); and (iii) the addition of a Ty element to our strain, which affected it by disrupting UBC4 (Figure 7).
PHO81-R701S and vacuole inheritance:
A review of the literature provides strong support for the notion that PHO81-R701S could have substantial impact on Pho81 function and vacuole inheritance. Pho81 is an inhibitor of the Pho80/85 CDK complex and mediates inactivation of the kinase in response to starvation for inorganic phosphate (Lenburg and O'Shea 1996). Unlike many CDK inhibitors, Pho81 remains constitutively bound to Pho80/85 (Schneider et al. 1994). CDK inhibition instead depends on binding of inositol heptakisphosphate (IP7) to the complex (Lee et al. 2007). Dissection of the molecular interaction between Ph081 and Pho80/85 in vitro suggested that “minimum domain” segment 3, from residues 665 to 701, binds constitutively to Pho80/85 while minimum domain segment 1, from residues 702 to 723, binds only in the presence of IP7 (Lee et al. 2008). It is thus plausible that Pho81 R701 is at the hinge point of a domain movement that occurs in response to IP7 binding and that the VAC6-1/PHO81-R701S mutation alters this function. Precedent for a role of Pho80/85 signaling in vacuole inheritance is provided by the vac5 mutant, which corresponds to a truncated allele of the Pho80 cyclin (Nicolson et al. 1995). Indeed, vac5 and VAC6 display similar vacuole morphologies. Precisely how alterations in Pho80/85 signaling lead to deregulation of vacuolar biogenesis is the subject of ongoing investigations.
Acknowledgments
We thank Emily Kaufman for assistance in recovering archived strains and scoring their phenotype and David Engelke, Anuj Kumar, Jun Li, and Thomas Glover for helpful comments on the manuscript. A. C. Ozdemir is a member of the Glover laboratory with whom the Wilson laboratory has closely collaborated in the development of human mate-pair sequencing approaches that greatly informed this project. We are grateful to the University of Michigan Depression Center for use of their computing cluster and support personnel including Fan Meng, Manhong Dai, Walter Mexner, and Tyler Brubaker. This work was supported by the University of Michigan Center for Genetics in Health and Medicine Pilot Feasibility Grant to T.E.W. and National Institutes of Health grant R01-GM050403 to L.S.W.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.123232/DC1.
References
- Araya, C. L., C. Payen, M. J. Dunham and S. Fields, 2010. Whole-genome sequencing of a laboratory-evolved yeast strain. BMC Genomics 11 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton, E. C., and J. D. Boeke, 2003. Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: a genomic point of view. Genome Res. 13 254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino, C. J., N. L. Catlett and L. S. Weisman, 1997. Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol. Cell. Biol. 17 6847–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer, M. J., C. M. Christianson, D. A. Pai and M. J. Dunham, 2006. Mapping novel traits by array-assisted bulk segregant analysis in Saccharomyces cerevisiae. Genetics 173 1813–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagna, D., A. Albiero, A. Bilardi, E. Caniato, C. Forcato et al., 2009. PASS: a program to align short sequences. Bioinformatics 25 967–968. [DOI] [PubMed] [Google Scholar]
- Cherry, J. M., C. Ball, S. Weng, G. Juvik, R. Schmidt et al., 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature 387 67–73. [PMC free article] [PubMed] [Google Scholar]
- Cordaux, R., and M. A. Batzer, 2009. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demianova, M., T. G. Formosa and S. R. Ellis, 1996. Yeast proteins related to the p40/laminin receptor precursor are essential components of the 40 S ribosomal subunit. J. Biol. Chem. 271 11383–11391. [DOI] [PubMed] [Google Scholar]
- Dew, I. M., B. Walenz and G. Sutton, 2005. A tool for analyzing mate pairs in assemblies (TAMPA). J. Comput. Biol. 12 497–513. [DOI] [PubMed] [Google Scholar]
- Ehrenreich, I. M., N. Torabi, Y. Jia, J. Kent, S. Martis et al., 2010. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464 1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr, S. D., R. Schekman, M. C. Flessel and J. Thorner, 1983. An MF alpha 1-SUC2 (alpha-factor-invertase) gene fusion for study of protein localization and gene expression in yeast. Proc. Natl. Acad. Sci. USA 80 7080–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel, D. J., 2005. Genome evolution mediated by Ty elements in Saccharomyces. Cytogenet. Genome Res. 110 63–69. [DOI] [PubMed] [Google Scholar]
- Gomes de Mesquita, D. S., H. B. van den Hazel, J. Bouwman and C. L. Woldringh, 1996. Characterization of new vacuolar segregation mutants, isolated by screening for loss of proteinase B self-activation. Eur. J. Cell Biol. 71 237–247. [PubMed] [Google Scholar]
- Kim, J. M., S. Vanguri, J. D. Boeke, A. Gabriel and D. F. Voytas, 1998. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8 464–478. [DOI] [PubMed] [Google Scholar]
- Korbel, J. O., A. E. Urban, J. P. Affourtit, B. Godwin, F. Grubert et al., 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318 420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. S., S. Mulugu, J. D. York and E. K. O'Shea, 2007. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. S., K. Huang, F. A. Quiocho and E. K. O'hea, 2008. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 4 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenburg, M. E., and E. K. O'Shea, 1996. Signaling phosphate starvation. Trends Biochem. Sci. 21 383–387. [PubMed] [Google Scholar]
- Liti, G., D. M. Carter, A. M. Moses, J. Warringer, L. Parts et al., 2009. Population genomics of domestic and wild yeasts. Nature 458 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., W. Sung, K. Morris, N. Coffey, C. R. Landry et al., 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, N., H. Nishiyori, M. Nakamura, C. Kawazu, M. Murata et al., 2008. Development of a DNA barcode tagging method for monitoring dynamic changes in gene expression by using an ultra high-throughput sequencer. Biotechniques 45 95–97. [DOI] [PubMed] [Google Scholar]
- Metzker, M. L., 2010. Sequencing technologies: the next generation. Nat. Rev. Genet. 11 31–46. [DOI] [PubMed] [Google Scholar]
- Mortimer, R. K., and D. C. Hawthorne, 1975. Genetic mapping in yeast. Methods Cell Biol. 11 221–233. [DOI] [PubMed] [Google Scholar]
- Mortimer, R. K., and D. Schild, 1985. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol. Rev. 49 181–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, S. B., K. J. Buckingham, C. Lee, A. W. Bigham, H. K. Tabor et al., 2010. Exome sequencing identifies the cause of a Mendelian disorder. Nat. Genet. 42 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson, T. A., L. S. Weisman, G. S. Payne and W. T. Wickner, 1995. A truncated form of the Pho80 cyclin redirects the Pho85 kinase to disrupt vacuole inheritance in S. cerevisiae. J. Cell Biol. 130 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. S., D. J. Klionsky, L. M. Banta and S. D. Emr, 1988. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8 4936–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M., and F. Winston, 1984. Identification of a Ty insertion within the coding sequence of the S. cerevisiae URA3 gene. Mol. Gen. Genet. 193 557–560. [DOI] [PubMed] [Google Scholar]
- Scannell, D. R., G. Butler and K. H. Wolfe, 2007. Yeast genome evolution: the origin of the species. Yeast 24 929–942. [DOI] [PubMed] [Google Scholar]
- Schneeberger, K., S. Ossowski, C. Lanz, T. Juul, A. H. Petersen et al., 2009. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6 550–551. [DOI] [PubMed] [Google Scholar]
- Schneider, K. R., R. L. Smith and E. K. O'Shea, 1994. Phosphate-regulated inactivation of the kinase PHO80–PHO85 by the CDK inhibitor PHO81. Science 266 122–126. [DOI] [PubMed] [Google Scholar]
- Seufert, W., and S. Jentsch, 1990. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 9 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, N. K., and M. D. Haimes, 1981. Molecular mechanisms of substitution mutagenesis. An experimental test of the Watson-Crick and topal-fresco models of base mispairings. J. Biol. Chem. 256 10671–10683. [PubMed] [Google Scholar]
- Tsai, A. G., and M. R. Lieber, 2010. Mechanisms of chromosomal rearrangement in the human genome. BMC Genomics 11(Suppl. 1): S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida, T. A., and S. D. Emr, 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas, D. F., and J. D. Boeke, 1993. Yeast retrotransposons and tRNAs. Trends Genet. 9 421–427. [DOI] [PubMed] [Google Scholar]
- Wang, Y. X., H. Zhao, T. M. Harding, D. S. Gomes de Mesquita, C. L. Woldringh et al., 1996. Multiple classes of yeast mutants are defective in vacuole partitioning yet target vacuole proteins correctly. Mol. Biol. Cell 7 1375–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger, J. W., K. Schwartz and G. Sherlock, 2010. Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet. 6 e1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., and M. Gerstein, 2003. Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucleic Acids Res. 31 5338–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]