Abstract
Phenovariance may be obscured when genetic mapping is performed using highly divergent strains, and closely similar strains are preferred if adequate marker density can be established. We sequenced the C57BL/10J mouse genome using the Applied Biosystems SOLiD platform and here describe a genome-wide panel of informative markers that permits the mapping of mutations induced on the closely related C57BL/6J background by outcrossing to C57BL/10J, and backcrossing or intercrossing. The panel consists of 127 single nucleotide polymorphisms validated by capillary sequencing: 124 spaced at ∼20-Mb intervals across the 19 autosomes, and three markers on the X chromosome. We determined the genetic relationship between four C57BL-derived substrains and used the panel to map two N-ethyl-N-nitrosourea (ENU)-induced mutations responsible for visible phenotypes in C57BL/6J mice through bulk segregation analysis. Capillary sequencing, with computation of relative chromatogram peak heights, was used to determine the proportion of alleles from each strain at each marker.
DESPITE the advent of massively parallel DNA sequencing, genetic mapping remains necessary to establish linkage between phenotypes and the mutations that cause them. Ideally, closely related strains are used for mapping, since phenotypes can be affected by modifier loci, which occur in rough proportion to the genetic distance between strains. Modifier loci can alter the inheritance, penetrance, and pleiotropy of mutant phenotypes (Montagutelli 2000).
C57BL/6J is the only inbred laboratory mouse strain for which a complete annotated genomic sequence has been published and is therefore the strain most commonly used for random germline mutagenesis and phenotypic screening. For mutagenesis centers around the world, including The Jackson Laboratory, Australian Phenomics Facility (Canberra, Australia), RIKEN Genomics Science Center (Saitama, Japan), Medical Research Council Harwell (Oxfordshire, United Kingdom), Genomic Institute of the Novartis Foundation (San Diego, CA), and the Mouse Mutagenesis Center at Baylor College of Medicine (Houston, TX), C57BL/6J has been mutagenized. However, a closely related strain has not been available for genetic mapping of phenotypes induced in C57BL/6J, precluding the mapping and study of some phenotypes.
In 1921, a single cross gave rise to the black subline C57BL and the brown sublines C57BR and C57L. Subsequently, the C57BL/6 and C57BL/10 strains were derived from the inbred C57BL colony and separated prior to 1937; no contribution from other strains to the parentage of either line is known (Festing 2010). In terms of phenotype, C57BL/6J and C57BL/10J have been extensively characterized; 82 and 51 phenotypes are listed, respectively, for these two strains in the Mouse Phenome Database (http://phenome.jax.org). The strains are highly similar in appearance, body size, and weight, coat color, metabolism, blood composition, immune function, and cardiovascular, renal, and liver physiology. Although the two strains are presumed to share a close genetic relationship due to their recent common origin, by 1995 genetic differences on chromosomes 2, 4, 11, 12, 13, and 16 had been reported (McClive et al. 1994; Slingsby et al. 1995). The Mouse Genome Informatics (MGI) database currently lists a total of 412 autosomal single nucleotide polymorphisms (SNPs), restriction fragment length polymorphisms (RFLPs), or PCR-based polymorphisms between C57BL/6J and C57BL/10J strains. However, at least 12 gaps exceeding 30 Mb in length are evident when the locations of these markers are examined, and not all of the differences could be validated when reexamined by capillary electrophoresis in our laboratory.
To construct a more useful set of informative markers distinguishing C57BL/10J and C57BL/6J genomes, we sequenced the C57BL/10J genome using the Applied Biosystems SOLiD sequencing technology. We validated 127 SNPs by capillary sequencing and then used them to map two N-ethyl-N-nitrosourea (ENU)-induced mutations, one causing pigment dilution and the other causing circling behavior. Mapping was accomplished by bulk segregation analysis (BSA), in which allele frequency was measured at each informative locus in pools of DNA from phenotypically affected and nonaffected F2 mice. These measurements depended upon software that measures the amplitude of individual nucleotide peaks in trace files generated by sequencing amplified DNA fragments from pooled F2 genomic DNA samples.
MATERIALS AND METHODS
Animals:
The C57BL/6J and C57BL/10J strains were obtained from The Jackson Laboratory (Bar Harbor, ME) and housed in The Scripps Research Institute Animal Facility (La Jolla, CA). All studies were performed in accordance with the guidelines established by the Institutional Animal Care and Use Committee of The Scripps Research Institute. The C57BL/6NCrl and C57BL/10SgSnJ strains were purchased, respectively, from the Charles River Laboratories International (Wilmington, MA) in March 2010 and The Jackson Laboratory in 2004, and maintained at Australian National University (Canberra, Australia).
ENU mutagenesis was performed on C57BL/6J mice ordered from The Jackson Laboratory as described previously (Hoebe et al. 2003a,b). Each mutagenized (G0) male was bred to a C57BL/6J female, and the resulting G1 males were crossed to C57BL/6J females to yield G2 mice. G2 females were backcrossed to their G1 sires to yield G3 mice. The june gloom phenotype was observed among G3 mice of a single pedigree and expanded to form a stock on the basis of the visible pigmentation phenotype. The mayday circler phenotype was identified in a screen for suppressors of the mayday phenotype caused by a Kcnj8 mutation (Croker et al. 2007). The june gloom, mayday circler, galak, gray goose, cardigan, sweater, and zuckerkuss strains are described at http://mutagenetix.scripps.edu. Their MGI accession nos. are respectively, 4420315, 4458386, 3784980, 3784982, 3784977, 3784986, and 4420306.
Whole genome DNA sequencing:
SOLiD sequencing was performed according to the Applied Biosystems SOLiD 2 System Library Preparation Guide, Templated Beads Preparation Guide, and Instrument Operation Guide. Purified genomic DNA from a tail section of a male C57BL/10J mouse was sheared into 60- to 90-bp fragments using the Covaris S2 System. The sheared DNA was end repaired and ligated to adapters P1 and P2 and the products separated by agarose gel electrophoresis. Products 150–200 bp in size were excised, purified, and amplified using library PCR primers. This fragment library was clonally amplified on P1-coupled beads by emulsion PCR. Templated beads were separated from nontemplated beads through capture on polystyrene beads coated with P2 and attached to the surface of a SOLiD slide via a 3′ modification of the DNA strands. A total of 326,034,743 template-bound beads were collected and loaded onto one glass slide and subjected to SOLiD sequencing.
SOLiD sequencing data were analyzed using a Linux computer cluster, with a total of 3936 central processing units (CPUs), and the Applied Biosystems software, Corona Lite. Raw data from SOLiD (in .csfasta file format) were matched to the mouse reference genome (NCBI reference assembly Build 37) allowing a maximum of three mismatches per read. Uniquely matched reads were sent to a SNP calling pipeline to identify discrepancies.
SNP validations:
Genomic DNA from the C57BL/10J mouse analyzed by SOLiD sequencing was amplified by PCR with primers that were designed using a Perl script embedded with the Prime program from the GCG DNA software analysis package. The PCR products were purified on a Biomek FX using AMPure beads (Agencourt) and sequenced using Big Dye Terminator v3.1 (Applied Biosystems) on an ABI 3730 XL capillary sequencer. Evaluation of validation sequencing data was performed using a Perl script embedded with PhredPhrap, and discrepant base pair calls were visualized with consed.
For the first round of validation, two SNPs were selected within each of 138 windows an average size of 1.6 Mb (maximum and minimum windows 21.5 Mb and 387 bp, respectively) and spaced at an average distance of 20.4 Mb (maximum and minimum distances 29.3 Mb and 12.6 Mb, respectively) across the 19 autosomes and the X chromosome. For the second and third rounds of validation, candidate SNPs were selected for sequencing on the basis of (1) a position ∼20 Mb from the nearest successfully validated SNP, (2) number of reads by SOLiD sequencing and the score/confidence values assigned to the SNP, and (3) whether none of the SOLiD sequencing reads agreed with the published reference sequence. Sixteen discrepancies representing 16 sites validated in the laboratories of C. C. Goodnow and S. M. Vidal were included in the third round of validation and of these, eight were included in the final SNP panel. Of a total of 131 validated SNPs, 3 autosomal SNPs were excluded from the final panel because they were either polymorphic among mice of our inbred C57BL/6J stock or they were polymorphic for more than two different nucleotides. Three SNPs were located on the X chromosome and were not used in mapping the june gloom or mayday circler phenotypes. All SNPs behaved as autosomal recessive traits. The final panel contained 127 SNPs: 124 on the 19 autosomes and 3 on the X chromosome.
Bulk segregation analysis:
Each SNP was amplified by PCR from the two pooled F2 DNA samples. The PCR products were purified on a Biomek FX using AMPure beads (Agencourt) and sequenced using Big Dye Terminator v3.1 (Applied Biosystems) on an ABI 3730 XL capillary sequencer. DNA sequencing trace peak heights were used to interpolate C57BL/6J (B6) and C57BL/10J (B10) allele frequencies at each SNP site in the F2 test samples from standard curves of normalized peak height vs. allele frequency, generated using DNA samples containing known ratios of B6:B10 DNA. To generate standard curves, each SNP was amplified by PCR from four DNA samples containing B6:B10 contribution ratios of 100:0, 75:25, 50:50, and 0:100, and PCR products were sequenced by capillary electrophoresis. For each SNP site, B6 or B10 allele percentage was plotted against normalized trace peak height, defined as [SNP peak height/trace basal signal level], where the trace basal signal level was calculated as the average height of 10 nucleotides flanking the SNP site (5 nts upstream and 5 nts downstream of the SNP). This normalization is necessary to correct for differences in overall efficiency of individual sequencing runs. Linear regression was used to fit the plotted data points to a line.
The concentration of genomic DNA from each F2 mouse was measured by qPCR of a single copy locus (Chr 12: 25,631,411–25,631,567 within the Mboat2 gene). Pooled F2 DNA samples were created by mixing equimolar quantities of DNA from each individual into the pool. To determine allele frequencies in the pooled F2 DNA samples, normalized peak height was calculated for B6 and B10 alleles at each SNP site as described above, and estimated B6 and B10 allele percentages were interpolated from the standard curve for each corresponding SNP. This method for measuring allele frequency corrects for differences in nucleotide incorporation (A vs. T vs. C vs. G) within a sequencing run. The total allele number (N) was taken to be twice the number of mice at all autosomal loci. The estimated B6 and B10 allele numbers for each SNP site were determined as [estimated allele percentage × N], and were used to calculate P-values separately for mice with mutant and normal phenotypes on the basis of the χ2 distribution and the expected frequencies of B6 and B10 alleles at each variant site (Table 1). In the mutant phenotype DNA pool, when the estimated B10 allele count was higher than the expected B10 allele number, the particular marker was considered to be not linked, and the P-value was set at 1, i.e., estimated allele number was assumed to equal expected allele number. Similarly, in the normal phenotype DNA pool, when the estimated B6 allele count was higher than the expected B6 allele number, the particular marker was also considered to be not linked, and the P-value was set at 1. In the converse situation (lower number of B10 alleles than expected in the mutant pool or lower number of B6 alleles than expected in the normal pool), P-values were calculated as described above.
TABLE 1.
B6:B10 allele frequencies in F2 mice with a recessive mutation
| Phenotype |
|||
|---|---|---|---|
| Marker | Mutant | Normal | |
| Intercross | Unlinked | 50:50 | 50:50 |
| Linked | 100:0 | 33.3:66.7 | |
| Backcross | Unlinked | 75:25 | 75:25 |
| Linked | 100:0 | 50:50 | |
The probabilities for the mutant phenotype sample and the normal phenotype sample were combined after calculation using Fisher's method of combining P-values with the formula χ2 = −2[ln(p1) + ln(p2)]; the P-value for χ2, pcomposite, was determined from a χ2 distribution with four degrees of freedom. Scores for linkage were calculated as −log10(p) separately for the mutant phenotype pool, the normal phenotype pool, and for all mice combined.
Calculation of the synthetic LOD score:
The synthetic LOD score was calculated on the basis of the estimated number of mice with genotypes concordant with either the mutant or normal phenotype, using the formula LOD = log10[p(linkage)/p(nonlinkage)]. The probability of linkage was calculated as [p(linkage) = 1 − p(nonlinkage)]. The probability of nonlinkage was calculated from a binomial distribution, where the probability of either of two possible outcomes (concordance or discordance with phenotype) for each unlinked marker in a given F2 mouse differs depending on the type of cross that has been made. For backcross progeny, the expected frequency of genotypic concordance with either the mutant or the normal phenotype is 50%. For intercross progeny, the expected frequencies of genotypic concordance with the mutant and normal phenotypes are 25 and 75%, respectively. Probabilities calculated independently for F2 intercross mice with mutant phenotype and mice with normal phenotype were combined using Fisher's method.
RESULTS AND DISCUSSION
To identify SNPs between C57BL/6J and C57BL/10J strains, we performed a single fragment library sequencing run on genomic DNA from a male C57BL/10J mouse using version 2 of Applied Biosystems SOLiD apparatus. A total of 145,423,150 reads were uniquely mapped to the C57BL/6J reference genome [National Center for Biotechnology Information (NCBI) version 37] using a criterion of three or fewer mismatches within the read. In aggregate, the uniquely mapped reads covered 55.8, 39.1, and 28.1% of all nucleotides in the genome with one or more, two or more, or three or more sequencing reads, respectively. Among 62,367 discrepancies from the reference sequence identified in three or more reads (File S1), we manually selected for validation two discrepancies within each of 138 windows an average of 1.6 Mb in size and spaced at an average distance of 20.4 Mb across the 19 autosomes and the X chromosome. At least one SNP at each of 87 sites was successfully validated upon PCR amplification and DNA sequencing by capillary electrophoresis. In a second round of validation, 227 discrepancies at 47 sites were chosen for DNA sequencing; at least one SNP at each of 24 sites was confirmed. In a final round of validation, 150 discrepancies representing 24 sites were sequenced, including 16 informative differences representing 16 sites known to the Goodnow and Vidal laboratories. At least one SNP at each of 20 sites was confirmed in this round.
From 131 SNPs, a final SNP panel was compiled, containing 127 SNPs spaced at ∼20-Mb intervals across the 19 autosomes and the X chromosome (Table S1). Of these, nine are listed in the SNP database at Mouse Genome Informatics (MGI). In all, among 641 discrepancies we identified by SOLiD sequencing, 489 were successfully analyzed by capillary sequencing and 216 were authenticated. Taking into account a validation rate of ∼44.2% and a total of 62,367 uniquely mapped discrepancies with three or more sequencing reads (28.1% of all nucleotides), we estimate that ∼98,000 single nucleotide differences exist between the C57BL/6J and C57BL/10J strains (i.e., the strains are 99.9963% identical, with mutations occurring at intervals averaging 27,000 bp in length). We note, however, that insertions and deletions are not detected by SOLiD sequencing, and also that repetitive sequences are underrepresented in the data captured by SOLiD.
To estimate the similarity between C57BL/6 and C57BL/10 sublines, we also sequenced the 127 SNP sites in our panel in DNA from the C57BL/6NCrl and C57BL/10SgSnJ substrains (Table S2). Of 127 sites distinguishing C57BL/6J and C57BL/10J, the two C57BL/6 substrains and the two C57BL/10 substrains differed at 44 and 42 sites, respectively, supporting a model (Figure S1) in which the ancestral C57BL strain diverged to C57BL/6 and C57BL/10 strains, with subsequent splitting of the C57BL/6 line to C57BL/6J and C57BL/6NCrl, and splitting of the C57BL/10 line to C57BL/10J and C57BL/10SgSnJ. There is no evidence of introgression between C57BL/6J and C57BL/10SgSnJ nor between C57BL/6NCrl and C57BL/10J, pairs which showed differences at 81 and 79 sites, respectively. In contrast, differences were found at only 36 sites on comparison of C57BL/6NCrl and C57BL/10SgSnJ, suggesting introgression of these lines.
To demonstrate the utility of the SNP panel in both intercross and backcross mapping, we used it to locate two mutations responsible for unrelated phenotypes in C57BL/6J G3 mice homozygous for random germline mutations induced by ENU. june gloom is a recessive phenotype characterized by a gray coat color that differs from the normally black coat of wild-type C57BL/6J mice. Mice with the recessive mayday circler phenotype exhibit bidirectional circling and head tilting. We used BSA to minimize the cost of mapping. BSA has been applied to plants, where it was first developed (Michelmore et al. 1991), to Saccharomyces cerevisiae (Brauer et al. 2006), Danio rerio (Rawls et al. 2003), and Caenorhabditis elegans (Wicks et al. 2001); it has been employed for mapping traits in mice using simple sequence length polymorphisms (SSLPs) as markers (Collin et al. 1996; Taylor and Phillips. 1996). After either intercross or backcross of F1 mice to generate F2 offspring, we performed BSA by sequencing 124 autosomal markers in two pools of DNA from F2 animals grouped by phenotype. Each pool was created by measuring genomic DNA from each F2 mouse at a single copy locus (the Mboat2 gene) by qPCR and then mixing equimolar quantities of DNA from each individual into the pool.
For the june gloom phenotype, we outcrossed homozygous males to C57BL/10J females and intercrossed the resulting F1 hybrids. DNA from 13 F2 offspring with gray coat color, or 12 F2 offspring with black coat color was pooled and amplicons of ∼200 bp encompassing each of the 124 SNP sites were sequenced in both forward and reverse directions by capillary electrophoresis. For the mayday circler phenotype, we outcrossed homozygous males to C57BL/10J females and backcrossed the resulting female F1 hybrids to a homozygous male. Genomic DNA from 13 F2 offspring with circling behavior, or 13 F2 offspring with normal locomotor behavior, was pooled and sequenced across all markers. At each informative locus, enrichment of the C57BL/6J allele in the pool from mice with the mutant phenotype, and depletion of the C57BL/6J allele in the pool from mice with the normal phenotype, were used to establish linkage.
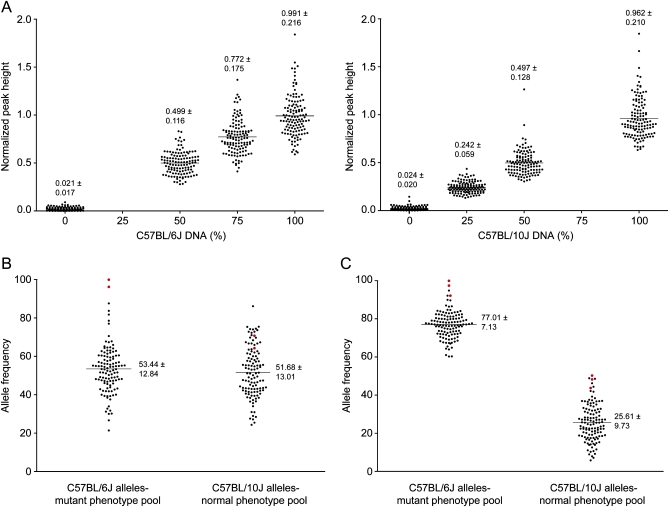
Capillary sequencing chromatogram peak heights were used to infer C57BL/6J and C57BL/10J allele frequencies at each variant site in the pooled DNA samples (Figure 1). Allele frequencies were estimated by linear interpolation, using a standard curve generated using DNA samples containing known amounts of C57BL/6J and C57BL/10J DNA (Figure 2A) (see materials and methods). Marker peak heights in standards and in test samples were normalized to the average signal within each sequencing reaction, calculated as the average height of 10 nucleotides flanking the SNP site (five peaks proximal and five peaks distal to the informative site). This method corrects SNP peak height measurements for differences between individual sequencing runs in overall efficiency and in the incorporation of particular nucleotides, which would be overlooked if peak heights were used directly in estimating allele frequency. As expected for pooled DNA from june gloom F2 intercross progeny, C57BL/6J and C57BL/10J allele frequencies were ∼50% at unlinked loci (Figure 2B). C57BL/6J and C57BL/10J allele frequencies were ∼75 and 25%, respectively, at unlinked loci in pooled DNA from mayday circler F2 backcross progeny (Figure 2C).
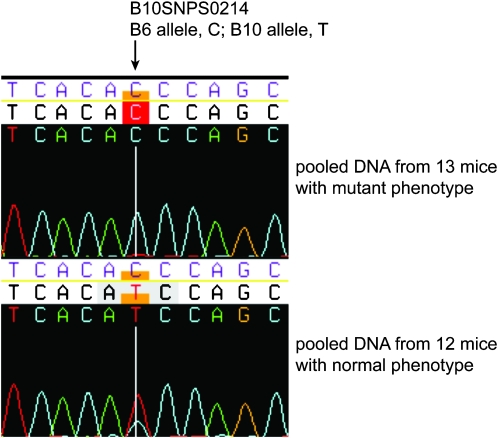
Figure 1.—
Sample DNA sequence trace chromatograms. The region containing the SNP with strongest linkage to the june gloom phenotype from pooled DNA of 12 mice with normal phenotype (black coat) and 13 mice with the mutant phenotype (gray coat). Note that because this marker is linked to the phenotype, the sample from mice with gray coats is expected to contain the C57BL/6J allele exclusively, whereas the sample from mice with black coats is expected to contain C57BL/6J and C57BL/10J alleles at a ratio of 33.3:66.7.
Figure 2.—
Estimated C57BL/6J and C57BL/10J allele frequencies in june gloom and mayday circler mutant and normal phenotype pools at 124 SNP sites. (A) Normalized peak heights, calculated as described in materials and methods, for each of 124 markers in samples with known ratios of C57BL/6J:C57BL/10 genomic DNA. For each individual marker, a standard curve was generated and fitted to a line by linear regression. Values, mean ± SD; lines represent mean. (B and C) C57BL/6J and C57BL/10J allele frequencies at each of 124 markers in the mutant and normal phenotype DNA pools, respectively. Shown in red are data points representing the marker with the highest linkage score and those flanking it. Values, mean ± SD; lines represent mean. june gloom samples (B) and mayday circler samples (C) are shown.
Because BSA does not determine the genotypes of individual mice, it is not possible to formally calculate LOD scores for each marker. Therefore, the χ2 distribution was used to calculate the significance of departure from the expectation that for intercross and backcross progeny, respectively, C57BL/6J:C57BL/10J allele frequencies should approximate 50:50 and 75:25 at unlinked loci. In these calculations, the total number of alleles was taken to be twice the number of mice at all autosomal loci; the number of C57BL/6J or C57BL/10J alleles was calculated as the interpolated allele frequency multiplied by the total number of alleles. P-values were determined separately for the mutant phenotype and normal phenotype pools and then combined using Fisher's method to give a P-value for linkage reflective of data from both pools. Finally, a linkage score, defined as −log10(p), was calculated for each marker.
In addition, a “synthetic LOD score” was calculated for each marker on the basis of the nearest integer approximation of concordance between marker genotype and phenotype and different assumptions were applied, depending upon whether a backcross or intercross had been performed. For example, given 12 F2 mice with the june gloom phenotype (where an intercross was performed) and an estimated allele frequency of 80% C57BL/6J (=P), 20% C57BL/10J (=Q) at a particular marker, we assume genotype frequencies would correspond to P2 B6/B6; 2PQ B6/B10; and Q2 B10/B10, or 0.64 (B6/B6), 0.32 (B6/B10), and 0.04 (B10/B10). The most probable numbers of mice of each genotype, rounded to the nearest integer, would then correspond to 0.64 × 12 = 8 (B6/B6), 0.32 × 12 = 4 (B6/B10), and 0.04 × 13 = 0 (B10/B10) mice (8:4:0). Calculation of the synthetic LOD score would then be based on 8 instances of concordance out of 12, with the expectation of 25% concordance for each event. A synthetic LOD score of 2.55 would be calculated.
In the second example, given 13 F2 mice with mayday circler phenotype (where a backcross was performed) and an estimated allele frequency of 80% C57BL/6J, 20% C57BL/10J at a particular marker, 26 alleles would be assumed to exist in the pool of DNA. This would correspond to a nearest estimate of 21 B6 alleles and 5 B10 alleles and thus to five heterozygous calls at the locus and eight C57BL/6J homozygous calls at the locus. Calculation of LOD would be based on 8 instances of concordance out of 13 events, with the expectation of a 50% concordance for each event. Hence a synthetic LOD score of 0.39 would be calculated.
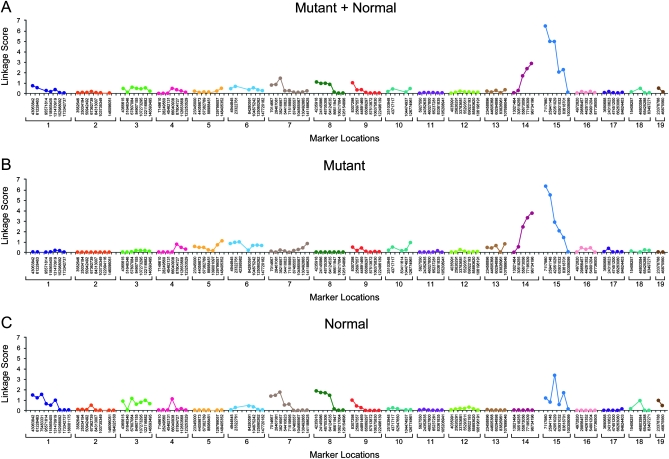
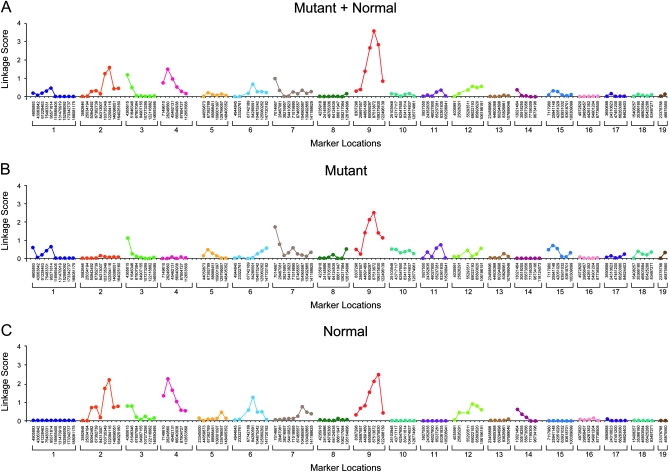
For june gloom and mayday circler mapping, linkage scores for all markers are graphed in Figures 3 and 4, respectively. The june gloom phenotype showed strongest linkage with marker B10SNPS0212 at position 7,117,980 bp on chromosome 15 (P < 2.91 × 10−7). In support of bona fide linkage with the june gloom phenotype, a peak at the same locus was also observed when the mutant phenotype and normal phenotype samples were analyzed separately (Figure 3, B and C). Although an upward trend in the composite linkage score was observed at the distal end of chromosome 14, it does not achieve significance. The allele frequencies and linkage scores from the normal phenotype sample do not support linkage of these loci to the mutant phenotype, i.e., C57BL/10J allele frequencies close to the expected value of 50% and linkage scores near zero were observed. The mayday circler phenotype showed peak linkage with marker B10SNPS0144 at position 87,513,872 bp on chromosome 9 (P < 2.48 × 10−4). The same locus exhibited the strongest linkage with the mutant phenotype when mutant phenotype and normal phenotype samples were analyzed separately (Figure 4, B and C). A lower peak was also observed on chromosome 2, but here allele frequencies and linkage scores for the normal phenotype sample, but not those of the mutant phenotype sample, supported linkage of these loci to the mutation. Smaller linkage score peaks may indicate the presence of modifier loci.
Figure 3.—
Linkage scores for june gloom (intercross genetic mapping). Scores for all mice combined (A), for the mutant phenotype pool (B), and for the normal phenotype pool (C) are shown.
Figure 4.—
Linkage scores for mayday circler (backcross genetic mapping). Scores for all mice combined (A), for the mutant phenotype pool (B), and for the normal phenotype pool (C) are shown.
The synthetic LOD scores of the top-scoring markers for june gloom and mayday circler phenotypes were 7.3 (for B10SNPS0212) and 7.8 (for B10SNPS0144), respectively (Figure S2 and Figure S3). Although these values are more familiar to workers who routinely map mutations and are strongly suggestive of linkage, we consider the synthetic LOD score to be less reliable than the P-values calculated on the basis of the χ2 distribution, since the number of mice with each genotype is not directly assessed in BSA.
On the basis of sequencing data collected for >30 G3 mice, an average of 21 mutations alter coding sense in each G3 mouse, about one-third of them homozygous. Therefore a mutation found close to the marker with peak linkage is very likely to be causative for the phenotype in question. If necessary, the genotypes of individual F2 mice at markers near the linkage peak may be examined to define the boundaries of a critical region.
In the case of june gloom, we identified Slc45a2 (GenBank: NM_053077; MGI: 2153040), located 3.8 Mb from B10SNPS0212, as a candidate gene for the phenotype that when mutated causes varied degrees of hypopigmentation of the eyes, skin, and hair, as observed in mice homozygous for the spontaneous underwhite (uw) mutation (Newton et al. 2001), and the cardigan, galak, gray goose, sweater, and zuckerkuss mutations generated by ENU mutagenesis in our laboratory (see http://mutagenetix.scripps.edu). A T-to-C transition was identified at position 22,723 of the Slc45a2 sequence in june gloom, corresponding to nucleotide 1228 of the mRNA transcript. The mutation causes a serine-to-proline substitution at amino acid 378 of SLC45A2, predicted to be possibly damaging by the PolyPhen-2 SNP effect prediction tool (Adzhubei et al. 2010).
Myo6 (GenBank: NM_001039546; MGI: 104785), located 7.5 Mb from B10SNPS0144, was identified as a candidate gene for the mayday circler phenotype and was directly sequenced. Mutations in Myo6, such as the classical Snell's waltzer (sv) mutation (Avraham et al. 1995), result in circling, head tossing, and hyperactive behavior. A T-to-A transversion at position 929 of the Myo6 transcript was found in mayday circler mice. The mutation converts codon 236, normally encoding cysteine, to a premature stop codon.
The SNP panel we have described should be sufficient to map any robust trait induced on the C57BL/6J background through outcross to C57BL/10J followed by backcrossing or intercrossing, although embryonic lethal mutations may be localized more efficiently using a balancer strategy (Boles et al. 2009). Phenotypes with reduced penetrance can be mapped using BSA by analyzing only animals with the mutant phenotype. BSA is more time and cost efficient than traditional mapping, with a cost of approximately $200 in reagents for each DNA pool subjected to genotypic analysis by capillary sequencing. However, BSA demands accurate assessment of phenotype, because individual mice with “questionable” phenotypes cannot be excluded in post hoc analysis.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (to S.M.V.), the National Institutes of Health grant 5 PO1 AI070167 (B.B.), and broad agency announcement (BAA) contract HHSN272200700038C (B.B. and C.C.G.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.121160/DC1.
References
- Adzhubei, I. A., S. Schmidt, L. Peshkin, V. E. Ramensky, A. Gerasimova et al., 2010. A method and server for predicting damaging missense mutations. Nat. Methods 7 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham, K. B., T. Hasson, K. P. Steel, D. M. Kingsley, L. B. Russell et al., 1995. The mouse Snell's waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat. Genet. 11 369–375. [DOI] [PubMed] [Google Scholar]
- Boles, M. K., B. M. Wilkinson, A. Maxwell, L. Lai, A. A. Mills et al., 2009. A mouse chromosome 4 balancer ENU-mutagenesis screen isolates eleven lethal lines. BMC Genet. 10 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer, M. J., C. M. Christianson, D. A. Pai and M. J. Dunham, 2006. Mapping novel traits by array-assisted bulk segregant analysis in saccharomyces cerevisiae. Genetics 173 1813–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin, G. B., Y. Asada, D. S. Varnum and J. H. Nadeau, 1996. DNA pooling as a quick method for finding candidate linkages in multigenic trait analysis: An example involving susceptibility to germ cell tumors. Mamm. Genome 7 68–70. [DOI] [PubMed] [Google Scholar]
- Croker, B., K. Crozat, M. Berger, Y. Xia, S. Sovath et al., 2007. ATP-sensitive potassium channels mediate survival during infection in mammals and insects. Nat. Genet. 39 1453–1460. [DOI] [PubMed] [Google Scholar]
- Festing, M. F. W., 2010 Inbred strains of mice and rats. Mouse Genome Database (MGD), Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, Maine. (http://www.informatics.jax.org/external/festing/search_form.cgi).
- Hoebe, K., X. Du, J. Goode, N. Mann and B. Beutler, 2003. a Lps2: a new locus required for responses to lipopolysaccharide, revealed by germline mutagenesis and phenotypic screening. J. Endotoxin Res. 9 250–255. [DOI] [PubMed] [Google Scholar]
- Hoebe, K., X. Du, P. Georgel, E. Janssen, K. Tabeta et al., 2003. b Identification of Lps2 as a key transducer of MyD88-independent TIR signaling. Nature 424 743–748. [DOI] [PubMed] [Google Scholar]
- McClive, P. J., D. Huang and G. Morahan, 1994. C57BL/6 and C57BL/10 inbred mouse strains differ at multiple loci on chromosome 4. Immunogenetics 39 286–288. [DOI] [PubMed] [Google Scholar]
- Michelmore, R. W., I. Paran and R. V. Kesseli, 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagutelli, X., 2000. Effect of the genetic background on the phenotype of mouse mutations. J. Am. Soc. Nephrol. 11(Suppl 16): S101–S105. [PubMed] [Google Scholar]
- Newton, J. M., O. Cohen-Barak, N. Hagiwara, J. M. Gardner, M. T. Davisson et al., 2001. Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am. J. Hum. Genet. 69 981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls, J. F., M. R. Frieda, A. R. McAdow, J. P. Gross, C. M. Clayton et al., 2003. Coupled mutagenesis screens and genetic mapping in zebrafish. Genetics 163 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slingsby, J. H., M. B. Hogarth, E. Simpson, M. J. Walport and B. J. Morley, 1995. New microsatellite polymorphisms identified between C57BL/6, C57BL/10, and C57BL/KsJ inbred mouse strains. Immunogenetics 43 72–75. [DOI] [PubMed] [Google Scholar]
- Taylor, B. A., and S. J. Phillips, 1996. Detection of obesity QTLs on mouse chromosomes 1 and 7 by selective DNA pooling. Genomics 34 389–398. [DOI] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. Plasterk, 2001. Rapid gene mapping in caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28 160–164. [DOI] [PubMed] [Google Scholar]