Abstract
In this study we profiled spatial and temporal transcriptional changes during asexual sporulation in the filamentous fungus Neurospora crassa. Aerial tissue was separated from the mycelium to allow detection of genes specific to each tissue. We identified 2641 genes that were differentially expressed during development, which represents ∼25% of the predicted genes in the genome of this model fungus. On the basis of the distribution of functional annotations of 1102 of these genes, we identified gene expression patterns that define key physiological events during conidial development. Not surprisingly, genes encoding transcription factors, cell wall remodeling proteins, and proteins involved in signal transduction were differentially regulated during asexual development. Among the genes differentially expressed in aerial tissues the majority were unclassified and tended to be unique to ascomycete genomes. This finding is consistent with the view that these genes evolved for asexual development in the Pezizomycotina. Strains containing deletions of several differentially expressed genes encoding transcription factors exhibited asexual development-associated phenotypes. Gene expression patterns during asexual development suggested that cAMP signaling plays a critical role in the transition from aerial growth to proconidial chain formation. This observation prompted us to characterize a deletion of the gene encoding a high-affinity cAMP phosphodiesterase (NCU00478). NCU00478 was determined to be allelic to aconidiate-2, a previously identified genetic locus controlling conidiation.
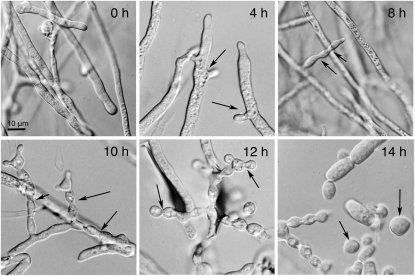
IN the filamentous fungus Neurospora crassa, there are two forms of asexual sporulation: macroconidiation, which produces multinucleate conidia that are 5–10 μm in diameter and microconidiation, which produces mononucleate conidia that are 1 μm in diameter. In this study, we assessed expression profiles in N. crassa during macroconidiation (hereafter conidiation). The sequence of events during asexual development in N. crassa is well documented (Springer and Yanofsky 1989). Briefy, macroconidiophores grow by apical budding from aerial hyphae. In initial budding growth the septum that forms the junction between cells has a diameter that is nearly as wide as the cell and produces a chain of barrel-shaped cells termed minor constriction chains. As the chain continues to grow at the tip, the septum between newly budded cells is of smaller diameter, producing chains of cells that resemble beads on a string called major constriction chains. Approximately 12 hr postinduction, growth ceases and nuclei migrate into the chains. Subsequently, cross-walls are laid down at each conidial junction, they undergo thickening, and starting at 14 hr the cross-walls are cleaved so that conidia (barrel to spherical in shape) can separate as individual spores (Springer and Yanofsky 1989) (Figure 1).
Figure 1.—
Time course of N. crassa asexual sporulation. 0 hr, hyphae of the mycelial mat at the initiation of the experiment; 4 hr, nascent aerial hyphae with hyphal branching (arrows); 8 hr, minor constriction chains present (arrows); 10 hr, minor constriction chains common (arrows); 12 hr, major constriction chains abundant (arrows); 14 hr, free conidia present (arrows). Bar, 10 μm.
Previous investigations to identify differentially expressed genes include both subtractive hybridization (Berlin and Yanofsky 1985a) and microarrays (Rerngsamran et al. 2005; Kasuga and Glass 2008). Specifically, Berlin and Yanofsky (1985a) identified several conidiation-specific genes, termed “con” genes. Rerngsamran et al. (2005) investigated the conidiation mutant fluffy, using a cDNA microarray with partial coverage of the genome. Kasuga and Glass (2008) tracked gene expression across defined sections from a growing colony that also exhibited asexual development using the same full genome microarray used in this study. No matter what the approach, the con genes remain among the most highly expressed genes in all analyses, including the present study.
Conidiation in N. crassa is induced by exposure of the mycelium to an air interface (Berlin and Yanofsky 1985a). The current hypothesis is that exposure of the mycelium to air induces a hyperoxidant state, which leads to the activation of cAMP and MAP kinase signal transduction pathways that trigger formation of aerial hyphae, which grow perpendicular to the substrate mycelium (Aguirre et al. 1989, 2006; Michan et al. 2003). Aerial hyphae are thought to be physiologically different from the vegetative mycelium, and aerial hyphae-specific genes have been identified, which are not expressed in the substrate mycelium or in conidiophores (Li et al. 2005). Since adenylate cyclase (cr-1, crisp) and protein kinase A (pkac-1) mutants are induced for conidiophore development but lack nonconidiogenous aerial hyphae (Rosenberg and Pall 1979; Banno et al. 2005), it is thought that the formation of aerial hyphae is promoted by cAMP signaling. Strains containing mutations in genes encoding Gα subunits (gna-3 and to a lesser extent gna-1, guanine nucleotide-a) have a similar phenotype and are known to regulate cr-1 activity (Rosenberg and Pall 1979; Kays and Borkovich 2004; Banno et al. 2005). Likewise, homologs of Saccharomyces cerevisiae ras1 (N. crassa smco-7, semicolonial) and ras2 (bd, band) affect vegetative growth rate and conidial development (Garnjobst and Tatum 1967; Sargent and Woodward 1969) and have been implicated as regulators of conidiation, possibly through effects on cAMP signaling (Hasunuma and Shinohara 1985; Belden et al. 2007). N. crassa strains containing a mutation in mak-2 (mitogen-activated protein kinase) display a colony and conidiation phenotype similar to cAMP pathway mutants (Li et al. 2005). Thus, the activities of these signaling pathways are likely to be regulated during conidiation.
Tissue-specific gene expression has been demonstrated for several genes expressed during conidiation (Berlin and Yanofsky 1985a; Springer and Yanofsky 1992; Lee and Ebbole 1998; Bailey-Shrode and Ebbole 2004). However, a global analysis of tissue-specific (aerial hyphae and conidiophores vs. substrate mycelium) has not been reported. For example, the role of the substrate mycelium during conidial morphogenesis is poorly understood. One possibility is that the substrate mycelium is metabolically inactive and is simply digested to provide material for aerial development. Alternatively, it is possible that the mycelial mat remains viable and contributes metabolic products required for aerial hyphae formation.
In this study, we used 70-mer oligonucleotide microarrays to examine N. crassa gene expression during synchronous development of conidia across nine time points. A unique aspect of this study was the separate profiling of aerial and mycelial tissue. Thus, we examined tissue specificity as well as temporal control of gene expression during development. Gene expression differences were most striking at the time in development when major constriction chains were beginning to mature.
MATERIALS AND METHODS
Time course:
N. crassa cultures at discrete stages during the development of conidia were prepared as previously described (Berlin and Yanofsky 1985b). Briefly, an 8-liter carboy containing 4 liters of Vogel's medium (2% sucrose) was inoculated with conidia to make a suspension with a final concentration of 106 conidia/ml. Filter sterilized air was vigorously bubbled through the suspension to ensure aeration and agitation for 20 hr at room temperature, resulting in a logarithmic phase suspension of mycelial growth.
Subsequently, 200 ml aliquots of the mycelial culture were harvested by vacuum filtration onto 9-cm Whatman no. 1 filter paper discs. One sample, time point “0,” was immediately frozen in liquid nitrogen. The remaining 18 mycelial mats on Whatman paper discs were placed onto the surface of 40 ml of Vogel's agar (2.0% sucrose, 0.75% agar) in 9-cm glass Petri plates. Plates were numbered and placed in randomized positions under constant fluorescent light in a biosafety cabinet with a lowered glass sash and no airflow.
At hours 1–4, 6, 8, 10, 12, 14, 16, 18, 20, 22, and 24, the mycelial mat was harvested from the Whatman paper and immediately frozen in liquid nitrogen. At the 12-, 14-, 16-, and 18-hr time points the aerial hyphae and conidiophores were easily separated from the more firmly adhered substrate mycelium by peeling the surface with a razor blade (Toledo et al. 1986). This was done to generate a “top” and “bottom” sample for the 12-hr time point (12T and 12B), and top samples for 14- and 18-hr time points (14T and 18T) for RNA isolation. For these 4 time points, each tissue type (aerial and mycelial) was flash frozen and stored separately. Simultaneous to the harvesting of tissue, microscopic analysis was performed using designated parallel cultures to assess the developmental stage at each time point. On the basis of the morphological characteristics monitored, 9 of the 14 time points were selected for microarray analysis. Both the aerial and mycelial tissues of 1 time point (12 hr) were used in our analysis, and only the aerial tissue was used for 14 and 18 hr.
Transcriptional profiling:
Each tissue sample was ground in a mortar and pestle with liquid nitrogen, generating a fine powder. RNA was isolated using TRIzol reagent (Invitrogen) following the manufacturer's protocol. The RNA was purified using an RNAeasy kit (Qiagen) following the manufacturer's protocol.
Synthesis of cDNA, labeling, hybridization, and image acquisition were performed as described in Dunlap et al. (2007). For cDNA synthesis and labeling, the ChipShot indirect labeling and clean-up system (Promega) was used. Briefly, 10 μg of total RNA was used for cDNA synthesis using the oligo(dT) primer according to the manufacturer's suggested protocol. The cDNA was purified using a ChipShot membrane column. The Cy3 or Cy5 cyanine dyes were incorporated into cDNA by adding Cy3 or Cy5 mono-n-hydroxysuccinimide ester dye (Amersham) to the cDNA solution for 1 hr at 25°. The cDNA was then cleaned using a ChipShot membrane column, vacuum dried, and subsequently used for hybridization.
Experimental design and statistical analysis:
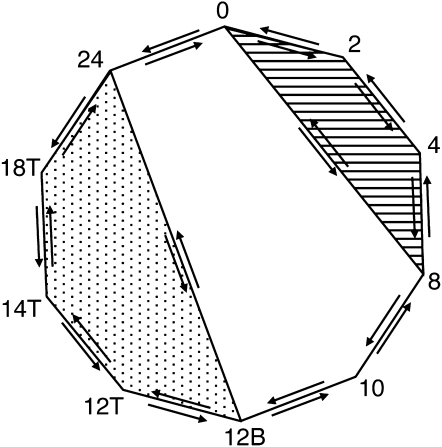
A “cross circuit” design was used to compare samples across different time points (Figure 2). Cross circuit designs increase the robustness of comparisons on spotted microarrays (Townsend 2003). Hybridized spots with one or more mean fluorescence intensities of Cy3 or Cy5, that were higher than mean background intensity plus three standard deviations and with >2% of the pixels saturated, were used for further analysis (Yang and Speed 2002). Bayesian analysis of gene expression levels (BAGEL) software was used to generate an expression profile and confidence intervals for each gene (Meiklejohn and Townsend 2005). We recognized that during asexual development, certain genes might be expressed at low levels at the early or late time points. Since there might be missing hybridizations associated with these genes, we reasoned that these data could be excluded during analysis of the entire circuit. To recover additional information about these genes whose expression was not detected in all samples, we conducted separate circuit analyses of the “early” and “late” subsets of the data using BAGEL (Figure 2, striped and dappled areas, respectively). Following BAGEL analysis, we classified genes as “significantly” regulated if they contained nonoverlapping 95% confidence intervals. These genes were then used for subsequent analyses.
Figure 2.—
Microarray experimental design for N. crassa conidiation time course. Each arrow represents one hybridization, with arrows pointing in opposite orientations indicating dye swap experiments. The striped area represents comparisons of the earlier time points only; the dappled area represents comparisons of late time points only.
Gene accession numbers were annotated according to version 7 of the N. crassa genome assembly (http://www.broadinstitute.org/annotation/genome/neurospora/). Genes showing significantly different relative expression levels were classified according to their major annotation in the MIPS Functional Catalogue (FUNCAT; FUNCAT annotations obtained from N. crassa Functional Genomics Database http://www.yale.edu/townsend; gene list updated March 2006) (Ruepp et al. 2004). To determine whether there were differences in the functional categories in each cluster, the distribution within each cluster was compared to the total distribution of all the annotated genes using independent χ2 tests. We used JMP 8.01 for cluster analysis (hierarchical, Ward's minimum variance method) and to produce the heat map. In graphs, relative expression levels are ratios of a given time point relative to the zero time point for that gene. Where indicated, these data have been log2 transformed. In tables, data are expression levels relative to the time point with the lowest expression for that gene. Microarray data are available at the Neurospora Functional Genomics Website (experiment ID 64; http://bioinfo.townsend.yale.edu/browse.jsp).
Analysis of NCU00478 and acon-2 loci:
A cross between the temperature-sensitive acon-2 mutant [Fungal Genetics Stock Center (FGSC) 3262] and the NCU00478 deletion strain (FGSC 11431) was performed on synthetic cross-medium (Davis and De Serres 1970). FGSC 11431 served as the female parent with the acon-2 mutant overlaid as the male. Progeny were grown for 6 days at 34° in constant light and scored for conidiation phenotype.
Sequencing of the NCU00478.4 locus in the acon-2 strain (FGSC 3262) was performed at the Gene Technologies Laboratory (GTL) at Texas A&M University. Briefly, the coding region for the NCU00478.4 locus of acon-2 was amplified using primers ACON2for2 and NCU00477Rev from the acon-2 strain (supporting information, Table S1) using Phusion Flash DNA polymerase (New England Biolabs, Ipswich, MA). The PCR product was inserted into pCR8 TOPO vector using the manufacturer's protocol (Invitrogen, Carlsbad, CA) and transformed into chemically competent Escherichia coli. The resulting transformants were screened for the presence of the insert via EcoRI digestion. The vector and insert DNA was purified using a QiaPrep Spin miniprep kit (Qiagen, no. 27104). Similarly, the NCU00478.4 locus of N. crassa was amplified from wild-type N. crassa using primers ACON2for1 and ACON2rev1 and cloned into pCR8 TOPO. The 3791-bp EcoRI-digested fragment was gel purified and ligated into vector pCB1004 (Carroll et al. 1994) to produce pCG2. In both cases, the inserts were sequenced using the primers listed in Table S1.
To generate transformable conidia, a culture of the acon-2 mutant was grown in a 250-ml flask containing 50 ml Vogel's agar (2% agar) supplemented with 1.0% yeast extract for 3 days at 25° in constant light. The resulting conidia were harvested with 50 ml sterile 1 m sorbitol and filtered through sterile miracloth (Calbiochem, 475855). The conidia were quantified using a hemocytometer and suspended at 8 × 108 conidia/ml in 1 m sorbitol. Lysing enzyme (Sigma L-1412) was added at a concentration of 20 mg/ml and the solution was allowed to incubate for 1 hr at 28° with shaking at 200 rpm. All subsequent manipulations of these partially digested conidia were performed on ice with solutions also maintained on ice. The conidial suspension was centrifuged for 5 min at 500 × g and 4°. The conidia were washed twice using 1 m sorbitol and resuspended to a concentration of 108 conidia/ml in 1 m electroporation buffer (1 m sorbitol, 1% PEG 4000). A 400-μl aliquot of conidia in electroporation buffer was mixed with 2 μg pCG2. This mixture was transferred to a prechilled electroporation cuvette (0.2-cm gap, Bio-Rad, 165-2086). Electroporation was performed using three pulses in rapid succession (∼1 sec) (750 V, 600 ohms, 25 μF). The cuvette was incubated on ice for 10 min. Outgrowth medium (1× Vogel's salts, 1 m sorbitol, 2% sorbose, 2% sucrose) was added to the reaction and incubated at 30° for 1 hr. A total of 50 ml of top agar (outgrowth media with 0.7% SeaPlaque LMP agarose; Lonza, 50101) maintained at 40° was mixed with the electroporation reaction and overlaid on FGS bottom agar (Davis and De Serres 1970) containing 250 μg/mL hygromycin B (Calbiochem, 400052). The resulting transformants were screened for presence or absence of conidia after growth at 34° for 3–5 days. Homokaryons of the conidiating cultures were generated by three rounds of streaking for single colonies on FGS plates (Ebbole and Sachs 1990).
RESULTS AND DISCUSSION
Developmental time course analysis of well-characterized conidiation-specific genes:
The development of conidiophores and conidia from mycelial tissue proceeded as observed in previous studies (Springer 1993). At 2-hr postinduction aerial hyphae emerged from the substrate mycelium and by 6 hr a uniform layer was observed. Rare minor constrictions were visible at 4 hr and common from 6 to 10 hr of development. Major constriction chains were first observed at 10 hr and uniformly distributed by 12 hr. Free conidia were observed starting at 14 hr (Figure 1).
Of the 9348 features on the microarray representing predicted protein coding genes in N. crassa, 5393 yielded mRNA profiles with the statistical support necessary for the inference of relative expression levels by BAGEL. In the initial comparison of all 10 samples, 2263 genes were developmentally regulated, exhibiting significant differences (nonoverlapping confidence intervals) across time points or tissue types. For some genes, data were only available for early time points (0–8 hr) or late time points (12–24 hr). Analysis of subsets of data for the early and late time points identified an additional 378 genes with nonoverlapping confidence intervals within these time points. This 2641-gene total is ∼25% of the predicted genes in the N. crassa genome exhibiting developmental regulation.
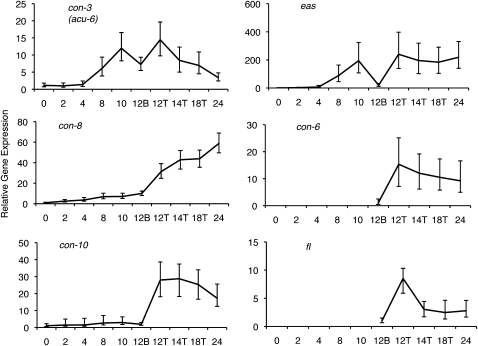
The expression of previously characterized conidiation-specific genes (Figure 3) was consistent with established patterns (Berlin and Yanofsky 1985a). Expression of con-3 was induced at 8 hr, consistent with previous Northern blot data showing weak induction as early as 2 hr with increasing expression peaking at 8 and 12 hr. In our microarray analysis, the induction of con-3 occurred at 8 hr and reached a maximum at 10 and 12 hr. The con-6 gene is typically induced at 8–10 hr, slightly earlier than con-10 and con-8 (Berlin and Yanofsky 1985a). In our time course, con-6 did not pass filtering prior to the 12-hr time point, likely due to a lack of detectable expression in the early time points. Similarly, mRNA from fl, a transcription factor required for conidiation (Bailey and Ebbole 1998), was only reliably detected at the 12-hr and later time points (Figure 3). On the basis of the data available for many of the known conidiation genes, our experiment represents a typical developmental time course of conidiation.
Figure 3.—
Expression of conidiation-associated genes. Gene expression values relative to the lowest expression value across the time course. The relative expression of con-3, con-8, con-10, eas, con-6, and fl are shown. The x-axis represents the time in hours and specific tissues where appropriate. The error bars indicate the BAGEL-generated 95% confidence interval for each time point.
Classification of gene expression patterns:
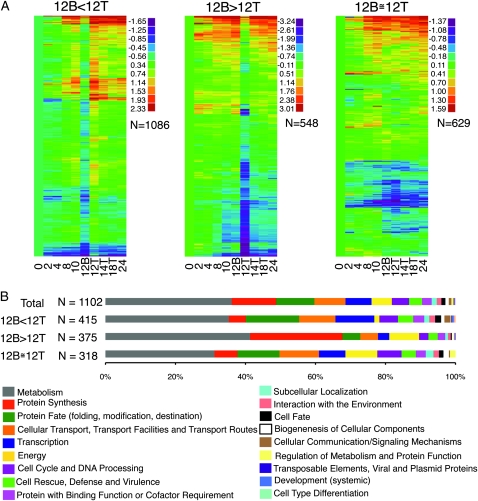
A cluster analysis was performed on the 2263 differentially expressed genes whose expression was detected at all time points (Figure S1). From these data, we observed differential gene expression for many genes in the 12-hr top (12T) and 12-hr bottom (12B) samples within the major clusters. These observations prompted us to reanalyze the full data set by dividing the genes into three subgroups: genes with lower relative expression levels in 12B compared to 12T (12B < 12T; N = 1086), genes with greater relative expression at 12B vs. 12T (12B > 12T; N = 548), and genes with similar relative expression levels at 12B and 12T (12B ≅ 12T; N = 629); cluster analysis was then performed on each group independently (Figure 4A). For example, in the 12B < 12T group (Figure 4A, left), genes in the 12B sample displayed an expression pattern most similar to the 2- and 4-hr time points. In contrast, the pattern of gene expression in the 12T sample was most similar to the 14T, 18T, and 24-hr samples. The 8-hr sample and the 10-hr sample had a hybrid pattern (similar to what one might expect for a combined 12B and 12T sample) of gene expression, with 8 hr more similar to 12B and 10 hr more similar to 12T. Thus, in the 12B < 12T gene set, the greater the mass of aerial tissue relative to mycelial mass the more the expression pattern resembles the patterns exhibited by isolated aerial hyphae.
Figure 4.—
Expression profiles for significantly regulated genes and corresponding classified FUNCAT category distributions. The number of genes in each category (N) is indicated. (A) The 2263 genes identified as significantly regulated, on the basis of nonoverlapping 95% confidence intervals in a BAGEL analysis. Genes were grouped according to their tissue-specific relative expression at 12 hr. Heat maps representing cluster analyses are shown for each 12B vs. 12T category. The expression values are relative to 0 hr and log2 transformed. Thus, all data represent either an increase or decrease of gene expression relative to 0 hr for that gene. (B) Distribution of 1102 genes on the basis of their major FUNCAT annotation (not including categories “unclassified proteins” or “classification not yet clear-cut”). The bar labeled “total” shows the distribution of all 1102 BAGEL significant genes with FUNCAT annotations. The 12B vs. 12T bars (12B < 12T, 12B > 12T, and 12B ≅ 12T) show each subset of FUNCAT category distributions of BAGEL significant genes grouped according to their relative expression at 12 hr.
Genes in the 12B > 12T samples (Figure 4A, middle) displayed an expression pattern in which the 12T sample was quite different from the 14T and 18T samples. Surprisingly, the 14T and 18T aerial tissue samples had expression patterns that are more similar to the 12B sample. It is worth noting that the 12T sample represents the time point at which the transition to major constriction chains was in progress and conidia formation had begun. Also, genes induced early in the time course appeared to be expressed in a mycelium-specific manner up to 12 hr. The similarity of 12B, 14T, and 18T suggested that as development proceeds beyond 12 hr, the expression pattern for these genes was recapitulated in the aerial tissue. It is tempting to speculate that these genes are needed for vegetative growth upon conidial germination. For example, genes involved in glycolysis and ribosomal biogenesis are in this gene set (see below). The 12B ≅ 12T genes (Figure 4A, right) are differentially expressed during conidiation but do not show tissue specificity (aerial hyphae vs. mycelial mat) at the 12-hr time point. The gene lists corresponding to each of the gene sets in Figure 4A are indicated in File S1.
Of the 2263 significantly regulated genes, 1102 had a FUNCAT annotation other than “unclassified proteins” or “classification not yet clear-cut.” Figure 4B shows the distributions of classified FUNCAT annotations for the total gene set and each of the 12B relative to 12T gene sets defined in Figure 4A. We used independent χ2 analyses to compare the distribution of annotations in each group to the distribution of annotations in the total. In all cases, the groups were significantly different from the entire group of FUNCAT annotated genes (P < 0.05). The 12B < 12T gene set had 415 genes with classified FUNCAT annotations. This group was relatively reduced for “protein synthesis” and “energy” and elevated for genes involved in “transcription.” It is interesting to note that the majority of the genes in the 12B < 12T set is composed of genes for unclassified proteins (566 of the 986 annotated for FUNCAT). Homologs of the majority of genes for unclassified proteins in the N. crassa genome can be found only within the subphylum Pezizomycotina (filamentous ascomycetes) (Kasuga et al. 2009) and thus are hypothesized to have functions characteristic to the phylum, such as conidiogenesis (Kasuga and Glass 2008). Specifically, among the 566 unclassified genes differentially expressed in aerial hyphae in this study, 45.9% are Pezizomycotina specific and 34.4% are N. crassa specific (File S1). The 12B > 12T gene set had 375 genes with classified FUNCAT annotation. In contrast, this set was relatively enriched for genes in protein synthesis and energy and reduced for transcription. The 12B ≅ 12T gene set has 318 genes with classified FUNCAT annotations. In this set protein synthesis was reduced and energy was elevated. Genes classified as having metabolic functions are found evenly distributed across the three groups. To further elucidate the processes occurring in the aerial and mycelial tissue, we queried genes and gene networks with biological functions relevant to conidiation (Figure 5; see below).
Figure 5.—
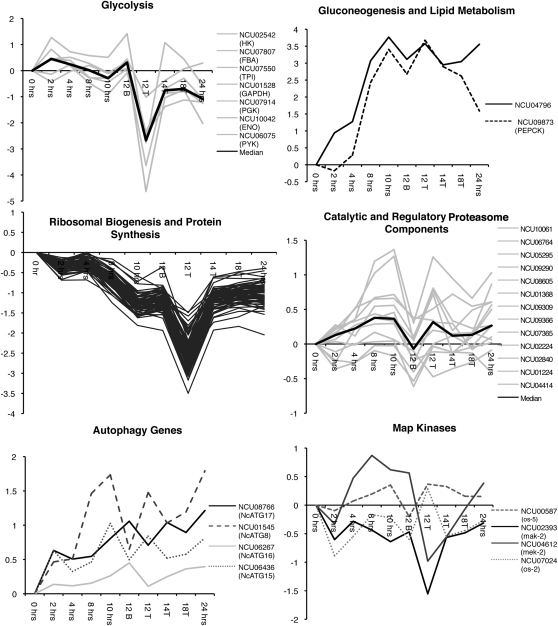
Expression levels of genes for metabolism and cell fate. Y-axis represents expression levels (log2) relative to time 0 h. Glycolysis: the bold line represents the median expression, based on the 7 individual genes (shaded lines). Gluconeogenesis and lipid metabolism: NCU04796, 3-ketoacyl-CoA thiolase, a key enzyme in lipid metabolism, is represented by a dashed line. NCU09873, PEPCK, a key enzyme in gluconeogenesis, is represented by a solid line. Ribosomal biogenesis and protein synthesis: solid lines represent individual coregulated genes. Catalytic and regulatory proteosome components: the bold line represents the median expression based on the 13 individual genes (shaded lines). Autophagy genes: two autophagy genes have a 12B < 12T pattern of expression (dashed lines) and two have a 12B > 12T expression pattern (solid lines). MAP kinases: the solid lines in the MAP kinase graph represent genes in the mak-2 MAP kinase pathway, while the dashed lines are genes in the os-2 MAP kinase pathway.
Carbon metabolism:
Previous studies suggested a potential relationship between physiological status with respect to carbon and the induction of asexual development in N. crassa. Nutrient conditions that promote glycolysis (liquid media with high glucose plus ammonia) repress conidiation, whereas, conditions that promote gluconeogenesis (liquid media with low glucose plus nitrate) are inductive (Turian and Bianchi 1972). Clearly nutrient status, in and of itself, influences induction of development. Thus, to tease apart the different factors contributing to induction of development in our experiment, we were interested in examining transcriptional profiles of genes for carbon metabolism. The median expression ratios of genes involved in glycolysis (Figure 5, glycolysis) remained relatively constant in samples up to 10 hr. At the 12-hr time point, the mycelial mat (12B) resembles the earlier time points, whereas, the aerial hyphae (12T) displayed reduced expression of these genes. The 12-hr time point appears to have captured a critical point in development, since recovery of a mycelial-like expression pattern for these genes was apparent in the 14- and 18-hr aerial hyphae. Both the gene for PEP carboxykinase (NCU09873), a rate-limiting step in gluconeogenesis, and 3-ketoacyl-CoA thiolase (NCU04796), a key enzyme in fatty acid degradation (Figure 5, gluconeogenesis and lipid metabolism), showed elevated expression levels in the aerial tissue relative to the mycelial mat. The increase in expression for these genes began at 4–8 hr and reached a maximum at ∼10–12 hr. These data suggest that fatty acid degradation in aerial tissue may supply substrates for gluconeogenesis. The reduction in glycolytic gene expression exhibited in 12T, combined with elevated expression of genes for gluconeogenesis, resembles the response of N. crassa to glucose starvation (Xie et al. 2004). Therefore, it appears that aerial tissue is limited for glucose. This starvation condition may serve as a signal for asexual morphogenesis; alternatively, signaling pathways associated with the developmental program may induce this pattern of carbon metabolism gene expression.
Surprisingly, the transcription pattern in the mycelial mat suggests that glycolysis (pyruvate kinase) and gluconeogenesis (PEP carboxykinase) were both active. This indicates tissue specificity within the mycelial mat to avoid the futile cycle of PEP synthesis by PEP carboxykinase and PEP degradation by pyruvate kinase. This suggestion is consistent with the finding of subspecialization of the layers of mycelial mat (Toledo et al. 1986). The hyphae of the upper layer of the mycelium are adhered to one another and give rise to aerial hyphae. The hyphae within the lower layer are not adhered to one another. In addition, enzymes for nitrogen metabolism were found to be differentially present in upper and lower layers (see below).
Genes involved in breakdown of starch (NCU01517, NCU07027, and NCU06877) were expressed more highly in the substrate mycelium, whereas genes for 1,3-β-glucan (NCU06871) and trehalose-6-P synthesis (NCU09715) were more abundantly expressed in aerial tissue at 12 hr (Table 1). Furthermore, two hexokinase homologs are expressed. NCU02542, was expressed at a relatively constant level across time and in different tissues, whereas, NCU04728 showed elevated expression levels in the 12-hr aerial hyphae (Table 1), and its role in development is unclear. NCU02542 is likely involved in phosphorylation of glucose produced from the breakdown of glycogen. Taken together, we propose that sugar is being mobilized in the mycelium and consumed for cell wall synthesis and carbohydrate storage in the aerial hyphae.
TABLE 1.
Relative gene expression of carbohydrate metabolism and sugar transporter genes
| Broad ID | Function/description | 0 hr | 2 hr | 4 hr | 8 hr | 10 hr | 12B | 12T | 14T | 18T | 24 hr |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NCU00821 | Sugar transporter | 1.71 | 1.00 | 2.15 | 4.10 | 4.37 | 5.49 | 4.75 | 6.37 | 6.81 | 7.51 |
| NCU01517 | Glycoamylase precursor | 1.00 | 1.00 | 1.57 | 3.28 | 5.01 | 9.29 | 2.43 | 3.94 | 4.35 | 4.10 |
| NCU01633 | Hexose transporter | 1.02 | 3.19 | 2.78 | 2.64 | 2.72 | 5.32 | 1.00 | 1.97 | 2.16 | 1.45 |
| NCU01813 | Sugar transporter | 1.18 | 1.00 | 1.41 | 2.33 | 3.95 | 1.51 | 3.36 | 3.37 | 3.03 | 4.58 |
| NCU02542 | Hexokinase | 2.03 | 1.84 | 2.38 | 2.05 | 1.65 | 2.75 | 1.00 | 1.56 | 2.13 | 2.48 |
| NCU04728 | Hexokinase | 1.02 | 1.06 | 1.14 | 1.10 | 1.03 | 1.00 | 2.68 | 1.55 | 1.38 | 1.21 |
| NCU04963 | Sugar transporter | 1.00 | 6.16 | 14.79 | 29.16 | 29.80 | 88.25 | 1.65 | 32.01 | 40.32 | 32.80 |
| NCU05597 | asd-3 | 1.06 | 1.12 | 1.19 | 1.97 | 2.92 | 1.00 | 6.39 | 2.95 | 2.97 | 3.53 |
| NCU05627 | Sugar transporter | 1.00 | 2.11 | 3.15 | 4.11 | 5.83 | 9.31 | 2.20 | 4.52 | 4.76 | 4.27 |
| NCU06871 | 1,3-β-glucan synthase | 2.54 | 1.09 | 1.00 | 1.06 | 1.36 | 1.42 | 3.77 | 1.86 | 1.38 | 1.41 |
| NCU06877 | Phosphatidylinositol transfer | 1.12 | 1.15 | 1.15 | 1.00 | 1.12 | 1.17 | 1.58 | 1.30 | 1.30 | 1.29 |
| NCU07027 | Glycogen phosphorylase | 2.26 | 4.62 | 5.82 | 4.53 | 3.72 | 6.33 | 1.00 | 3.07 | 3.60 | 3.03 |
| NCU09287 | Sugar transporter | 1.00 | 1.02 | 1.34 | 1.89 | 2.13 | 2.42 | 2.47 | 2.47 | 2.47 | 3.15 |
| NCU09715 | Trehalose phosphate synthase | 1.56 | 1.00 | 1.27 | 1.73 | 1.68 | 1.31 | 2.12 | 1.51 | 1.28 | 1.62 |
| NCU10021 | hgt-1 | 1.00 | 1.68 | 3.05 | 3.59 | 3.63 | 7.88 | 1.02 | 2.80 | 4.54 | 5.25 |
Several sugar transporter homologs were regulated during development (Table 1). Transcripts for the low-affinity glucose transporter homolog (NCU01633) (Forment et al. 2006) and two other sugar transporter homologs NCU00821 and NCU09287 were elevated in both the mycelium and the aerial tissue at 12 hr. Expression of the high-affinity glucose transporter, NCU10021, and one other sugar transporter homolog (NCU04963) was elevated in the mycelium at 12 hr relative to time 0; however, expression did not increase in the aerial tissue. Transcripts for sugar transporter homologs NCU05627, NCU01813, and NCU05597 were elevated in the aerial tissue but not in the substrate mycelium at 12 hr. The role of the induced sugar transporter genes and the NCU04728 hexokinase gene in aerial hyphae is unclear since we expect glucose and other sugars to be transported intracellularly, or synthesized via gluconeogenesis. Clearly, aerial hyphae extend beyond the substrate and thus are not exposed to extracellular glucose. Thus, aerial hyphae may either experience derepression of glucose repressed genes, such as sugar transporters, or another possible interpretation is that these transcripts were produced to prepare the nascent conidia for sugar uptake upon germination.
Nitrogen metabolism:
Nitrogen starvation is a strong inducer of conidiation (Plesofsky-Vig et al. 1983), thus, genes for nitrogen metabolism may also be regulated during development. Cárdenas and Hansberg (1984) suggested that glutamine is the main compound used to transfer nitrogen from the mycelial mat to the aerial tissues. Furthermore, glutamine synthetase activity and protein levels have been measured in the mycelial mat and aerial hyphae during a time course of development similar to ours (Cárdenas and Hansberg 1984) allowing us to compare mRNA and protein levels. N. crassa is known to possess two glutamine synthetase polypeptides that are encoded by different genes (Lara et al. 1982). NCU06724 is gln-1 (glutamine-1), encoding the β-glutamine synthetase polypeptide that forms an octameric enzyme required for growth in the absence of glutamine. NCU04856 encodes a close paralog to gln-1 (81% identical at the amino acid level) and thus likely is gln-2, encoding the α-glutamine synthetase polypeptide. The purified α-glutamine synthetase polypeptide forms a homotetramer with lower activity than the homooctameric β-glutamine synthetase. When both proteins are present they form a heteromeric enzyme and the overall activity of the enzyme is thought to be determined by the ratio of the polypeptides (Cárdenas and Hansberg 1984; Aguirre and Hansberg 1986). Under general conditions of nitrogen deficiency mRNA and protein levels of the β-glutamine synthetase are low, whereas the levels for the α-glutamine synthetase are high.
The expression level of gln-1 (NCU06724) declined 2.4-fold at 2 hr (Table 2) and remained relatively constant after that time in all tissues. Glutamine synthetase activity was found to decline 8-fold in the upper layer of the mycelial mat, but increase 2-fold in the lower layer over the 12-hr time course (Cárdenas and Hansberg 1984). In our case, the 2.4-fold decrease in transcript is consistent with combined changes in enzyme activity in the two layers. In 12-hr aerial hyphae, glutamine synthetase activity was half of the level of the preharvested culture. The transcript level for NCU04856 was similar in the mycelium during the 12-hr time course, but showed a 14-fold reduction in expression level specifically in the 12-hr aerial hyphae. This is consistent with the finding that the relative proportion of α-polypeptide was lower in the aerial hyphae than in the mycelial mat (Cárdenas and Hansberg 1984). Thus, the relative amount and ratio of α- and β-polypeptide appears to parallel their transcript levels. Furthermore, the near absence of NCU04856 (α-polypeptide) transcript suggests that nitrogen levels in aerial hyphae may be sufficiently high to repress transcription of NCU04856. Thus, although we concluded that carbon source was limiting in aerial hyphae on the basis of the observed induction of gluconeogenesis, we conclude that nitrogen transport into the aerial tissue is sufficient to repress nitrogen-regulated genes.
TABLE 2.
Relative gene expression of glutamine synthetases
| Broad ID | Gene name/description | 0 hr | 2 hr | 4 hr | 8 hr | 10 hr | 12B | 12T | 14T | 18T | 24 hr |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NCU04856 | gln-2 | 15.28 | 12.59 | 9.38 | 9.27 | 8.28 | 14.28 | 1.00 | 9.32 | 9.48 | 6.99 |
| NCU06724 | gln-1 | 4.06 | 1.68 | 1.64 | 1.70 | 1.25 | 1.00 | 1.70 | 1.61 | 1.00 | 1.22 |
Ribosome biogenesis, proteasome components, and autophagy:
During conidiation, a decline in ribosomal protein synthesis has been observed for Aspergillus fumigatus (Twumasi-Boateng et al. 2009). In our analysis, transcripts expressed at a higher level in the mycelium vs. the aerial hyphae at 12 hr (Figure 4A, middle) were enriched for genes encoding ribosome components. These data are consistent with the hypothesis that the mycelium is active during morphogenesis and provides needed substrates for aerial hyphae formation. It is important to note that the ribosomal protein synthesis genes were downregulated in the aerial tissue at 12 hr. Transcription of ribosomal protein genes declines during carbon starvation in fungi (Klein and Struhl 1994; Yin et al. 2003). This is consistent with our findings for carbon metabolism genes discussed above, suggesting that aerial hyphae of N. crassa are subject to carbon starvation. A decline in ribosomal protein synthesis during carbon- or nitrogen starvation-induced conidiation is known for A. fumigatus (Twumasi-Boateng et al. 2009). Interestingly, the key regulator of conidiation in A. fumigatus, brlA, is required for the reduction in ribosomal protein synthesis under nitrogen starvation but not carbon starvation. Thus, in these two fungi there seems to be a complex interplay of development and physiology.
In eukaryotes, the proteasome is important in the recycling of proteins (Coux et al. 1996). One model for development is that protein recycling occurs in the mycelial mat and that protein turnover provides substrates for transport into aerial hyphae to support growth. However, we did not observe an induction of genes for proteasome synthesis in the mycelium; rather, expression levels for some proteasome components were higher in the aerial tissue (Figure 5, proteasome components). Also, we did not find evidence for induction of genes involved in autophagy in the mycelium (Figure 5, autophagy genes). While we recognize that transcript levels may not reflect the activity of the proteasome or autophagy processes, Toledo et al. (1986) observed that the hyphae in both the upper and lower layers of the mycelial mat maintained viability over the time course of development.
Cell wall synthesis and membrane modification:
As expected for growing aerial hyphae and developing conidiophores, the expression of glucan synthase (NCU06871) along with several glucanases, glucosidases, glucanosyltransferases, chitinases, and proteases were elevated in aerial tissue (Table 3). These genes are likely involved in direct synthesis and modification of cell wall and polysaccharides.
TABLE 3.
Relative gene expression of cell wall and membrane modification genes
| Broad ID | Gene name/description | 0 hr | 2 hr | 4 hr | 8 hr | 10 hr | 12B | 12T | 14T | 18T | 24 hr |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NCU00471 | Carboxypeptidase | 1.18 | 1.00 | 1.51 | 2.90 | 3.58 | 1.36 | 5.04 | 2.58 | 2.61 | 4.01 |
| NCU00586 | ncw-6, SUR7a | 1.00 | 1.31 | 1.13 | 1.26 | 1.45 | 1.22 | 3.43 | 5.68 | 3.82 | 4.64 |
| NCU00673 | spr-4 | 1.00 | 1.72 | 2.04 | 3.23 | 4.22 | 1.76 | 6.05 | 2.93 | 3.34 | 4.48 |
| NCU01068 | MUG137a | 1.28 | 1.03 | 1.04 | 1.36 | 1.76 | 1.00 | 3.15 | 1.50 | 1.38 | 2.65 |
| NCU01366 | cpr-5 | 1.10 | 1.13 | 1.00 | 1.55 | 3.76 | 1.16 | 5.16 | 2.75 | 2.16 | 2.52 |
| NCU01510 | C2 domain protein | 2.68 | 1.00 | 1.37 | 1.87 | 1.66 | 2.28 | 8.28 | 4.93 | 4.96 | 5.84 |
| NCU02322 | m-6-p isomerase | — | — | — | — | — | 1.00 | 5.50 | 5.41 | 3.81 | 4.59 |
| NCU02540 | LSP1a | 1.00 | 1.32 | 1.24 | 2.19 | 2.15 | 1.46 | 4.49 | 4.59 | 4.08 | 4.53 |
| NCU02956 | Aspartic proteinase | 1.13 | 1.05 | 1.11 | 1.11 | 1.05 | 1.00 | 3.23 | 1.64 | 1.50 | 1.84 |
| NCU03254 | Cellulase | — | — | — | — | — | 1.00 | 7.62 | 5.64 | 3.39 | 3.58 |
| NCU04421 | anx-14 | 2.37 | 1.61 | 2.30 | 4.33 | 4.23 | 1.00 | 3.75 | 1.80 | 1.44 | 4.08 |
| NCU04431 | Glucanase | — | — | — | — | — | 1.00 | 5.16 | 2.25 | 1.49 | 2.07 |
| NCU04727 | Glucosamine deaminase | 1.00 | 1.25 | 1.40 | 1.29 | 1.65 | 1.02 | 5.48 | 3.46 | 1.70 | 1.58 |
| NCU04798 | Alpha-1,2-mannosidase | 1.21 | 1.39 | 1.43 | 1.65 | 2.01 | 1.00 | 3.06 | 2.11 | 2.04 | 1.71 |
| NCU04883 | gh18-4 | — | — | — | — | — | 1.00 | 16.42 | 3.77 | 1.39 | 1.51 |
| NCU04924 | cut-1 | 1.36 | 1.04 | 1.10 | 2.35 | 2.95 | 1.00 | 3.71 | 2.58 | 2.27 | 2.82 |
| NCU05858 | Fatty acid oxygenase | — | — | — | — | — | 1.00 | 12.45 | 5.97 | 4.62 | 6.38 |
| NCU06020 | Chitinase | 1.17 | 1.00 | 1.46 | 1.47 | 1.57 | 1.22 | 3.63 | 2.83 | 1.92 | 2.32 |
| NCU06871 | Glucan synthase | 2.54 | 1.09 | 1.00 | 1.06 | 1.36 | 1.42 | 3.77 | 1.86 | 1.38 | 1.41 |
| NCU07253 | Glucanosyltransferases | 2.77 | 2.08 | 1.55 | 2.40 | 1.99 | 1.00 | 7.35 | 3.28 | 2.19 | 3.29 |
| NCU07495 | PIL1a | — | — | — | — | — | 1.00 | 3.83 | 4.90 | 2.65 | 2.99 |
| NCU07523 | Glucanase | 1.04 | 1.14 | 1.37 | 1.43 | 1.20 | 1.00 | 8.62 | 2.42 | 1.20 | 1.72 |
| NCU07923 | Peptidase S41 family | 1.10 | 1.20 | 1.35 | 1.55 | 1.85 | 1.00 | 3.69 | 2.52 | 1.25 | 1.37 |
| NCU08087 | Glucanase | 1.05 | 1.03 | 1.10 | 2.31 | 3.47 | 1.00 | 6.70 | 4.16 | 4.38 | 5.02 |
| NCU08600 | KAR9a | 1.03 | 1.06 | 1.17 | 1.54 | 1.85 | 1.00 | 3.06 | 1.94 | 1.92 | 1.78 |
—, no data.
Orthologs in Saccharomyces cerevisiae.
Similarly the expression level of cut-1 (NCU04924) was induced in aerial tissue. The cut-1 gene encodes a member of the haloacid dehalogenase family and resembles members of the NagD subfamily (COG0647) of sugar phosphatases. It is induced by osmotic stress or heat shock (Youssar et al. 2005). The N. crassa cut-1 mutant is osmotically sensitive, has altered aerial hyphae development, and does not properly release conidia (Youssar et al. 2005). We hypothesize that cut-1 is involved in membrane structure or cell wall biosynthesis, since many members of this gene family are involved in phosphatidyl or carbohydrate metabolism.
Several orthologs to yeast genes involved in morphogenesis were regulated during N. crassa asexual sporulation (Table 3). In yeast, Sur7p, Lsp1p and Pil1p physically interact with each other at the bud tip to regulate cellular morphogenesis (Young et al. 2002; Zhang et al. 2004; Walther et al. 2006). The orthologs to yeast SUR7 (NCU00586), LSP1 (NCU02540), and PIL1 (NCU07495) showed increased expression levels in the aerial hyphae tissue. NCU00586 is a member of the Sur7p/Pal1p family, which is involved in signal transduction with roles in pH sensing, endocytosis, sporulation, and plasma membrane organization. NCU08600 has sequence similarity to the yeast KAR9 gene and is coregulated with the Sur7p/Pal1p homolog NCU00586. This finding is interesting because in S. cerevisiae, Kar9p is required for karyogamy, positioning of the mitotic spindle, and organization of cytoplasmic microtubules. Kar9p has been shown to localize to the tip of the shmoo projection, the cortical dot in mitotic cells, and to the tip of growing buds (Miller and Rose 1998). In conidiation, nuclei migrate into proconida chains prior to septation (Springer and Yanofsky 1989). Thus we hypothesize these orthologs to yeast morphogenesis genes are essential to conidia formation. A gene encoding a BAR domain containing protein NCU01068 also showed increased expression levels in aerial tissue. BAR domains are found in protein families that are membrane binding and are involved in membrane curvature sensing (Peter et al. 2004). NCU01068 is most similar to S. pombe meiotically upregulated gene 137 protein (Mug137). Mug137 contains a predicted N-terminal BAR domain and a C-terminal SH3 domain, similar to endophilins. A further connection to the cytoskeleton was found in the induction of NCU04421, an annexin (anx-14). Annexins are a family of calcium-dependent phospholipid-binding proteins. The annexin family has been linked with many intracellular and extracellular activities including membrane scaffolding, organization and trafficking of vesicles, endocytosis, exocytosis, signal transduction, inhibition of phospholipase activity, resistance to oxygen species, DNA replication, and calcium ion channel formation (Vishwanatha and Kumble 1993; Mira et al. 1997; Gerke et al. 2005). In addition, NCU01510, a gene encoding a protein that contains C2 domains was induced. C2 domains are Ca2+-dependent membrane targeting modules that bind to a wide variety of substances including phopholipids, inositol polyphosphates, and intracellular proteins (Davletov and Südhof 1993). Since NCU01510 contains two C2 domains, it is likely to function as a membrane trafficking protein, similar to the role of the calcium sensor synaptotagmin 1, which controls exocytosis (Tang et al. 2006). Overall, there was a striking tissue-specific induction of genes involved in cell wall and membrane modification and signaling.
Signal transduction:
Many, but not all, of the genes encoding transcription factors of N. crassa have been deleted (Colot et al. 2006). Of the regulated genes encoding transcription factors with phenotypic characterizations, eight exhibited asexual development-specific phenotypes, two exhibited sexual development phenotypes, two exhibited colony growth phenotypes, one exhibited colony growth and asexual phenotypes, one exhibited asexual and sexual phenotypes, and three exhibited all development-altered phenotypes (Colot et al. 2006) (Table 4). Many of the genes encoding transcription factors were only moderately regulated, indicating that either a small change in expression is sufficient, or post-translational modifications of these factors may also be important for their activity. Of these characterized transcription factor mutants, we found that orthologs of asexual developmental regulators in A. nidulans corresponding to flbC (NCU03043) and medA (NCU07617) were differentially expressed during asexual development in N. crassa (Table 4). The NCU03043 and NCU07617 deletion mutants are defective in conidiation (Colot et al. 2006), suggesting that they play a central role in regulating this developmental process.
TABLE 4.
Relative expression of N. crassa transcription factors with developmental phenotypes in deletion mutants
| Broad ID | Gene name | 0 hr | 2 hr | 4 hr | 8 hr | 10 hr | 12B | 12T | 14T | 18T | 24 hr |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NCU00499 | ada-1 | 1.00 | 1.63 | 1.65 | 2.29 | 2.97 | 3.93 | 2.21 | 2.35 | 2.68 | 4.66 |
| NCU01459 | asl-2 | 1.00 | 1.17 | 1.24 | 1.64 | 2.05 | 1.07 | 2.49 | 1.79 | 1.81 | 1.67 |
| NCU02713 | csp-1 | 1.00 | 1.73 | 1.17 | 1.38 | — | — | — | — | — | — |
| NCU02713 | csp-1 | — | — | — | — | — | 1.00 | 6.05 | 2.43 | 2.17 | 3.00 |
| NCU03043 | flbCa | — | — | — | — | — | 1.00 | 6.05 | 2.43 | 2.17 | 3.00 |
| NCU03489 | col-21 | 1.25 | 1.30 | 1.33 | 1.44 | 1.59 | 1.00 | 1.69 | 1.53 | 1.58 | 1.38 |
| NCU03593 | kal-1 | 1.34 | 1.98 | 1.47 | 1.43 | 1.38 | 1.45 | 1.04 | 1.01 | 1.00 | 1.09 |
| NCU03686 | lah-3 | 1.80 | 1.32 | 1.32 | 1.46 | 1.61 | 1.38 | 1.00 | 1.01 | 1.39 | 1.35 |
| NCU03931 | ada-5 | 1.20 | 1.07 | 1.07 | 1.02 | 1.00 | 1.08 | 1.52 | 1.10 | 1.18 | 1.15 |
| NCU04179 | sah-1 | 1.00 | 2.16 | 1.73 | 2.63 | 2.39 | 1.41 | 1.84 | 2.01 | 1.94 | 2.10 |
| NCU04731 | sah-2 | — | — | — | — | — | 1.00 | 1.34 | 1.08 | 1.05 | 1.01 |
| NCU06656 | acu-15 | 1.00 | 1.43 | 1.15 | 1.72 | 1.85 | 2.00 | 1.37 | 1.61 | 1.60 | 2.60 |
| NCU07374 | sah-3 | 1.33 | 1.23 | 1.27 | 1.36 | 1.51 | 1.00 | 1.76 | 1.45 | 1.55 | 1.26 |
| NCU07617 | medAa | 1.37 | 1.25 | 1.34 | 1.63 | 2.17 | 1.00 | 3.56 | 2.00 | 1.93 | 1.75 |
| NCU08307 | C6-Zn2b | — | — | — | — | — | 1.00 | 5.12 | 1.92 | 1.23 | 1.00 |
| NCU08651 | col-27 | 1.38 | 1.73 | 1.95 | 2.30 | 2.73 | 1.00 | 1.48 | 1.56 | 2.17 | 2.31 |
| NCU08726 | fl | — | — | — | — | — | 1.00 | 8.52 | 3.03 | 2.47 | 2.79 |
—, no data.
Ortholog in Aspergillus nidulans.
An uncharacterized binuclear zinc finger transcription factor.
Other genes with aerial hyphae-specific induction included NCU04272, NCU04379, NCU04493, and NCU05667 (Table 5). NCU04272 encodes a protein containing four ZZ-type zinc finger domains. ZZ zinc finger proteins are known to bind DNA, RNA, protein, and lipid substrates (Klug 1999; Legge et al. 2004; Brown 2005; Hall 2005; Gamsjaeger et al. 2007). NCU04379 encodes a protein with similarity to Ca2+-binding proteins (EF-Hand superfamily). These proteins are involved in signal transduction mechanisms related to cell division and chromosome partitioning (Day et al. 2002). NCU04493 and its paralog NCU05667 (acw-3) encode predicted secreted proteins with similarity to fungal proteins that are anchored in the cell wall (GPI anchors). The functions of these proteins in the Ascomycota are unclear, but in some Basidiomycota, they appear to be involved in both fruiting body formation and infection cell development (Link and Voegele 2008). One member of this family from the Shitake mushroom interacts with a fruiting body formation-induced MAP kinase in the yeast two-hybrid system (Szeto et al. 2007). Conceivably these proteins could interact to link information about cell wall surface architecture to calcium or MAP kinase signaling pathways.
TABLE 5.
Relative gene expression of signal transduction genes
| Broad ID | Gene name/description | 0 hr | 2 hr | 4 hr | 8 hr | 10 hr | 12B | 12T | 14T | 18T | 24 hr |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NCU00587 | os-5 | 1.15 | 1.07 | 1.21 | 1.31 | 1.47 | 1.00 | 1.48 | 1.44 | 1.28 | 1.28 |
| NCU01166 | mcb | 1.20 | 1.00 | 1.22 | 1.36 | 1.52 | 1.45 | 2.11 | 1.91 | 2.21 | 2.53 |
| NCU02393 | mak-2 | 2.94 | 1.93 | 2.41 | 2.17 | 1.88 | 2.12 | 1.00 | 1.98 | 2.10 | 2.37 |
| NCU04272 | ZZ-type zinc finger | 1.00 | 1.44 | 1.29 | 2.19 | 3.36 | 1.79 | 5.14 | 3.15 | 2.91 | 3.91 |
| NCU04379 | EF-Hand superfamily | — | — | — | — | — | 1.00 | 3.85 | 2.12 | 1.39 | 1.64 |
| NCU04493 | MAPK interacting | 6.71 | 1.18 | 1.03 | 1.60 | 1.82 | 1.00 | 13.30 | 9.52 | 2.45 | 3.86 |
| NCU05667 | acw-3 | 2.96 | 1.96 | 1.47 | 1.05 | 1.43 | 1.00 | 3.46 | 1.37 | 1.27 | 1.06 |
| NCU06240 | pkac-1 | 1.34 | 1.25 | 1.36 | 1.63 | 1.83 | 1.00 | 2.40 | 1.68 | 1.70 | 1.64 |
| NCU07024 | os-2 | 1.85 | 1.00 | 1.27 | 1.65 | 1.59 | 1.22 | 2.26 | 1.27 | 1.36 | 1.54 |
| NCU08377 | cr-1 | 1.01 | 1.02 | 1.00 | 1.48 | — | — | — | — | — | — |
At 12 hr, expression of mak-2 (sexual development MAP kinase; NCU02393) was significantly lower in the aerial tissue as compared to the mycelial mat, and os-2 (osmolarity MAP kinase; NCU07024) exhibited an opposite pattern in the 12B vs. 12T comparison. The MAPKKs from the MAK-2 (NCU02393) and the OS-2 (NCU00587) pathways were coregulated with their respective MAPK partners (Figure 5, MAP kinases, Table 5). Strains with mutations in mak-2 lack aerial hyphae and are activated for conidiation similar to cr-1 (Li et al. 2005). Therefore a decline in activity of the mak-2 pathway is expected in aerial tissue and a decline in the mak-2 transcript levels is consistent with this view (Pandey et al. 2004). Once aerial hyphae are produced, a decline in mak-2 transcript levels may correspond to a decrease in the activity of the pathway, which would promote conidial morphogenesis at the expense of continued growth of the aerial hyphae. On the basis of the phenotypes of the cr-1 and pkac-1 mutants (Rosenberg and Pall 1979; Banno et al. 2005), we expect the cAMP signaling pathway to be upregulated until the elongation of aerial hyphae ceases and then downregulated at the onset of conidiogenesis (∼12 hr in the aerial tissue). A decline in cAMP synthesis would be consistent with the decline in expression of the genes for glycolysis and ribosomal proteins observed in the 12T tissue. However, we did not detect a decrease in expression of genes associated with the cAMP pathway (NCU06240, NCU01166, and NCU08377; Table 5). Rather for pkac-1, we observed a twofold increase in expression levels in the aerial hyphae at 12 hr. Thus, we speculate that transcription of pkac-1 is subject to feedback activation when cAMP levels are low.
In S. cerevisiae, the high-affinity phosphodiesterase (Pde2p) is responsible for the breakdown of cAMP (Sass et al. 1986). Expression for the N. crassa PDE2 ortholog (NCU00478) was constitutive. The NCU00478 deletion mutant was preliminarily characterized as aconidial (Colot et al. 2006). We obtained the NCU00478 deletion mutant and confirmed that it was aconidial, blocked prior to minor constriction chain formation. Therefore, in absence of cAMP turnover, the transition from aerial hyphae formation to proconidial chain formation is blocked. We noted the morphological defect of NCU00478 was similar to the phenotype of a previously identified aconidial temperature-sensitive mutant (acon-2) (Matsuyama et al. 1974). Furthermore, NCU00478 is located on linkage group IIIR, close to the map location of acon-2 (Perkins et al. 2001). Therefore, we performed the crosses and direct sequencing necessary to determine whether NCU00478 and acon-2 were allelic.
Crosses between acon-2 and the NCU00478 deletion strain were fertile. We scored 316 progeny for conidiation at 34°, and all were aconidial, suggesting acon-2 and NCU00478 are allelic. Amplification of the NCU00478 region from the acon-2 mutant generated a PCR product that was 1693 bp smaller than expected. DNA sequencing revealed that 757 bp of the C terminus of NCU00478 were deleted in the acon-2 mutant. This deletion creates a loss of 252 amino acids in the predicted C terminus and shifts the stop codon to within the intergenic region with the addition of 23 amino acids. Transformation of the acon-2 mutant strain with a plasmid containing the wild-type version of NCU00478 (pCG2) restored conidiation at 34°, thus, confirming that acon-2 is an allele of NCU00478.
Ivey et al. (2002) showed that the GNA1 Gα protein activates adenlyate cyclase and that the gna-1 mutant is derepressed for conidiation. Interestingly, the cr-1; gna-1 double mutant is synergistic in the degree of derepression of conidiation and addition of exogenous cAMP restores repression of conidiation in the cr-1 mutant, but does not fully restore repression of conidiation in the cr-1; gna-1 double mutant. This suggests that gna-1 regulates conidiation by its effect on cAMP signaling and by a cAMP-independent pathway (Ivey et al. 2002). Li et al. (2005) showed that a MAP kinase (encoded by mak-2) is derepressed for conidiation and that repression of conidiation is not restored by addition of exogenous cAMP (Li et al. 2005). Thus, gna-1 may be involved in regulating both the MAP kinase and cAMP signaling pathways. We hypothesize that the acon-2; mak-2 double mutant may display some partial restoration of conidiation compared to the acon-2 mutant.
In conclusion, this microarray analysis captured an important time point (12 hr) in asexual development, which corresponded to the transition from aerial hyphae formation to proconidial chain development. In particular, by separating the mycelial and aerial tissue we were able to observe spatial differentiation of gene expression, including genes involved in transcriptional regulation, signal transduction, carbon and nitrogen metabolism, ribosomal protein synthesis, cytoskeleton structure, and cell wall differentiation. Further analysis of transcription factors identified in this study and which, when deleted give aconidial phenotypes (Colot et al. 2006), will serve to further elucidate the molecular genetic control of N. crassa asexual development. Finally, transcriptional downregulation of the mak-2 MAP kinase pathway in aerial hyphae is consistent with our model of MAK2 function (Li et al. 2005). The mak-2 mutant is induced for conidiation and has reduced aerial hyphae. Thus, we expect MAK2 to be active during the growth of aerial hyphae. MAK2 activity is expected to decline at the onset of conidial morphogenesis and transcriptional downregulation of mak-2 and NCU04612 the MAPKK of the MAK2 pathway may be the means of reducing the activity of this MAP kinase pathway. As with the mak-2 pathway, a block in the cAMP pathway causes induction of conidiation at the expense of aerial hyphae growth (Li et al. 2005). However, no transcriptional downregulation of components of the cAMP pathway was observed, and indeed an increase in transcripts for the cAMP-dependent protein kinase catalytic subunit, pkac-1, was found. We reasoned that the pathway may be regulated by cAMP levels during conidiation, rather than transcriptional regulation of genes encoding components of the cAMP pathway. Since mutations that block the cAMP pathway induce conidiation, we characterized a mutant in the high-affinity phosphodiesterase that should lead to elevated cAMP levels. As expected, we found that this mutant was blocked in conidial morphogenesis. Furthermore, we showed that this gene corresponds to the previously identified temperature-sensitive conidiation gene, acon-2.
Acknowledgments
We thank the Gene Technology Lab at Texas A&M and the Fungal Genetic Stock Center. This research was funded by National Science Foundation IOS-0716894 (to B.D.S., H.H.W., and D.J.E.) and a National Institutes of Health multi-institutional program project grant (GM068087) (to N.L.G.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.121780/DC1.
Microarray data are available at the Neurospora Functional Genomics Website (experiment ID 64; http://bioinfo.townsend.yale.edu/browse.jsp).
References
- Aguirre, J., and W. Hansberg, 1986. Oxidation of Neurospora crassa glutamine synthetase. J. Bacteriol. 166 1040–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre, J., R. Rodriguez and W. Hansberg, 1989. Oxidation of Neurospora crassa NADP-specific glutamate dehydrogenase by activated oxygen species. J. Bacteriol. 171 6243–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre, J., W. Hansberg and R. Navarro, 2006. Fungal responses to reactive oxygen species. Med. Mycol. 44 101–107. [DOI] [PubMed] [Google Scholar]
- Bailey, L. A., and D. J. Ebbole, 1998. The fluffy gene of Neurospora crassa encodes a Gal4p-Type C6 zinc cluster protein required for conidial development. Genetics 148 1813–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Shrode, L., and D. J. Ebbole, 2004. The fluffy gene of Neurospora crassa is necessary and sufficient to induce conidiophore development. Genetics 166 1741–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno, S., N. Ochiai, R. Noguchi, M. Kimura, I. Yamaguchi et al., 2005. A catalytic subunit of cyclic AMP-dependent protein kinase, PKAC-1, regulates asexual differentiation in Neurospora crassa. Genes Genet. Syst. 80 25–34. [DOI] [PubMed] [Google Scholar]
- Belden, W. J., L. F. Larrondo, A. C. Froehlich, M. Shi, C.-H. Chen et al., 2007. The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev. 21 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin, V., and C. Yanofsky, 1985. a Isolation and characterization of genes differentially expressed during conidiation of Neurospora crassa. Mol. Cell. Biol. 5 849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin, V., and C. Yanofsky, 1985. b Protein changes during the asexual cycle of Neurospora crassa. Mol. Cell. Biol. 5 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, R. S., 2005. Zinc finger proteins: getting a grip on RNA. Curr. Opin. Struct. Biol. 15 94–98. [DOI] [PubMed] [Google Scholar]
- Cárdenas, M. E., and W. Hansberg, 1984. Glutamine metabolism during aerial mycelium growth of Neurospora crassa. J. Gen. Microbiol. 130 1733–1741. [DOI] [PubMed] [Google Scholar]
- Carroll, A. M., J. A. Sweigard and B. S. Valent, 1994. Improved vectors for selecting resistance to hygromycin. Fung. Genet. Newslett. 41 20–21. [Google Scholar]
- Colot, H. V., G. Park, G. E. Turner, C. Ringelberg, C. M. Crew et al., 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux, O., K. Tanaka and A. L. Goldberg, 1996. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65 801–847. [DOI] [PubMed] [Google Scholar]
- Davis, R. H., and F. J. de Serres, 1970. Genetic and microbiological research techniques for Neurospora crassa. Meth. Enzymol. 17A 79–143. [Google Scholar]
- Davletov, B. A., and T. C. Südhof, 1993. A single C2 domain from synaptotagmin I is sufficient for high affinity Ca2+/phospholipid binding. J. Biol. Chem. 268 26386–26390. [PubMed] [Google Scholar]
- Day, I., V. Reddy, G. Shad Ali and A. Reddy, 2002. Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol. 3 56.51–56.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap, J. C., K. A. Borkovich, M. R. Henn, G. E. Turner, M. S. Sachs et al., 2007. Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv. Genet. 57 49–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole, D. J., and M. S. Sachs, 1990. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fung. Genet. Newslett. 37 17–18. [Google Scholar]
- Forment, J. V., M. Flipphi, D. Ramon, L. Ventura and A. P. MacCabe, 2006. Identification of the mstE gene encoding a glucose-inducible, low affinity glucose transporter in Aspergillus nidulans. J. Biol. Chem. 281 8339–8346. [DOI] [PubMed] [Google Scholar]
- Gamsjaeger, R., C. K. Liew, F. E. Loughlin, M. Crossley and J. P. Mackay, 2007. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 32 63–70. [DOI] [PubMed] [Google Scholar]
- Garnjobst, L., and E. L. Tatum, 1967. A survey of new morphological mutants in Neurospora crassa. Genetics 57 579–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke, V., C. E. Creutz and S. E. Moss, 2005. Annexins: linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 6 449–461. [DOI] [PubMed] [Google Scholar]
- Hall, T. M. T., 2005. Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 15 367–373. [DOI] [PubMed] [Google Scholar]
- Hasunuma, K., and Y. Shinohara, 1985. Characterization of cpd-1 and cpd-2 mutants which affect the activity of orthophosphate regulated cyclic phosphodiesterase in Neurospora. Curr. Genet. 10 197–203. [DOI] [PubMed] [Google Scholar]
- Ivey, F. D., A. M. Kays and K. A. Borkovich, 2002. Shared and independent roles for a Gαi protein and adenylyl cyclase in regulating development and stress responses in Neurospora crassa. Eukaryotic Cell 1 634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga, T., and N. L. Glass, 2008. Dissecting colony development of Neurospora crassa using mRNA profiling and comparative genomics approaches. Eukaryot. Cell 7 1549–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga, T., G. Mannhaupt and N. L. Glass, 2009. Relationship between phylogenetic distribution and genomic features in Neurospora crassa. PLoS ONE 4 e5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kays, A. M., and K. A. Borkovich, 2004. Severe impairment of growth and differentiation in a Neurospora crassa mutant lacking all geterotrimeric Gα proteins. Genetics 166 1229–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, C., and K. Struhl, 1994. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol. Cell. Biol. 14 1920–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug, A., 1999. Zinc finger peptides for the regulation of gene expression. J. Mol. Biol. 293 215–218. [DOI] [PubMed] [Google Scholar]
- Lara, M., L. Blanco, M. Campomanes, E. Calva, R. Palacios et al., 1982. Physiology of ammonium assimilation in Neurospora crassa. J. Bacteriol. 150 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., and D. J. Ebbole, 1998. Tissue-specific repression of starvation and stress responses of the Neurospora crassa con-10 gene is mediated by RCO1. Fungal Genet. Biol. 23 269–278. [DOI] [PubMed] [Google Scholar]
- Legge, G. B., M. A. Martinez-Yamout, D. M. Hambly, T. Trinh, B. M. Lee et al., 2004. ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module. J. Mol. Biol. 343 1081–1093. [DOI] [PubMed] [Google Scholar]
- Li, D., P. Bobrowicz, H. H. Wilkinson and D. J. Ebbole, 2005. A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics 170 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, T. I., and R. T. Voegele, 2008. Secreted proteins of Uromyces fabae: similarities and stage specificity. Mol. Plant Pathol. 9 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama, S. S., R. E. Nelson and R. W. Siegel, 1974. Mutations specifically blocking differentiation of macroconidia in Neurospora crassa. Dev. Biol. 41 278–287. [DOI] [PubMed] [Google Scholar]
- Meiklejohn, C. D., and J. P. Townsend, 2005. A Bayesian method for analysing spotted microarray data. Brief. Bioinformatics 6 318–330. [DOI] [PubMed] [Google Scholar]
- Michan, S., F. Lledias and W. Hansberg, 2003. Asexual development is increased in Neurospora crassa cat-3- null mutant strains. Eukaryot. Cell 2 798–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R. K., and M. D. Rose, 1998. Kar9p Is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira, J.-P., T. Dubois, J.-P. Oudinet, S. Lukowski, F. o. Russo-Marie et al., 1997. Inhibition of cytosolic phospholipase A2 by annexin V in differentiated permeabilized HL-60 cells. J. Biol. Chem. 272 10474–10482. [DOI] [PubMed] [Google Scholar]
- Pandey, A., M. G. Roca, N. D. Read and N. L. Glass, 2004. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot. Cell 3 348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, D. D., A. Radford and M. S. Sachs, 2001. The Neurospora Compendium: Chromosomal Loci. Academic Press, San Diego, CA.
- Peter, B. J., H. M. Kent, I. G. Mills, Y. Vallis, P. J. G. Butler et al., 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303 495–499. [DOI] [PubMed] [Google Scholar]
- Plesofsky-Vig, N., D. Light and R. Brambl, 1983. Paedogenetic conidiation in Neurospora crassa. Exp. Mycol. 7 283–286. [Google Scholar]
- Rerngsamran, P., M. B. Murphy, S. A. Doyle and D. J. Ebbole, 2005. Fluffy, the major regulator of conidiation in Neurospora crassa, directly activates a developmentally regulated hydrophobin gene. Mol. Microbiol. 56 282–297. [DOI] [PubMed] [Google Scholar]
- Rosenberg, G., and M. L. Pall, 1979. Properties of two cyclic nucleotide-deficient mutants of Neurospora crassa. J. Bacteriol. 137 1140–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp, A., A. Zollner, D. Maier, K. Albermann, J. Hani et al., 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32 5539–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent, M. L., and D. O. Woodward, 1969. Genetic determinants of circadian rhythmicity in Neurospora. J. Bacteriol. 97 861–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass, P., J. Field, J. Nikawa, T. Toda and M. Wigler, 1986. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 83 9303–9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, M. L., 1993. Genetic control of fungal differentiation: the three sporulation pathways of Neurospora crassa. BioEssays 15 365–374. [DOI] [PubMed] [Google Scholar]
- Springer, M. L., and C. Yanofsky, 1989. A morphological and genetic analysis of conidiophore development in Neurospora crassa. Genes Dev. 3 559–571. [DOI] [PubMed] [Google Scholar]
- Springer, M. L., and C. Yanofsky, 1992. Expression of con genes along the three sporulation pathways of Neurospora crassa. Genes Dev. 6 1052–1057. [DOI] [PubMed] [Google Scholar]
- Szeto, C. Y., G. S. Leung and H. S. Kwan, 2007. Le.MAPK and its interacting partner, Le.DRMIP, in fruiting body development in Lentinula edodes. Gene 393 87–93. [DOI] [PubMed] [Google Scholar]
- Tang, J., A. Maximov, O.-H. Shin, H. Dai, J. Rizo et al., 2006. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell 126 1175–1187. [DOI] [PubMed] [Google Scholar]
- Toledo, I., J. Aguirre and W. Hansberg, 1986. Aerial growth in Neurospora crassa: characterization of an experimental model system. Exp. Mycol. 10 114–125. [Google Scholar]
- Townsend, J., 2003. Multifactorial experimental design and the transitivity of ratios with spotted DNA microarrays. BMC Genomics 4 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turian, G., and D. E. Bianchi, 1972. Conidiation in Neurospora. Bot. Rev. 38 119. [Google Scholar]
- Twumasi-Boateng, K., Y. Yu, D. Chen, F. N. Gravelat, W. C. Nierman et al., 2009. Transcriptional profiling identifies a role for BrlA in the response to nitrogen depletion and for StuA in the Regulation of Secondary Metabolite Clusters in Aspergillus fumigatus. Eukaryot. Cell 8 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanatha, J., and S. Kumble, 1993. Involvement of annexin II in DNA replication: evidence from cell-free extracts of Xenopus eggs. J. Cell. Sci. 105 533–540. [DOI] [PubMed] [Google Scholar]
- Walther, T. C., J. H. Brickner, P. S. Aguilar, S. Bernales, C. Pantoja et al., 2006. Eisosomes mark static sites of endocytosis. Nature 439 998–1003. [DOI] [PubMed] [Google Scholar]
- Xie, X., H. H. Wilkinson, A. Correa, Z. A. Lewis, D. Bell-Pedersen et al., 2004. Transcriptional response to glucose starvation and functional analysis of a glucose transporter of Neurospora crassa. Fungal Genet. Biol. 41 1104–1119. [DOI] [PubMed] [Google Scholar]
- Yang, Y. H., and T. Speed, 2002. Design issues for cDNA microarray experiments. Nat. Rev. Genet. 3 579–588. [DOI] [PubMed] [Google Scholar]
- Yin, Z., S. Wilson, N. C. Hauser, H. Tournu, J. D. Hoheisel et al., 2003. Glucose triggers different global responses in yeast, depending on the strength of the signal, and transiently stabilizes ribosomal protein mRNAs. Mol. Microbiol. 48 713–724. [DOI] [PubMed] [Google Scholar]
- Young, M. E., T. S. Karpova, B. Brugger, D. M. Moschenross, G. K. Wang et al., 2002. The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol. Cell. Biol. 22 927–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssar, L., T. J. Schmidhauser and J. Avalos, 2005. The Neurospora crassa gene responsible for the cut and ovc phenotypes encodes a protein of the haloacid dehalogenase family. Mol. Microbiol. 55 828–838. [DOI] [PubMed] [Google Scholar]
- Zhang, X., R. L. Lester and R. C. Dickson, 2004. Pil1p and Lsp1p negatively regulate the 3-phosphoinositide-dependent protein pinase-like kinase Pkh1p and downstream signaling pathways Pkc1p and Ypk1p. J. Biol. Chem. 279 22030–22038. [DOI] [PubMed] [Google Scholar]