Abstract
Aneuploid cells are characterized by incomplete chromosome sets. The resulting imbalance in gene dosage has phenotypic consequences that are specific to each karyotype. Even in the case of Down syndrome, the most viable and studied form of human aneuploidy, the mechanisms underlying the connected phenotypes remain mostly unclear. Because of their tolerance to aneuploidy, plants provide a powerful system for a genome-wide investigation of aneuploid syndromes, an approach that is not feasible in animal systems. Indeed, in many plant species, populations of aneuploid individuals can be easily obtained from triploid individuals. We phenotyped a population of Arabidopsis thaliana aneuploid individuals containing 25 different karyotypes. Even in this highly heterogeneous population, we demonstrate that certain traits are strongly associated with the dosage of specific chromosome types and that chromosomal effects can be additive. Further, we identified subtle developmental phenotypes expressed in the diploid progeny of aneuploid parent(s) but not in euploid controls from diploid lineages. These results indicate long-term phenotypic consequences of aneuploidy that can persist after chromosomal balance has been restored. We verified the diploid nature of these individuals by whole-genome sequencing and discuss the possibility that trans-generational phenotypic effects stem from epigenetic modifications passed from aneuploid parents to their diploid progeny.
THE genome of aneuploid individuals contains incomplete chromosome sets. The balance between chromosome types, and the genes they encode, is compromised, resulting in altered expression of many genes, including genes with dosage-sensitive effects on phenotypes. In humans, only a few types of aneuploid karyotypes are viable (Hassold and Hunt 2001), highlighting the deleterious effect of chromosome imbalance. The most commonly known viable form of aneuploidy in humans is Down syndrome, which results from a trisomy of chromosome 21 in an otherwise diploid background. Down syndrome patients exhibit many specific phenotypes, sometimes visible only in a subset of patients (Antonarakis et al. 2004). For phenotypes found in all Down syndrome patients, the penetrance of each phenotype varies between patients (Antonarakis et al. 2004). Despite the increasing amount of information available about the human genome and the availability of a mouse model for Down syndrome (O'Doherty et al. 2005), the genes responsible for most of the phenotypes associated with Down syndrome are still unknown (Patterson 2007; Korbel et al. 2009; Patterson 2009). Recently, detailed phenotypic analyses of as many as 30 aneuploid patients have allowed the identification of susceptibility regions for several specific phenotypes (Patterson 2007, 2009; Korbel et al. 2009; Lyle et al. 2009), but the specific genes remain to be identified. Understanding the physiology of aneuploidy is not only relevant to those individuals with aneuploid genomes but also to understanding cancer since most cancerous cells are aneuploid (Matzke et al. 2003; Pihan and Doxsey 2003; Storchova and Pellman 2004; Holland and Cleveland 2009; Williams and Amon 2009) or the consequences of copy number variation and dosage sensitivity (Dear 2009; Henrichsen et al. 2009).
Plants are more tolerant of aneuploidy than animals (Matzke et al. 2003) for reasons that remain unclear. Since the discovery of the Datura trisomic “chromosome mutants” by Blakeslee (1921, 1922), viable trisomics of each chromosome type have been described in numerous species. Trisomics exhibit phenotypes specific to the identity of the triplicated chromosome (Blakeslee 1922; Khush 1973; Koornneef and Van der Veen 1983; Singh 2003). More complex aneuploids, i.e., individuals carrying more than one additional chromosome, can be viable as well and have been observed in many plants species, especially among the progeny of triploid individuals (McClintock 1929; Levan 1942; Johnsson 1945; Khush 1973). Some species appear to be more tolerant of complex aneuploidies than others, suggesting a genetic basis for aneuploidy tolerance (Satina and Blakeslee 1938; Khush 1973; Ramsey and Schemske 2002; Henry et al. 2009). Aneuploid individuals frequently appear spontaneously within polyploid plant populations, presumably due to a failure to equally partition the multiple chromosome sets at meiosis (Randolph 1935; Doyle 1986). These aneuploids exhibit few or subtle phenotypic abnormalities and can often compete with their euploid progenitors (Ramsey and Schemske 1998). Plants therefore provide an excellent opportunity for a genome-wide investigation of aneuploid syndromes: sample size is not limited, phenotypes can be described and assessed in detail, and plant aneuploid populations provide a complex mixture of viable karyotypes.
In this article, we report our investigation of the relationship between phenotype and karyotype in populations of aneuploid Arabidopsis thaliana plants. All simple trisomics of A. thaliana have been previously isolated and phenotypically characterized (Steinitz-Sears 1962; Lee-Chen and Steinitz-Sears 1967; Steinitz-Sears and Lee-Chen 1970; Koornneef and Van der Veen 1983), demonstrating that they are tolerated in A. thaliana. We previously reported that aneuploid swarms—populations of aneuploid individuals of varying aneuploid karyotypes—could be obtained from the progeny of triploid A. thaliana individuals (Henry et al. 2005, 2009). Using a combination of a quantitative PCR-based method and flow cytometry, we were able to derive the full aneuploid karyotype of each of these individuals (Henry et al. 2006). We further crossed triploid A. thaliana to diploid or tetraploid individuals and demonstrated that at least 44 of the 60 possible aneuploid karyotypes that could result from these crosses (aneuploid individuals carrying between 11 and 19 chromosomes) were viable and successfully produced adult plants. Taken together, these populations and methods make it possible to explore the basis of aneuploid syndromes in A. thaliana. In this study, we were able to phenotypically characterize at least one individual from 25 different aneuploid karyotypes falling between diploidy and tetraploidy. We demonstrated that specific phenotypes are affected by the dosage of specific chromosome types. The effect of the dosage of specific chromosome types on traits was additive and could be used to predict the observed phenotype. The availability of multiple generations of aneuploid and euploid individuals allowed us to investigate potential long-term effects of aneuploidy as well as parent-of-origin effects on aneuploid phenotypes.
MATERIALS AND METHODS
Plant materials:
All plants were grown in soil (Sunshine Professional Peat-Lite mix 4, SunGro Horticulture, Vancouver, BC) in a growth room lit by fluorescent lamps (model TL80; Phillips, Sunnyvale, CA) at 22° ± 3° with a 16 hr:8 hr light:dark photoperiod or in a greenhouse at similar temperatures and light regimes, with supplemental light provided by sodium lamp illumination as required.
Tetraploid lines were produced as previously described (Henry et al. 2005). Col-0 represents the diploid ecotype Columbia; 4x-Col represents tetraploidized Col-0, and Wa-1 represents the naturally occurring tetraploid ecotype Warschau-1 [The Arabidopsis Information Resource (TAIR) accession no. CS6885]. C and W are used to represent the basic genomes of Col-0 and Wa-1, respectively. Crosses are always represented with the seed parent mentioned first and the pollen parent next. For example, CWW plants are triploids generated by crossing Col-0 as the seed parent to Wa-1. All CCC triploid plants studied here were generated by crossing Col-0 as the seed parent to 4x-Col.
The different populations used in this study were the following (see Table 1 for details). The CCWW F1 population was generated by crossing Wa-1 to 4x-Col tetraploid plants in either direction and contained tetraploid and aneuploid individuals (Henry et al. 2006). The CWW F2 population is the selfed progeny of a CWW triploid plant. It contains diploid, triploid, and tetraploid individuals and a swarm of aneuploid individuals of intermediate genome content. Four pseudo-backcross populations (pBCs) were generated by crossing the CWW triploid as pollen parent or egg parent to either diploid Col-0 or tetraploid 4x-Col (Henry et al. 2006, 2009). All individuals in the CWW F2, CCWW F1, and pBC populations were analyzed for genome content using flow cytometric analysis of nuclear DNA content. In addition, the CCWW F1 and pBC populations were fully karyotyped using quantitative fluorescent PCR (QF-PCR) as previously described (Henry et al. 2005, 2006).
TABLE 1.
Origin of the different plants, lines and populations used in this study.
| Name | Cross (seed parent first) | N | Ploidy | Note |
|---|---|---|---|---|
| CWW | Col-0 x Wa-1 | n/a | 3x | – |
| CCWW F1 | 4x-Col x Wa-1 | 46 | 4x, aneuploids | Fully karyotyped |
| CCWW F1 | Wa-1 x 4x-Col | 47 | 4x, aneuploids | Fully karyotyped |
| CWW F2 | CWW selfed | 109 | 2x, 3x, 4x, aneuploids | Genome content determined but no full karyotypes |
| pBC | Col-0 x CWW | 80 | 2x, 3x, aneuploids | Fully karyotyped |
| pBC | CWW x Col-0 | 102 | 2x, 3x, aneuploids | Fully karyotyped |
| pBC | 4x-Col x CWW | 33 | 2x, 3x, aneuploids | Fully karyotyped |
| pBC | CWW x 4x-Col | 47 | 2x, 3x, aneuploids | Fully karyotyped |
Analysis of qualitative phenotypes:
Aneuploid phenotypes were described and defined through the observation of individual aneuploid plants from the pBCs (N = 57) and comparison with diploid Col-0 individuals. In all cases, the observers were unaware of the karyotype of the plants and recorded visible phenotypic variation as compared to diploid individuals. After all plants had been described, 12 traits were selected for further study on the basis of the recurrent observation of specific non-wild-type phenotypes in at least five independent plants.
Phenotypic traits:
Qualitative phenotypic traits were divided into three scoring categories, depending on the number of phenotypic states associated with them. Eight traits were binary. If the phenotype was observed, the value of “1” was assigned to that plant. If the phenotype was not observed, it was scored “0.” These phenotypes are described in Table 2. Three traits for which two opposite phenotypes, in addition to the wild-type phenotype, were observed in the aneuploid individuals, were identified. For these traits, the value of “1” was assigned to one of the two phenotypes and “−1” was assigned to the opposite phenotype. If neither phenotype was observed, the individual was assigned the score “0.” The effect of chromosome dosage on these traits was determined as described below. In addition, the effect of chromosome dosage on each of the two opposite phenotypes was tested separately by excluding the individuals exhibiting the opposite phenotype. Finally, in the last category, fertility of aneuploids was assessed qualitatively (associated quantitative scores in parentheses): fertile (3), moderately fertile (2), low fertility (1), and sterile (0). Seeds were considered as “plump” if they contained a visible embryo structure at least 20% the size of a wild-type seed. Each aneuploid plant was selfed and a few siliques were collected from each plant. Plants were considered fertile if siliques were well formed and contained many seeds, most of which (>95%) were plump. Plants were considered as moderately fertile if siliques contained at least 20 plump seeds, which is ∼40–50% of a seed set. A low-fertility score was assigned to plants carrying siliques either that contained few seeds or in which most of the seeds were shriveled. Finally, plants were recorded as sterile if no plump seeds were observed.
TABLE 2.
Phenotypes observed
| Trait | Description |
|---|---|
| Branchy | Secondary branching and loss of apical dominance. |
| Curly leaves | Rolled blades of cauline leaves as illustrated in Figure 1N. |
| Empty axils | Apparent lack of axillary buds at the basis of a cauline leaf (Figure 1Q). In the qualitative analysis (Table 4), a plant was scored as a “yes” for the presence of empty axils on the basis of a preponderance of empty axils as compared to control plants. |
| Fan-shaped vasculature | Unusual vein pattern on the longitudinal leaf axis resulting in a fan-shaped pattern. |
| Fasciation | Gross morphological evidence for radial stem growth resulting in divergence of the vasculature and bifurcation of the meristem (Figure 1O). |
| Flower in axil | Direct conversion of an axillary meristem to a floral meristem (Figure 1I). |
| Hairy | Presence of higher density of trichomes on both the adaxial and abaxial leaf surfaces as well as on the stem (Figure 1, R and S). |
| Irregularities | Instances of meristematic reversion, fasciation, triple branches, and double-headed flowers. |
| Irregular spacing | Periods of failed elongation resulting in disorganized and compacted nodes followed by longer-than-normal internodes (Figure 1, L–M). |
| Meristematic reversion | Meristem fate switching from a later to an earlier developmental state. For example, reversion from a floral meristem to an inflorescence meristem or from an inflorescence meristem to a vegetative meristem. Evidenced by out-of-order placement of organs (flowers and leaves). |
| Nubbin | Angular projection/bend in stem often with light irregular growth at position. Frequently found at the base of a secondary stem or immediately following or preceding a node (Figure 1P) (Iltis 2000). |
| Triple branches | Three-way branching, typically at an axil where the axillary meristem produced a bifurcated shoot. |
Statistical analysis:
For each individual plant, a complete karyotype had previously been determined (Henry et al. 2005). From this karyotype, the total number of chromosomes was calculated, as well as the average number of copies per chromosome type. For example, a double trisomic of chromosomes 3 and 5 contained three copies of chromosomes 3 and 5 and two copies of chromosomes 1, 2, and 4. The total number of chromosomes was thus 12, and the average number of copies per chromosome type was 12/5 = 2.4. Next, for each chromosome type, dosage was expressed as the number of copies relative to the average [relative chromosome dosage (rChrX)]. In the previous example, the dosage of chromosome 1 would be rChr1 = 2/2.4 = 0.83 while the dosage of chromosome 3 would be rChr3 = 3/2.4 = 1.25. Using this method, rChrX values >1 thus indicate an over-representation relative to at least one other chromosome type while rChrX values <1 indicate an under-representation. Finally, the relationship between each phenotype and the relative chromosome dosage was calculated by regression analysis. P-values were corrected for five independent tests due to five chromosome types (i.e., Bonferroni corrected) such that a P-value < 0.01 was regarded as significant.
Analysis of quantitative phenotypes:
Quantitative measurements were recorded for individual aneuploid plants from the pBC populations for three traits: stem diameter, percentage of empty axils, and rosette size. Stem diameter (SD) was measured ∼1 cm above the rosette on each individual using a caliper. The percentage of empty axils was calculated by dividing the number of empty axils (Table 2) by the total number of leaves. Finally, for each population of plants, individual pictures were taken of each plant. Rosette-size measurements were obtained from photos by recording the length of the smallest possible rectangle that included the totality of the rosette. At the time the photos were taken, the CCWW F1 plants (no. of euploids = 62 and no. of aneuploids = 31) were 29 days old, the CWW F2 plants (no. of euploids = 13 and no. of aneuploids = 70) were 28 days old, and the progeny of trisomic plants were 21 days old.
Statistical analysis:
For all three traits, the association between phenotype and relative chromosome dosage values was analyzed by regression analysis, as presented above. In the case of rosette size, each population was analyzed separately to eliminate the effect of rosette age. Finally, aneuploid and euploid groups were compared using Student's t-tests.
Test of additivity:
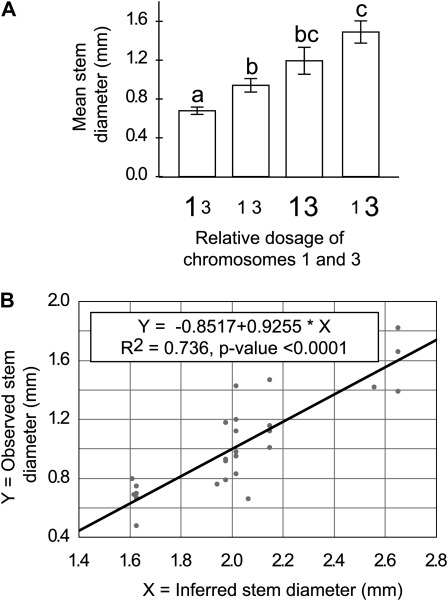
The relative dosage of chromosomes 1 and 3 was found to significantly affect SD. Specifically, the effect of dosage of chromosomes 1 and 3 followed, respectively, the following models: SD = 1.9033 – 0.8616 × rChr1 and SD = −0.3731 + 1.3956 × rChr3. To test if the effects of chromosomes 1 and 3 were additive, the observed stem diameter was compared to an inferred stem diameter on the basis of the dosage of chromosomes 1 and 3 and calculated using the following fully additive model: SD = 1.5302 + 1.3956 × rChr3 – 0.8616 × rChr1 (Figure 2B).
Figure 2.—
Additive chromosomal dosage effects on stem diameter. (A) Mean stem diameter as a function of the relative dosage of chromosomes 1 and 3. For ease of illustration, all aneuploid plants were classified into four categories, symbolized by the size of the chromosome type character, and depending on whether the dosage of chromosomes 1 and 3 was over-represented (rChrX > 1, large characters) or under-represented (rChrX < 1, small characters). Different letters above the two bars indicate significantly different means for these two populations of aneuploid individuals. Values were considered significantly different when the t-test P-values were <0.05. Standard errors are indicated. (B) Relationship between observed mean stem diameter and stem diameter values calculated using a model that assumes full additivity of the single effects of chromosomes 1 and 3 on stem diameter (see materials and methods for details).
A similar analysis was performed to assess the additivity of the effects of the dosage of chromosomes 3 and 5 on the percentage of empty axils (%EA). Specifically, the effect of dosage of chromosomes 3 and 5 was expressed as follows: %EA = −0.8216 + 1.0790 × rChr3 and %EA = 1.2333 − 0.9878 × rChr5, respectively. The fully additive model was as follows: inferred %EA = 0.4117 + 1.0790 × rChr3 − 0.9878 × rChr5.
Quantitative measure of seed viability:
Siliques were harvested into individual tubes, and all the seeds from each fruit were counted using a dissecting microscope. Seeds were characterized as “plump” if they contained a visible embryo structure at least 20% the size of wild-type seed or “shriveled” if they did not.
Origin and phenotypic characterization of the pseudo-diploid plants:
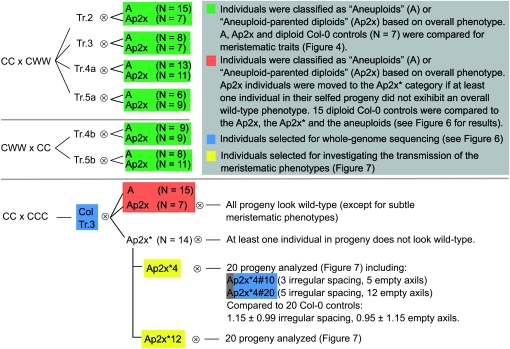
From the progeny of the pBCs, six plants were selected that were trisomic for one chromosome type and diploid for all other chromosome types. The trisomic chromosome is indicated in the name of the line: Tr.2, Tr.3, Tr.4a, and Tr.5a originated from CC × CWW crosses and Tr.4b and Tr.5b originated from CWW × CC crosses (see Figure 4). In addition, ColTr.3 was an all-Col-0 individual originating from a CC × CCC cross. It was originally determined to be trisomic for chromosome 3 in an otherwise diploid background on the basis of its phenotype, which was later confirmed using whole-genome sequencing (see below). Each of these trisomic plants was allowed to self, and some of the produced seeds were planted (see Figure 4). Rosette sizes were measured as described above. Additionally, a variety of meristematic abnormalities (defined in Table 2) were repeatedly observed in these populations, both in the aneuploid individuals and in the supposed diploid individuals. For each of the resulting progeny, the number of instances of each of these traits was recorded.
Figure 4.—
Summary of the populations used to investigate a possible long-term effect of aneuploidy on meristematic traits. Seed parents are indicated first, and “⊗” indicates a self-cross.
Karyotyping using whole-genome sequencing:
Genomic DNA was extracted using the Fast DNA kit (MP Biomedicals, Solon, OH) from six selected individuals. For each individual, between 1.2 and 2.0 μg of DNA was further processed. First, water was added to the DNA to reach a total of 200 μl in 1.5-ml Eppendorf tubes. The DNA was then fragmented by sonication (Bioruptor UCD-200; Diagenode) for 15 min (pulses of 30 sec on the high setting and 30 sec off) and cleaned using MinElute columns (Qiagen Sciences) as recommended by the manufacturer. End-repair and “A”-base addition were carried out using the Next DNA Sample Prep kit (New England Biolabs, Ipswich, MA) according to the manufacturer's recommendations. Reactions were cleaned using MinElute columns after each step, with a final elution volume of 10 μl. Adaptors for Illumina GAIIX sequencing were ligated by combining the following: 10 μl of DNA, 15 μl of quick ligation reaction buffer (2×), 1.2 μl of DNAse-free water, 1.8 μl of adaptor mix (50 μm), and 2 μl of quick T4 DNA ligase for a total of 30 μl. Each of the six samples was ligated to different 5-bp barcoded adaptors (see Table S1for adaptor sequences). The reaction mixes were again purified using MinElute columns before being run on a 1.5% agarose gel for size selection. DNA of the desired size range (in this case, 300–400 bp) was extracted from the gel using the Qiagen Gel Extraction Kit (Qiagen Sciences) and eluted in 30 μl. The resulting libraries were amplified by PCR by mixing 13.5 μl of template DNA, 15 μl of 2× Phusion High-Fidelity PCR Master Mix (Finnzymes Oy, Espoo, Finland), and 1.5 μl of 5 μm paired-end primer mix (PE-PrimerA + PE-PrimerB; see Table S1); we used the following protocol: 3 sec at 98° followed by 12 cycles of 10 sec at 98°, 30 sec at 65°, and 30 sec at 72°, ending with 5 min at 72°. The PCR products were purified using MinElute columns and eluted in 10 μl before being submitted for 41-bp sequencing using Solexa Sequencing technology on an Illumina Genome Analyzer II (Illumina, San Diego). The original sequence file has been deposited in the National Center for Biotechnology Information Sequence Read Archive under accession no. SRP003606.1.
Sequencing reads were divided into individual pools according to their barcode, and the barcodes were truncated from each read. The resulting read sequences were aligned to the A. thaliana TAIR 9.0 genome using the Efficient Local Alignment of Nucleotide Data (ELAND) software (Illumina, San Diego). Only reads that matched perfectly to a single location in the reference genome were processed further. For each individual, reads were pooled into 100,000-bp nonoverlapping bins covering the whole A. thaliana genome using two custom Python scripts (see File S1 and File S2). In short, coverage across the genome was calculated by counting the number of reads in each bin. Col-0 #1 (diploid Col-0) was used as the control sample. For each of the other five individuals, relative coverage was derived for each bin using the following formula: read coverage = [no. of reads in bin (sample)] × (total no. of reads from Col-0 control)/(total no. of reads from sample). Using this formula, coverage values along chromosomes oscillated around the values for relative chromosome dosage (rChr#X) described above. The coverage value of all chromosomes from a diploid individual oscillated around 1.0.
RESULTS
We phenotypically characterized A. thaliana aneuploids either isolated from tetraploid populations or resulting from crosses between triploid and diploid or tetraploid individuals (Henry et al. 2006). We focused our analysis on aneuploids of genome content ranging between diploidy and tetraploidy (aneuploids containing between 11 and 19 chromosomes) for two reasons: they can most accurately be karyotyped using QF-PCR (Henry et al. 2006) and they exhibit more severe phenotypes than aneuploids of higher ploidy backgrounds. Indeed, the consequences of chromosomal dosage imbalance may be buffered by the presence of more copies of the genome in aneuploid individuals of higher ploidy backgrounds (Khush 1973; Ramsey and Schemske 1998; Vizir and Mulligan 1999; Birchler et al. 2001).
Aneuploid individuals exhibited diverse phenotypes affecting a wide variety of traits (Figure 1). Individuals of the same karyotype exhibited similar phenotypic traits, but the degree of severity of each phenotype was variable. The overall severity of the aneuploid phenotype did not appear to be correlated with the number of unbalanced chromosomes but rather with the identity of the unbalanced chromosome. For example, plants trisomic for chromosome 1 (Tr.1) were much more severely affected than Chr.1, Chr.3 double trisomics (data not shown). Finally, one haploid plant that originated from pollination of a diploid plant by pollen from a triploid was observed. No specific phenotype was observed with the exception that the plant was smaller overall and had narrower stems than its diploid counterpart. This haploid plant produced very few seeds, which all gave rise to diploid plants, similar to a recent report (Ravi and Chan 2010).
Figure 1.—
Aneuploid phenotypes in A. thaliana. Photos are of aneuploid individuals with the exception of E and R. (A–E) Whole rosettes. (F–H and J) Whole plants. (I and K–T) Specific phenotypes. (I) Flower in axil. (K) Aerial rosette. (L) Severe irregular spacing. (M) Moderate irregular spacing. (N) Curly leaves. (O) Fasciation. (P) Nubbin. (Q) Empty axils. (R) Diploid cauline leaf showing wild-type stem and leaf trichomes. (S) Hairy stem and cauline leaf. Karyotypes: (A) 3x + chromosomes 3 and 5; (B) 3x – chromosome 5; (C) 2x + chromosome 2; (D) 2x + chromosomes 3 and 5; (E) 2x-Col-0; (F) 3x – chromosome 5; (G) 2x + chromosome 5; (H) 2x + chromosomes 1, 2, and 3; (I) 2x + chromosome 3 and 4; (J) 2x + chromosomes 1 and 2 (K) 2x + chromosome 3; (L) 2x + chromosomes 1, 2, and 3; (M) 2x + chromosome 5; (N) 2x + chromosome 5; (O) 2x + chromosome 3; (P) 4x + chromosome 4; (Q) 2x + chromosome 3; (R) 2x-Col-0; (S) 2x + chromosome 4.
Can specific phenotypes be associated with specific chromosome types?
Fifty-seven karyotyped aneuploid individuals, representing 25 different karyotypes and carrying chromosome numbers between 11 and 19 (Table 3), were characterized phenotypically. Phenotypic abnormalities were recorded after close observation of each plant and comparison with Col-0 diploid controls. Twelve traits were selected for further study on the basis of the recurrent observation of similar phenotypes in at least five plants. Traits were divided into three groups depending on how the data were gathered or analyzed: “single” traits, “opposite” traits, and fertility.
TABLE 3.
Karyotype distribution of the phenotyped plants
| Chromosome no. | No. of individuals | No. of different karyotypes represented |
|---|---|---|
| 11 | 24 | 5 |
| 12 | 6 | 4 |
| 13 | 1 | 1 |
| 14 | 1 | 1 |
| 16 | 7 | 3 |
| 17 | 7 | 4 |
| 18 | 7 | 4 |
| 19 | 4 | 3 |
| Total | 57 | 25 |
The first group contained eight binary traits (see materials and methods for details). With the exception of “branchy,” all binary traits were altered by the relative dosage of one or two chromosome types (Table 4). Half of the effects observed were very strong (P-values < 0.0001), suggesting the existence of dosage-sensitive genes on those chromosomes responsible for the observed phenotype. Additional observations could be made from these results. For example, over-representation of chromosome 5 increases the occurrence of triple branches while under-representation of chromosome 5 increases the occurrence of empty axils. This increase and decrease in axillary meristem number could result from a single dosage-sensitive mechanism encoded by chromosome 5 that controls the production of axillary meristems.
TABLE 4.
Effect of relative chromosome dosage on binary phenotypes
| No. of individuals |
Effect of rChrX on phenotype occurrence (correlation P-values)a |
||||||
|---|---|---|---|---|---|---|---|
| Phenotype | No | Yes | Chromosome 1 | Chromosome 2 | Chromosome 3 | Chromosome 4 | Chromosome 5 |
| Hairy | 47 | 10 | 0.555 | 0.014 | 0.686 | <0.0001+ | 0.016 |
| Curly leaves | 50 | 7 | 0.646 | 0.319 | 0.475 | 0.260 | 0.0044+ |
| Empty axils | 35 | 22 | 0.126 | 0.572 | 0.00011+ | 0.372 | 0.0034− |
| Nubbins | 48 | 9 | <0.0001+ | 0.045 | 0.237 | 0.316 | 0.115 |
| Branchy | 51 | 6 | 0.710 | 0.189 | 0.484 | 0.175 | 0.638 |
| Triple branches | 43 | 14 | 0.520 | 0.291 | 0.035 | 0.038 | <0.0001+ |
| Fasciation | 47 | 10 | 0.0057+ | 0.287 | 0.707 | 0.893 | 0.570 |
| Fan-shaped vasculature | 51 | 6 | 0.213 | 0.218 | 0.137 | 0.151 | <0.0001+ |
For each chromosome type, the correlation between phenotype occurrence and rChrX was calculated. Regressions were considered significant when P-values were <0.01 (in boldface type) to control for independent testing on five chromosome types. “+” and “−” indicate positive and negative correlations, respectively.
To further address this possibility, traits that exhibited opposite phenotypes were analyzed. For example, some individuals exhibited leaves that were a lighter green than wild type, while others exhibited leaves that were a darker green than wild type. Results for this group of opposite phenotypes were similar to those obtained for the first group with the dosage of none to two chromosome types influencing each trait, as compared to the wild-type group (Table 5). To determine if opposite traits were influenced by the same chromosome types, karyotypes in the two phenotypic groups were compared to the wild-type group, while individuals exhibiting the opposite phenotype were excluded. In our analysis, none of the opposite traits were influenced by opposite changes in the dosage of the same chromosome type (Table 5).
TABLE 5.
Effect of chromosome dosage on opposite phenotypes
| Phenotype | Effecta on the incidence of: | Chromosome 1 | Chromosome 2 | Chromosome 3 | Chromosome 4 | Chromosome 5 | No. of plants | |
|---|---|---|---|---|---|---|---|---|
| Leaf width | Both | 0.354 | 0.00016 | 0.089 | 0.583 | 0.00034 | Wild type | 38 |
| Narrow | 0.151 | 0.63718 | 0.818 | 0.169 | 0.0061+ | Narrow | 7 | |
| Wide | 0.868 | 0.000030+ | 0.025 | 0.546 | 0.08498 | Wide | 12 | |
| Apical dominance | Both | 0.094 | 0.776 | 0.013 | 0.035 | 0.050 | Wild type | 47 |
| Weak | 0.0068+ | 0.468 | 0.118 | 0.576 | 0.764 | Weak | 4 | |
| Strong | 0.810 | 0.328 | 0.081 | 0.033 | 0.031 | Strong | 6 | |
| Color | Both | 0.00033 | 0.898 | 0.012 | 0.724 | 0.384 | Wild type | 34 |
| Dark green | 0.00055+ | 0.635 | 0.647 | 0.552 | 0.231 | Dark | 9 | |
| Light green | 0.29353 | 0.800 | 0.012 | 0.341 | 0.926 | Light | 14 | |
For each chromosome type, the correlation between phenotype and rChrX was calculated. Regressions were considered significant when P-values were <0.01 (in boldface type) to control for independent testing on five chromosome types. “+”indicates a positive correlation. When testing the effect of chromosome dosage on one of the two opposite phenotypes, individuals exhibiting that phenotype were compared to the wild-type group, and individuals exhibiting the opposite phenotype were excluded from the analysis.
Finally, the fertility of selfed aneuploid individuals was assessed in a semiquantitative manner (see materials and methods for details), and the effect of dosage of each chromosome type was assessed by regression analysis. No euploid individuals were included in this analysis, as they exhibited no variation from the maximum fertility score. Within the context of our aneuploid swarm, the dosage of chromosome 2 was found to significantly affect fertility. Specifically, over-representation of chromosome 2 increased fertility (regression P-value = 0.0032).
Are the phenotypic effects of chromosome dosage additive?
The results presented above suggested that specific aneuploid phenotypes can be associated with the relative dosage of one or more specific chromosome types. To test whether the effects of the dosage of two chromosome types were additive, we quantitatively phenotyped a subset of the plants for two traits: stem diameter (N = 28) and percentage of empty axils (N = 25).
First, the effect of each chromosome type was assessed by regression analysis (see materials and methods for details). The relative dosage of chromosomes 1 and 3 was found to significantly affect stem diameter. Specifically, a relative over-representation of chromosome 1 was associated with a decrease in stem diameter (regression P-values = 0.0049) while a relative over-representation of chromosome 3 was associated with an increase in stem diameter (regression P-value < 0.0001). Next, for each individual, stem diameter was calculated on the basis of the relative dosage of chromosomes 1 and 3 in these individuals, assuming fully additive effects of these two chromosome types (see materials and methods for details). These values were compared to the observed values by regression analysis. The regression was significant (P-value < 0.0001), and the goodness of fit (R2 = 0.7362) indicated that the two effects were sufficient to explain most of the observed variability (Figure 2B).
A similar analysis was performed for the empty axil phenotype, with similar results. As observed in our initial nonquantitative analysis (Table 4), the dosage of chromosomes 3 and 5 influenced the percentage of empty axils. Specifically, an over-representation of copies of chromosome 3 resulted in an increase in the percentage of empty axils (regression P-value < 0.0001). An under-representation of chromosome 5 had a similar effect (regression P-value = 0.00012). Comparison of the inferred percentage of empty axils assuming a fully additive model with the observed percentage of empty axils resulted in a significant regression (P-value < 0.0001; R2 = 0.7162).
Can rosette size serve as a phenotypic marker for aneuploid severity?
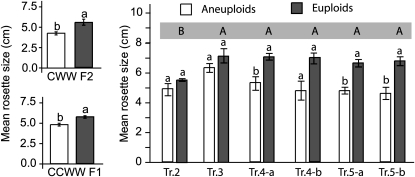
In general, aneuploid individuals often appeared less vigorous than euploid individuals, suggesting a general reduction of growth due to aneuploidy. This observation is confirmed by the measurement of the size of young rosettes in two of the populations characterized earlier (Figure 3): the CWW F2 and the CCWW F1 populations. In both populations, mean rosette diameter was significantly smaller for the aneuploid individuals than for the euploid individuals (Student's t-test P-value < 0.0001 for the CCWW F1 population and 0.0058 for the CWW F2 population). Interestingly, rosette size in the CWW F2 population was not associated with fertility (percentage of plump seed) or with our quantitative measure of aneuploid selection (calculated as the ratio of the observed number of individuals in a particular genome content class to the expected number), which provides a measure of selection against specific genome content classes (Henry et al. 2007). This suggests that rosette diameter is influenced by aneuploidy but not necessarily indicative of selection against a particular type of aneuploid karyotype.
Figure 3.—
Effect of aneuploidy on rosette diameter. Mean rosette diameter values were compared between euploid (gray) and aneuploid (white) groups within the CWW F2 population (left, top), the CCWW F1 population (left, bottom), and the progeny of trisomic individuals (right). Means were compared using Student's t-tests. Lowercase letters directly above the two columns indicate significantly different means between the corresponding populations. Uppercase letters in the blue-gray rectangle above the graph represent significantly different means between the different populations of Ap2x individuals. Values were considered significantly different when the Student's t-test P-values were <0.05, except for the analysis of the trisomic populations where significant P-values were <0.0083 to compensate for multiple testing on six independent populations. Standard errors are indicated.
Next, the effect of dosage of each chromosome type on rosette diameter was examined. In the CCWW F1 population, individuals carrying an additional copy of chromosome 1 (N = 12) were on average smaller than the rest of the individuals (regression P-value = 0.00088). Over-representation of chromosome 5 (N = 13) did not influence rosette diameter, and low numbers of aneuploid individuals prevented the analysis of the effect of dosage of the other chromosome types. This was consistent with our general observation that individuals carrying extra copies of chromosome 1 were weaker overall than all other aneuploid types observed (data not shown).
Next, rosette diameter was measured in the progeny of selfed trisomics of each chromosome type, with the exception of the trisomics of chromosome 1 from which seeds could not be obtained. For each population of trisomic progeny, plants were qualified as aneuploid or diploid on the basis of the overall phenotype of the plants once they had reached reproductive stage (see Figure 3). Indeed, selfed trisomics produce a mixture of diploid and trisomic individuals, and the rate of transmission of the trisomic chromosome depends on the chromosome type (Khush 1973). Trisomics of each chromosome type can easily be recognized from the diploids through the observation of specific phenotypes, as reported previously (Steinitz-Sears 1962; Lee-Chen and Steinitz-Sears 1967; Steinitz-Sears and Lee-Chen 1970; Koornneef and Van der Veen 1983). Next, we correlated this information with the rosette size of each of the plants as recorded when they were still vegetative. Comparison of the mean rosette size of the trisomic and diploid populations demonstrated a deleterious effect of an additional copy of chromosome 5 in both lines and of chromosome 4 in one of the two lines but not of chromosomes 2 or 3 (Figure 3). Altogether, our results suggest that decreased rosette size is not a general response to aneuploidy per se but merely another trait influenced by the dosage of specific chromosome types.
Are there long-term effects of aneuploidy?
The analysis of rosette diameter in the progeny of selfed trisomics highlighted an unexpected phenomenon. While the mean rosette diameter of most wild-type subpopulations was similar, that of the wild-type population produced by Tr.2 was significantly smaller than all others (Figure 3; Student's t-test P-values ≤ 0.0062). This suggested that diploid individuals originating from aneuploid parents might not necessarily be phenotypically wild type and that aneuploid ancestry might have phenotypic consequences. We therefore refer to diploid individuals originating from at least one aneuploid parent as “Aneuploid-parented diploid” (Ap2x) (Figure 4).
To test this hypothesis, the progenies of the selfed trisomics were characterized for a number of meristematic traits that had been recurrently observed among these populations. For each trait, the mean values obtained for the aneuploid and Ap2x subpopulations were compared to each other as well as to mean values obtained from a set of control Col-0 diploid plants (Figure S1). As previously observed, comparison of the aneuploid populations with the control diploid Col-0 suggested that most traits are affected by aneuploidy in a chromosome-dependent manner (Figure S1). Similarly, the Ap2x populations were phenotypically different from the control population for several traits.
To rule out the possibility that this effect originated from the fact that the aneuploid plants described above are hybrids of the Col-0 and Wa-1 genomes, we analyzed the progeny of an all-Col-0 aneuploid. Aneuploid Col-0 plants were produced by crossing a CCC triploid to a Col-0 diploid. Among the progeny, one (ColTr.3) was identified as a likely trisomic of chromosome 3 on the basis of its phenotype. This karyotype was confirmed by whole-genome sequencing (see below). This individual was selfed and 36 progeny were characterized in detail, along with 15 Col-0 control individuals (Figure 4). The progeny were divided into aneuploids and Ap2x individuals on the basis of overall phenotype, as described above. At a glance, these Ap2x individuals could not be distinguished from a wild-type plant whereas the trisomic individuals were immediately apparent.
Selfed trisomics normally produce diploids or parental trisomics (i.e., trisomics of the same chromosome type as the parents), but a low percentage of secondary trisomics (trisomics for a chromosomal arm instead of a whole chromosome) can also be found in the progeny of trisomics and, rarely, unrelated aneuploids can be produced as well (Khush 1973). It is therefore possible that some of the Ap2x individuals were segmental aneuploids and not visually recognizable as such. Therefore, all of the Ap2x individuals were again selfed and their progeny were observed for trueness to type. Specifically, only individuals that were phenotypically close to wild type and exclusively produced progeny that were also phenotypically close to wild type were labeled as Ap2x (as opposed to Ap2x* in Figure 4). After this very conservative selection, only seven individuals could be unambiguously labeled as Ap2x.
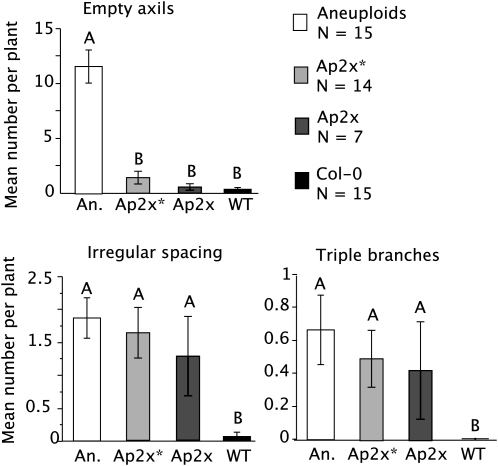
Meristematic traits from the Ap2x, the Ap2x*, and the aneuploids were compared to the control Col-0 using Student's t-tests (Figure 5). Neither the Ap2x nor the Ap2x* differed significantly from the Col-0 for the number of empty axils, which was significantly higher in the aneuploid individuals. Both the number of triple branches and irregular spacing were higher in the Ap2x, Ap2x*, and aneuploid individuals than in the Col-0 control plants (P-values = 0.042 and 0.0080, respectively). This confirms the observations recorded in the progeny of the Col-0/Wa-1 trisomics and suggests that the meristematic phenotypes observed do not stem from the presence of a hybrid background. This provides strong support to the idea of a long-term effect of parental aneuploidy.
Figure 5.—
Effect of direct and parental aneuploidy on meristematic traits in an all-Col-0 population. An all-Col-0 trisomic of chromosome 3 was selfed and its progeny divided into aneuploids (A, in open bar) and aneuploidy-parented diploid (Ap2x or Ap2x*; see Figure 4 for details). Each individual was scored for meristematic phenotypes, and the mean trait values of the different subpopulations were compared to each other as well as to a set of 15 control Col-0 plants (WT solid bar) on a pair-wise basis using Student's t-tests. Different letters above two columns indicate significantly different means (P-values < 0.01 to compensate for multiple testing on six independent populations) for these two measurements. Standard errors are indicated.
To further verify that the Ap2x individuals were not segmental aneuploids, two of them were karyotyped using whole-genome sequencing. The individuals selected for this purpose were among the progeny of Ap2x*4, an individual that was phenotypically wild type but that produced at least one progeny exhibiting phenotypes that did not conform to the wild type (Figure 4). Among the 20 individuals analyzed in the progeny of Ap2x*4, two (#10 and #20) were selected for whole-genome sequencing on the basis of their high incidence of meristematic abnormalities (see Figure 4 for details). We reasoned that, if segmental aneuploidy were present in Ap2x*4 and its progeny, these individuals would be most likely to carry them. The lack of secondary aneuploidy in these progeny and the persistence of meristematic abnormalities indicate a grandparental effect of aneuploid ancestry.
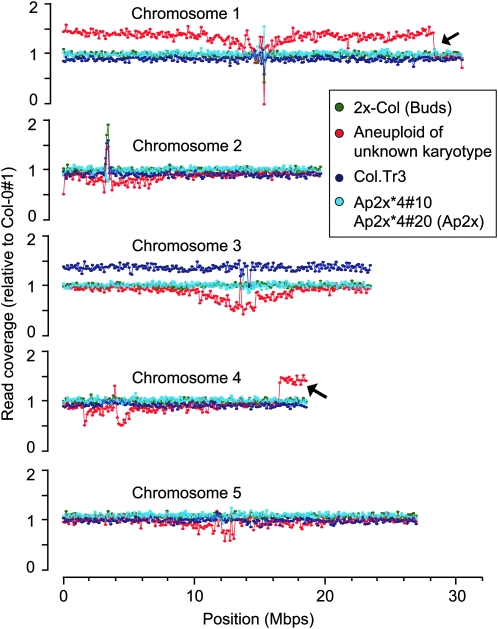
Two diploid Col-0 controls, the ColTr.3 trisomic individual and an aneuploid individual of unknown karyotype, were also subjected to whole-genome sequencing. Between ∼1.4 million and 3.6 million reads that matched unambiguously and perfectly to the A. thaliana TAIR 9.0 genomic sequence were obtained from each of the six individuals. Reads were pooled within bins of 100,000 bp along all five chromosome types, and coverage was measured as the number of reads mapping to each bin. Read counts in each bin were normalized to those obtained for one of the diploid Col-0 individuals (Col-0 #1), such that the coverage of all chromosomes from diploid individuals oscillated around 1.0 (Figure 6). For aneuploid individuals, chromosome coverage is expected to oscillate around the rCHr value described previously. The second diploid Col-0 control exhibited no deviation from the expected two copies of each chromosome. Similarly, no aneuploidy could be detected in the two Ap2x individuals (Ap2x*4#10 or Ap2x*4#20). The karyotype of the ColTr.3 individual was confirmed. Indeed, for a trisomic individual, rChr = 0.91 for the disomic chromosomes and rChr = 1.36 for the trisomic chromosome. This is consistent with what we observed for Col.Tr.3: the average read coverage of the four disomic chromosomes was between 0.9062 and 0.9135, and the average read coverage for chromosome 3 was 1.3595. Finally, the coverage of the aneuploid of unknown karyotype was interesting in several ways. This aneuploid individual had been produced by a Col-0 × Wa-1 cross. It therefore carried both Col-0 and Wa-1 genomes, which resulted in much more variable relative coverage curves, especially in the areas surrounding the centromeres. This was presumably caused by a higher frequency of SNPs in these regions, leading to a lower number of perfectly matched reads when aligning Wa-0 sequences to the Col-0 reference genome. Nevertheless, this variability was not important enough to mask variation in chromosome number. Indeed, trisomy of chromosome 1 for this individual was easily identified by the higher average read coverage for that chromosome (Figure 6). Moreover, our data suggest that the terminal 2.1 Mbp of one of the copies of chromosome 1 had been replaced by a fragment of similar size from the terminal end of chromosome 4 (arrows in Figure 6). Investigating the mechanisms leading to this translocation is beyond the goal of this report, but its detection perfectly illustrates that changes in copy number of small chromosomal fragments, <2% of the genome in this case, are readily identifiable using this method.
Figure 6.—
Karyotyping using whole-genome sequencing. Genomic DNA from six individuals was prepared for whole-genome sequencing: diploid Col-0 #1 (used to normalize data from the other individuals), diploid Col-0 #2 (control), ColTr.3 (presumed trisomic of chromosome 3, Col-0 genotype), two of the progeny of the Ap2x*4 individual, and finally an aneuploid of unknown karyotype carrying Col-0 and Wa-1 chromosomes. The number of reads obtained were pooled by bins of 100,000 bp and counted. Numbers of reads per bin were normalized to the Col-0 #1 control. For ease of visualization, bin coverage was expressed such that the average bin coverage = 1. Chromosomes or chromosomal fragments present in more or less than two copies would therefore deviate from this average. Arrows point at two examples of such deviations.
What is the pattern of inheritance of the meristematic phenotypes?
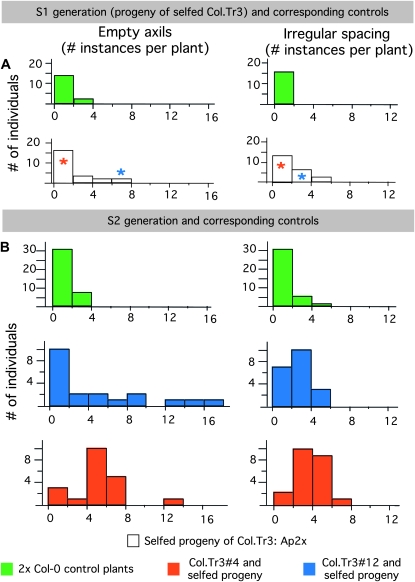
To determine whether the severity of a given meristematic phenotype was stably inherited from one plant by its progeny, we recorded the number of empty axils and irregular spacings in the Ap2x* individuals (first selfing generation from the Col.Tr3 trisomic; see Figure 4) as well as in 20 individuals from the selfed progeny of two of Ap2x* (S2 generation). Both for the Ap2x* and for their progeny, a set of diploid Col-0 plants were grown alongside as controls. For both traits, the number of instances in the S2 populations was higher than for the control Col-0 populations (Figure 7B). Both Ap2x#4, which exhibited a low number of meristematic abnormalities, and Ap2x#12, which exhibited a high number of meristematic abnormalities (see the asterisks in Figure 7A), produced progeny with high and low numbers of abnormalities (Figure 7B). These results suggest that the number of instances of abnormalities recorded in the S1 plants was not predictive of the number of instances of abnormalities in the corresponding S2 plants.
Figure 7.—
Pattern of inheritance of two meristematic traits upon selfing. Distributions of the number of instances of two meristematic traits (empty axils and irregular spacing) in two subsequent selfing generations of Ap2x plants. (A) Distributions in the Ap2x individuals that originated from the selfed Col.Tr3 (white bars) and in control plants grown simultaneously (green bars). Red and blue asterisks indicate the number of instances of empty axils and irregular spacing in Col.Tr3#4 and in Col.Tr3#12, respectively. (B) Distributions in the selfed progeny of two of the Ap2x individuals (red, progeny of Col.Tr3#4; blue, progeny of Col.Tr3#12). For both Ap2x individuals, 20 progeny were grown along with 20 control individuals (green bars).
Does the parental origin of aneuploidy affect the phenotype of the progeny?
To address this question, the phenotype of individuals of the same karyotype, but differing in the origin of the aneuploidy (maternal or paternal), needed to be compared. Due to a high number of possible aneuploid karyotypes, only two karyotypic classes contained enough individuals for this purpose: trisomics of chromosome 2 (N = 21) and trisomics of chromosome 5 (N = 16). Each group of trisomics was divided into two pools, depending on whether the trisomic chromosome had been inherited through the ovule (individuals produced from CWW × Col-0 crosses) or the pollen (individuals produced from Col-0 × CWW crosses). Trisomics from both groups were selfed, and percentages of plump seeds produced were recorded. Mean percentages of plump seeds were compared between individuals with maternal and paternal aneuploidy. For both karyotypes tested, trisomics for which the extra chromosomal copy had been inherited through the ovule produced a lower percentage of plump seed than trisomics that had inherited the extra chromosomal copy through the pollen. For trisomics of chromosome 5, this effect was not significant (85.46% vs. 90.0% plump seed; Student's t-test P-value = 0.31) but it was significant for trisomics of chromosome 2 (65.6% vs. 96.2% plump seed; Student's t-test P-value = 0.0078). This parent-of-origin effect on the fertility of the next generation suggests epigenetic effects capable of influencing plant performance and seed fitness during reproduction. Future investigations of multiple phenotypes on larger populations will be required to test this hypothesis.
DISCUSSION
We have analyzed phenotypic syndromes in populations of A. thaliana aneuploids and investigated the correlation of specific phenotypes to the relative dosage of each chromosome type. The relatively small number of observed phenotypes suggests that they might be caused by the imbalance of only a few dosage-sensitive gene products.
Taken together, our results suggest that the mechanisms underlying aneuploid syndromes in A. thaliana are similar to those operating in Down syndrome patients: similar phenotypes are observed in individuals of the same karyotype, but the severity of the phenotype varies from individual to individual. The mechanism behind the observed variation in phenotype severity between individuals carrying the same karyotype remains unclear. In the case of our population, different growth conditions could contribute to this effect. For example, some of the plants were grown in a growth chamber and others in a greenhouse. In addition, we cannot exclude the possibility of karyotype mosaicism in some of the plants, i.e., that not all cells of the individuals carry the same chromosome number. This type of situation is thought to be responsible for some of the phenotypic variation observed in aneuploid syndromes in humans (Papavassiliou et al. 2009), but it is unclear whether such mosaicism exists in plants. In our populations of aneuploid individuals, we did not observe any phenotype that could be identified as a clear example of cellular mosaicism.
Specific phenotypes were linked to the relative dosage of specific chromosome types (Tables 4 and 5 and Figure 2), even in the context of a highly heterogeneous population of karyotypes in the population (Table 3). Most traits were strongly associated with a single chromosome type. These results contrast with previous data suggesting that multiple aneuploidies each have small effects on the same traits (Birchler et al. 2001; Birchler 2010). For example, a series of growth-related traits were quantified in a dosage series of 18 of the 20 chromosomal arms of maize (Lee et al. 1996). All traits investigated in this study were affected by most of the chromosome arms (13 of 18 on average). The nature of the traits studied may be responsible for the difference between these results in maize and those reported here. In addition, we focused on the appearance of non-wild-type phenotypes while the traits investigated in maize related to plant vigor (plant height, leaf width, etc.) and reproductive potential (days to anthesis, etc.), which had been previously reported as quantitatively inherited (Lee et al. 1996). Nevertheless, our measurement of rosette size, which more closely matches the type of trait investigated in maize, was also affected by the dosage of specific chromosome types rather than aneuploidy as a general phenomenon. Similarly, we found that stem diameter was also associated with specific chromosome types and that the relative doses of these chromosomes were good predictors of stem diameter, irrespective of the dosage of the other chromosome types (Figure 2). Moreover, aneuploid individuals exhibited stem diameters that were both smaller and bigger than those of control plants (data not shown), indicating that aneuploidy could be associated with both reduced and increased trait values, as opposed to the traits investigated in maize, which were all either reduced or unchanged in aneuploid individuals (Lee et al. 1996). The mechanisms behind this differential response remain to be investigated. Maize differs from Arabidopsis by being both a highly domesticated species and an outcrosser. Unknown genetic features related to these characteristics might underlie the difference.
Toward the identification of specific gene products:
Studies in both plants (Huettel et al. 2008; Makarevitch et al. 2008) and humans (FitzPatrick 2005 and references therein) have demonstrated that, in trisomic individuals, the overall expression level of genes located on the triplicated chromosome is increased according to gene dosage. However, these studies have also highlighted many cases of dosage compensation as well as secondary dosage effects resulting in altered expression of genes located on the other chromosome types. The genes responsible for aneuploid syndromes are not easily inferred from such gene expression data. Furthermore, recent studies in maize have demonstrated that changes in gene expression due to aneuploidy were tissue-specific and changed across developmental stages (Makarevitch and Harris 2010). Analysis of candidate genes will therefore require an in-depth analysis of gene expression both temporally and spatially.
The data presented here do not allow identification of candidate genes, but specific hypotheses can be put forth. For example, two of the phenotypes analyzed exhibited an interesting pattern: the number of empty axils was associated with the relative dosage of chromosomes 3 and 5, in both the qualitative (Table 4) and the quantitative (see results) analyses performed. Similarly, the presence of “triple branches” was associated with the relative dosage of chromosomes 3 and 5 but in the opposite direction. Considering the nature of these phenotypes, it is possible that they are associated with the same factors and that variations in the dosage of these two chromosome types determine their incidence. In Arabidopsis, the class III homeodomain leucine zipper transcription (HD-ZIP III) factors function as polarity determinants (Husbands et al. 2009) and as a determinant of the fate of the apical meristem (Smith and Long 2010). Among them, REVOLUTA (REV) is located on chromosome 5. Plants carrying a loss-of-function mutation in REV (rev-1) exhibit a high frequency of empty axils (Talbert et al. 2002); the action of REV is dosage-sensitive (Ann J. Slade, personal communication); and REV is regulated by the microRNA MIR165/166 family, one of which, MIR166b, is located on chromosome 3 (Floyd and Bowman 2004; Mallory et al. 2004; Jung and Park 2007; Smith and Long 2010). REV therefore constitutes a plausible candidate for the observation of the empty-axil and triple-branch traits. It will be interesting to determine if the empty-axil phenotype can be rescued in, for example, plants trisomics for chromosome 5 but carrying a mutant allele of REV.
Long-term effect of aneuploidy:
We have observed meristematic abnormalities in the progeny of aneuploid individuals even after genomic balance had been restored in these individuals. One possible explanation is that the observed meristematic abnormalities result from dosage imbalance between the two gametes rather than from aneuploidy per se. We have worked extensively with various interploidy crosses (2x by 4x, 2x by 3x, etc.) and have obtained diploid or triploid individuals from these crosses in previous experiments (Henry et al. 2005, 2006, 2007, 2009). We have never noted irregularities such as those observed in the Ap2x individuals. This suggests that parental whole-genome imbalance is insufficient to produce the observed phenotypic abnormalities.
Our results therefore raise the possibility that aneuploidy may have a long-term effect, visible in subsequent euploid generations, which we have referred to as Ap2x. The mechanism behind the phenotypes of these Ap2x's is unknown, but several observations suggest a role for epigenetic modifications. First, this phenomenon was observed in three independent populations (Figure 4), involving either Col-0 and Wa-1 or only Col-0 genotypes, suggesting that they are reproducible and not linked to possible negative interactions between the Col-0 and Wa-1 genomes. Second, the phenotypes observed in the Ap2x individuals are similar, irrespective of the karyotype and phenotype of the aneuploid parent, suggesting that they are not linked to dosage alterations of specific chromosomal fragments but rather to a more general genomic state associated with aneuploidy. Third, the number of meristematic abnormalities in an individual Ap2x does not predict whether or not that individual's progeny will carry many or few of these abnormalities (Figure 7). Furthermore, plants exhibiting strong meristematic abnormalities early in development sometimes reverted to normal development later on (our unpublished data). Finally, we have observed that fertility of trisomic individuals varied depending on the parental origin of the trisomic chromosome. Taken together, our results suggest that aneuploidy might result in epigenetic modifications in the aneuploid parent, which are passed on to the next generation but are unstable.
Similar dosage-sensitive epigenetic phenomena have been reported previously. For example, in tobacco, a transgene was spontaneously methylated when present on the triplicated chromosome of an aneuploid line (Papp et al. 1996). A change in gene dosage caused by polyploidy, even without aneuploidy, has also been shown to result in changes in epigenetic silencing of a specific transgene (Mittelsten Scheid et al. 1996). The fact that similar phenotypes are observed in the Ap2x individuals, irrespective of the origin or type of parental aneuploidy, suggests that a similar set of genes is targeted or perhaps that the actions of the genes associated with these phenotypes are more sensitive to subtle epigenetic changes. A study of fission yeast demonstrated that the additional presence of a minichromosome, even devoid of a protein-coding gene, was sufficient to result in altered binding of the heterochromatic protein Swi6 at the telomeres (Chikashige et al. 2007). This resulted in increased expression of some of the genes located in these regions. Imbalance in chromosome number can thus alter chromatin structure, possibly via a change in the ratio of heterochromatic DNA to the enzymes responsible for the establishment and/or maintenance of the heterochromatic state. An in-depth analysis of the epigenome of some of the Ap2x individuals might help to shed light on the mechanisms underlying this potential long-term effect of aneuploidy.
The power of whole-genome sequencing as a karyotyping and cytogenetic tool:
Our data (Figure 6) confirm the power of whole-genome sequencing as a cytogenetics tool. Trisomy was detected with high reliability, as was a translocation event covering <2% of the genome (2.1 Mbp). Our data indicate that deletions or duplications of much smaller fragments would be easily detectable as well and that genomes of up to 100 individuals could be analyzed on a single Illumina flow cell lane, thereby drastically reducing the cost per individual.
Interestingly, the aneuploid individual karyotyped using whole-genome sequencing (in red in Figure 6) and determined to be trisomic for all but the tail end of chromosome 1 exhibited phenotypes consistent with chromosome 1 trisomy (dark green leaves, shorter leaves, shorter stature), but the plant overall was much sturdier than other trisomics of chromosome 1 (data not shown). This raises the possibility that the stunting associated with trisomy of chromosome 1 could be linked to the tail end of chromosome 1. The combination of whole-genome sequencing and the possibility of obtaining partial aneuploids or aneuploids containing chromosome abnormalities such as the putative translocation observed in aneuploid #1 raise the possibility of an in-depth mapping of loci linked to specific phenotypes. Indeed, studies similar to those performed on humans (Korbel et al. 2009; Lyle et al. 2009), in which phenotypes are correlated with karyotypes using mapping of chromosomal dosage, could easily be performed here on much larger populations and encompassing variation in the dosage of all chromosome types.
Acknowledgments
We thank the Biology Greenhouse (Biology Department, University of Washington) for material support; the University of California at Davis Genome Center Analysis Core for whole-genome sequencing services; Jennifer Monsoon-Miller for technical advice; and Kathie Ngo for assistance with the bioinformatics analysis of the whole-genome sequencing data. We also thank Ravi Maruthachalam and Simon Chan for providing us with aneuploid material.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.121079/DC1.
References
- Antonarakis, S. E., R. Lyle, E. T. Dermitzakis, A. Reymond and S. Deutsch, 2004. Chromosome 21 and Down syndrome: from genomics to pathophysiology. Nat. Rev. Genet. 5 725–738. [DOI] [PubMed] [Google Scholar]
- Birchler, J. A., 2010. Reflections on studies of gene expression in aneuploids. Biochem. J. 426 119–123. [DOI] [PubMed] [Google Scholar]
- Birchler, J. A., U. Bhadra, M. P. Bhadra and D. L. Auger, 2001. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev. Biol. 234 275–288. [DOI] [PubMed] [Google Scholar]
- Blakeslee, A., 1921. The globe mutant in the Jimson weed (Datura stramonium). Genetics 6 241–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee, A., 1922. Variation in Datura due to changes in chromosome number. Am. Natur. 56 16–31. [Google Scholar]
- Chikashige, Y., C. Tsutsumi, K. Okamasa, M. Yamane, J. Nakayama et al., 2007. Gene expression and distribution of Swi6 in partial aneuploids of the fission yeast Schizosaccharomyces pombe. Cell Struct. Funct. 32 149–161. [DOI] [PubMed] [Google Scholar]
- Dear, P. H., 2009. Copy-number variation: The end of the human genome? Trends Biotechnol. 27 448–454. [DOI] [PubMed] [Google Scholar]
- Doyle, G., 1986. Aneuploidy and inbreeding depression in random mating and self-fertilization autotetraploid populations. Theor. Appl. Genet. 72 799–806. [DOI] [PubMed] [Google Scholar]
- FitzPatrick, D., 2005. Transcriptional consequences of autosomal trisomy: primary gene dosage with complex downstream effects. Trends Genet. 21 249–253. [DOI] [PubMed] [Google Scholar]
- Floyd, S. K., and J. L. Bowman, 2004. Gene regulation: ancient microRNA target sequences in plants. Nature 428 485–486. [DOI] [PubMed] [Google Scholar]
- Hassold, T. J., and P. Hunt, 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2 280–291. [DOI] [PubMed] [Google Scholar]
- Henrichsen, C. N., N. Vinckenbosch, S. Zollner, E. Chaignat, S. Pradervand et al., 2009. Segmental copy number variation shapes tissue transcriptomes. Nat. Genet. 41 424–429. [DOI] [PubMed] [Google Scholar]
- Henry, I. M., B. P. Dilkes, K. Young, B. Watson, H. Wu et al., 2005. Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics 170 1979–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, I. M., B. P. Dilkes and L. Comai, 2006. Molecular karyotyping and aneuploidy detection in Arabidopsis thaliana using quantitative fluorescent polymerase chain reaction. Plant J. 48 307–319. [DOI] [PubMed] [Google Scholar]
- Henry, I. M., B. P. Dilkes and L. Comai, 2007. Genetic basis for dosage sensitivity in Arabidopsis thaliana. PLoS Genet. 3 e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, I. M., B. P. Dilkes, A. P. Tyagi, H. Y. Lin and L. Comai, 2009. Dosage and parent-of-origin effects shaping aneuploid swarms in A. thaliana. Heredity 103 458–468. [DOI] [PubMed] [Google Scholar]
- Holland, A. J., and D. W. Cleveland, 2009. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel, B., D. P. Kreil, M. Matzke and A. J. Matzke, 2008. Effects of aneuploidy on genome structure, expression, and interphase organization in Arabidopsis thaliana. PLoS Genet. 4 e1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands, A. Y., D. H. Chitwood, Y. Plavskin and M. C. Timmermans, 2009. Signals and prepatterns: new insights into organ polarity in plants. Genes Dev. 23 1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iltis, H. H., 2000. Homeotic sexual translocations and the origin of maize (Zea mays, poaceae): a new look at an old problem. Econ. Bot. 54 7–42. [Google Scholar]
- Johnsson, H., 1945. The triploid progeny of the cross diploid x tetraploid Populus tremula. Hereditas 31 411. [DOI] [PubMed] [Google Scholar]
- Jung, J. H., and C. M. Park, 2007. MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta 225 1327–1338. [DOI] [PubMed] [Google Scholar]
- Khush, G., 1973. Cytogenetics of Aneuploids. Academic Press, New York.
- Koornneef, M., and J. H. Van der Veen, 1983. Trisomics in Arabidopsis thaliana and the location of linkage groups. Genetica 61 41–46. [Google Scholar]
- Korbel, J. O., T. Tirosh-Wagner, A. E. Urban, X. N. Chen, M. Kasowski et al., 2009. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc. Natl. Acad. Sci. USA 106 12031–12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E. A., L. L. Darrah and E. H. Coe, 1996. Dosage effects on morphological and quantitative traits in maize aneuploids. Genome 39 898–908. [DOI] [PubMed] [Google Scholar]
- Lee-Chen, S., and L. Steinitz-Sears, 1967. The location of linkage groups in Arabidopsis thaliana. Can. J. Genet. Cytol. 9 381–384. [Google Scholar]
- Levan, A., 1942. The effect of chromosomal variation in sugar beets. Hereditas 28 345–399. [Google Scholar]
- Lyle, R., F. Bena, S. Gagos, C. Gehrig, G. Lopez et al., 2009. Genotype-phenotype correlations in Down syndrome identified by array CGH in 30 cases of partial trisomy and partial monosomy chromosome 21. Eur. J. Hum. Genet. 17 454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch, I., and C. Harris, 2010. Aneuploidy causes tissue-specific qualitative changes in global gene expression patterns in maize. Plant Physiol. 152 927–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch, I., R. L. Phillips and N. M. Springer, 2008. Profiling expression changes caused by a segmental aneuploid in maize. BMC Genomics 9 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A. C., B. J. Reinhart, M. W. Jones-Rhoades, G. Tang, P. D. Zamore et al., 2004. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 23 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke, M. A., M. F. Mette, T. Kanno and A. J. Matzke, 2003. Does the intrinsic instability of aneuploid genomes have a causal role in cancer? Trends Genet. 19 253–256. [DOI] [PubMed] [Google Scholar]
- McClintock, B., 1929. A cytological and genetical study of triploid maize. Genetics 14 180–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelsten Scheid, O., L. Jakovleva, K. Afsar, J. Maluszynska and J. Oaszkowski, 1996. A change of ploidy can modify epigenetic silencing. Proc. Natl. Acad. Sci. USA 93 7114–7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Doherty, A., S. Ruf, C. Mulligan, V. Hildreth, M. L. Errington et al., 2005. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science 309 2033–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavassiliou, P., T. P. York, N. Gursoy, G. Hill, L. V. Nicely et al., 2009. The phenotype of persons having mosaicism for trisomy 21/Down syndrome reflects the percentage of trisomic cells present in different tissues. Am. J. Med. Genet. A 149A 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp, I., V. A. Iglesias, E. A. Moscone, S. Michalowski, S. Spiker et al., 1996. Structural instability of a transgene locus in tobacco is associated with aneuploidy. Plant J. 10 469–478. [DOI] [PubMed] [Google Scholar]
- Patterson, D., 2007. Genetic mechanisms involved in the phenotype of Down syndrome. Ment. Retard. Dev. Disabil. Res. Rev. 13 199–206. [DOI] [PubMed] [Google Scholar]
- Patterson, D., 2009. Molecular genetic analysis of Down syndrome. Hum. Genet. 126 195–214. [DOI] [PubMed] [Google Scholar]
- Pihan, G., and S. J. Doxsey, 2003. Mutations and aneuploidy: Co-conspirators in cancer? Cancer Cell 4 89–94. [DOI] [PubMed] [Google Scholar]
- Ramsey, J., and D. W. Schemske, 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29 467–501. [Google Scholar]
- Ramsey, J., and D. W. Schemske, 2002. Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33 589–639. [Google Scholar]
- Randolph, L., 1935. Cytogenetics of tetraploid maize. J. Agric. Res. 50 591–605. [Google Scholar]
- Ravi, M., and S. W. Chan, 2010. Haploid plants produced by centromere-mediated genome elimination. Nature 464 615–618. [DOI] [PubMed] [Google Scholar]
- Satina, S., and A. Blakeslee, 1938. Chromosome behavior in triploid Datura. III. The seed. Am. J. Bot. 25 595–602. [Google Scholar]
- Singh, R., 2003. Plant Cytogenetics. CRC Press, Boca Raton, FL.
- Smith, Z. R., and J. A. Long, 2010. Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464 423–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinitz-Sears, L., 1962. Chromosome studies in Arabidopsis thaliana. Genetics 48 483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinitz-Sears, L., and S. Lee-Chen, 1970. Cytogenetic studies in Arabidopsis thaliana. Can. J. Genet. Cytol. 12 217–223. [Google Scholar]
- Storchova, Z., and D. Pellman, 2004. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 5 45–54. [DOI] [PubMed] [Google Scholar]
- Talbert, P. B., R. Masuelli, A. P. Tyagi, L. Comai and S. Henikoff, 2002. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizir, I., and B. Mulligan, 1999. Genetics of gamma-irradiation-induced mutations in Arabidopsis thaliana: large chromosomal deletions can be rescued through the fertilization of diploid eggs. J. Hered. 90 412–417. [DOI] [PubMed] [Google Scholar]
- Williams, B. R., and A. Amon, 2009. Aneuploidy: Cancer's fatal flaw? Cancer Res. 69 5289–5291. [DOI] [PMC free article] [PubMed] [Google Scholar]