Abstract
Slk19p is a member of the Cdc-14 early anaphase release (FEAR) pathway, a signaling network that is responsible for activation of the cell-cycle regulator Cdc14p in Saccharomyces cerevisiae. Disruption of the FEAR pathway results in defects in anaphase, including alterations in the assembly and behavior of the anaphase spindle. Many phenotypes of slk19Δ mutants are consistent with a loss of FEAR signaling, but other phenotypes suggest that Slk19p may have FEAR-independent roles in modulating the behavior of microtubules in anaphase. Here, a series of SLK19 in-frame deletion mutations were used to test whether Slk19p has distinct roles in anaphase that can be ascribed to specific regions of the protein. Separation-of-function alleles were identified that are defective for either FEAR signaling or aspects of anaphase spindle function. The data suggest that in early anaphase one region of Slk19p is essential for FEAR signaling, while later in anaphase another region is critical for maintaining the coordination between spindle elongation and the growth of interpolar microtubules.
ACCURATE separation of chromatids at anaphase is dependent on both the coordination of cell cycle events through signaling pathways and the assembly of a robust apparatus for separating sister chromatids. The Cdc14p phosphatase is a key regulator of the final stages of mitosis. Cdc14p activity modulates the structure and behavior of the anaphase spindle (Pereira and Schiebel 2003; Woodbury and Morgan 2007) and is necessary for exit from mitosis and entry into the next cell cycle (Visintin et al. 1998). During early parts of the cell cycle Cdc14p is sequestered in the nucleolus; two signaling pathways modulate its release. At the onset of anaphase the FEAR network (Cdc fourteen early anaphase release) triggers a nonessential, transient release of Cdc14p (Stegmeier et al. 2002) predominantly into the nucleus. In late anaphase, the mitotic exit network (MEN) is activated, triggering a complete release of Cdc14p from the nucleolus into the nucleus and cytoplasm, and subsequent exit from mitosis (Shou et al. 1999; Visintin et al. 1999).
The FEAR network includes a number of proteins required for the timely release of Cdc14p from the nucleolus (reviewed in Stegmeier and Amon 2004; Queralt and Uhlmann 2008). These include Esp1p/Separase (Stegmeier et al. 2002; Sullivan and Uhlmann 2003; Stegmeier and Amon 2004; Queralt et al. 2006), Spo12p, a protein of unknown function (Stegmeier et al. 2002; Tomson et al. 2009), and Slk19p (Stegmeier et al. 2002; Sullivan and Uhlmann 2003). Slk19p and Esp1p physically interact, Slk19p is a cleavage substrate of Esp1p, and Slk19p is necessary for Esp1p to perform its role in FEAR (Sullivan et al. 2001; Sullivan and Uhlmann 2003; Tomson et al. 2009). Cdc14p is necessary for multiple functions in cell division including the localization of several proteins to the spindle midzone (e.g., Sli15p–Ipl1p, Ase1p, and Cin8p) (Pereira and Schiebel 2003; Khmelinskii et al. 2007). These proteins act to modify spindle behavior by reducing tubulin turnover, stabilizing the spindle midzone to prevent collapse of the spindle as it elongates, and providing force generation for spindle elongation (Bouck and Bloom 2005; Khmelinskii et al. 2007; Kotwaliwale et al. 2007). Thus, because of the defect in localizing these proteins to the spindle midzone, FEAR mutants exhibit defects in anaphase spindle behavior.
SLK19 was first identified in a screen for genes necessary for growth in budding yeast strains lacking KAR3, a gene encoding a minus-end directed microtubule motor (Zeng et al. 1999). Kar3p localizes to microtubule plus ends and promotes depolymerization of microtubules in vitro (Manning et al. 1999; Zeng et al. 1999). Slk19–GFP was found to localize to the kinetochores before anaphase and to both the spindle midzone and kinetochores during anaphase (Zeng et al. 1999). Strains lacking SLK19 exhibit multiple spindle defects including short mitotic spindles, increased numbers of astral microtubules, and fragile anaphase spindles (Zeng et al. 1999; Sullivan et al. 2001; Gardner et al. 2008). These and other data suggest that Slk19p is a plus-end microtubule stabilizer (Zeng et al. 1999; Movshovich et al. 2008).
The fact that the abrogation of the FEAR pathway leads to aberrant anaphase spindle dynamics (Pereira and Schiebel 2003; Higuchi and Uhlmann 2005; Khmelinskii et al. 2007, 2009) raises the question of whether the spindle-related defects of slk19 deletion mutants are due to a loss of only FEAR signaling or to a loss of both FEAR signaling and additional functions. The results from a number of experiments touch on, but do not resolve, this issue. Localization of Esp1p and Slk19p to the anaphase spindle midzone is interdependent and both are required for midzone localization of the spindle stabilizing protein, Ase1p (Khmelinskii et al. 2007). However, induction of pGAL1–CDC14, which leads to dephosphorylation of Ase1p and stabilization of anaphase interpolar microtubule (ipMT) turnover (Higuchi and Uhlmann 2005), is insufficient to rescue midzone organization of Ase1p in esp1 or slk19 localization mutants (Khmelinskii et al. 2007). A nonphosphorylatable form of Ase1p (one that does not require Cdc14p to be dephosphorylated) can rescue the midzone assembly defects of slk19Δ mutants, but not their spindle stability defects. These findings suggested to Schiebel and colleagues that the midzone assembly defects of SLK19 mutants are due to loss of FEAR (Khmelinskii and Schiebel 2008) and raised the possibility that the spindle stability defects of SLK19 mutants are due to loss of a FEAR-independent function. This work tests these ideas.
We have constructed SLK19 separation-of-function alleles, which were used to dissect the roles of Slk19p in anaphase. This analysis demonstrates, first, that Slk19p has roles in anaphase beyond the FEAR pathway, and second, that one region of the protein is essential for FEAR signaling and a separate region is essential for anaphase spindle dynamics. Slk19p was found to have roles, independent of FEAR, in promoting a transition between fast and slow anaphase B spindle elongation and in coordinating the length of interpolar microtubules with the length of the elongating anaphase spindle. Therefore, Slk19p participates in two mechanisms to stabilize and strengthen the anaphase spindle: the FEAR pathway and a separate process that affects the zone of overlap of antiparallel ipMTs.
MATERIALS AND METHODS
Yeast strains and media:
Strains used for the majority of experiments were derivatives of S288C (dc48-5.1c and dc49-7.1c are the parent strains) (Nicolas et al. 1989); anaphase elongation experiments were performed both in this strain background and a separate strain that was a gift from Mike Dresser (X strain) (Dresser et al. 1994). Strains and genotypes are listed in Table S1. Synthetic complete medium, YPAD, and YPAc were prepared as described in Amberg et al. (2005).
Gene disruption, truncation, and fusions:
In-frame slk19 deletion alleles A through F were obtained using a PCR-based mutagenesis approach; the template used was a pRS416 vector (Sikorski and Hieter 1989) containing the SLK19 ORF with 895 bp of the promoter region and a 3′ GFP:KANMX tag. Deletion derivatives of the parent plasmid were sequenced. A total of 770 bp of the 3′-UTR was added downstream of the KANMX tag using in vivo cloning (Oldenburg et al. 1997). This allowed the SalI and DraIII digestion product of these plasmids with deletion alleles to be targeted to the chromosome, where it replaced a slk19∷URA3 allele, using the lithium acetate transformation method described in Gietz and Woods (2002). The GFP:KANMX tag was replaced with the pRS404 (Christianson et al. 1992) TRP1 allele through transformation of a PCR product. These plasmids were also used as templates for PCR-mediated gene replacement of the slk19∷URA3 allele for isolation of untagged alleles. To build slk19–ΔG∷TRP1, with pRS406 as the template for PCR-mediated gene replacement, we integrated a premature stop codon and the TRP1 ORF at the 3′ end of SLK19. This template was also used for TRP1 replacement of slk19 and spo12 ORFs.
To construct SLK19–GFP∷TRP1 alleles a single C-terminal GFP fusion of wild-type SLK19 was created using a PCR product from pFA6a–GFP–TRP1 (Longtine et al. 1998). Next a URA3 cassette was inserted into the SLK19–GFP ORF. SLK19 deletion derivatives were obtained by transforming this strain with PCR products corresponding to SLK19 deletion alleles and selecting for 5-FOA resistant transformants. slk19–ΔG–GFP was isolated with a single transformation of a PCR product using the GFP∷TRP1 cassette.
The kar3∷loxP–KANMX–loxP allele was described previously (Shanks et al. 2004), and MR820[KAR3, URA3, CEN4] was a gift from M. Rose. The bim1∷KAN and cla4∷KAN alleles were generated with PCR-mediated gene replacement using pFA6a–KANMX6 (Longtine et al. 1998).
pRK44 is a cen plasmid expressing SLK19 with 895 bp of promoter region, created through PCR-based in vivo cloning into BamHI–NotI digested pRS416 (Sikorski and Hieter 1989).
SPC42–mCHERRY∷HIS3 was created with a PCR product using the template pKT355 (a gift from K. Thorn). PHIS3–mCHERRY–TUB1∷URA3 was integrated into the URA3 locus after ApaI digestion of pAK011 (a gift from E. Schiebel). The GFP–TUB1∷URA3 cassette was introduced transforming strains with StuI digestion of pAFS125 (a gift from A. Straight). A strain expressing SPC42–DsRED∷URA3, X400, was a gift from M. Conrad and was backcrossed into slk19 and spo12 deletion strains.
The KanMX–PGAL1NLS–slk19709-822–GFP∷TRP1 locus was created by transforming a strain with a SLK19–GFP∷TRP1 locus with a KanMX–PGAL1NLS PCR product. pFA6a–KAN–PGAL was used as a template and the downstream primer contained the NLS sequence GGCAACCTTTCTCTTCTTCTTTGGTGGAGTACA along with homology to SLK19 starting at bp +2127.
To make a CFP–SLK19 allele TKH16 was transformed with a PCR product using template pBS5 from the Yeast Resource Center, resulting in loxP∷KAN∷loxP immediately upstream of CFP–SLK1977–822. The majority of this selection cassette was removed (and the original 5′ sequence restored) with CRE recombinase. This method results in one loxP site remaining between the original promoter and the CFP allele.
Sporulation:
Sporulation was induced by first growing diploid strains to a density of 5 × 107 cells/ml at 30° in YPAD medium. Cells were then washed once in sporulation medium (1% potassium acetate supplemented with 1 mg/ml adenine) and resuspended in sporulation medium at the same density. Sporulation efficiency and the frequencies of dyads, tryads, and tetrads were determined with a light microscope.
Synthetic lethality assay:
To determine synthetic lethality, haploid strains were grown to stationary phase in YPAD medium, allowing for loss of the URA3-expressing plasmid. Tenfold serial dilutions in ddH2O were prepared in a microtiter dish, ending with a final concentration of two cells/μl. Using a 48-pin “frogger,” the cells were then transferred to both YPAD plates and plates containing 5-FOA. Using a similar method, strains were streaked to single colonies on YPAD plates and replica plated onto 5-FOA-containing plates.
Imaging of live cells (Slk19p–GFP localization analysis):
Images of Slk19–GFP fusions in live cells were obtained after cells at a density of 0.5 × 107 cells/ml had been arrested in 20 μg/ml α-factor (dissolved in 0.1 n HCL) for 3 hr and then released into SC medium. A total of 3 μl of concentrated cells were placed onto a coverslip, and then a thin (∼1 mm) 1% agarose pad was placed over the cells and pressure applied with a Kimwipe to both remove excess moisture and produce a single layer of cells below the pad. The coverslip was then inverted and placed over a rubber gasket on a glass slide, creating an enclosed chamber [method described in Dresser (2009)]. Images were obtained with a Zeiss AxioImager using a ×100 Plan-Apo 1.4 NA objective, a Roper HQ2 CCD, and Axiovision software. At ∼4-min intervals a new field of each strain was imaged in three channels (GFP, mCherry, and DIC) over 5–7 Z-planes, 0.5 μm apart. Of the stacks, the clearest single plane image was used for displaying the localization pattern.
Anaphase elongation rates:
Cells expressing Spc42–DsRed (or GFP–Tub1) were grown to 0.5 × 107 cells/ml, arrested in 20 μg/ml α-factor in SC medium for 2 hr, and loaded into a CellASIC Y2 microfluidic plate. Cells were trapped in 4- to 5-μm thick chambers while fresh SC medium was pumped around them at 4 psi for the duration of the experiment. Time-lapse images were collected every minute from multiple regions using a Nikon Eclipse TE2000 inverted microscope with an enclosed incubator, ×60 Plan-Apo 1.4 NA objective, Perfect Focus System attachment, a Roper HQ2 CCD, a Prior-Proscan II stage controller, and Nikon NES software. The distance between spindle poles over time was measured using OMRFQANT (Kateneva et al. 2005) and analyzed using Excel and Prism.
Relative GFP–Tub1 intensity:
Strains expressing GFP–Tub1 were grown to 0.5 × 107 cells/ml and arrested in 20 μg/ml α-factor in SC medium for 3 hr, then released into SC medium for 30 min before being placed on a coverslip and covered with an agarose pad. Images were collected on a Zeiss AxioObserver inverted microscope using a ×100 Plan-Apo 1.4 NA objective, Hamamatsu Orca-ER camera and controller with a Yokosawa CSU22 spinning disk confocal scanner, using Slidebook imaging software. A Z-series of 15 images 0.25 μm apart was collected with 2 × 2 binning and then a sum intensity projection exported for analysis in Metamorph. To measure tubulin intensity along the spindle, the Metamorph Linescan tool, set at a 5-pixel width, was traced over each anaphase spindle and the values exported to Excel for analysis. Each spindle was normalized to 24 bins in length and then the two halves of the spindle averaged together; spindles with similar length were also averaged.
Modeling GFP–Tub1 distribution:
The method and software used were as described previously (Gardner et al. 2007, 2008). Briefly, images were collected using the same Zeiss Axiovert spinning-disk apparatus described above, but with no binning, and from strains expressing both GFP–Tub1 and DsRed–Spc42. The DsRed signal was used to define the position of the SPBs. GFP intensity distributions were measured using custom MATLAB software, which also normalized the length of each spindle to 24 bins. Kinetochore microtubule (kMT) length distributions were fit to an exponential model as previously described. The distribution of interpolar microtubule (ipMT) lengths were fit to a Gaussian distribution, such that the mean and the standard deviation of the length distribution were free-fitting parameters and were adjusted to match experimental GFP–Tub1 fluorescence distributions for wild-type and mutant spindles.
Model-convolution simulations:
Model convolution of simulated fluorescence distributions was completed by convolving the experimentally observed microscope point spread function with the simulated distribution of fluorescent proteins, as previously described. The convolution algorithm was applied to simulated spindles, and then a custom MATLAB measurement package was used to determine the GFP–Tub1 distribution in the simulated images. A total of 360 model spindles were evaluated for each of the 17 wild-type and 28 mutant spindle lengths measured experimentally. Profiles between experimental and simulation were compared to determine best-fit models as described previously (Sprague et al. 2003; Gardner et al. 2007).
RESULTS
Synthetic lethality of slk19Δ kar3Δ is not due to the FEAR defect:
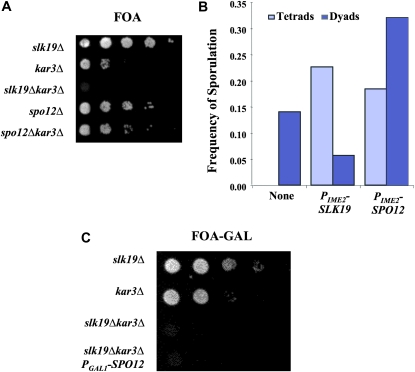
To test whether the synthetic lethality observed between SLK19 and KAR3 deletion alleles is due to the simultaneous loss of FEAR activity and Kar3p function, we asked whether other FEAR mutants are also synthetic lethal with the kar3Δ allele. We created double mutant strains carrying the kar3Δ allele and deletions of either SLK19 or SPO12, which are both required for FEAR activity (Stegmeier et al. 2002); these strains also contained a copy of KAR3 on a plasmid expressing URA3. The viability of slk19Δkar3Δ strains was dependent on the presence of the KAR3 plasmid as demonstrated by the lack of growth on medium containing 5-fluoroorotic acid (5-FOA), which is toxic to cells expressing the URA3 gene product (Figure 1A) (Boeke et al. 1984). spo12Δ kar3Δ strains were viable without the plasmid. Thus, loss of the FEAR pathway in the kar3Δ background is not lethal. In fact, the spo12Δ allele seemed to increase the viability of kar3Δ strains (Figure 1A). The reason for this is not at all clear—perhaps the prolonged time in anaphase or the altered spindle dynamics in FEAR mutants enhances spindle integrity in kar3Δ mutants, increasing the number of mutant cells that can assemble a functional spindle.
Figure 1.—
slk19Δ kar3Δ synthetic lethality is independent of the FEAR pathway. (A) Cells were grown to stationary phase in YPAD. Tenfold serial dilutions in water were spotted onto 5-FOA to test for growth in the absence of the plasmid MR820 [YCp–KAR3∷URA3]. Strains: slk19Δ (TRK27), kar3Δ (TRS107), slk19Δ kar3Δ (DRK212.2D), spo12Δ (DRK215.1D), and spo12Δ kar3Δ (DRK215.4). (B) slk19Δ/slk19Δ strains were sporulated and the number of tetrad and dyad asci determined with a light microscope (n > 100 cells). Strain slk19Δ/slk19Δ (DRK9) was transformed with empty vector pRS424, pRK50 (PIME2–SLK19), or pRK52 (PIME2–SPO12). (C) Cells were grown to stationary phase in YP–GAL, serially diluted 10-fold in water, and spotted onto 5-FOA–GAL medium to test for growth in the absence of the plasmid MR820. Strains: slk19Δ (TRK27), kar3Δ (TRS107), slk19Δ kar3Δ (DRK212.2D), and slk19Δ kar3Δ, PGAL1–SPO12 (TRK201).
To confirm that loss of FEAR activity was not the cause of the slk19Δ kar3Δ synthetic lethality, we tested whether suppression of the FEAR defect could rescue viability of slk19Δ kar3Δ mutants. Overexpression of SPO12 was previously demonstrated to be sufficient for the exit from mitosis when the essential MEN gene, CDC15, is disrupted (Shirayama et al. 1996; Jaspersen et al. 1998), suggesting that SPO12 overexpression might also rescue other Cdc14p release defects. Indeed, expression of SPO12 from a high-copy plasmid with the PIME2 promoter, which is induced early in meiosis (Smith and Mitchell 1989), substantially suppressed the FEAR-related dyad phenotype of slk19Δ mutants; while the slk19Δ mutant produced only dyads, the PIME2–SPO12 plasmid resulted in the production of many tetrads. This rescue was not complete (the slk19Δ PIME2–SPO12 strain did not produce as many tetrads as the slk19Δ strain carrying a complementing SLK19 plasmid), suggesting that the SPO12 overexpression did not completely restore the FEAR pathway or that there is an additional role for SLK19 in tetrad formation beyond FEAR (Figure 1B). While overexpression of SPO12 does provide some FEAR activity (above), overexpression of SPO12 with the PGAL1 promoter did not rescue the slk19Δ kar3Δ synthetic lethality (Figure 1C). Thus, we conclude that although slk19Δ mutants do have a defect in FEAR signaling, this is not the cause of the synthetic lethality of slk19Δ kar3Δ double mutants, and therefore this synthetic lethality must be attributable to some other functional defect in SLK19 deletion mutants.
Slk19p has functionally separable roles in chromosome segregation:
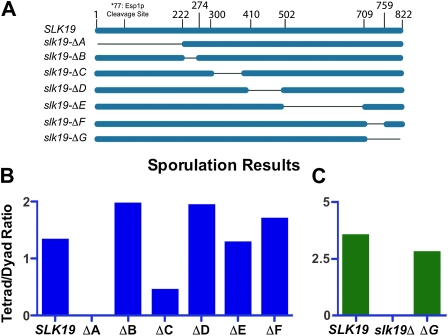
Structural predictions suggest Slk19p is composed of a series of coiled-coil domains and an N-terminal globular domain. We reasoned that loss of specific coiled coils might lead to loss of individual functions of Slk19p while limiting disruption of the other functions. We created a series of in-frame deletions, designated A through G, each eliminating either a predicted coiled-coil domain or the N-terminal globular domain (Figure 2A). These deletions were then tested in two assays, one assessing FEAR network activity and one identifying alleles synthetically lethal with the KAR3 deletion.
Figure 2.—
Only the N-terminal region of Slk19p is necessary for FEAR signaling. (A) A diagram depicting each of five predicted coiled-coil regions (denoted B through F) individually deleted from the endogenous locus, along with the N-terminal globular region (deletion A). (B and C) Homozygous diploid slk19 deletion strains were induced to sporulate and the ratio of tetrads to dyads scored using light microscopy (n > 100 cells). (C) Strains were grown in YP-acetate prior to sporulation. Strains: (B) (mixed parents; dc48-9.1c and dc49-7.1c derivatives) SLK19/SLK19 (DKH60), slk19–ΔA/slk19–ΔA (DKH54), slk19–ΔB/slk19–ΔB (DKH55), slk19–ΔC/slk19–ΔC (DKH56), slk19–ΔD/slk19–ΔD (DKH57), slk19–ΔE/slk19–ΔE (DKH58), and slk19–ΔF/slk19–ΔF (DKH59); (C) (both parents dc49-7.1c derivatives) SLK19/SLK19 (DD812), slk19–ΔG/slk19–ΔG (DD802), and slk19Δ/slk19Δ (DD810).
We tested the ability of these deleted versions of Slk19p to activate the FEAR network by taking advantage of the fact that FEAR mutants form only dyads after meiosis (Klapholz and Esposito 1980; Kamieniecki et al. 2000; Zeng and Saunders 2000; Buonomo et al. 2003; Marston et al. 2003). We created diploid strains homozygous for each deletion allele and scored for the ability of the strain to form tetrads after meiosis. The slk19–ΔG allele was constructed (and tested for FEAR function) after the others, following the demonstration that this region of Slk19p shares homology with the TACC (transforming acidic coiled-coil) domain of Alp7 from Schizosaccharomyces pombe (Sato et al. 2003) (reviewed in Peset and Vernos 2008). Our laboratory strain background (an S288C derivative; Nicolas et al. 1989) exhibited a tetrad-to-dyad ratio >1, while isogenic slk19Δ/slk19Δ diploid strains produced dyads but not tetrads (Figure 2, B and C). Homozygous diploid strains expressing deletions B, D, E, F, and G also produced a ratio >1 after meiosis, indistinguishable from the wild-type (WT) control. These FEAR assays show that large regions of Slk19p corresponding to these deletions B, D, E, F, and G (i.e., much of the carboxyl half of the protein) are not necessary for FEAR activity. Deletion of the globular domain encompassing amino acids 1–202 (i.e., slk19–ΔA) produced only dyads after meiosis, demonstrating a loss of FEAR activity that was indistinguishable from the complete deletion (Figure 2B). Deletion of region C also reduced the FEAR activity somewhat (Figure 2B).
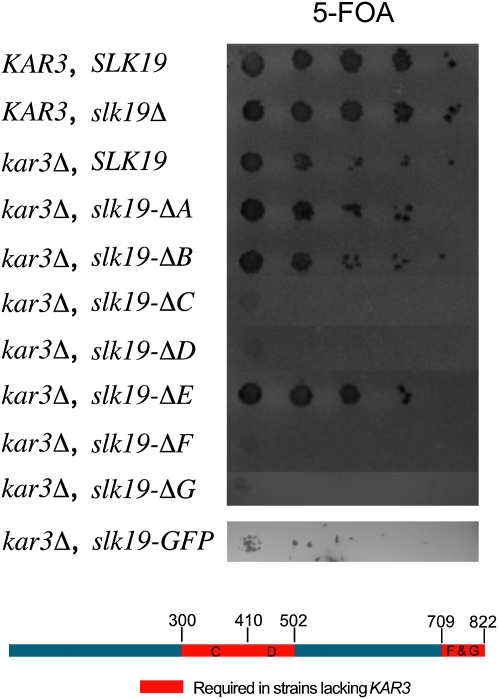
After identifying a region essential for producing a FEAR signal (region A), we wanted to determine which regions of Slk19p are necessary for viability in a kar3Δ background. To test this, we created double mutants lacking endogenous KAR3, expressing one of the SLK19 deletions, and also containing a CEN plasmid carrying the KAR3 and URA3 genes. These strains were then assayed for viability on medium containing 5-FOA. SLK19 alleles lacking regions A, B, or E are not synthetically lethal with a KAR3 deletion, while the slk19–ΔC, slk19–ΔD, slk19–ΔF, and slk19–ΔG alleles are all synthetically lethal with kar3Δ (Figure 3). Thus region A, while essential for a FEAR signal, is not required for viability in the kar3 deletion background. Conversely, the central D region, and the C-terminal F and G regions, are dispensable for FEAR signaling but essential for viability if KAR3 is deleted. Fusion of GFP to the C terminus of Slk19p also resulted in reduced viability in strains lacking KAR3.
Figure 3.—
Multiple coiled-coil regions of Slk19p are required for viability in strains lacking KAR3. Haploid strains lacking KAR3, containing a deletion allele of SLK19, and also carrying the plasmid MR820 [YCp–KAR3∷URA3] were first grown in YPAD, serially diluted 10-fold in water, and then spotted onto 5-FOA medium. Below is a diagram of Slk19p showing the regions essential for viability in a KAR3 deletion strain. KAR3 strains: wild type (DC49-7.1c) and slk19Δ (TRK28). KAR3 deletion strains: SLK19 (dKH46-2.1a), slk19–ΔA (dKH53-1.1c), slk19–ΔB (dKH42-1.7c), slk19–ΔC (dKH43-1.7d), slk19–ΔD (dKH61-1.5d), slk19–ΔE (dKH44-1.6a), slk19–ΔF (dKH45-1.10d), slk19–ΔG (dKH75-1.8d), and slk19–GFP (dKH186-2.1c).
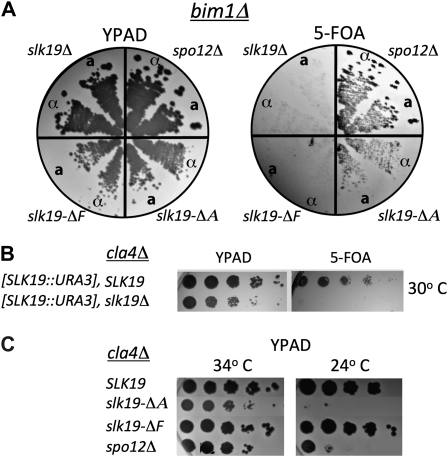
Large-scale yeast genetic screens have identified genes involved in spindle positioning, cohesin function, and cytokinesis that exhibit synthetic interactions with slk19Δ (Tong et al. 2004; Ye et al. 2005; Collins et al. 2007). We used the separation-of-function alleles to test which genetic interactions require which functions of Slk19p. We first tested bim1Δ, cla4Δ, ctf8Δ, and kip3Δ as representative alleles involved in the different pathways identified to have synthetic interactions with slk19Δ. Of those tested only bim1Δ and cla4Δ were synthetically lethal with slk19Δ in our strain background (Figure 4, A and B). We then tested these deletions for synthetic defects with slk19–ΔA (no FEAR signal) and slk19–ΔF (synthetic lethal with kar3Δ).
Figure 4.—
Separation-of-function slk19 alleles show consistent interactions with BIM1 or CLA4 deletions. (A) Haploid strains lacking BIM1, containing a deletion allele of slk19 or spo12Δ for a FEAR− control and also carrying pRK44 [YCp–SLK19∷URA3] were streaked to single colonies on YPAD and replica plated onto 5-FOA–containing plates. Strains: slk19Δ, MATa (dKH274-1.4b) and MATα (dKH274-1.9d), slk19–ΔF, MATa (dKH268-7.7d) and MATα (dKH268-2.8b), slk19–ΔA, MATa (dKH262-6.4b) and MATα (dKH262-6.10b), and spo12Δ, MATa (dKH280-5.3b) and MATα (dKH280-5.7a). (B) Haploid cla4Δ strains carrying pRK44 [YCp-SLK19∷URA3] were grown to stationary phase in YPAD medium, serially diluted 10-fold in water, spotted onto 5-FOA medium, and grown at 30°. Strains: SLK19 (TKH223) and slk19Δ (dKH272-3.3a). (C) Ura− (therefore they do not contain the SLK19 expressing plasmid), cla4Δ strains also containing a slk19 partial deletion (or spo12Δ, FEAR− control) were isolated from tetrad dissection. These strains were serially diluted 10-fold in water, spotted onto YPAD medium, and allowed to grow at 34° or 24°. Strains: SLK19 (TKH223), slk19–ΔA (dKH260-5.1a), slk19–ΔF (dKH266-6.2c), and spo12Δ (dKH278-5.3b).
Bim1p is a microtubule plus-end tracking protein that associates with both cytoplasmic and nuclear microtubules (Tirnauer et al. 1999). During anaphase, Bim1p is required to maintain a large overlap zone of ipMTs; loss of the protein results in spindles with a reduced ability to withstand the pulling apart of sister chromatids in anaphase B (Gardner et al. 2008). We found that like the complete SLK19 deletion, slk19–ΔF was synthetically lethal with bim1Δ, while the FEAR mutant slk19–ΔA was not (Figure 4A).
CLA4 and SLK19 complete deletions exhibit synthetic growth defects (Goehring et al. 2003). In our strain background the slk19Δ cla4Δ double mutant combination renders cells cold sensitive. We found that the A region, but not the F region, of Slk19p was essential for growth of cla4Δ mutants at 24° (Figure 4C). This pattern of synthetic interaction between slk19 deletion alleles and cla4Δ was the opposite of that observed for bim1Δ (Figure 4A). To test whether the synthetic interaction observed between slk19–ΔA and cla4Δ is recapitulated when cla4Δ is combined with other FEAR mutants, we assayed the viability of the spo12Δ cla4Δ combination. These double mutants were also cold sensitive.
The above results reinforce the notion that Slk19p has two genetically separable roles and each role is dependent on a separate region of the protein. As TACC proteins have been shown to associate with and promote the growth of microtubules (Bellanger and Gonczy 2003), we chose the slk19–ΔG allele to investigate the FEAR-independent functions of Slk19p.
The C terminus of Slk19p is essential for localization to the mitotic spindle:
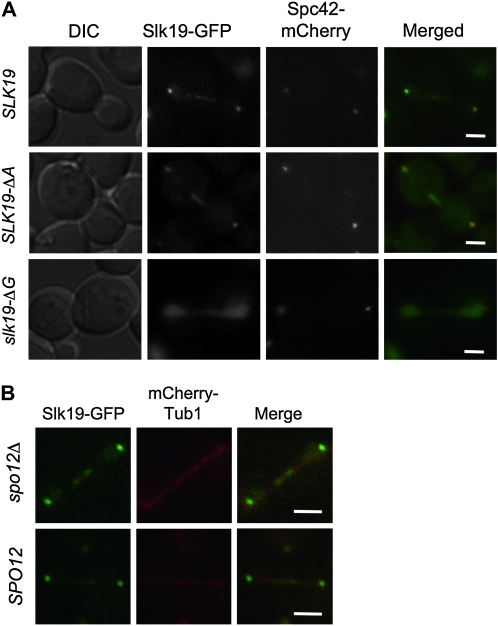
Slk19p–GFP fusion proteins have been shown to localize near the spindle pole body (SPB) in G1, the kinetochores in metaphase and anaphase, and at the spindle midzone also during anaphase (Zeng et al. 1999; Pereira and Schiebel 2003; Higuchi and Uhlmann 2005; Pagliuca et al. 2009). The FEAR and kar3Δ synthetic lethality assays (Figures 2 and 3), above, showed that the Slk19–GFP fusion protein is functional for FEAR signaling but partially defective for spindle function, raising the possibility that our (and other) localization studies with this protein might be misleading. To address this, localization experiments were also performed with a CFP–Slk19 protein. This allele does not exhibit a synthetic defect with kar3Δ and exhibits localization patterns that are indistinguishable from those seem with Slk19–GFP (though with less signal intensity) (supporting information, Figure S1). Thus we believe the general localization patterns obtained with the Slk19–GFP construct to accurately reflect the localizations of Slk19p. To determine whether the slk19–ΔA or slk19–ΔG separation-of-function mutants disrupted these localization patterns, we created C-terminal GFP fusions of these alleles. In these cells Spc42p was tagged with mCherry to visualize the spindle pole bodies. Cells were arrested in G1 with α-factor, released from the arrest, and imaged at 5-min intervals. At early time points after release into the cell cycle a focus of Slk19p–GFP was observed to colocalize with the spindle pole body (Figure S2). Frequently, in cells that had yet to reach metaphase, a second focus of Slk19p–GFP was also observed (Figure S2). This second Slk19p focus frequently colocalized at the ends of, or along, microtubule projections from the aster as well as with kinetochores, consistent with an association of Slk19p with kinetochores at the time of early attachment to the microtubules. Metaphase cells exhibited a bi-lobed GFP structure between the two SPBs (not shown), consistent with kinetochore localization, as previously observed (Zeng et al. 1999). Anaphase cells exhibited GFP foci at both SPBs as well as on the middle of the spindle (Figure 5A, top row), as previously observed (Zeng et al. 1999).
Figure 5.—
Known Slk19–GFP localizations do not require the presence of the A region, but do depend on the G region. (A) Strains expressing SPC42–mCherry and a SLK19 allele with a C-terminal GFP fusion were arrested in G1 with α-factor and then released by washing with SC media. Cells were concentrated, put onto a cover slip, and covered with an agarose pad. Multiple Z-plane images were obtained as the cell progressed through mitosis; single planes from example cell cycle stages are shown. Strains: SLK19–GFP (dKH202-2.1b), slk19–ΔA–GFP (dKH204-2.4b), and slk19–ΔG–GFP (dKH203-1.3a). (B) Using strains expressing SLK19–GFP and mCHERRY–TUB1, cells were prepared as above, maximum intensity projections are shown. Strains: SPO12 (dKH305-2.20a) and spo12Δ (dKH305-2.20c). Bars, 2 μm.
The N terminus (region A) of Slk19p is not essential for the observed localization pattern of Slk19–GFP: in most slk19–ΔA anaphase cells Slk19–ΔA–GFP was present at the midzone in a distribution that was not clearly different from that seen in wild-type cells (Figure 5A second row). Previous studies have shown that Cdc14p is required for visualization of Slk19p–GFP at the spindle midzone (Pereira and Schiebel 2003; Khmelinskii et al. 2007). Since slk19–ΔA strains are FEAR defective, the localization of Slk19–ΔA–GFP to the midzone must occur independently of a FEAR signal. To test this, we assayed localization of Slk19–GFP to the spindle midzone in SPO12 deletion strains. GFP was visible at the spindle midzone in both spo12Δ and SPO12 cells with no clear difference in localization patterns (Figure 5B). These data show that Slk19p midzone localization in anaphase is independent of the FEAR pathway through two separate mutations, spo12Δ and slk19-ΔA, while previous work showed dependence on Cdc14p for localization (Pereira and Schiebel 2003; Khmelinskii et al. 2007). Slk19p is initially recruited to the early anaphase midzone in FEAR deficient cells (Higuchi and Uhlmann 2005). Further, other studies have demonstrated Cdc14p activity without a functional FEAR pathway (Stegmeier et al. 2002; Tomson et al. 2009). Together these data suggest that Slk19p midzone localization requires Cdc14p activity and that FEAR is not necessary to provide sufficient Cdc14p activity for this localization. However, in wild-type cells, it is possible that the FEAR pathway can contribute to the Cdc14p activity that mediates midzone localization of Slk19p.
The C terminus (region G) of Slk19p is essential for its localization to the spindle. In cells expressing slk19–ΔG–GFP, we could not detect any specific localization of GFP to the kinetochores, spindle poles, or anaphase midzone (Figure 5A). Instead, Slk19–ΔG–GFP exhibited a uniform distribution of GFP throughout the nucleus during the entire cell cycle. As all kinetochore- and spindle-related localizations were dependent on the G region, we tested whether a G region–GFP fusion, under the control of the GAL1 promoter (and with an added nuclear localization signal), could recapitulate the localization pattern of Slk19–GFP. While we found weak association with the SPB and kinetochores in G1 and metaphase, respectively, this construct did not fully recapitulate the localization of the wild-type protein (not shown). Thus the G region is essential, but not sufficient, for spindle localization of the protein.
SLK19 mutants lose the transition between fast and slow anaphase B:
Several previous observations suggest SLK19 plays a role in mitotic spindle dynamics and some of these defects are almost certainly due to loss of the FEAR pathway. For example, slk19Δ mutants (and esp1 mutants) show aberrant organization of Ase1p at the spindle midzone (Khmelinskii et al. 2007). This phenotype can be suppressed, but not completely rescued, by a nonphosphorylatable form of Ase1p that does not require the action of the FEAR pathway to be dephosphorylated, suggesting that this slk19Δ spindle-related defect is partially due to the loss of the FEAR pathway (Khmelinskii et al. 2007, 2009).
There are other slk19Δ spindle-related phenotypes, however, for which it is not clear whether the defect is related to a loss of FEAR activity. First, Slk19p mediates a pause during anaphase spindle elongation in kinesin-5 motor mutants (cin8–FA kip1Δ) (Movshovich et al. 2008). Second, deletions of SLK19 result in more fragile or less forceful spindles; in slk19Δkar3–ts double mutants (but less so in kar3–ts single mutants) the spindles collapse after a shift to the nonpermissive temperature (Zeng et al. 1999), and slk19 mutants, like bim1, kar3, and ase1 mutants, are less able than wild-type cells to break a dicentric chromosome (Gardner et al. 2008). Therefore, anaphase spindle dynamics in the slk19 separation-of-function mutants were evaluated using multiple assays.
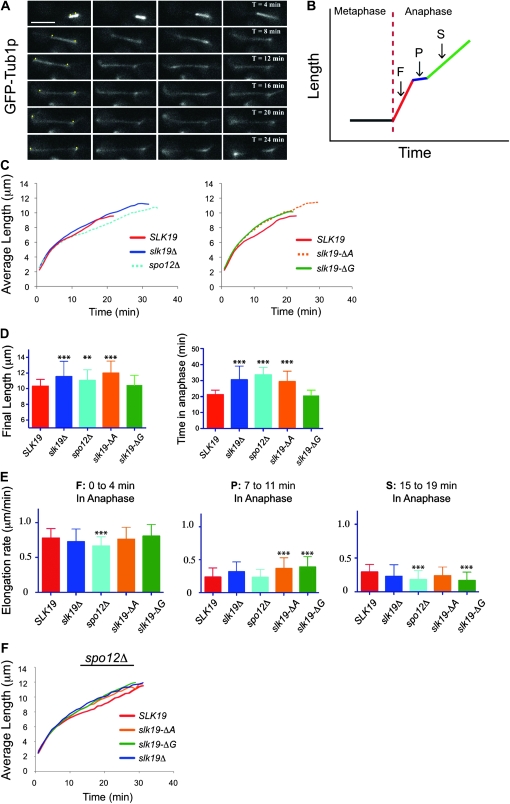
The first approach was to measure anaphase spindle elongation rates in wild-type and slk19 mutant strains. Living cells expressing either GFP–Tub1p or Spc42p–DsRed (a SPB marker) were imaged every minute after release from an α-factor arrest (Figure 6A), and the distance between the spindle poles was measured to produce spindle elongation traces for individual cells. For each genotype, the elongation traces from 40 to 100 cells were averaged. Results were similar for each fluorescent marker and in two independent strain backgrounds.
Figure 6.—
slk19 separation-of-function mutants display disrupted anaphase spindle elongation independent of FEAR defects. (A) Example of WT elongation: an otherwise wild-type strain expressing GFP–TUB1 (dKH160-1.2a) was arrested in α-factor, trapped in a microfluidic plate from CellASIC, and imaged every 30 sec while flowing complete medium through the chamber. Yellow dots in the left column were added to mark the location of SPBs. Bar, 5 μm. (B) A cartoon representing a WT trace, highlighting the four observed stages; M, metaphase; F, fast phase; P, paused state; S, slow phase. (C) Individual elongation traces were aligned at anaphase initiation (time = 0 min) and the average distance determined at each timepoint. (D) Comparison of time spent in anaphase and final length of the spindle. Anaphase was deemed completed at the timepoint prior to the rapid movement of SPBs that occurs when the spindle breaks and disassembly begins. (E) The rate of elongation during three 4-minute windows, corresponding to early, mid, or late anaphase, were determined from individual elongation traces and then averaged. Strains and number of cells counted: SLK19 (dKH257-1.2d), n = 25; slk19Δ (dKH256-5.5a), n = 40; spo12Δ (dKH282-5.10c), n = 40; slk19–ΔA (dKH283-1.2b), n = 40; and slk19–ΔG (dKH255-7.6a), n = 100. [D and E, bars show standard deviation (SD) **P < 0.01 and ***P < 0.001 when compared to SLK19 using a two-tailed unpaired t-test.] (F) Anaphase elongation rates of slk19 spo12Δ double mutants. Strains: SLK19 (dKH295-1.9b), n = 32; slk19Δ (dKH296-1.1a), n = 43; slk19–ΔA (dKH297-1.19c), n = 36; and slk19–ΔG (dKH299-1.16d), n = 40.
Wild-type spindles exhibit two phases of elongation, a fast phase and a slow phase (Kahana et al. 1995). Between these two elongation phases is a short pause, the molecular basis of which is unknown (Kahana et al. 1995; Yeh et al. 1995). An idealized anaphase elongation plot is shown in Figure 6B. The averaged elongation plots from multiple wild-type cells exhibited the reported phases of elongation (Figure 6C, SLK19): a fast phase with a rate of 0.85 ± 0.16 μm/min lasting 3.9 ± 1.35 min, a short pause state, and a slow phase of 0.29 ± 0.11 μm/min lasting ∼17 min. The endpoint of each trace indicates the average time at which the spindles started to collapse in each population (see below).
The previously published tendency of FEAR mutants to persist in anaphase was clear when the spindle elongation profiles from spo12Δ mutants were compared to those from wild-type cells (Figure 6C, spo12Δ) (Stegmeier et al. 2002). spo12Δ mutants spent a longer time in anaphase than wild-type cells and finished with significantly longer spindles (Figure 6D). The slk19 FEAR-defective mutants slk19Δ and slk19–ΔA exhibited these same phenotypes (Figure 6, C and D). However, slk19Δ, slk19–ΔA, and slk19–ΔG mutants exhibit phenotypes distinct from spo12Δ mutants: first, the fast phase is more rapid in the slk19 mutants, and second, there is loss of the pause between the fast and slow elongation phases in each of the slk19Δ mutants, but not in the spo12Δ mutant (Figure 6C). At the time wild-type spindle elongation pauses, the slk19 spindles continue elongation at a faster rate (Figure 6E, 7- to 11-min window), which gradually becomes slower over time (Figure 6E, 15–19 min). The rate of spindle elongation in the pause period was statistically indistinguishable between wild-type cells and spo12Δ mutants (Figure 6E), while slk19–ΔA and slk19–ΔG mutants both exhibited significantly faster spindle elongation in this interval than wild type (Figure 6E). This pause phase variation is seen in the average curves (Figure 6C), as well as in individual elongation traces (Figure S3), which exhibit similarly smooth profiles and frequently lack a transition.
Figure 7.—
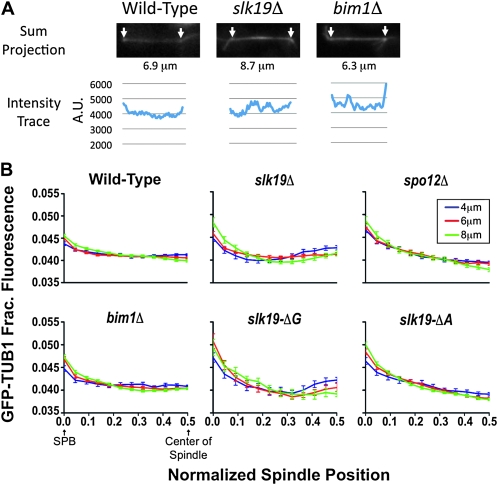
slk19 C-terminal mutants have a disrupted ipMT distribution. Cells expressing GFP–TUB1 were trapped under an agarose pad following release from an α-factor arrest, and a Z-series of 15 images, 0.25 μm apart, was obtained with a spinning-disk confocal microscope. Relative intensity was determined starting with a sum projection of the stack and using Metamorph's linescan tool to measure the intensity of a 5-pixel column at each point along the spindle. (A) Examples of sum projections of observed spindles and the corresponding intensity traces are shown. Arrows mark the GFP intensity peaks denoting the SPBs, and the length between the poles is given. (B) Each spindle was divided into 24 bins, split into half, and then spindles of lengths 4–5.9 μm, 6–7.9 μm, and 8–9.9 μm were averaged. Graphs show relative intensity and standard error of the mean (SEM) for each bin. Strains used and number of half spindles (4 μm), (6 μm), (8 μm); SLK19 (dKH160-1.2a), n = 68, 80, and 64; slk19Δ (TKH198), n = 40, 62, and 26; spo12Δ (TKH206), n = 74, 66, and 30; bim1Δ (dKH291-2.7c), n = 42, 78, and 74; slk19–ΔG (TKH194), n = 16, 20, and 14; and slk19–ΔA (TKH196), n = 60, 78, and 84.
The spindle elongation characteristics of the slk19 mutants suggest that slk19Δ has two (or more) defects in anaphase: (1) a defect in anaphase exit, due to loss of a FEAR signal; and (2) loss of the pause between the fast and slow phases of anaphase spindle elongation. The anaphase elongation profiles of slk19Δ mutants should reflect the combination of these two defects. By this model a spo12Δ (FEAR defect) slk19–ΔG (pause defect) double mutant should phenocopy the complete SLK19 deletion. To test this, the spindle elongation profile of a spo12Δ slk19–ΔG double mutant was evaluated. Indeed, loss of SPO12 from a slk19–ΔG mutant also led to longer spindles and an anaphase elongation profile similar to slk19Δ cells (Figure 6F).
Slk19p's C terminus is essential for normal anaphase microtubule dynamics:
The anaphase elongation studies above did not reveal clear differences between the slk19–ΔA and slk19–ΔG mutants other than the known anaphase FEAR defects specific to slk19–ΔA, yet genetic assays suggest that a unique microtubule role is disrupted in the slk19–ΔG mutant. To more closely examine potential differences between the slk19–ΔA and slk19–ΔG anaphase spindles, we adopted methods to evaluate the distributions of ipMTs in these strains. Previous studies have shown that anaphase spindles in slk19Δ mutants are fragile and other mutants with this phenotype have defects in ipMT dynamics (Gardner et al. 2008). Yeast anaphase spindles are spanned by about eight ipMTs (four from each pole), which are cross-linked by microtubule-associated proteins (Winey et al. 1995). This lattice provides the structural support that prevents the spindle from collapsing during elongation (anaphase B), and as sister chromatids are pulled to the poles by kinetochore MTs (anaphase A) (O'Toole et al. 1999). In wild-type cells these ipMTs extend from their origin nearly to the opposite pole, on average spanning 78% of the length of the spindle (O'Toole et al. 1999). However, in bim1Δ mutants the ipMTs only span 52% of the spindle, which results in a shorter zone of overlap and less cross-bridging of the ipMTs by the remaining cross-bridging proteins; this likely explains why bim1Δ spindles are prone to collapse (Gardner et al. 2008). The fragility of slk19 mutant anaphase spindles suggests that, like bim1Δ mutants, these spindles might also have an aberrant ipMT distribution.
The distribution of ipMTs can be estimated by evaluating the fluorescence intensity of GFP-tubulin across the anaphase spindle (Gardner et al. 2007, 2008). To determine the distribution of ipMTs in anaphase spindles we created isogenic strains that expressed GFP–Tub1p and carried mutant alleles of SLK19, BIM1, or SPO12. Strains were released into a synchronous cell cycle, images of developing spindles were collected, and tubulin density along the spindle was determined. Figure 7A shows example images of wild-type and mutant spindles and the corresponding intensity traces.
Anaphase spindle distributions were pooled according to spindle length and the data plotted as a function of GFP–Tub1p intensity from the SPB (brightest GFP point) to the center of the spindle. Therefore, each spindle contributes two half-spindles to the average curve. As has been described previously (Gardner et al. 2008), wild-type spindles exhibited GFP–Tub1p intensity distributions with a slight peak near the pole, corresponding to the short kMTs that have pulled the centromeres to the poles, and then a relatively flat distribution across the middle of the spindle (Figure 7B). This flat tubulin intensity distribution across the central region reflects the known distribution of ipMTs in wild-type spindles, which extend across the majority of the spindle (Gardner et al. 2008; O'Toole et al. 1999). In wild-type spindles this distribution pattern was found to be largely invariant among all spindle lengths though there is a slight shift in intensity away from the midzone toward the poles in longer spindles (Figure 7B, 8 μm). Overall the amount of ipMT overlap remains constant throughout anaphase. Thus, in wild-type cells, the growth of ipMTs is coordinated with microtubule sliding such that the ipMTs continue to span the majority of the length of the spindle as it elongates. Evaluation of bim1Δ mutants demonstrates the ability of the assay to reveal defects in ipMT distribution, as was described previously (Gardner et al. 2008). The higher proportion of tubulin intensity at the poles is indicative of shorter and fewer ipMTs in bim1Δ mutants (Figure 7B) (Gardner et al. 2008).
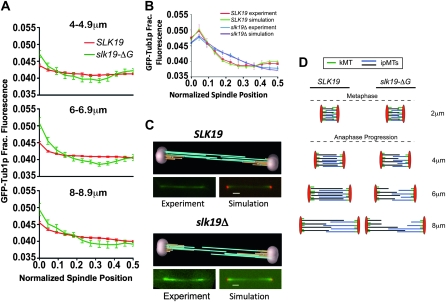
Figure 8.—
slk19Δ mutants have fewer ipMTs extending through the midzone. (A) Tubulin intensity of anaphase spindles as a function of spindle length. Data are the same as those shown in another format in Figure 7B. (B) Modeling tubulin density in slk19Δ spindles. Modeling software was used to simulate features of anaphase spindles that would yield tubulin distributions like those observed experimentally with SLK19 and slk19Δ strains (Gardner et al. 2007, 2008). The tubulin distribution that would be produced by the simulated spindles was compared to the observed distributions. The distribution of tubulin in SLK19 and slk19Δ strains was obtained by measuring spindles of 5–9 μm in length (average length 6.6 μm for each strain) using SPBs labeled with Spc42–DsRed as the endpoints. Strains and number of half spindles measured: SLK19 (dKH300-1-1d), n = 34; slk19Δ (dKH303-1.6a), n = 56. (C) Example animations produced by the modeling software used to create the simulated distributions in B. (D) A model of the defects observed in slk19–ΔG mutant spindles including shorter ipMTs from the onset of anaphase and an inability to coordinate growth of the spindle with growth of ipMTs.
Loss of the FEAR pathway (spo12Δ) results in a slight redistribution of GFP–Tub1p (Figure 7B). spo12Δ mutant cells produce spindles with a greater proportion of intensity at the poles, suggesting slightly fewer ipMTs that extend past the midzone. However, as in wild-type cells, the tubulin distribution is constant as the spindle grows. So while FEAR mutants may have a slightly different ipMT set point, they are able to maintain the balance between MT sliding and ipMT elongation. The tubulin distribution from slk19–ΔA mutant spindles quite closely resembled that seen in the spo12Δ mutants, suggesting that the A region does not have a dramatic role in ipMT distribution beyond its role in FEAR (Figure 7B).
Anaphase ipMT distributions in the slk19–ΔG and slk19Δ mutants were greatly disrupted (Figure 7B). In these mutants, short anaphase spindles (4–4.9 μm) had a high proportion of GFP–Tub1 in the center (Figure 8A), indicative of a population of ipMTs that extend just past the midzone. Long spindles (8–8.9 μm) in these mutants had a smaller proportion of GFP–Tub1 in the center of the spindle (Figure 8A), suggesting that there are fewer ipMTs that reach the midzone. In summary the GFP–Tub1 intensity distribution is not constant in slk19Δ mutants, changing as the spindles lengthen (Figure 7B). This suggests that in these mutants ipMT growth is not coordinated with spindle elongation as it is in wild-type cells.
A modeling approach was used to predict the ipMT characteristics of slk19Δ spindles that could result in the observed GFP–Tub1p intensity profiles. This approach was used previously to model ipMT distributions in bim1Δ and kar3Δ mutant spindles (Gardner et al. 2008). Spindles from wild-type and slk19Δ strains 5–9 μm in length (average 6.6 μm for both populations) were analyzed to determine the relative distribution of GFP–Tub1 (Figure 8B). Briefly, simulated images were produced from model spindles with varied ipMT distributions. Comparison of these simulated images to experimentally obtained GFP–Tub1 distributions allowed for the identification of model ipMT distributions that best fit the observed data (Figure 8B) (Sprague et al. 2003; Pearson et al. 2006; Gardner et al. 2007, 2008).
This modeling approach suggests that the spindles produced by wild-type cells contain ipMTs of relatively similar lengths with several regions of overlap between antiparallel MTs (Figure 8C). In contrast, the best-fit model spindles for slk19Δ mutants feature ipMTs that are on average 25% shorter than the wild-type ipMTs and are much more variable in length than the ipMTs in wild-type cells (average length ± SD = 3.46 ± 0.71 μm for SLK19 and 2.53 ± 1.08 μm for slk19Δ). The result of this variation in ipMT length is a midzone with fewer antiparallel ipMTs and shorter overlaps between antiparallel ipMTs (Figure 8C). Thus, this modeling predicts fewer potential contacts for cross-linking proteins to bridge the antiparallel MTs in slk19 mutants.
DISCUSSION
Using a collection of functional assays we have determined that Slk19p is composed of at least two separable functional domains. First, the N-terminal region is essential for FEAR signaling, but dispensable for certain aspects of microtubule behavior. Second, the C-terminal region is necessary for localization of Slk19p to the spindle midzone and contributes to the coordination of spindle elongation and the growth of ipMTs during anaphase, but is not required for FEAR signaling.
FEAR pathway:
Slk19p has at least two roles in the FEAR signaling pathway, inhibiting PP2A and activating Spo12p (Stegmeier et al. 2002; Queralt et al. 2006; Tomson et al. 2009); both roles appear to depend on the association of Slk19p with Esp1p (Sullivan et al. 2001; Rahal and Amon 2008). We determined that the N-terminal A region of Slk19p (amino acids 1–202) is essential to provide enough of a FEAR signal to promote two successive divisions in meiosis. As the A region (amino acids 1–222) contains the site of cleavage by Esp1p at amino acid 77 (Sullivan et al. 2001), it seems likely that the loss of FEAR activity in slk19–ΔA mutants is attributable to a loss in the interaction of Slk19p and Esp1p that is essential for FEAR signaling.
In addition to being incapable of producing a FEAR signal in meiosis, the slk19–ΔA allele showed a synthetic interaction with the MEN-defective cla4Δ allele. Cla4p is a kinase involved in septin ring formation and the completion of cytokinesis (Goehring et al. 2003) and is required for the localization of the MEN activator Lte1p to the daughter cortex (Hofken and Schiebel 2002). FEAR mutants are dependent on the MEN pathway for viability (Stegmeier et al. 2002). Thus, combining a FEAR defect with a MEN defect likely causes the observed synthetic interactions between either slk19-ΔA or spo12Δ and cla4Δ.
Slk19p and the midanaphase pause:
In a variety of assays the slk19–ΔA and slk19–ΔG alleles yield distinctly different phenotypes, but they both exhibit a loss of the pause that occurs between the fast and slow phases of anaphase spindle elongation. Slk19p, but not Spo12p, was previously demonstrated to be required for a midanaphase pause that could be detected in kinesin-5 mutants (Movshovich et al. 2008). This is probably the same anaphase pause that is apparent in the analysis of our wild-type strains and wild-type strains used by others (Kahana et al. 1995; Yeh et al. 1995). How might both the slk19–ΔA and slk19–ΔG alleles exhibit a similar phenotype that is not seen in FEAR mutants? Slk19–ΔAp and Slk19–ΔGp are both likely defective in localizing Slk19p/Esp1p to the spindle midzone, which is essential for midzone organization of Ase1p (Khmelinskii et al. 2007); Slk19–ΔAp localizes to the midzone but is likely defective in its ability to bind Esp1p (since it is missing a known Esp1p interaction site and is FEAR−) and Slk19–ΔGp can likely bind Esp1p (since it is FEAR+) but cannot localize to the midzone.
Though the molecular basis for the anaphase pause is not known, these results suggest parallels in the requirements for the anaphase pause and for the organization of Ase1p at the midzone: slk19 and esp1 mutants are defective in organizing Ase1p at the midzone in a manner which, along with the pause defect (this article), is independent of FEAR signaling (Khmelinskii et al. 2009). The fast phase of anaphase appears to be mainly dependent on the sliding of antiparallel ipMTs, while slow phase elongation is probably tied to the rate of tubulin subunit addition to these ipMTs (Kahana et al. 1995; Straight et al. 1998; Maddox et al. 2000; Schuyler et al. 2003). Ase1p is a crucial regulator of the transition to slow elongation. Deletion of ase1 prevents progression past the fast elongation phase (Schuyler et al. 2003), and ase1–7A, an unphosphorylatable mutant, has an anaphase elongation profile similar to the slk19 mutants, lacking a transition from fast to slow anaphase (Khmelinskii et al. 2009). An explanation for both ase1 and slk19 mutant elongation profiles is that both have uncoupled ipMT polymerzation from antiparallel sliding during the slow phase, a consequence of a malformed spindle midzone structure.
Microtubule dynamics:
Four phenotypes of slk19 mutants demonstrate a FEAR-independent role for Slk19p in microtubule dynamics. (1) The slk19–ΔG allele, which is proficient for FEAR signaling, is synthetically lethal with deletions of the spindle regulators KAR3 and BIM1. (2) SLK19 deletion alleles, but not spo12Δ mutants, eliminate the pause between the fast and slow anaphase elongation phases. (3) Deletion of the C terminus of Slk19p prevents localization of the protein to the spindle midzone, consistent with a FEAR-independent role on the spindle. (4) SLK19 alleles lacking the C-terminal region disrupt anaphase ipMT distribution.
In mutants lacking the C terminus of Slk19p, ipMTs fail to extend far past the center of the early anaphase spindle (Figure 8D, 4-μm spindles). This spindle morphology might arise if the elongation of the metaphase spindle occurs without an accompanying growth of the plus ends of ipMTs. This failure of plus-end growth would be predicted to result in early anaphase spindles with a concentration of tubulin at the midzone, flanked by regions of lesser density between the midzone and the poles (Figure 8D), just as we have seen on short spindles (Figure 8A). The coordination of spindle elongation and plus-end ipMT growth in wild-type spindles results in an even distribution of tubulin density between the poles as anaphase spindles elongate (Figure 8A,D). In wild-type cells, MT growth appears to fall behind near the end of anaphase, resulting in a lower proportion of tubulin between the midzone and the poles (Figure 8, B and D). The late anaphase reduction in MT growth is exacerbated in slk19 mutants, which results in longer spindles that have fewer overlaps between antiparallel MTs and a subsequent decrease in cross-linking strength (Gardner et al. 2008). This is similar to what has been observed in bim1Δ mutants (Gardner et al. 2008).
Slk19p may be working directly to regulate ipMT length, through modifying plus-end dynamics, or indirectly through its action on midzone regulators and motors such as Ase1p and Cin8p. Numerous observations suggest a role for Slk19p in stabilizing the plus ends of microtubules or promoting their growth. These include (1) the localization of Slk19p–GFP at the plus ends of MTs and kinetochores throughout the cell cycle (this article) (Higuchi and Uhlmann 2005; Fridman et al. 2009; Pagliuca et al. 2009); (2) premature chromatin stretching during metaphase in slk19Δ mutants (Zhang et al. 2006), which could be due to increased catastrophic shortening of kMTs leading to greater pulling force on kinetochores and excess stress on centromeric cohesion; (3) synthetic lethality of the slk19–ΔG allele with deletions of the MT regulators BIM1 and KAR3; and (4) shortened ipMTs in SLK19 carboxy-terminus deletions. Since there is no direct evidence of Slk19p binding to microtubules (data not shown), it is likely Slk19p modulates other known regulators. The recent demonstration that Slk19p shares a similar outer kinetochore localization with the plus-end tracking proteins Bim1p, Bik1p, and Kar3p, as well as Cin8p (Pagliuca et al. 2009), is consistent with the notion that Slk19p works with these proteins to modulate MT dynamics.
Acknowledgments
The authors acknowledge and thank members of the Dawson lab and the Cell Cycle and Cancer Biology Department at the Oklahoma Medical Research Foundation for helpful discussion of the work, H. Coffin for assistance building the deletion mutants, M. Conrad for strain construction advice, M. Rose for the MR820 plasmid, E. Schiebel for the mCHERRY-TUB1 construct, and K. Thorn for the SPC42-mCHERRY construct. We also thank G. Gorbsky and members of his lab for the use of, and assistance with, spinning-disk microscopy image acquisition and the use of Metamorph; C. Lee for advice on the agarose pad method for flattening yeast cells for live-cell image acquisition; S. Rankin for assistance with the Nikon perfect focus system; and A. Mok from CellASIC. This research was supported by a grant from the Oklahoma Center for the Advancement of Science and Technology (OCAST) (to D.S.D.) and National Institutes of Health grant GM087516 (to M.E.D.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.123257/DC1.
References
- Amberg, D., D. Burke and J. Strathern, 2005. Methods in Yeast Genetics, a Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bellanger, J. M., and P. Gonczy, 2003. TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr. Biol. 13 1488–1498. [DOI] [PubMed] [Google Scholar]
- Boeke, J. D., F. LaCroute and G. R. Fink, 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197 345–346. [DOI] [PubMed] [Google Scholar]
- Bouck, D. C., and K. S. Bloom, 2005. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc. Natl. Acad. Sci. USA 102 5408–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo, S. B., K. P. Rabitsch, J. Fuchs, S. Gruber, M. Sullivan et al., 2003. Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12, and SLK19. Dev. Cell 4 727–739. [DOI] [PubMed] [Google Scholar]
- Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero and P. Hieter, 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110 119–122. [DOI] [PubMed] [Google Scholar]
- Collins, S. R., K. M. Miller, N. L. Maas, A. Roguev, J. Fillingham et al., 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446 806–810. [DOI] [PubMed] [Google Scholar]
- Dresser, M. E., 2009. Time-lapse fluorescence microscopy of Saccharomyces cerevisiae in meiosis. Methods Mol. Biol. 558 65–79. [DOI] [PubMed] [Google Scholar]
- Dresser, M. E., D. J. Ewing, S. N. Harwell, D. Coody and M. N. Conrad, 1994. Nonhomologous synapsis and reduced crossing over in a heterozygous paracentric inversion in Saccharomyces cerevisiae. Genetics 138 633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman, V., A. Gerson-Gurwitz, N. Movshovich, M. Kupiec and L. Gheber, 2009. Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep. 10 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, M. K., D. J. Odde and K. Bloom, 2007. Hypothesis testing via integrated computer modeling and digital fluorescence microscopy. Methods 41 232–237. [DOI] [PubMed] [Google Scholar]
- Gardner, M. K., J. Haase, K. Mythreye, J. N. Molk, M. Anderson et al., 2008. The microtubule-based motor Kar3 and plus end-binding protein Bim1 provide structural support for the anaphase spindle. J. Cell Biol. 180 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R. D., and R. A. Woods, 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350 87–96. [DOI] [PubMed] [Google Scholar]
- Goehring, A. S., D. A. Mitchell, A. H. Tong, M. E. Keniry, C. Boone et al., 2003. Synthetic lethal analysis implicates Ste20p, a p21-activated potein kinase, in polarisome activation. Mol. Biol. Cell 14 1501–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, T., and F. Uhlmann, 2005. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature 433 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofken, T., and E. Schiebel, 2002. A role for cell polarity proteins in mitotic exit. EMBO J. 21 4851–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen, S. L., J. F. Charles, R. L. Tinker-Kulberg and D. O. Morgan, 1998. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell 9 2803–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana, J. A., B. J. Schnapp and P. A. Silver, 1995. Kinetics of spindle pole body separation in budding yeast. Proc. Natl. Acad. Sci. USA 92 9707–9711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamieniecki, R. J., R. M. Shanks and D. S. Dawson, 2000. Slk19p is necessary to prevent separation of sister chromatids in meiosis I. Curr. Biol. 10 1182–1190. [DOI] [PubMed] [Google Scholar]
- Kateneva, A. V., A. A. Konovchenko, V. Guacci and M. E. Dresser, 2005. Recombination protein Tid1p controls resolution of cohesin-dependent linkages in meiosis in Saccharomyces cerevisiae. J. Cell Biol. 171 241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii, A., and E. Schiebel, 2008. Assembling the spindle midzone in the right place at the right time. Cell Cycle 7 283–286. [DOI] [PubMed] [Google Scholar]
- Khmelinskii, A., C. Lawrence, J. Roostalu and E. Schiebel, 2007. Cdc14-regulated midzone assembly controls anaphase B. J. Cell Biol. 177 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii, A., J. Roostalu, H. Roque, C. Antony and E. Schiebel, 2009. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev. Cell 17 244–256. [DOI] [PubMed] [Google Scholar]
- Klapholz, S., and R. E. Esposito, 1980. Isolation of SPO12–1 and SPO13–1 from a natural variant of yeast that undergoes a single meiotic division. Genetics 96 567–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwaliwale, C. V., S. B. Frei, B. M. Stern and S. Biggins, 2007. A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Dev. Cell 13 433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Maddox, P. S., K. S. Bloom and E. D. Salmon, 2000. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, B. D., J. G. Barrett, J. A. Wallace, H. Granok and M. Snyder, 1999. Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J. Cell Biol. 144 1219–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston, A. L., B. H. Lee and A. Amon, 2003. The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Dev. Cell 4 711–726. [DOI] [PubMed] [Google Scholar]
- Movshovich, N., V. Fridman, A. Gerson-Gurwitz, I. Shumacher, I. Gertsberg et al., 2008. Slk19-dependent mid-anaphase pause in kinesin-5-mutated cells. J. Cell Sci. 121 2529–2539. [DOI] [PubMed] [Google Scholar]
- Nicolas, A., D. Treco, N. P. Schultes and J. W. Szostak, 1989. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature 338 35–39. [DOI] [PubMed] [Google Scholar]
- O'Toole, E. T., M. Winey and J. R. McIntosh, 1999. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 10 2017–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg, K. R., K. T. Vo, S. Michaelis and C. Paddon, 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25 451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca, C., V. M. Draviam, E. Marco, P. K. Sorger and P. De Wulf, 2009. Roles for the conserved spc105p/kre28p complex in kinetochore-microtubule binding and the spindle assembly checkpoint. PLoS One 4 e7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, C. G., M. K. Gardner, L. V. Paliulis, E. D. Salmon, D. J. Odde et al., 2006. Measuring nanometer scale gradients in spindle microtubule dynamics using model convolution microscopy. Mol. Biol. Cell 17 4069–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., and E. Schiebel, 2003. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science 302 2120–2124. [DOI] [PubMed] [Google Scholar]
- Peset, I., and I. Vernos, 2008. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 18 379–388. [DOI] [PubMed] [Google Scholar]
- Queralt, E., and F. Uhlmann, 2008. Cdk-counteracting phosphatases unlock mitotic exit. Curr. Opin. Cell Biol. 20 661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queralt, E., C. Lehane, B. Novak and F. Uhlmann, 2006. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell 125 719–732. [DOI] [PubMed] [Google Scholar]
- Rahal, R., and A. Amon, 2008. The Polo-like kinase Cdc5 interacts with FEAR network components and Cdc14. Cell Cycle 7 3262–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M., N. Koonrugsa, T. Toda, L. Vardy, S. Tournier et al., 2003. Deletion of Mia1/Alp7 activates Mad2-dependent spindle assembly checkpoint in fission yeast. Nat. Cell Biol. 5 764–766, author reply 766. [DOI] [PubMed] [Google Scholar]
- Schuyler, S. C., J. Y. Liu and D. Pellman, 2003. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J. Cell Biol. 160 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks, R. M., C. Bascom-Slack and D. S. Dawson, 2004. Analysis of the kar3 meiotic arrest in Saccharomyces cerevisiae. Cell Cycle 3 363–371. [PubMed] [Google Scholar]
- Shirayama, M., Y. Matsui and A. Toh-e, 1996. Dominant mutant alleles of yeast protein kinase gene CDC15 suppress the lte1 defect in termination of M phase and genetically interact with CDC14. Mol. Gen. Genet. 251 176–185. [DOI] [PubMed] [Google Scholar]
- Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed et al., 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97 233–244. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H. E., and A. P. Mitchell, 1989. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 9 2142–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague, B. L., C. G. Pearson, P. S. Maddox, K. S. Bloom, E. D. Salmon et al., 2003. Mechanisms of microtubule-based kinetochore positioning in the yeast metaphase spindle. Biophys. J. 84 3529–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier, F., and A. Amon, 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38 203–232. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., R. Visintin and A. Amon, 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108 207–220. [DOI] [PubMed] [Google Scholar]
- Straight, A. F., J. W. Sedat and A. W. Murray, 1998. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 143 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, M., and F. Uhlmann, 2003. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat. Cell Biol. 5 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, M., C. Lehane and F. Uhlmann, 2001. Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nat. Cell Biol. 3 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer, J. S., E. O'Toole, L. Berrueta, B. E. Bierer and D. Pellman, 1999. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J. Cell Biol. 145 993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomson, B. N., R. Rahal, V. Reiser, F. Monje-Casas, K. Mekhail et al., 2009. Regulation of Spo12 phosphorylation and its essential role in the FEAR network. Curr. Biol. 19 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu et al., 2004. Global mapping of the yeast genetic interaction network. Science 303 808–813. [DOI] [PubMed] [Google Scholar]
- Visintin, R., K. Craig, E. S. Hwang, S. Prinz, M. Tyers et al., 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2 709–718. [DOI] [PubMed] [Google Scholar]
- Visintin, R., E. S. Hwang and A. Amon, 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398 818–823. [DOI] [PubMed] [Google Scholar]
- Winey, M., C. L. Mamay, E. T. O'Toole, D. N. Mastronarde, T. H. Giddings, Jr. et al., 1995. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 129 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury, E. L., and D. O. Morgan, 2007. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat. Cell Biol. 9 106–112. [DOI] [PubMed] [Google Scholar]
- Ye, P., B. D. Peyser, X. Pan, J. D. Boeke, F. A. Spencer et al., 2005. Gene function prediction from congruent synthetic lethal interactions in yeast. Mol. Syst. Biol. 1 0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, E., R. V. Skibbens, J. W. Cheng, E. D. Salmon and K. Bloom, 1995. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, X., and W. S. Saunders, 2000. The Saccharomyces cerevisiae centromere protein Slk19p is required for two successive divisions during meiosis. Genetics 155 577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, X., J. A. Kahana, P. A. Silver, M. K. Morphew, J. R. McIntosh et al., 1999. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 146 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T., H. H. Lim, C. S. Cheng and U. Surana, 2006. Deficiency of centromere-associated protein Slk19 causes premature nuclear migration and loss of centromeric elasticity. J. Cell Sci. 119 519–531. [DOI] [PubMed] [Google Scholar]