Abstract
In Schizosaccharomyces pombe, Etd1 is a positive regulator of the septation initiation network (SIN), a conserved GTPase-regulated kinase cascade that triggers cytokinesis. Here we show that a mutation in the pab1 gene, which encodes the B-regulatory subunit of the protein phosphatase 2A (PP2A), suppresses mutations in the etd1 gene. Etd1 is required for the function of the GTPase Spg1, a key regulator of SIN signaling. Interestingly, the loss of Pab1 function restored the activity of Spg1 in Etd1-deficient cells. This result suggests that PP2A-Pab1–mediated dephosphorylation inhibits Spg1, thus antagonizing Etd1 function. The loss of pab1 function also rescues the lethality of mutants of other genes in the SIN cascade such as mob1, sid1, and cdc11. Two-hybrid assays indicate that Pab1 physically interacts with Mob1, Sid1, Sid2, and Cdc11, suggesting that the phosphatase 2A B-subunit is a component of the SIN complex. Together, our results indicate that PP2A-Pab1 plays a novel role in cytokinesis, regulating SIN activity at different levels. Pab1 is also required to activate polarized cell growth. Thus, PP2A-Pab1 may be involved in coordinating polar growth and cytokinesis.
THE fission yeast Schizosaccharomyces pombe is a leading experimental model for eukaryotic cytokinesis (Bathe and Chang 2009; Pollard and Wu 2010). Fission yeast cells grow in a polarized manner by elongation at the cell ends and divide during cytokinesis by the action of a contractile actomyosin ring assembled in the middle of the cell (Snell and Nurse 1993). At the end of mitosis, when nuclear separation has been completed, actomyosin ring constriction is triggered by the septation initiation network (SIN). This signal transduction cascade is composed of the GTPase Spg1 and three protein kinases—Cdc7, GC-kinase Sid1, and NDR-kinase Sid2 in their presumed order of action—and the associated proteins Cdc14 with Sid1 and Mob1 with Sid2. These proteins are all located at the spindle pole body (SPB) during mitosis on a scaffold composed of the coiled-coil proteins Sid4 and Cdc11 (Krapp et al. 2004). The Sid2-Mob1 protein kinase complex is thought to transmit the division signal from the SPB to the actomyosin ring since it also associates at the division site during septation (Krapp and Simanis 2008). The SIN triggers actomyosin ring contraction coordinated with the synthesis of the primary and secondary septa that will form the new cell wall (Krapp et al. 2004; Wolfe and Gould 2005). The small GTPase Rho1 is known to promote cell-wall formation at the division site by stimulation of Cps1p/Drc1 1,3-β-glucan synthase (Le Goff et al. 1999), but the mechanism remains unclear.
SIN activity is tightly regulated during the cell cycle to ensure proper coordination of mitosis and cytokinesis. Mutants that negatively affect SIN function undergo nuclear division in the absence of septation, while increased SIN activity induces septation in interphase cells (Krapp and Simanis 2008). Regulation of the SIN is complex, involving multiple, partially redundant mechanisms, but the nucleotide status of the Ras superfamily small GTPase, Spg1, represents a key step in SIN activity (Lattmann et al. 2009). Cdc16 and Byr4 form a two-component GTPase-activating protein (GAP) for Spg1 that inhibits its activity (Furge et al. 1998; Cerutti and Simanis 1999). Proteins acting as a guanine nucleotide-exchange factor (GEF) for this GTPase have not been identified. In the budding yeast Saccharomyces cerevisiae, the pathway analogous to the SIN is known as the mitotic exit network (MEN) (reviewed in Krapp and Simanis 2008). Contact between the SPB-localized GTPase Tem1 (the Spg1 homolog) with its putative GEF Lte1, which is present only within the bud, has been proposed as a mechanism to ensure that mitotic exit occurs only after the spindle has oriented correctly (Bardin et al. 2000; Pereira et al. 2000). Bfa1-Bub2 (the Cdc16-Byr4 equivalent) are negative regulators of the MEN, acting as a two-component GAP for Tem1 (Geymonat et al. 2002).
Etd1 was identified in a genetic screen searching for new regulators of the S. pombe cell division cycle (Jimenez and Oballe 1994). Further characterization indicated that Etd1 acts as a positive regulator of the SIN (Daga et al. 2005). A recent study has established a key role for Etd1 in the timing of cytokinesis via the regulation of Spg1, acting as a potential homolog of budding yeast Lte1 (Garcia-Cortes and McCollum 2009). Loss of Etd1 function can be suppressed by mutations in a number of genes, some of which are involved in morphogenesis (Jimenez and Oballe 1994). Here we show that one of the mutations that bypass the requirement for etd1 in cytokinesis affects the activity of pab1, which encodes the protein phosphatase 2A (PP2A) regulatory subunit B. The characterization of Pab1 and pab1 mutants described in this study reveals a novel role for PP2A-Pab1 in SIN regulation and provides new insight into the mechanism by which Etd1 might regulate SIN signaling. We also show that Pab1 participates in activation of the morphological pathway, suggesting a role for PP2A-Pab1 in the coordination of cytokinesis and morphogenesis.
MATERIALS AND METHODS
Media and general methods:
Growth conditions and strain manipulations were described previously (Moreno et al. 1991). Experiments in liquid culture were carried out in minimal medium (EMM), supplemented as required, with a starting cell density of 2–4 × 106 cells/ml, corresponding to the mid-exponential growth phase. For regulated nmt expression, described methods were used (Maundrell 1993). Conditional deficiency for etd1 was assessed by using the etd1-1 allele (37° or 6% v/v ethanol are permissive conditions), the null etd1Δ allele (37°, permissive temperature), or the etd1Δ nmt81x-etd1 strain at any growth temperature, where etd1 expression is regulated by thiamine under the weak repressible nmt81x promoter (Daga et al. 2005). This later strain was used to produce etd1Δ mutants with the desired genetic background, removing the nmt81x-etd1 marker by tetrad dissection.
Genetic and cytological techniques were performed according to Moreno et al. (1991). Double mutants were constructed by tetrad dissection. Transformation was achieved by using the lithium acetate transformation protocol (Norbury and Moreno 1997).
pab1 cloning and deletion:
To clone the pab1 gene, we looked for a hypersensitive condition of the ret1-4/pab1-4 mutant. We found pab1-4 lethality when 0.005% of SDS was added to the growth media; pab1-4 mutants were transformed with a cDNA library controlled by a thiamine-repressible nmt1 promoter (plasmid pREP3x) to obtain 300,000 colonies. Only one clone rescued both lethality and shape defects under low levels of expression (in thiamine-containing medium). We purified and sequenced this clone, which encoded for protein phosphatase 2A regulatory subunit B, Pab1. The pab1 sequence, including its promoter and terminator, was amplified using ATGTCACTTTTATGCAGAGC and AGAAAAATACTGTGGATAGC primers from a wild-type (wt) strain and cloned into the pUR19 plasmid. This construct was transformed into a pab1-4 strain to check whether the phenotype was suppressed.
Deletion of pab1 in the desired strain (wt and etd1Δ) was made by using the described PCR-based gene-targeting method (Bahler et al. 1998). The oligonucleotides used for that purpose were 5′-TCCCGGAAGTTCAATAAAAAAGGAACTATGTTCAGTCTATCCGTAGATCATAAAAACAAACCAAACTCCACATAAGGACAGAATTCGAGCTCGTTTAAAC-3′ and 5′-AAGATCAAGATCTTCGTATACAATTGAAATTGGCAAAGTGTACCAACGGGAACGAAATCTTAATATAATACGAGTAGAAGCGGATCCCCGGGTTAATTAA-3′. The pab1 deletion in the respective genetic backgrounds was checked by PCR.
Plasmids:
Gene fragments were obtained by PCR amplification from either S. pombe genomic DNA or the cDNA library, as appropriate. Overexpression assays were achieved using the pREP3x plasmid (expression driven by the nmt1 promoter). The open reading frame of the desired gene was amplified by PCR from a cDNA library using appropriate primers and cloned into the cloning sites of the pREP3x vector (Maundrell 1993). For moderated overexpression, the pREP41x vector was used (nmt41x-driven expression). To construct the plasmids pREP41x-GFP-pab1 and pREP41x-MyHIspab1, pab1 was obtained by PCR amplification from genomic DNA and cloned into the NdeI-BamHI sites of the pREP41x-EGFPN vector and the SmaI-SalI sites in pREP41x-MycHis vectors, respectively (Craven et al. 1998). The GFP-Pab1 construct yields a GFP fused to the N-terminal end of Pab1 (C-terminal fusions were nonfunctional). A pJK148nmt41x-GFP-pab1 strain was created by PCR amplification from pREP41x-GFP-pab1 and subcloned into pJK148. The resulting plasmid was linearized with NruI and integrated into the S. pombe leu1 locus by homologous recombination. A strain containing a GFP-Pab1 construct under the expression of the endogenous pab1 promoter was not obtained.
Pab1 and Mob1p copurification:
Protein extracts were prepared from exponentially growing cells, collected by centrifugation, washed with stop buffer (0.9% NaCl, 1 mm NaN3, 10 mm EDTA, 50 mm NaF), and frozen on dry ice. All subsequent manipulations were done on ice or in the cold room (4°). For Western blotting, total protein extracts were prepared by Fast-prep vortexing with glass beads (Sigma) (Moreno et al. 1991).
To purify HisMyc-Pab1 and GFP-Mob1 protein, a cobalt resin was used (Talon Metal Affinity, Clontech). Soluble protein extracts were prepared in 100 mm NaCl, 0.05% NP-40, 20 mm Tris–HCl (pH 8), 1 mm 2-β-Mercaptoethanol, and 10% glycerol buffer with 1 mg/ml pepstatin, 10 mg/ml leupeptin, and 10 mg/ml aprotinin. Cells extracts were clarified at 14,000 × g for 15 min. Protein concentration was measured using a bicinchoninic acid protein assay kit (Sigma). For each purification, 500 μg/1 mg of soluble protein was incubated for 2 hr with 500 μg–1 mg of cobalt resin. Complex proteins were eluted by addition of 50 mm of imidazol and precipitated with trichloroacetic acid.
Crude extracts and purified proteins were separated on SDS–PAGE (10%). Blots were probed with anti-His (Qiagen) at 1:10,000 dilution or anti-GFP (Roche) at 1:20,000 followed by anti-mouse IgG conjugates (Sigma) at 1:1000 dilution. The Amersham Life Science ECL system was used to detect Pab1 while Supersignal (Pierce) was used to detect Mob1.
Two-hybrid analyses:
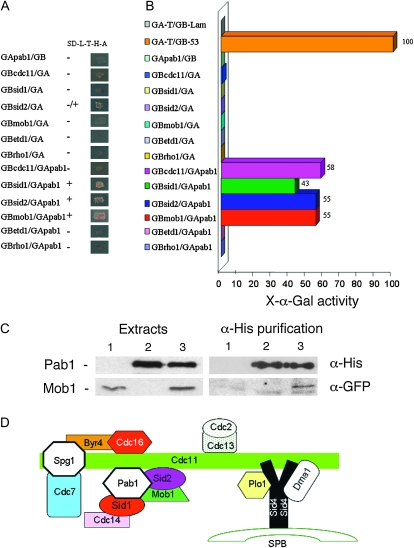
Two-hybrid system assays were developed using pGADT7 and pGBKT7 plasmids with the AH109 strain as the Matchmaker Gal4 Two-Hybrid System 3 (Clontech). As pGBKT7-pab1 was able to activate the reporter genes, we used pGADT7-pab1 to check interaction with Cdc11, Sid1, Sid2, Mob1, Etd1, and Rho1. We checked the interactions with expressions of both Gal1 and Gal2 in a plate assay and also in a colorimetric assay checking for expression of MEL1 reporter using X-α-Gal as substrate. All the procedures were developed according to the manufacturer's (Clontech) instructions.
Microscopy:
F-actin was visualized by rhodamine-phalloidin staining (Sigma) of formaldehyde-fixed cells. Cell-wall and chromatin regions were visualized by calcofluor white (Sigma) and 4,6-diamidino-2-phenylindole (DAPI; Sigma), respectively. Cells were examined using a Leica fluorescence microscope equipped with a plan Apo ×100 lens. Confocal images were obtained with a confocal Leica microscope. For time-lapse experiments in strains harboring GFP-Pab1 and Atb2-tomato constructs, cells were imaged in z-series with a step size of 0.3 μm between focal planes every 5 min.
Live-cell imaging was performed under Deltavision wide-field microscope systems (Applied Precision, Issaquah, WA). For time-lapse experiments, exponentially growing cells were concentrated by centrifugation and suspended in 100 μl of EMM medium, and the cell suspension (5 μl) was placed in 35-mm glass-bottom culture dishes (P35-1.5-10-C; MatTek) with 5 μl of 1 mg/ml soybean lectin (Sigma-Aldrich) and immersed in 3 ml of medium. Time-lapse experiments were performed at 25°, acquiring images every 5 min as an image stack of 10- × 0.5-μm z-planes with 2 × 2 binning, with the exception of pab1-4 and pab1-4 etd1Δ where acquisition was performed using 20 × z-planes per image stack.
Image analysis:
Fluorescence intensity measurements for Cdc7-GFP were made using ImageJ software (National Institutes of Health, Bethesda, MD) by placing a circle around SPB-localized Cdc7 and measuring the maximal fluorescence (Daga et al. 2005). A nearby cytosolic region was used as background.
RESULTS AND DISCUSSION
A mutation in the regulatory subunit B of PP2A, Pab1, suppresses the requirement for Etd1 in cytokinesis:
Both the etd1-1 mutant and the etd1Δ null strain are lethal at normal S. pombe growth temperatures, ranging from 20° to 35°, but are viable at 36°–37°. At restrictive growth temperatures, these etd1-deficient mutants exhibit a conventional sin-deficient phenotype, yielding elongated and multinucleate cells without a septum (Jimenez and Oballe 1994; Daga et al. 2005; Garcia-Cortes and McCollum 2009).
The lethal phenotype of etd1-1 can be spontaneously reverted by a large number of extragenic suppressors defective in polar cell growth (ret mutants), suggesting for the first time a connection between pathways regulating cytokinesis and morphogenesis (Jimenez and Oballe 1994). One of these mutants, ret1-4, gives rise to round or pear-shaped cells and is viable at any temperature but grows optimally in the range of 30°–35°. This mutant was also able to suppress the etd1Δ null allele at any temperature, indicating that the ret1-4 mutation activates septation and counteracts the requirement for etd1 function in cytokinesis (Figure 1A). Binucleate cells were occasionally observed (enhanced in an etd1-deficient background) (Figure 1A), suggesting a role for the etd1-1 suppressor gene in the coordination of polar growth and cell division, as proposed for genes of the morphogenesis pathway (Verde et al. 1998).
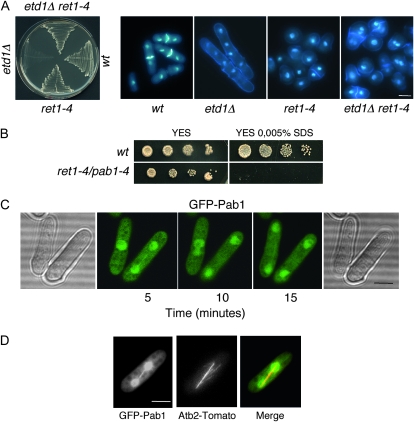
Figure 1.—
Identification of the etd1-1 suppressor gene, pab1. (A) Growth at 25° of wild-type (wt), ret1-4, etd1Δ (etd1∷ura4), and etd1Δ ret1-4 strains. These strains were grown at 37° (permissive temperature for etd1Δ) and replica-plated to assay for growth at 25° (restrictive temperature for etd1Δ). The results show that ret1-4 is able to suppress growth lethality of a null etd1Δ allele (left) and septation deficiencies of these mutant cells, as determined by DAPI-calcofluor staining (right panels). (B) Growth sensitivity of ret1-4 to 0.005% SDS assessed by serial dilution drop tests on plates (10-fold dilutions starting with 104 cells). Wild-type strain (wt) was used as a control. This property allowed complementation cloning using a cDNA library driven by the thiamine-repressible nmt1 promoter at low levels of expression (with thiamine) in the pREP3x plasmid. The corresponding cDNA coded for the pab1 gene, and the ret1-4 mutation was renamed pab1-4. (C) In vivo localization of Pab1 using a GFP-pab1 construct in pab1Δ cells. GFP-Pab1 was imaged in living cells at 25° by time-lapse microscopy at 5-min intervals in the same focal plane. GFP-Pab1 expression was monitored from time 0 to full expression after nmt41x-GFP-pab1 induction, and GFP-Pab1 localization was the same in the wide range of expression levels assessed. (D) Association of GFP-Pab1 with the mitotic spindle. The Acb2-tomato construct was used for microtubule localization. Bars, 5 μm.
To know more about this gene and its connection to etd1, we decided to characterize ret1 and further analyze its function in morphogenesis and its relationship with etd1 and the SIN cascade. SDS sensitivity of ret1-4 cells was used to clone the ret1 gene by complementation with a S. pombe cDNA library driven by the thiamine-repressible nmt1 promoter at low expression level (with thiamine) (see materials and methods and Figure 1B). This screen yielded only one rescuing plasmid. Sequence analysis of the corresponding cDNA indicated that the complementing gene encodes Pab1, the B-regulatory subunit of PP2A. DNA sequence also determined that ret1-4 contained a point mutation at nucleotide 694 that introduces a premature stop codon, thereby yielding a truncated, nonfunctional Pab1 protein. The truncation is predicted to occur adjacent to a conserved central region of subunit B that is thought to perform a substrate recognition function (Mayer-Jaekel et al. 1993). This demonstrates that the ret1-4 mutant is an allele of pab1 and therefore we renamed it pab1-4. The discovery that a mutation in pab1, the only B-type regulatory subunit found in the entire S. pombe genome, can suppress etd1 deficiencies indicates that the PP2A-Pab1 complex is involved in the control of cytokinesis.
To localize Pab1 in vivo, the protein was tagged by the addition of GFP to the N terminus and expressed at low levels (in the presence of thiamine) under the nmt41x promoter. The nmt41x-GFP-pab1 construct was cloned into the plasmid pJK148 and integrated by homologous recombination into the leu1 locus in pab1-deleted cells (see materials and methods). These cells had normal morphology, indicating that the tagged Pab1 protein is functional. Examination of living cells indicated that GFP-Pab1 is found in the cytoplasm, but the PP2A B-subunit is more concentrated in the nucleus and also localizes at the mitotic spindle (Figure 1, C and D). As described below this section, Pab1 interacts with SPB proteins, suggesting that this PP2A B-subunit might be associated with this subcellular structure. However, to date we have not observed Pab1 at the SPB or at the medial ring, as occurs for different SIN components (Krapp and Simanis 2008) and the PP2A B′ regulatory subunit Par1 (Le Goff et al. 2001). Curiously, the localization of Pab1 is similar to that described for the microtubule-associated kinase-like MAST-L (hGwl) in human cells, a protein that directly inhibits PP2A activity (Burgess et al. 2010).
Construction of diploid strains with the phenotype pab1-4/pab1+ indicated that pab1-4 is recessive (according to a pab1 loss-of-function mutation) with respect to cell morphology and etd1-1 suppression phenotypes. In agreement with this analysis, we found that a pab1Δ null allele also suppressed lethality of etd1-deficient cells (data not shown). Since Etd1 is a positive regulator of SIN (Daga et al. 2005; Garcia-Cortes and McCollum 2009), the ability of pab1-4 and pab1Δ to suppress etd1 mutants suggests that Pab1 might act as an inhibitor of the SIN pathway.
A molecular function for PP2A-Pab1 in negative regulation of Spg1:
In S. pombe, Cdc16 functions as a GAP that inhibits SIN via negative regulation of Spg1 (Furge et al. 1998). Since Pab1 might be acting as a negative regulator of SIN signaling, we wondered whether the effects of the mutants pab1-4 and cdc16-116 could be additive. As shown in Figure 2A, the double mutant cdc16-116 pab1-4 had a reduced nonpermissive temperature as compared to the single cdc16-116 mutant (30–32° in Figure 2A). At 32°, these cdc16-116 pab1-4 cells were round, according to morphogenesis defects conferred by the pab1-4 mutation, and enhanced the cdc16-116 multi-septated phenotype, consistent with a negative role for Pab1 in the regulation of SIN (Figure 2B).
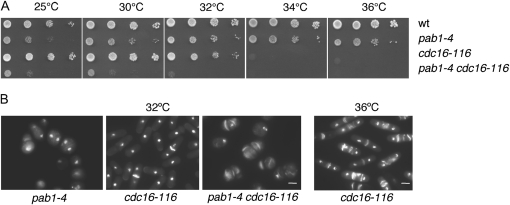
Figure 2.—
Negative regulation of cytokinesis by Pab1. (A) Serial dilution of cdc16-116 pab1-4 double mutant and the single mutants to test for potential additive effects of SIN inhibitors growing at 25°, 30°, 32°, 34°, and 36°. (B) pab1-4, cdc16-116, and pab1 cdc16-116 cells were DAPI- and calcofluor-stained after 8 hr incubation at 32° (first three panels from left). DAPI-calcofluor staining of cdc16-116 cell at 36° (restrictive temperature for this strain) is also shown (far right panel).
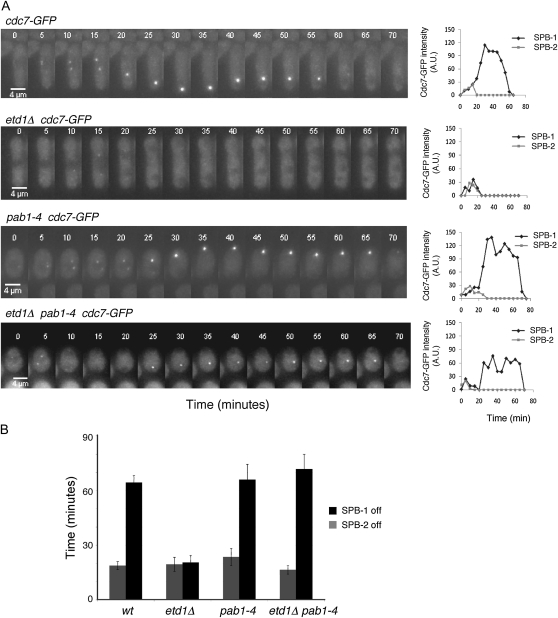
Etd1 is an essential component of the SIN (Daga et al. 2005). A recent study has shown that this protein is a positive regulator of Spg1 and a potential homolog of the budding yeast Lte1 (Garcia-Cortes and McCollum 2009). To determine the mechanisms by which pab1-4 suppresses etd1 deficiencies, we studied Spg1 activity in etd1Δ, pab1-4, and etd1Δ pab1-4 mutant cells. Cdc7 binds only the active (GTP-bound) form of Spg1 (Sohrmann et al. 1998). Thus, the nucleotide-binding state of Spg1 can be indirectly monitored through the binding of Cdc7 to the SPB. Cdc7-GFP localization by time-lapse microscopy was used for this purpose. In wild-type cells (used as a control), Cdc7-GFP localized to both SPBs in early mitosis and then to only one SPB during anaphase B. Similar results were observed in pab1-4 cells (Figure 3A). Etd1 is required to maintain Spg1 activity during late anaphase (Daga et al. 2005; Garcia-Cortes and McCollum 2009). Accordingly, Spg1 activity decreased prematurely in etd1 mutant cells (Figure 3A). Interestingly, the pab1-4 mutation restored Spg1 activity during anaphase B in cells lacking Etd1 (etd1Δ pab1-4 mutant in Figure 3A and in supporting information, Figure S1), counteracting the defect in Spg1 activation caused by the inactivity of Etd1. Examination of 10 cells for each strain by time-lapse microscopy confirmed this observation (average time of SPB activation is represented in Figure 3B). Thus, PP2A-Pab1 might function as a negative regulator of Spg1, antagonizing Etd1 activity.
Figure 3.—
Effects of pab1-4 in Spg1 activity. (A) Cdc7-GFP was imaged by time-lapse microscopy at 5-min intervals on a single focal plane in living wild-type cells and in etd1Δ, pab1-4, and etd1Δ pab1-4 mutant cells as indicated. Fluorescence intensity was quantified (arbitrary units) and represented for each SPB (SPB-1 and SPB-2). Bar, 4 μm. (B) Average of the time that SPBs remained active. Time (minutes) from initial SPB activation until the SPB was switched off was determined for each SPB (SPB-1 and SPB-2) in 10 cells per strain. Error bars represent standard deviations.
We cannot exclude that PP2A-Pab1 could directly inhibit Spg1; however, we failed to detect physical interactions between Pab1 and this GTPase (data not shown). Phosphorylation and dephosphorylation have been shown to regulate GAP function in a number of different eukaryotic cells (Sopko et al. 2007; Zheng et al. 2007; Toure et al. 2008; McAvoy et al. 2009; Mori et al. 2009; Wolfe et al. 2009). Therefore, we hypothesize that PP2A-Pab1 inhibition of Spg1 is more likely to operate indirectly, perhaps through activating dephosphorylation of the Cdc16-Byr4 two-component GAP. Under this hypothesis, Etd1 might antagonize PP2A-Pab1 function through Spg1 GAP(s) inactivation and delocalization rather than by GEF activity stimulating Spg1 nucleotide exchange. In agreement with this possibility, Lte1 has been shown to activate Tem1 by regulating the localization of the GAP Bfa1 at the SPBs (Geymonat et al. 2009).
Suppression analysis of SIN mutants by pab1-4:
Protein phosphatase 2A plays a role in controlling cytokinesis and septum formation through its regulatory subunits. PP2A is a heterotrimer composed of a catalytic subunit (C), a scaffold (A), and a regulatory or targeting subunit (B or B′) (Mayer-Jaekel and Hemmings 1994; Virshup and Shenolikar 2009). In S. pombe, there are two genes encoding the B′-regulatory subunits, par1 and par2. The double null mutant is viable, but is sensitive to temperature (high and low) and stress and shows defects in septation and cleavage (Jiang and Hallberg 2000; Tanabe et al. 2001). Curiously, a mutation in the regulatory subunit B′ Par1 is able to suppress the septation defects of cdc11-136, cdc7-24, spg1-B8 (Le Goff et al. 2001), and sid2-250 mutants (Jin et al. 2006).
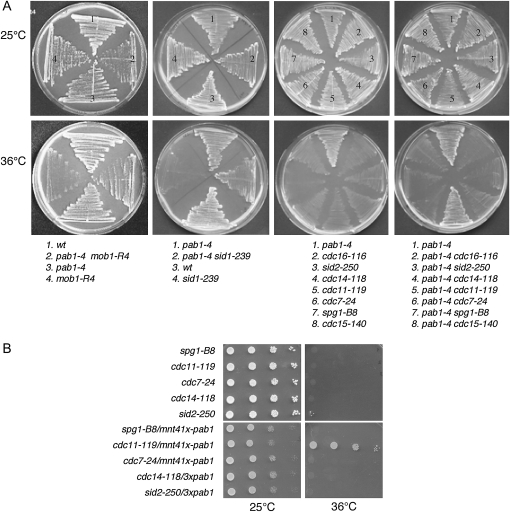
Since pab1-4 was isolated as a suppressor of the etd1-1 mutant, we also examined the effect of pab1-4 on other SIN components. To this end, we assessed the ability of pab1-4 to suppress the growth defects of other SIN mutants when grown at restrictive temperatures. As shown in Figure 4A, the double mutants of pab1-4 with mob1-R4, sid1-239, or cdc11-119 were able to grow at 36° (nonpermissive growth temperature for these SIN mutants), although the cytokinesis defects associated with the sin− phenotype of these cells were only partially rescued. Double mutants of pab1-4 with spg1-B8, cdc7-24 cdc14-118, or sid2-250 were unable to grow at 36° (Figure 4A). In these cases, the double mutants retained the round shape of pab1-4 cells but accumulated several nuclei as is typical of SIN mutants (data not shown). Therefore, unlike the par1 mutant, pab1-4 was able to suppress the growth defects of mob1-R4 and sid1-239, indicating that different regulatory subunits of PP2A might regulate different components of the SIN pathway. Interestingly, mutations in pab1 (Figure 4A) as well as in par1 (Le Goff et al. 2001) can rescue the lethality of cdc11 thermosensitive mutants, suggesting that the scaffold Cdc11 protein might be a common target for these PP2A B-and B′-subunits. PP2A-Par1 is required for dephosphorylation of Cdc11 at the end of mitosis, but in par1 mutant cells, not all Cdc11 protein remains hyperphosphorylated, suggesting that Cdc11 dephosphorylation is due to the action of PP2A-Par1 and to another protein phosphatase(s) yet to be identified (Krapp et al. 2004). Since both par1 and pab1 mutants rescue cdc11 mutants, we are currently testing whether PP2A-Pab1 could be also involved in Cdc11 dephosphorylation.
Figure 4.—
Genetic interactions for pab1-4 with SIN mutants. (A) Growth of cdc15-140, spg1-B8, cdc7-24, cdc11-119, cdc14-118, sid2-250, cdc16-116, mob1-R4, and sid1-239 septation mutants at their permissive (25°) and restrictive (36°) temperature. Double mutants were used to assay for suppression of lethality of these septation mutants by pab1-4, as indicated. Wild type (wt) and pab1-4 strains were used as a control. (B) Growth of spg1-B8, cdc11-119, cdc7-24, cdc14-11, and sid2-250 septation mutants at their permissive (25°) and restrictive (36°) temperature, assessed by serial dilution drop tests on plates (10-fold dilutions starting with 104 cells). Moderated overexpression of pab1 (nmt41x-driven expression in the absence of thiamine) was used to assay for suppression of lethality by an excess of Pab1 in these septation mutants, as indicated.
It has been suggested that the B- and B′-subunits compete for binding to the core A-C dimer (Shu et al. 1997; Evans and Hemmings 2000). To test whether an excess of Pab1 could also suppress the lethality of sin− mutants, expression of pab1 was induced from the moderated mnt41x promoter (high expression level is toxic and pleiotropic) in spg1-B8, cdc11-119, cdc7-24 cdc14-118, or sid2-250 mutant strains, and cells shifted to 36°. As observed in Figure 4B, elevated expression of Pab1 failed to suppress lethality of all these mutants, except cdc11-119. On the basis of a binding competition model, increased expression of Pab1 would favor accumulation of PP2A with mostly B- rather than B′-subunits, mimicking Par1-deficient cells (Le Goff et al. 2001).
Physical interactions between Pab1 and SIN components:
Loss of Pab1 function efficiently suppresses etd1 mutations, suggesting that Etd1 and Pab1 are antagonistic regulators of a common target, namely Spg1 activity or its regulators. Partial rescue of cdc11-119, mob1-R4, and sid1-239 mutants by pab1-4 was also observed. Sid1-Cdc14 and Sid2-Mob1 complexes are assembled at the Cdc11-Sid4 scaffold and transduce SIN signaling downstream of Spg1. Thus, pab1-4–mediated hyper-activation of Spg1 could increase the residual SIN activity of these mutants, accounting for the incomplete suppression of their septation defects. However, we cannot discard the possibility that PP2A-Pab1 could regulate SIN signaling at different levels of the cascade. NDR/DBF2 family protein kinases, of which Sid2 is a member, are negatively regulated by PP2A in human cells (Millward et al. 1999), and PP2A complexes regulate the class II mMOB1, a homolog of the yeast protein Mob1, to modulate changes in the cytoskeleton (Moreno et al. 2001). Thus, the rescue of cdc11-119, mob1-R4, and sid1-239 mutants could also result from PP2A-Pab1 being a negative regulator of the Sid1-Cdc14 and Sid2-Mob1 complexes and/or of their assembly at the Cdc11-Sid4 scaffold.
According to the above genetic integrations (see also Figure 4A), we analyzed whether a physical interaction occurs between Pab1 and different components of the SIN. Consistently, two-hybrid assays allowed us to detect in vivo associations between the full-length version of Pab1 and Mob1 and Sid1 and Sid2 after growth selection in media without histidine and adenine (Figure 5A). By using a sensitive colorimetric method for the detection and quantification of yeast α-galactosidase activity, an interaction between Pab1 and Cdc11 was also detected (Figure 5B). Therefore, Pab1 might be a component of the SIN complex, suggesting that protein dephosphorylation mediated by PP2A-Pab1 might regulate SIN at different levels.
Figure 5.—
Physical interaction between Pab1 and SIN proteins. (A) Protein–protein interactions were examined using the yeast two-hybrid system. S. cerevisiae AH109 cells transformed with Gal4-binding domain-fused and Gal4 activation domain-fused genes were grown on SD without leucine, tryptophan, histidine, and adenine. (B) Interactions were also tested using a colorimetric assay to test for α-galactosidase activity. The constructions GA-T/GB-Lam and GA-T/GB-53 were used as negative and positive controls, respectively. (C) Western blot analysis of Pab1 (MycHis-pab1construct) and Mob1 (mob1-GFP construct) with anti-α-His and anti-α-GFP antibodies, respectively. Yeast extracts were purified using immobilized cobalt affinity from a Mob1-GFP strain expressing Myc6His-Pab1 protein from a plasmid. Lane 1: mob1-GFP strain; lane 2: wild-type strain expressing pREPMycHis-pab1; and lane 3: mob1-GFP strain expressing pREPMycHis-pab1. In lane 3, Mob1-GFP is copurified with Pab1 protein. (D) Model for the physical assembly of different SIN components and regulators in the SPB from Morrell et al. (2004). The Sid2-Cdc14 complex is shown to interact with Pab1.
In mammalian cells, PP2A complexes physically interact with the class II mMOB1 (Moreno et al. 2001). To further analyze the interaction of the PP2A-Pab1 complex with Mob1 in fission yeast, we studied this association using a histidine purification procedure. As shown in Figure 5C, Mob1-GFP copurified with Myc6His-Pab1 protein, suggesting that the Pab1 interactions described here in S. pombe might be of relevance in all eukaryotic cells.
Components and regulators of the SIN cascade are organized at the spindle pole body by the scaffold proteins Sid4 and Cdc11. On the basis of our observations described here, we propose how we believe Pab1 fits into the model of SIN component organization at the SPB (Morrell et al. 2004) (Figure 5D).
Coordination of the SIN cascade and the morphogenesis pathway:
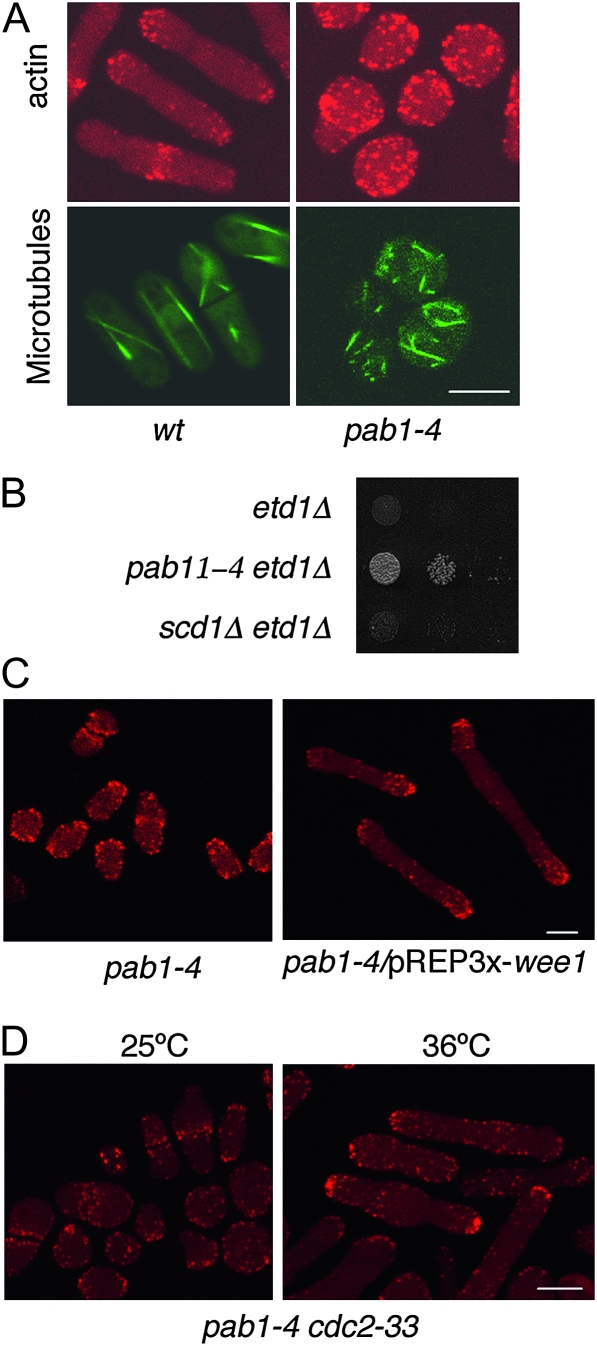
As mentioned above, the absence of Pab1 (pab1-4 and pab1Δ mutants) triggered septation even in the absence of Etd1 (Figure 1A), indicating that PP2A-Pab1 might act as a negative regulator of cytokinesis. At the same time, S. pombe pab1-4 cells lose growth polarity and manifest disorganized microtubule arrays and delocalized cortical actin patches (Figure 6A), a phenotype in agreement with previous studies of the pab1 null allele, which established a positive role for PP2A-Pab1 in morphogenesis (Kinoshita et al. 1996; Jiang and Hallberg 2000; Tanabe et al. 2001). Thus, PP2A-Pab1 seems to play a dual function: negative regulation of cell division with a positive role in morphogenesis.
Figure 6.—
Role for Pab1 in morphogenesis. (A) Actin staining using rhodamine-conjugated phalloidin (in red) and microtubule localization using Atb2-GFP fusion protein (in green) in wild-type (wt) and pab1-4 cells at 30°. (B) Suppression assay of etd1Δ cells lethality by scd1Δ, a null allele of the scd1 gene encoding a Cdc42 GEF required for morphogenesis. Growth of etd1Δ, etd1Δ pab1-4 (positive control), and etd1Δ scd1Δ cells was assessed by serial dilution drop tests on plates (10-fold dilutions starting with 104 cells). (C) Microscopic images of F-actin structures (rhodamine-conjugated phalloidin staining) in pab1-4 mutant cells (control) and pab1-4 cells overexpressing wee1 (pREP3x-wee1, nmt1-driven expression after 18 hr of derepression at 25°). (D) Microscopic images of F-actin (rhodamine-conjugated phalloidin staining) in pab1-4 cdc2-33 cells at 25° (permissive temperature as a control) and after 6 hr incubation at 36° (restrictive temperature for cdc2-33). Bars, 5 μm.
In fission yeast, cell polarity and morphogenesis are thought to be regulated by the product of the Ras homolog, ras1, which acts upstream of the Cdc42 GTPase and activates the serine/threonine kinase Pak1p/Shk1p/Orb2. This kinase maintains the cell in a polarized state during interphase in an (NDR-kinase) Orb6-dependent manner (Verde et al. 1998; Das et al. 2009). We observed that the scd1Δ mutation was unable to rescue the lethality of etd1Δ mutant cells (Figure 6B). The scd1 gene encodes a Cdc42 GEF (Das et al. 2009), and scd1Δ cells are round, indicating that suppression of etd1 mutants by pab1-4 is not indirectly caused by loss of cell polarity.
Polar growth and cell division seem to be inter-regulated at different levels. A number of regulatory proteins of the morphogenesis pathway have been reported to interact with members of the SIN cascade. For example, a possible role in S. pombe cytokinesis for Cdc42, which also localizes to the medial region, has been described (Coll et al. 2007). Similarly, phosphorylation of the myosin regulatory light chain by Orb2 has been shown to delay cytokinesis (Loo and Balasubramanian 2008). Pmo25 is essential for Orb6 kinase activity. The localization of Pmo25 at the SPBs and the kinase activities of Nak1-Orb6 during interphase are under the control of the Cdc7 and Sid1 SIN kinases, suggesting a functional linkage between SIN and the network for cell morphogenesis (Kanai et al. 2005). The antagonistic role of Etd1 and PP2A-Pab1 in SIN regulation described in this study represents a new connection between cytokinesis and morphogenesis. This dual function of Pab1 in activating polar growth and inhibiting septation suggests that PP2A-Pab1 might be involved in coordinating morphogenesis and cytokinesis.
The kinase Orb6 is proposed to act through the Wee1-Cdc2 pathway (Verde et al. 1998). Interestingly, an excess of Wee1 restored cell polarity to pab1-4 mutant cells. As shown in Figure 6, C and D, F-actin patches, a marker of the growing zones in the cell (Marks et al. 1986), localized to the cell tips in pab1-4 mutant cells lacking Cdc2 activity either by overexpression of wee1 or by direct inactivation in a cdc2-33 background at the restrictive temperature. Communication between the universal cell cycle regulator Cdc2 and cell morphology factors is essential to coordinate the cell cycle and cell morphology in eukaryotic cells (Moseley and Nurse 2009). This result suggests that PP2A-Pab1 phosphatase is involved in morphogenesis by inactivating Cdc2 (Kinoshita et al. 1996; Janssens and Goris 2001; Forester et al. 2007) and/or by counteracting its activity through dephosphorylation of Cdc2-phosphorylated substrates (Sopko et al. 2007; Zheng et al. 2007; Burgess et al. 2010; Lorca et al. 2010).
The genetic and molecular evidence described in this study suggest that PP2A-Pab1 and Etd1 antagonistically regulate Spg1 GTPase activity. Pab1 interacts with other components of the SIN, suggesting that PP2A-Pab1 may also act at other levels of the SIN cascade. During the cell cycle, eukaryotic cells alternate cell growth and cytokinesis. The dual function of Pab1 in activating polar growth and in inhibiting septation shown here suggests that PP2A-Pab1 might be involved in the molecular mechanisms that coordinate these two events. Further studies are required to determine the role of PP2A-Pab1 complexes and Etd1 in GTPase regulation and to provide new insights into the crosstalk established here between pathways controlling cell morphology and cell division.
Acknowledgments
We are grateful to Paul Nurse and Pilar Perez for providing strains and plasmids. We thank members of the Centro Andaluz de Biología del Desarrollo cell cycle group, Viesturs Simanis, John R. Pearson, and Ann Yonetani for useful comments and discussions. This work was supported by grants from the Ministerio de Ciencia e Innovación of the Spanish Government (grants BFU2009-13565 and BFU2010-21310) and by the Consejería de Economía, Innovación y Ciencia of the Junta de Andalucía.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.121368/DC1.
References
- Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie, III et al., 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14 943–951. [DOI] [PubMed] [Google Scholar]
- Bardin, A. J., R. Visintin and A. Amon, 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102 21–31. [DOI] [PubMed] [Google Scholar]
- Bathe, M., and F. Chang, 2009. Cytokinesis and the contractile ring in fission yeast: towards a systems-level understanding. Trends Microbiol. 18 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, A., S. Vigneron, E. Brioudes, J. C. Labbé, T. Lorca et al., 2010. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc. Natl. Acad. Sci. USA 107 12564–12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti, L., and V. Simanis, 1999. Asymmetry of the spindle pole bodies and Spg1 GAP segregation during mitosis in fission yeast. J. Cell Sci. 112(Pt. 14): 2313–2321. [DOI] [PubMed] [Google Scholar]
- Coll, P. M., S. A. Rincon, R. A. Izquierdo and P. Perez, 2007. Hob3p, the fission yeast ortholog of human BIN3, localizes Cdc42p to the division site and regulates cytokinesis. EMBO J. 26 1865–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven, R. A., D. J. Griffiths, K. S. Sheldrick, R. E. Randall, I. M. Hagan et al., 1998. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene 221 59–68. [DOI] [PubMed] [Google Scholar]
- Daga, R. R., A. Lahoz, M. J. Munoz, S. Moreno and J. Jimenez, 2005. Etd1 is a novel protein that links the SIN cascade with cytokinesis. EMBO J. 24 2436–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, M., D. J. Wiley, X. Chen, K. Shah and F. Verde, 2009. The conserved NDR kinase Orb6 controls polarized cell growth by spatial regulation of the small GTPase Cdc42. Curr. Biol. 19 1314–1319. [DOI] [PubMed] [Google Scholar]
- Evans, D. R., and B. A. Hemmings, 2000. Important role for phylogenetically invariant PP2Acα active site and C-terminal residues revealed by mutational analysis in Saccharomyces cerevisiae. Genetics 156 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forester, C. M., J. Maddox, J. V. Louis, J. Goris and D. M. Virshup, 2007. Control of mitotic exit by PP2A regulation of Cdc25C and Cdk1. Proc. Natl. Acad. Sci. USA 104 19867–19872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furge, K. A., K. Wong, J. Armstrong, M. Balasubramanian and C. F. Albright, 1998. Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr. Biol. 8 947–954. [DOI] [PubMed] [Google Scholar]
- Garcia-Cortes, J. C., and D. McCollum, 2009. Proper timing of cytokinesis is regulated by Schizosaccharomyces pombe Etd1. J. Cell Biol. 186 739–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat, M., A. Spanos, S. J. Smith, E. Wheatley, K. Rittinger et al., 2002. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J. Biol. Chem. 277 28439–28445. [DOI] [PubMed] [Google Scholar]
- Geymonat, M., A. Spanos, G. de Bettignies and S. G. Sedgwick, 2009. Lte1 contributes to Bfa1 localization rather than stimulating nucleotide exchange by Tem1. J. Cell Biol. 187 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens, V., and J. Goris, 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353 417–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W., and R. L. Hallberg, 2000. Isolation and characterization of par1(+) and par2(+): two Schizosaccharomyces pombe genes encoding B′ subunits of protein phosphatase 2A. Genetics 154 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez, J., and J. Oballe, 1994. Ethanol-hypersensitive and ethanol-dependent cdc- mutants in Schizosaccharomyces pombe. Mol. Gen. Genet. 245 86–95. [DOI] [PubMed] [Google Scholar]
- Jin, Q. W., M. Zhou, A. Bimbo, M. K. Balasubramanian and D. McCollum, 2006. A role for the septation initiation network in septum assembly revealed by genetic analysis of sid2–250 suppressors. Genetics 172 2101–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai, M., K. Kume, K. Miyahara, K. Sakai, K. Nakamura et al., 2005. Fission yeast MO25 protein is localized at SPB and septum and is essential for cell morphogenesis. EMBO J. 24 3012–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, K., T. Nemoto, K. Nabeshima, H. Kondoh, H. Niwa et al., 1996. The regulatory subunits of fission yeast protein phosphatase 2A (PP2A) affect cell morphogenesis, cell wall synthesis and cytokinesis. Genes Cells 1 29–45. [DOI] [PubMed] [Google Scholar]
- Krapp, A., and V. Simanis, 2008. An overview of the fission yeast septation initiation network (SIN). Biochem. Soc. Trans. 36 411–415. [DOI] [PubMed] [Google Scholar]
- Krapp, A., E. Cano and V. Simanis, 2004. Analysis of the S. pombe signalling scaffold protein Cdc11p reveals an essential role for the N-terminal domain in SIN signalling. FEBS Lett. 565 176–180. [DOI] [PubMed] [Google Scholar]
- Lattmann, E., A. Krapp and V. Simanis, 2009. Cytokinesis: closure resets your SIN. Curr. Biol. 19 R1040–R1042. [DOI] [PubMed] [Google Scholar]
- Le Goff, X., A. Woollard and V. Simanis, 1999. Analysis of the cps1 gene provides evidence for a septation checkpoint in Schizosaccharomyces pombe. Mol. Gen. Genet. 262 163–172. [DOI] [PubMed] [Google Scholar]
- Le Goff, X., S. Buvelot, E. Salimova, F. Guerry, S. Schmidt et al., 2001. The protein phosphatase 2A B'-regulatory subunit par1p is implicated in regulation of the S. pombe septation initiation network. FEBS Lett. 508 136–142. [DOI] [PubMed] [Google Scholar]
- Loo, T. H., and M. Balasubramanian, 2008. Schizosaccharomyces pombe Pak-related protein, Pak1p/Orb2p, phosphorylates myosin regulatory light chain to inhibit cytokinesis. J. Cell Biol. 183 785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca, T., C. Bernis, S. Vigneron, A. Burgess, E. Brioudes et al., 2010. Constant regulation of both the MPF amplification loop and the Greatwall-PP2A pathway is required for metaphase II arrest and correct entry into the first embryonic cell cycle. J. Cell Sci. 123 2281–2291. [DOI] [PubMed] [Google Scholar]
- Marks, J., I. M. Hagan and J. S. Hyams, 1986. Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J. Cell Sci. Suppl. 5 229–241. [DOI] [PubMed] [Google Scholar]
- Maundrell, K., 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123 127–130. [DOI] [PubMed] [Google Scholar]
- Mayer-Jaekel, R. E., and B. A. Hemmings, 1994. Protein phosphatase 2A: a ‘menage a trois’. Trends Cell Biol. 4 287–291. [DOI] [PubMed] [Google Scholar]
- Mayer-Jaekel, R. E., H. Ohkura, R. Gomes, C. E. Sunkel, S. Baumgartner et al., 1993. The 55 kd regulatory subunit of Drosophila protein phosphatase 2A is required for anaphase. Cell 72 621–633. [DOI] [PubMed] [Google Scholar]
- McAvoy, T., M. M. Zhou, P. Greengard and A. C. Nairn, 2009. Phosphorylation of Rap1GAP, a striatally enriched protein, by protein kinase A controls Rap1 activity and dendritic spine morphology. Proc. Natl. Acad. Sci. USA 106 3531–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward, T. A., S. Zolnierowicz and B. A. Hemmings, 1999. Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem. Sci. 24 186–191. [DOI] [PubMed] [Google Scholar]
- Moreno, C. S., W. S. Lane and D. C. Pallas, 2001. A mammalian homolog of yeast MOB1 is both a member and a putative substrate of striatin family-protein phosphatase 2A complexes. J. Biol. Chem. 276 24253–24260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., A. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194 795–823. [DOI] [PubMed] [Google Scholar]
- Mori, K., M. Amano, M. Takefuji, K. Kato, Y. Morita et al., 2009. Rho-kinase contributes to sustained RhoA activation through phosphorylation of p190A RhoGAP. J. Biol. Chem. 284 5067–5076. [DOI] [PubMed] [Google Scholar]
- Morrell, J. L., G. C. Tomlin, S. Rajagopalan, S. Venkatram, A. S. Feoktistova et al., 2004. Sid4p-Cdc11p assembles the septation initiation network and its regulators at the S. pombe SPB. Curr. Biol. 14 579–584. [DOI] [PubMed] [Google Scholar]
- Moseley, J. B., and P. Nurse, 2009. Cdk1 and cell morphology: connections and directions. Curr. Opin. Cell Biol. 21 82–88. [DOI] [PubMed] [Google Scholar]
- Norbury, C., and S. Moreno, 1997. Cloning cell cycle regulatory genes by transcomplementation in yeast. Methods Enzymol. 283 44–59. [DOI] [PubMed] [Google Scholar]
- Pereira, G., T. Hofken, J. Grindlay, C. Manson and E. Schiebel, 2000. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell 6 1–10. [PubMed] [Google Scholar]
- Pollard, T. D., and J. Q. Wu, 2010. Understanding cytokinesis: lessons from fission yeast. Nat. Rev. Mol. Cell Biol. 11 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, Y., H. Yang, E. Hallberg and R. Hallberg, 1997. Molecular genetic analysis of Rts1p, a B' regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol. Cell. Biol. 17 3242–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell, V., and P. Nurse, 1993. Investigations into the control of cell form and polarity: the use of morphological mutants in fission yeast. Dev. Suppl. 1993 289–299. [PubMed] [Google Scholar]
- Sohrmann, M., S. Schmidt, I. Hagan and V. Simanis, 1998. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev. 12 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko, R., D. Huang, J. C. Smith, D. Figeys and B. J. Andrews, 2007. Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J. 26 4487–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, O., D. Hirata, H. Usui, Y. Nishito, T. Miyakawa et al., 2001. Fission yeast homologues of the ‘;subunit of protein phosphatase 2A: multiple roles in mitotic cell division and functional interaction with calcineurin. Genes Cells 6 455–473. [DOI] [PubMed] [Google Scholar]
- Toure, A., R. Mzali, C. Liot, L. Seguin, L. Morin et al., 2008. Phosphoregulation of MgcRacGAP in mitosis involves Aurora B and Cdk1 protein kinases and the PP2A phosphatase. FEBS Lett. 582 1182–1188. [DOI] [PubMed] [Google Scholar]
- Verde, F., D. J. Wiley and P. Nurse, 1998. Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc. Natl. Acad. Sci. USA 95 7526–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virshup, D. M., and S. Shenolikar, 2009. From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell 33 537–545. [DOI] [PubMed] [Google Scholar]
- Wolfe, B. A., and K. L. Gould, 2005. Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 15 10–18. [DOI] [PubMed] [Google Scholar]
- Wolfe, B. A., T. Takaki, M. Petronczki and M. Glotzer, 2009. Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 7 e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X. D., R. T. Lee, Y. M. Wang, Q. S. Lin and Y. Wang, 2007. Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 26 3760–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]