Abstract
In Drosophila, where females mate multiply, sperm competition contributes strongly to fitness variability among males. Males transfer “Acp” seminal proteins to females during mating, and these proteins influence the outcome of sperm competition. Because Acps function within the female, male proteins can directly interact with female molecules in a manner that affects reproductive fitness. Here we begin to dissect the genetic architecture of male × female interactions underlying reproductive phenotypes important to sperm competition. By utilizing chromosome extraction lines, we demonstrate that the third and X chromosomes each have large effects on fertility phenotypes, female remating rate, and the sperm competition parameter, P1. Strikingly, the third and X chromosomes harbor genetic variation that gives rise to strong male × female interactions that modulate female remating rate and P1. Encoded on these chromosomes are, respectively, sex peptide (SP) and sex peptide receptor (SPR), the only pair of physically interacting male Acp and female receptor known. We identified several intriguing allelic interactions between SP and SPR. The results of this study begin to elucidate the complex genetic architecture of reproductive and sperm competition phenotypes and have significant implications for the evolution of male and female characters.
IN most organisms, extensive variation in reproductive success exists. The male and female effects of this variation are often strongly nonadditive. In Drosophila, where multiple matings occur frequently (Harshman and Clark 1998; Imhof et al. 1998), one major source of variation in male reproductive success is sperm competition. Sperm competition occurs when sperm from multiple males are present within a female. In the laboratory, sperm competition trials often involve mating a female with two different males in timed succession and measuring the proportion of progeny from each male. The magnitude of among-male variation in sperm competition is enormous, with some males appearing >10 times as successful as other males at competing (Fiumera et al. 2007). To maintain such a high level of fitness variation in a population (Hughes 1997), there must be high levels of functional polymorphism in genes underlying sperm competition.
Drosophila accessory gland proteins (Acps) are critical components of the seminal fluid, mediating many physiological and behavioral changes in the mated female. These changes include effects on egg production and egg laying rate, sperm storage patterns, expression of antimicrobial peptides, feeding rate, remating rate, longevity, locomotion, and sleep patterns (reviewed in Wolfner 2009; see also Isaac et al. 2010). Over 100 Acps have been identified (e.g., see Ravi Ram and Wolfner 2007a; Findlay et al. 2008, 2009). Of particular relevance to this study, Acps have been shown to be critical to sperm competitive success (Harshman and Prout 1994; Chapman et al. 2000; Wong et al. 2008a; Avila and Wolfner 2009), and Acp alleles are associated with sperm competition outcomes (Clark et al. 1995; Fiumera et al. 2005, 2007).
Sperm competition is subject to sexual selection due to the differing reproductive “interests” of the sexes (Parker 1970). For evolution by sexual selection to occur, population level variation must exist in alleles for Acps and other reproductive proteins influencing success in sperm competition. Indeed, studies have demonstrated that some Acps have high levels of variation (Aguadé et al. 1992; Tsaur and Wu 1997; Tsaur et al. 1998; Swanson et al. 2001; Swanson and Vacquier 2002; Haerty et al. 2007) that show nonneutral patterns of evolution (Civetta and Singh 1998; Swanson et al. 2001; Swanson and Vacquier 2002; Begun and Lindfors 2005; Mueller et al. 2005; Wong et al. 2008b; Kelleher and Markow 2009). Extraordinarily high levels of sequence variation have been found in male reproductive proteins across many taxa (Swanson and Vacquier 2002). It remains a mystery why such important genes controlling reproductive fitness maintain such high levels of variation. One possible mechanism for maintaining polymorphism is antagonistic pleiotropy. This phenomenon results in a trade-off between effects of alleles where, as second males, a particular allele results in more offspring sired, but as a first male, the same allele results in reduced ability to prevent remating. This trade-off may maintain genetic variation that benefits one sperm competition trait at the expense of another. Antagonistic pleiotropy has been demonstrated for several Acp genes (Fiumera et al. 2007). Furthermore, some of these genes harbor natural polymorphisms that are associated with large differences in sperm competition outcomes (Harshman and Prout 1994; Clark et al. 1995; Fiumera et al. 2005, 2007), and some of this variation in outcome might be explained by genetic interactions between the sexes.
Sperm competition is characterized by complex genetic interactions, some of which are readily quantified experimentally as male × female genotypic interaction (Clark and Begun 1998; Clark et al. 1999). A previous small study demonstrated that a male's sperm competitive ability as a first male (his P1 score) or as a second male (his P2 score) depends on the genotype of the female with whom he is mating (Clark et al. 1999). Rank order of male sperm competitive ability depends on female genotype. While male × female genotypic interactions are known to influence sperm competition outcomes, the generality, magnitude, and specific molecular genetic basis of these interactions are unknown.
In addition to complex male × female genotypic interactions, both male and female genotypes individually affect sperm competition parameters and other reproductive phenotypes (Clark et al. 1995, 1999; Clark and Begun 1998; Fiumera et al. 2005, 2007; Civetta et al. 2008). Male genotypic effects have been documented for female remating rate and male P1 score (Clark et al. 1995, 1999; Clark and Begun 1998; Fiumera et al. 2005, 2007; Civetta et al. 2008). Functional studies of specific Acps are providing us with a good molecular understanding of at least some male contributions to the male × female interaction (Harshman and Prout 1994; Chapman et al. 2000; Wong et al. 2008a; Avila and Wolfner 2009; Fricke et al. 2009). However, while female genotypic effects have been documented for the female's egg laying rate and the male's P1 and P2 scores (Clark et al. 1995, 1999; Clark and Begun 1998; Fiumera et al. 2005, 2007; Civetta et al. 2008), the female molecular contributions to the interactions that underlie sperm competition are unknown.
Only one female protein is known to interact with an Acp: the sex peptide receptor (SPR) (FBgn0029768) interacts with the 36-amino-acid sex peptide (SP) (Acp70A; FBgn0003034) (Yapici et al. 2008). SPR is a G-protein–coupled receptor expressed mainly in the female reproductive tract, in fru+ neurons in the brain, and in fru+ppk+ neurons of the female reproductive tract. SPR expression in the fru+ppk+ neurons innervating the female reproductive tract is necessary and sufficient for egg laying and receptivity changes induced in the female by receipt of SP in the male's seminal fluid (Hasemeyer et al. 2009; Yang et al. 2009).
In this study, we begin to dissect the genetic architecture of male × female interactions by examining the interaction between the third chromosome in males and the X chromosome in females. We identify novel male effects, female effects, and male × female interactions on important measures of reproductive fitness and sperm competition. More importantly, we demonstrate that the third chromosome and the X chromosome have strong individual and interactive effects on the phenotypes measured. We tested for interaction between SP and SPR and found several intriguing allelic interactions. Our results have important implications for the evolution of male and female reproductive fitness.
MATERIALS AND METHODS
Drosophila melanogaster fly cultures:
We used 90 third chromosome extraction lines previously reported (Fiumera et al. 2007) and 51 X chromosome extraction lines previously reported (Hill-Burns and Clark 2009). Briefly, each line captures a unique, wild-derived chromosome (3 or X, respectively) and renders it homozygous in a genetic background that is co-isogenic across all lines. Thus, genomes of the X chromosome extraction lines differ only by the X chromosome, and the genomes of the third chromosome extraction lines differ only by the third chromosome. Flies in all chromosome extraction lines have wild-type, red eyes. We sequenced SP or SPR in the third and X chromosome extraction lines, respectively, and, on the basis of the results, chose 10 lines of each for this study. The experimental lines were chosen to maximize genetic and protein diversity in SP and SPR. Males from the third chromosome extraction lines and females from the X chromosome extraction lines were used in the sperm competition assays. In the double matings described, second males were from a bwD stock previously described (Clark and Begun 1998). All flies were collected under CO2 anesthesia and aged 4–7 days in single-sex vials of 20–30 flies. All flies were maintained on standard agar–dextrose–yeast media and housed at 24° on a 12-hr light/dark cycle.
Sperm competition assays:
A 10 × 10 crossing scheme was employed. Males from 10 third chromosome extraction lines were tested against females from 10 X chromosome extraction lines, for a total of 100 separate cross combinations. This design differs from a standard diallel because we are not testing attributes of F1 flies. Instead this design tests the genetic contribution of the male (third chromosome line) and the female (X chromosome line) in each of the 100 pairwise mating combinations. The progeny are scored for eye color to ascertain the sperm competition and progeny phenotypes of the parental lines. All 100 crosses were replicated 18 times in three separate blocks. For each of the 100 crosses, six males were mass mated to six females for 12 hr (overnight) on day 0 (first mating). On day 1, males were discarded and each female was aspirated into an individual vial (vial 1) and allowed to lay eggs for 24 hr. On day 2, each female was aspirated into a new vial (vial 2) and allowed to lay eggs for 24 hr. On day 3, each female was aspirated into a new vial (vial 3), and 2 bwD males were placed with each female for 12 hr (overnight; second mating). On day 4, bwD males were discarded and each female was aspirated into a new vial (vial 4) and allowed to lay eggs for 48 hr. On day 6, each female was aspirated into a new vial (vial 5) and allowed to lay eggs for 48 hr. On day 8, females were discarded.
Egg laying rates were scored at 24 and 48 hr after the first mating. Eggs were counted in vials 1 and 2 within 8 hr of removal of females. Total egg count reported is the total number of eggs laid in vials 1 and 2. Progeny were allowed to eclose in vials 1–5 and were scored for eye color. Red-eyed progeny were sired by the third chromosome extraction line male (first male). Brown-eyed progeny were sired by the bwD male (second male). Hatchability/viability is the proportion of eggs that hatched and survived to adulthood (progeny no./egg no. in vials 1 and 2). Remating rate is the proportion of females that remate (of a total of 18 replicates), determined by the presence of one or more brown-eyed progeny in vials 3–5. P1 is the proportion of progeny from the first male after the second mating, calculated as the proportion of red-eyed progeny after the second mating (red/(red + brown)). P1 scores were calculated from vials 4 and 5 of the crosses that remated. Because remating rate was calculated from vials 3–5, a P1 score can be 100% if red-eyed flies were present in vial 3, but not in vials 4 and 5. We did not perform the corresponding P2 experiments for third chromosome extraction line males because no previous studies had implicated a role for SP or SPR in P2.
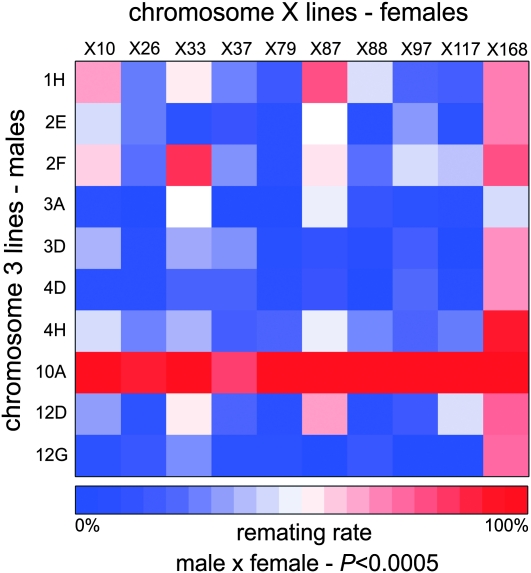
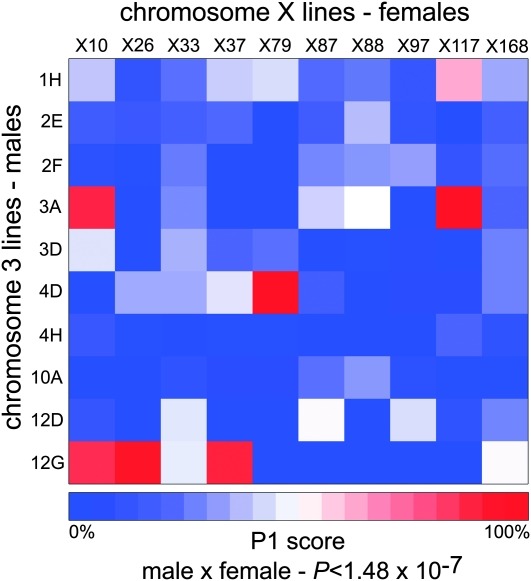
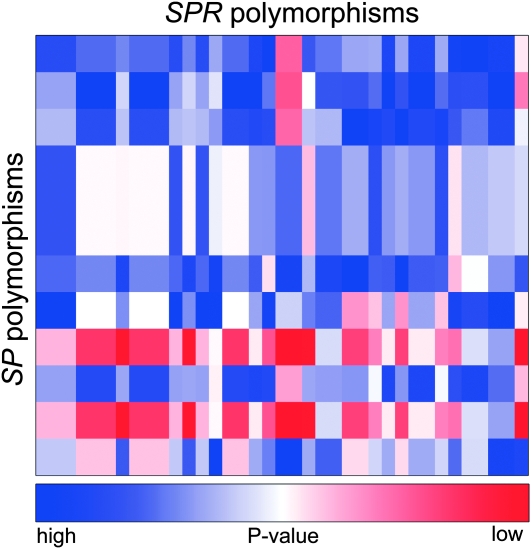
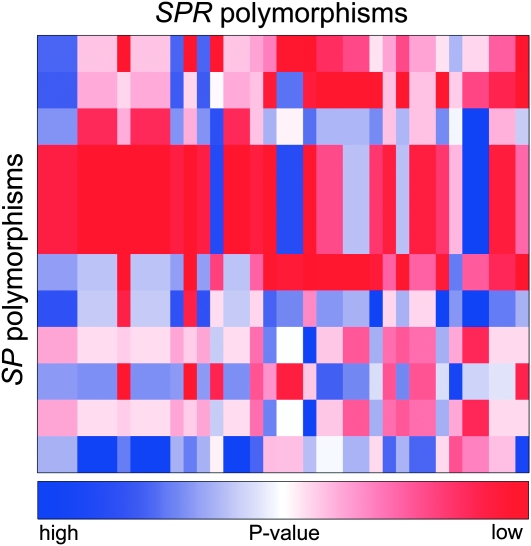
We constructed a heat map to visualize the remating rates and P1 scores from the 100 crosses. This provided a quick visualization of the quantitative data. These heat maps are generated in R with the standard heatmap() function in the package MASS (Venables and Ripley 2002). The colors represent remating rates in Figure 1, average P1 scores in Figure 2, and P-values for allelic interactions in Figures 3 and 4. We found this to be an effective way to convey differences between genotype combinations, particularly when multiple genotypes are involved in a large sperm competition experiment.
Figure 1.—
Remating rates are characterized by extensive male × female interactions. Significant male × female interactions ultimately determine the female remating rate. Male lines are represented by each row. Female lines are represented by each column. Colors represent remating rates. Significance was tested by a log-linear model.
Figure 2.—
P1 scores are driven by male × female interactions. Significant male × female interactions ultimately determine the success of the first male. Male lines are represented by each row. Female lines are represented by each column. Colors represent P1 scores. Significance was tested by a linear model.
Figure 3.—
Female remating rates are determined by SP × SPR polymorphism interactions. Polymorphisms in SP interact with polymorphisms in SPR to give the female remating rate. Male SP polymorphisms are represented by rows and female SPR polymorphisms are represented by columns. Several SP polymorphisms are in linkage disequilibrium and several SPR polymorphisms are in linkage disequilibrium, reducing the total number of interactions possible. Colors represent the uncorrected P-value of each interaction. Dark blue represents higher P-value, less significant interactions. Dark red represents lower P-value, more significant interactions. Significance was tested by a log-linear model.
Figure 4.—
SP × SPR polymorphism interactions in P1. Polymorphisms in SP interact with polymorphisms in SPR to give the final P1 score of each genotypic pair. Male SP polymorphisms are represented by rows and female SPR polymorphisms are represented by columns. Several SP polymorphisms are in linkage disequilibrium and several SPR polymorphisms are in linkage disequilibrium, reducing the total number of interactions possible. Colors represent the uncorrected P-value of each interaction. Dark blue represents higher P-value, less significant interactions. Dark red represents lower P-value, more significant interactions. Significance was tested by a log-linear model.
Sequencing:
Genomic PCR was performed as in Fiumera et al. (2007). Briefly, genomic DNA was extracted from flies using a standard phenol/chloroform extraction. PCR was performed on the genomic DNA, using gene-specific primers, and products were visualized on 1.5% agarose gels. PCR products were purified with shrimp alkaline phosphatase and exonuclease I (Promega, Madison, WI). The BigDye Termination kit and appropriate sequencing primers were used for automated sequencing (Applied Biosystems, Foster City, CA). The samples were filtered through Sephadex columns (Amersham Biosciences, Piscataway, NJ). Sequencing reaction products were separated and scored on ABI 3730 sequencers by the sequencing facility at the Cornell University Life Sciences Core Laboratories Center.
Primers for the SP gene were designed to flank the entire genomic locus to yield an ∼500-bp product. The amplified product included both exons, UTRs, and the single intron. Primers for the SPR gene were designed to flank the coding exons (exons 3–6). Primers were placed 50–100 bp upstream and downstream of exon/intron boundaries. Exons 1 and 2 of SPR are noncoding exons, AT-rich, and difficult to amplify; thus these exons were excluded from analysis. All amplicons were sequenced from both directions.
Sequence traces were manually assembled and examined by eye (Sequencher). Samples containing singleton variants were reamplified and resequenced to ensure that the variant was not a PCR-induced error.
Polymorphisms are denoted by standard methods where a polymorphism in the coding region of a gene is indicated by a ‘c’ and its nucleotide position from the transcription start site (e.g., c234). Intron polymorphisms are indicated by the number of nucleotides from the start of the intron (e.g., intron 1 + 23). UTR polymorphisms are denoted as the number of nucleotides upstream or downstream of the start or the stop site of transcription, respectively (e.g., 3′-UTR − 15 or 5′-UTR + 15). All polymorphism notations include the major allele followed by the minor allele (e.g., A > T).
Statistical analysis:
All statistical analysis was performed in R (version 2.8.1; R Development Core Team). To identify male line, female line, and male × female effects, analysis of variance (ANOVA) was used to apply a simple linear model to each phenotype measured, similar to the analysis described in Clark et al. (1999). These models were applied for each of the phenotypes: egg laying, progeny hatchability, P1 score, and remating rate. The ANOVA model has both random and fixed effects, and so it is a mixed model. The mean of the phenotype (yijk) for the cross of the ith line of male with the jth line of female was
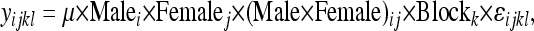 |
where the indexes for the male and the female genotypes are (1, 2, 3,…, 10) and there were three independent blocks. Recall that there were six replicates of each cross within each block.
To test the significance of interactions between SNPs within SP and SPR, the lines were recoded on the basis of their SNP genotypes, collapsing the 10 × 10 matrix of effects to 2 × 2 (homozygous lines for either of the two or three SNP alleles at each locus). From these collapsed data, the linear model to test for SNP interactions in egg laying, progeny hatchability, P1 score, and remating rate was
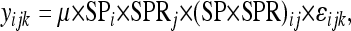 |
where yijk represents the phenotype of interest. Because there were 12 SNPs in SP and 37 in SPR, all nonredundant pairwise tests were performed, testing each SP polymorphic site for interaction with each polymorphic site in SPR. The SP × X chromosome and SPR × third chromosome tests were performed in a similar manner. Bonferroni correction was applied to infer significance in the face of multiple testing.
Each of the 10 × 10 crosses produces counts of females that successfully remated, and these counts were fitted by a log-linear model (Agresti 1990) using the R procedure loglm. Log-linear models are like an extension of a chi-square contingency table test, and terms in the model allow explicit testing for whether an interaction between lines or SP and SPR SNPs affects the counts of the response variable (remating rate). Bonferroni correction was applied to interaction P-values to correct for multiple testing. A total of 135 allele interaction tests were performed, rather than 444 tests (12 SP polymorphisms × 37 SPR polymorphisms) because there were several blocks of polymorphisms in strong linkage disequilibrium in SP and SPR (supporting information, Table S3 and Table S4).
qPCR:
Virgin male and female flies were collected and aged as virgins for 3–5 days. Fifteen flies per line were collected for RNA and flash frozen on dry ice. Total RNA was isolated with a standard phenol/chloroform extraction protocol and stored at −80°. cDNA was synthesized using a Promega kit. qPCR was performed with SYBR green reagents (Roche, Indianapolis). The qPCR reaction was performed on an ABI-7400 instrument and analysis of qPCR data was performed with the ABI Prism 7000 SDS software. SP qPCR primers span the exon 1–2 junction. SPR qPCR primers span the exon 5–6 junction. RP49 was used as a stably expressed control gene. Each line's expression was relative to a standard Canton-S laboratory strain. Six measurements were taken of each line: two biological replicates each containing three technical replicates.
Western blots:
Western blots were performed as previously reported (Liu and Kubli 2003). Male reproductive tracts were dissected from 3- to 5-day-old virgin flies. The SP antiserum (Liu and Kubli 2003) was kindly provided by Eric Kubli.
RESULTS
A 10 × 10 crossing scheme was employed for a total of 100 double-mating trials. All 100 crosses were replicated 18 times, and of these 1800 trials, 125 were removed because they produced no progeny. A total of 111,002 progeny were counted and scored, and from the progeny phenotypes, we inferred that 847 of the double-mating crosses in fact resulted in females producing progeny from both males. There were doubly mated females for all 100 of the distinct crosses.
Egg laying rate:
Egg laying rates after the first mating were scored 24 hr postmating (vial 1) and 48 hr postmating (vial 2). A total of 15,973 eggs (mean ± SD: 9.8 ± 5.4 per vial) were counted after the first 24 hr and 21,129 eggs (13.1 ± 7.1 per vial) were counted after 48 hr (Figure S1). For both 24 and 48 hr after mating, ANOVA tests indicated a significant male effect (24 hr, P < 0.0003; 48 hr, P < 2.2 × 10−16) and a significant female effect (24 hr, P < 2.2 × 10−16; 48 hr, P < 2.2 × 10−16) on egg laying. As expected from previous experience, there was a significant experimental block effect (24 hr, P < 0.0006; 48 hr, P < 0.001), but no significant male × female effect was detected for egg laying rate.
Progeny and hatchability/viability:
A total of 14,406 progeny (8.7 ± 5.3 per vial) resulted from the eggs counted at 24 hr postmating, and the corresponding count at 48 hr postmating was 18,427 (11.5 ± 7.2 per vial) (Figure S2). For both time points, there was a significant male effect (24 hr, P < 2.890 × 10−13; 48 hr, P < 2.2 × 10−16) and a significant female effect (24 hr, P < 2.2 × 10−16; 48 hr, P < 2.2 × 10−16) on progeny number. There was a marginally significant male × female effect on progeny number 24 hr after mating (P < 0.04), but not 48 hr after mating.
Hatchability/viability for the first mating was calculated from the number of eggs laid and the progeny eclosed for 24 and 48 hr postmating (0.85 ± 0.21 per vial; Figure S3). For 24 and 48 hr postmating, hatchability/viability showed a significant male effect (24 hr, P < 2.2 × 10−16; 48 hr, P < 2.2 × 10−16) and significant female effect (24 hr, P < 0.0002; 48 hr, P < 8.7 × 10−10). Similar to egg laying rate, there was a significant male × female effect for hatchability/viability at 24 hr (P < 0.021), but not at 48 hr.
Remating rate:
Remating rate was calculated as a proportion of the 18 replicate females that remated within a cross (0.35 ± 0.31 per cross; Figure S4). There was a significant male effect (P < 4.03 × 10−13) and significant female effect (P < 0.00038) on remating. Additionally, a significant male × female effect was also detected (P < 0.0005) (Figure 1).
P1 score:
P1 is the proportion of progeny from the first male after the second mating and was calculated from all females that remated with a bwD male. The P1 score (0.20 ± 0.31 per vial) was calculated from 4 days of egg laying, after the second mating (Figure S5). There was a significant male effect (P < 2.2 × 10−16) and significant female effect (P < 2.18 × 10−5). P1 also had the largest male × female interaction effect of all parameters measured (P < 1.48 × 10−7) (Figure 2).
Polymorphism in SP and SPR:
The entire SP genomic locus was sequenced in the 90 third chromosome extraction lines. Sequencing identified 14 sites with segregating polymorphism (Table S1 and Figure S6). One variant was a singleton and 13 variants occurred more than once. Twelve variants were SNPs and 2 variants were indels. One SNP was nonsynonymous and three SNPs were synonymous. The rest of the SNPs and indels were noncoding. There were 16 unique haplotypes represented among the 90 third chromosome extraction lines.
The four coding exons of SPR were sequenced in the 51 X chromosome extraction lines. There were a total of 37 polymorphic sites: 35 SNPs and two indels (Table S2 and Figure S7). Six SNPs were singletons and 29 variants occurred more than once. Two SNPs were nonsynonymous and 33 SNPs were synonymous. The rest of the SNPs were noncoding. Both indels occurred in the 5′-UTR. There were 42 unique haplotypes represented among the 51 X chromosome extraction lines.
Ten lines each from the third chromosome and X chromosome extraction lines were chosen for the double-mating experiments described above. The lines were selected to capture as much of the SP and SPR variation as possible. The third chromosome extraction lines represent nine unique SP haplotypes (Table S3). Twelve SP polymorphisms are segregating in the 10 third chromosome extraction lines that were used in the mating tests. The X chromosome extraction lines represent nine unique SPR haplotypes (Table S4). Thirty-seven SPR polymorphisms are segregating in the 10 X chromosome extraction lines that were used in the mating tests.
Associations of polymorphisms in SP and SPR with reproductive phenotypes:
Each polymorphism in SP and in SPR results in a partitioning of the homozygous lines into two or three groups, one for each of the alternative alleles (one SNP site was segregating with three different nucleotides). We asked whether there is evidence that each polymorphism is associated with differences in the measured phenotypes by performing simple t-tests for egg laying rate, progeny phenotypes, and P1 score and chi-square tests for female remating rate. In this study, with the exception of P1 score, approximately half of all SP and SPR polymorphisms have an effect on the phenotypes measured (Table 1, Table S5, and Table S6).
TABLE 1.
Proportion of individual SP and SPR polymorphisms with effects on reproductive phenotypes
| Gene | Egg total | Progeny total | Hatchability | Remating rate | P1 score |
|---|---|---|---|---|---|
| SP | 6/12 | 11/12 | 11/12 | 10/12 | 9/12 |
| SPR | 21/37 | 15/37 | 20/37 | 26/37 | 6/37 |
Specific SP × SPR interactions:
The above tests indicate that several aspects of sperm competition success show a signature of male × female interaction among the 10 × 10 lines tested. These lines were chosen to maximize genetic heterogeneity in SP and SPR and to assess whether SP and SPR allelic differences played a role in male × female interactions. We performed an analysis on the basis of the DNA sequence polymorphism within these two genes. By collapsing the lines into categories on the basis of their genotypes at these polymorphic sites, we could then determine the degree of departure from additivity of these genotypes as marginal tests of the full data and assess the likelihood that the SP and SPR genes are responsible for these effects.
As expected from the lack of male × female interaction, egg laying rates and the subsequent progeny phenotypes demonstrated no significant SP × SPR interactions (data not shown).
The significance of SP × SPR interactions on remating rate was assessed by fitting hierarchical log-linear models. Figure 3 shows the resulting uncorrected P-values for the interaction terms obtained from these remating tests. While there were 12 interactions that were significant (nominal P ≤ 0.05), none were significant after correcting for multiple testing.
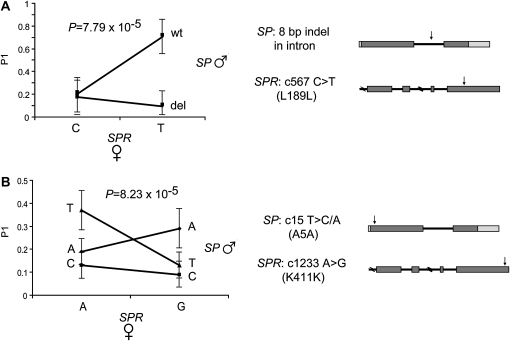
Figure 4 shows the resulting uncorrected P-values for the interaction terms from SNP interaction tests for P1 scores. There were 38 significant SP × SPR SNP interactions (nominal P ≤ 0.05) and two, in particular, remained significant after correction for multiple testing. The most significant interaction was between an 8-bp deletion in SP [intron 1 del(37–44)] and the SNP c567 C > T (L189L) in SPR (uncorrected P = 7.79 × 10−5, corrected P = 0.0105) (Figure 5A). The female SPR genotype at c567 does not have an effect on the P1 score of the deleted SP allele; rather, it affects only the P1 score of the wild-type SP allele. When the female carries the c567 C SPR allele, there is no difference between the P1 scores of males carrying the wild-type or the deleted SP allele. However, when females carry the c567 T SPR allele, males carrying the wild-type SP allele have an eightfold increase in P1 score as compared to males carrying the deleted allele.
Figure 5.—
Polymorphism interactions between SP and SPR. The two most significant interactions between SP and SPR are shown. (A) An intronic indel in SP interacts with a synonymous SNP in SPR. The P1 score of the male alleles is dependent on the female allele. Males have similar P1 scores when females carry the SPR C allele. However, there is an approximately eightfold difference in P1 scores between the male SP alleles when the female carries the SPR T allele. (B) A synonymous SNP in SP interacts with a synonymous SNP in SPR. The rank order of male SP allele changes dependent on the female SPR allele. Significance was tested by a log-linear model.
There was also a significant interaction effect on P1 scores between the SNP c15 T > C/A (A5A) in SP and the SNP c1233 A > G (K411K) in SPR (uncorrected P = 8.23 × 10−5, corrected P = 0.011) (Figure 5B). When females carry the c1233 A SPR allele, males carrying the c15 T SP allele have the highest P1 score, followed by males carrying the A allele and males carrying the C allele. However, when females carry the c1233 G SPR allele, there is a rank order change and males carrying the c15 A SP allele have the highest P1 score, followed by males carrying the T allele and males carrying the C allele. While the success of the male SP T and A alleles is affected by female genotype, the success of the C allele is unaffected by female genotype.
We tested whether the interactions observed between SP or SPR polymorphisms were spurious by testing for interaction with a set of SNPs not thought to be related to Acps or Acp processing. SPR polymorphisms were tested for interaction with SNPs in 20 immunity genes along the third chromosome (Hill-Burns and Clark 2009). There was a deficit of significant P-values between SPR and third chromosome SNPs. Seventy-seven of 1548 tests were significant with a nominal P < 0.05, but when correction for multiple testing was applied, none of the interactions remained significant. SP polymorphisms were also tested for interaction with SNPs in 25 immunity genes along the X chromosome (Sackton et al. 2010). Sixty-one of 636 tests between SP and X chromosome polymorphisms were significant with a nominal P < 0.05. Again, when correction for multiple testing was applied, none of these interactions remained significant. These data suggest that SP and SPR polymorphisms interact among themselves at a level that significantly exceeds their degree of interaction with unrelated SNPs.
SP and SPR expression levels:
Because the majority of polymorphisms identified in the tested lines did not affect the amino acid sequence of SP or SPR, we hypothesized that they might affect expression levels. Accordingly, we examined expression of SP and SPR. qPCR was performed for both, and Western blot analysis was performed for SP.
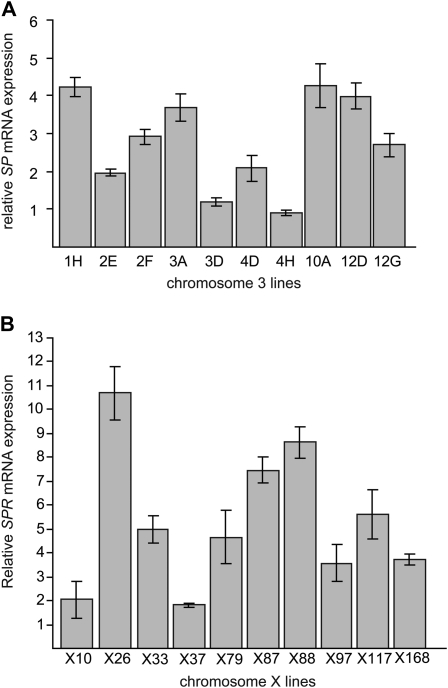
qPCR demonstrated that the 10 third chromosome extraction lines differed significantly in their SP mRNA levels (P < 2.2 × 10−16) (Figure 6A). There was greater than fourfold difference in normalized SP mRNA levels between the lowest-expressing line (4H) and the highest-expressing line (10A). There was no association between SP transcript abundance and any of the measured phenotypes, but with only 10 lines, these tests had low power (data not shown). Interestingly, Western blot analysis indicated that degree of variation in protein levels among lines is smaller than the variation in mRNA levels among lines (Figure S8). There is only a twofold difference in SP protein level between the lowest-expressing line (12D) and the highest-expressing line (10A). While line 10A displayed the highest SP mRNA and protein levels, different lines had the lowest SP mRNA levels (4H) and the lowest protein levels (12D). In fact, 12D has the lowest SP protein levels, but it displays one of the highest SP mRNA levels.
Figure 6.—
SP and SPR mRNA expression varies across lines. mRNA expression was quantified by qPCR. (A) SP expression in male third chromosome extraction lines significantly differs (P < 2.2 × 10−16). (B) SPR expression in female X chromosome extraction lines significantly differs (P < 2.2 × 10−16). Levels of mRNA are expressed relative to a standard Canton-S laboratory strain. All measurements consist of six measurements, two biological replicates containing three technical replicates each. Mean ± SD is shown.
SPR expression levels were evaluated by qPCR only since a suitable SPR antibody for Western blotting was not available. qPCR demonstrated that the 10 X chromosome extraction lines differed significantly in their levels of SPR mRNA (P < 2.2 × 10−16) (Figure 6B). There was greater than fivefold difference between the lowest-expressing line (X37) and the highest-expressing line (X26). Similar to the SP analysis, there was no significant correlation between SPR mRNA levels and the phenotypes measured.
To establish whether the polymorphisms identified in SP and SPR are associated with mRNA expression differences (eQTL), the lines were categorized according to mean mRNA expression levels and each SNP was tested for an effect of allele on expression level. All polymorphisms carried by only one extraction line were excluded from this analysis. The remaining 5 SP polymorphisms demonstrated a significant effect of allele on SP mRNA expression (Figure S9). Of the remaining 20 SPR polymorphisms, 19 showed a significant effect of allele on SPR mRNA expression levels (Figure S9).
DISCUSSION
To examine the genetic architecture of male × female interactions involved in phenotypes relevant to Drosophila reproductive fitness, we measured the interaction between the third chromosome in males and the X chromosome in females. We found that both the third and the X chromosomes have individual main effects on all the reproductive phenotypes measured in this study. In addition, we found that the third and X chromosomes have a large interaction effect on female remating rate and on the sperm competition parameter, P1.
Third chromosome effects:
Previous studies (Fiumera et al. 2007) demonstrated that there are multiple significant associations of polymorphisms in male reproductive proteins encoded on chromosome 3 and sperm competition. Our results support this observation. We identified a significant main effect of the third chromosome (male) on female remating rate and P1. Several male reproductive proteins encoded on the third chromosome were previously implicated in affecting either female remating rate or P1. These genes include CG6168, CG14560, and SP (Chapman et al. 2003; Liu and Kubli 2003; Fiumera et al. 2007). Our results suggest that natural variation in these and/or other male reproductive proteins on the third chromosome could contribute to differential sperm competition outcomes.
While whole-genome male effects on fertility phenotypes (egg laying rate, hatchability/viability, and progeny number) have been demonstrated (Civetta et al. 2008), individual chromosomes have not previously been examined for their contributions to this male effect. We demonstrate that the third chromosome (male) has significant effects on all these fertility phenotypes. The third chromosome harbors several male reproductive genes that have a demonstrated role in egg production, including DUP99B (Saudan et al. 2002), CG33943 (Ravi Ram and Wolfner 2007b), and SP (Chapman et al. 2003; Liu and Kubli 2003). Polymorphism in any of these genes could potentially contribute to the third chromosome effect we observe. To test this hypothesis, we sequenced these genes in the 10 third chromosome lines. We identified very few polymorphisms in DUP99B and CG33943, and all the variants we found were noncoding or synonymous, with no predicted functional consequences for the proteins (data not shown). The results for SP are discussed below. These data suggest that a large effect arising from DUP99B and CG33943 is not likely to underlie the variation in fertility phenotypes we observed. Instead, combined small effects of many genes or an as yet unidentified gene of large effect might explain the third chromosome effect. These data motivate the continued search for the functional role of male reproductive genes on the third chromosome.
X chromosome effects:
Past studies demonstrated a strong female effect on fertility phenotypes (Civetta et al. 2008) and P1 (Clark et al. 1999), but the female effect on female remating was not tested. Our results show a strong female effect on remating rate. Furthermore, we demonstrate a strong X chromosome female effect on all the phenotypes measured.
The X chromosome harbors at least one female gene with an important role in female remating rate and fertility phenotypes, SPR (Yapici et al. 2008). Variation in SPR might have a direct influence on these phenotypes. Although little is known about other female molecules important for female remating rate, SPR is likely not to be the only X chromosome gene to affect fertility phenotypes we measured; for example, numerous female molecules are known to affect egg laying. The process of egg production and egg laying takes place within the female reproductive tract, and although ovulation is stimulated by mating, the egg must progress through the female reproductive tract, encountering muscle contractions and neuroendocrine signals before fertilization can occur (reviewed in Bloch Qazi et al. 2003).
We observed an X chromosome female effect on P1, but it is not known which genes on the X chromosome might contribute to this effect. Variation in P1 requires that the female modulate her sperm usage from the first and the second male. This suggests that at least some female genes involved in sperm usage reside on the X chromosome. Identification of these genes would further elucidate the female's contribution to variation in P1. Candidate genes might include X-linked genes that are expressed in sperm storage organs in the female, including the spermathecae and the seminal receptacle (Allen and Spradling 2008; Prokupek et al. 2008, 2009, 2010).
Chromosome interactions:
In addition to separate male and female effects, we identified male × female genotypic interactions for female remating rate. This is the first documentation of an effect of interaction between the genotypes of both sexes on the female's willingness to remate. Two lines had strong individual sex effects on female remating rate. One line (third chromosome line 10A) demonstrated a male-only effect on remating rate, irrespective of female genotype. Males from this line are poor at preventing remating and must be carrying polymorphism on the third chromosome that affects one or more reproductive proteins. Another line (X chromosome line X168) showed a female-only effect on remating rate, suggesting that its X chromosome harbors polymorphisms that supersede effects of male genotype.
The strong male × female interactions that we identified are consistent with results from a smaller study (Clark et al. 1999), and we extend it by doubling the number of lines used, including lines from a different population, and, most importantly, examining the interaction between two specific chromosomes. Our results indicate that male × female interactions are not unique to the previously examined lines. The robust interactions identified in both studies show that interaction between the genotypes is critical to sperm competition outcomes.
While other studies reported male × female genotype interactions in fertility phenotypes (Civetta et al. 2008), we did not detect such an interaction. There are many possible explanations for the difference in findings across studies. First, environmental conditions can have a very large effect on egg laying rates (McGraw et al. 2007; Fricke et al. 2010), and it is almost certain the conditions of our study differed from those of the earlier study. Second, the experimental design we employed involved egg counts only on the first 2 days after the first mating, whereas the study reported egg counts over a longer period. Third, because the studies utilized very different experimental lines, we may not have captured polymorphisms that show interactive effects.
We limited our analysis to the third chromosome in males and the X chromosome in females, whereas Civetta et al. (2008) and Clark et al. (1999) did not focus on specific interchromosomal interactions. We found that the third and X chromosomes showed strong interaction for female remating rate and P1. However, genes on the second and fourth chromosomes, as well as male genes on the X chromosome and female genes on the third chromosome, could also potentially interact. Our design eliminated variation on the second and fourth chromosomes, and the primary goal was to determine interaction between the two chromosomes harboring SP and SPR. We expect there may be additional interactions involving genes throughout the genome.
SP × SPR interactions:
By examining the third chromosome in males and the X chromosome in females, we had the unique opportunity to test for potential interaction between two known physically interacting proteins. SP and SPR are the only two known male/female Drosophila reproductive genes whose protein products physically interact (Yapici et al. 2008). SP is on the third chromosome and its protein is transferred to the female during mating. In the female, SP binds its X-encoded receptor SPR, and this physical interaction results in many postmating changes in the female, potentially affecting all the phenotypes measured in this study (Yapici et al. 2008).
We tested the interaction of each SP polymorphism with each SPR polymorphism for all phenotypes measured in this study. We found no SP × SPR interactions for fertility phenotypes or remating rates, but this may be explained by the reasons discussed above (in Chromosome interactions). We found two very significant interactions between SP and SPR alleles for P1. In one case, the two male SP alleles display an eightfold difference in P1 depending on the female SPR allele. In another case, there is a rank-order change between the two male SP alleles in P1, dependent on the female SPR allele.
While these statistical observations are intriguing, they are only suggestive of biologically relevant allelic interactions given that the effects are detected from only 10 lines for each extracted chromosome, and the extracted chromosomes are segregating at many other sites. However, the observation of interaction among SP and SPR alleles makes sense in light of previous work. First, Fricke et al. (2009) presented evidence that SP might have a role in sperm competition. Second, SP function is required for proper release of stored sperm from female sperm storage organs (Avila et al. 2010). Inappropriate release of sperm from storage organs can have a direct effect on sperm usage and P1 scores (Avila and Wolfner 2009; Ram and Wolfner 2009). Furthermore, we demonstrate that SP alleles do not spuriously interact with unrelated polymorphisms along the X chromosome and similarly, SPR alleles do not interact with polymorphisms in non-reproduction-related genes along the third chromosome. Finally, in our tested lines, each SP and SPR allele is carried on multiple haplotypes (at least three for most alleles), reducing the potential for an interaction with a linked variant. We recognize that this is not definitive proof of allelic interactions between SP and SPR, but these observations motivate future functional testing of the interaction between these and other potential interacting SP and SPR alleles.
Evolutionary implications:
Interactive genotypic effects between the sexes described here and elsewhere (Clark et al. 1999; Civetta et al. 2008) could lead to alleles in interacting genes exhibiting dynamic cycling behavior that could protect and maintain polymorphism (Clark 2002). In agreement with this theory, some Acps display nonneutral patterns of evolution and high levels of polymorphism (Cirera and Aguade 1997, 1998; Tsaur and Wu 1997; Aguadé 1998, 1999; Clark and Begun 1998; Tsaur et al. 1998; Swanson et al. 2001; Haerty et al. 2007; Wong et al. 2008b; Kelleher and Markow 2009). Rapid evolution has been observed for some genes expressed in the female reproductive tract, which might potentially encode Acp receptors and proteins involved in postmating responses (Swanson et al. 2004; Prokupek et al. 2008, 2010). In a wild population, many male alleles may be segregating and their success will depend on the genotype of the female receptor/response gene.
Male × female genetic interactions might underlie the maintenance of variation in sperm competition success. Differences in sperm competition success likely reflect the interaction between allelic variation in critical genes. Rather than single, optimal alleles becoming fixed, this allelic variation could be maintained due to a variety of selective forces. For example, some aspects of sperm competition ability and female responses have been proposed to be under sexually antagonistic selection (Parker 1970). In this scenario, differing interests between the sexes may prevent fixation of an allele favorable to individuals of one sex but detrimental to individuals of the other sex. In another scenario, postcopulatory sexual selection (Eberhard 1996) could also maintain allelic variation, particularly in the context of cryptic female choice. If females of certain genotypes preferentially use sperm of select male genotypes, multiple combinations of male and female genotypes might have equivalent fitness and thus would all be maintained. It is unknown which of these or other scenarios are the driving force behind variation we see in sperm competition ability or in the allelic variation in SP and SPR, but the abundance of empirical evidence for male × female interaction leaves open these mechanisms as potentially critical aspects of the evolution of sperm competitive ability.
The X chromosome differs from autosomes in that it spends two-thirds (rather than half) of the time in females. This has permitted the “feminization” of the X chromosome (Rice 1984), in that the X chromosome carries a relative excess of female-expressed genes (Parisi et al. 2003; Ranz et al. 2003; Gurbich and Bachtrog 2008), whereas male-biased genes are skewed to the autosomes. Because female genes on the X chromosome are under stronger selection in the female, we might expect that sperm-competition-related female genes on the X chromosome are particularly poised to coevolve with male effects arising from male-expressed autosomal genes.
The data presented here motivate further study of the complex genetic architecture of sperm competition. Only when we identify the genes involved on both sides of the interaction, can we begin to understand the dynamic evolution of this important system.
Acknowledgments
We thank D. Castillo for his help with scoring flies. We thank L. Sirot, F. Avila, T. Connallon, and T. Greenberg for helpful comments. We thank A. Fiumera for discussions. We thank E. Kubli for kindly providing anti-SP and N. Yapici, Y.-J. Kim, and B. Dickson for helpful discussions and a sample of anti-SPR. We thank Montserrat Aguadé and four anonymous reviewers for suggestions that greatly improved this report. This work was funded by National Institutes of Health (NIH)/National Institute of Child Health and Human Development (NICHD) grant R01-HD059060 (to M.F.W. and A.G.C.), NIH/NICHD grant R01-HD038921 (to M.F.W.), National Science Foundation grant DEB-0743125 (to A.G.C.), and NIH/NICHD training grant in reproductive genomics T32-HD052471 (to C.Y.C.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.123174/DC1.
References
- Agresti, A., 1990. Categorical Data Analysis. JohnWiley & Sons, New York.
- Aguadé, M., 1998. Different forces drive the evolution of the Acp26Aa and Acp26Ab accessory gland genes in the Drosophila melanogaster species complex. Genetics 150 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M., 1999. Positive selection drives the evolution of the Acp29AB accessory gland protein in Drosophila. Genetics 152 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M., N. Miyashita and C. Langley, 1992. Polymorphism and divergence in the Mst26A male accessory gland gene region in Drosophila. Genetics 132 755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, A. K., and A. C. Spradling, 2008. The Sf1-related nuclear hormone receptor Hr39 regulates Drosophila female reproductive tract development and function. Development 135 311–321. [DOI] [PubMed] [Google Scholar]
- Avila, F. W., and M. F. Wolfner, 2009. Acp36DE is required for uterine conformational changes in mated Drosophila females. Proc. Natl. Acad. Sci. USA 106 15796–15800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila, F. W., K. Ravi Ram, M. C. Bloch Qazi and M. F. Wolfner, 2010. Sex peptide is required for the efficient release of stored sperm in mated Drosophila females. Genetics 186 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., and H. A. Lindfors, 2005. Rapid evolution of genomic Acp complement in the melanogaster subgroup of Drosophila. Mol. Biol. Evol. 22 2010–2021. [DOI] [PubMed] [Google Scholar]
- Bloch Qazi, M. C., Y. Heifetz and M. F. Wolfner, 2003. The developments between gametogenesis and fertilization: ovulation and female sperm storage in Drosophila melanogaster. Dev. Biol. 256 195–211. [DOI] [PubMed] [Google Scholar]
- Chapman, T., D. M. Neubaum, M. F. Wolfner and L. Partridge, 2000. The role of male accessory gland protein Acp36DE in sperm competition in Drosophila melanogaster. Proc. R. Soc. Lond. 267 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, T., J. Bangham, G. Vinti, B. Seifried, O. Lung et al., 2003. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc. Natl. Acad. Sci. USA 100 9923–9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirera, S., and M. Aguade, 1997. Evolutionary history of the sex-peptide (Acp70A) gene region in Drosophila melanogaster. Genetics 147 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirera, S., and M. Aguade, 1998. Molecular evolution of a duplication: the sex-peptide (Acp70A) gene region of Drosophila subobscura and Drosophila madeirensis. Mol. Biol. Evol. 15 988–996. [DOI] [PubMed] [Google Scholar]
- Civetta, A., and R. S. Singh, 1998. Sex-related genes, directional sexual selection, and speciation. Mol. Biol. Evol. 15 901–909. [DOI] [PubMed] [Google Scholar]
- Civetta, A., K. Rosing and J. Fisher, 2008. Differences in sperm competition and sperm competition avoidance in Drosophila melanogaster. Anim. Behav. 75 1739–1746. [Google Scholar]
- Clark, A. G., 2002. Sperm competition and the maintenance of polymorphism. Heredity 88 148–153. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., and D. J. Begun, 1998. Female genotypes affect sperm displacement in Drosophila. Genetics 149 1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., M. Aguadé, T. Prout, L. G. Harshman and C. H. Langley, 1995. Variation in sperm displacement and its association with accessory gland protein loci in Drosophila melanogaster. Genetics 139 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., D. J. Begun and T. Prout, 1999. Female x male interactions in Drosophila sperm competition. Science 283 217–220. [DOI] [PubMed] [Google Scholar]
- Eberhard, W. G., 1996. Female Control: Sexual Selection by Cryptic Female Choice. Princeton University Press, Princeton, NJ.
- Findlay, G. D., X. Yi, M. J. Maccoss and W. J. Swanson, 2008. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 6 e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay, G. D., M. J. MacCoss and W. J. Swanson, 2009. Proteomic discovery of previously unannotated, rapidly evolving seminal fluid genes in Drosophila. Genome Res. 19 886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera, A. C., B. L. Dumont and A. G. Clark, 2005. Sperm competitive ability in Drosophila melanogaster associated with variation in male reproductive proteins. Genetics 169 243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera, A. C., B. L. Dumont and A. G. Clark, 2007. Associations between sperm competition and natural variation in male reproductive genes on the third chromosome of Drosophila melanogaster. Genetics 176 1245–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke, C., S. Wigby, R. Hobbs and T. Chapman, 2009. The benefits of male ejaculate sex peptide transfer in Drosophila melanogaster. J. Evol. Biol. 22 275–286. [DOI] [PubMed] [Google Scholar]
- Fricke, C., A. Bretman and T. Chapman, 2010. Female nutritional status determines the magnitude and sign of responses to a male ejaculate signal in Drosophila melanogaster. J. Evol. Biol. 23 157–165. [DOI] [PubMed] [Google Scholar]
- Gurbich, T. A., and D. Bachtrog, 2008. Gene content evolution on the X chromosome. Curr. Opin. Genet. Dev. 18 493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerty, W., S. Jagadeeshan, R. J. Kulathinal, A. Wong, K. Ravi Ram et al., 2007. Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics 177 1321–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman, L. G., and A. G. Clark, 1998. Inference of sperm competition from broods of field-caught Drosophila. Evolution 52 1334–1341. [DOI] [PubMed] [Google Scholar]
- Harshman, L. G., and T. Prout, 1994. Sperm displacement without sperm transfer in Drosophila melanogaster. Evolution 48 758–766. [DOI] [PubMed] [Google Scholar]
- Hasemeyer, M., N. Yapici, U. Heberlein and B. J. Dickson, 2009. Sensory neurons in the Drosophila genital tract regulate female reproductive behavior. Neuron 61 511–518. [DOI] [PubMed] [Google Scholar]
- Hill-Burns, E. M., and A. G. Clark, 2009. X-linked variation in immune response in Drosophila melanogaster. Genetics 183 1477–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, K. A., 1997. Quantitative genetics of sperm precedence in Drosophila melanogaster. Genetics 145 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof, M., B. Harr, G. Brem and C. Schlotterer, 1998. Multiple mating in wild Drosophila melanogaster revisited by microsatellite analysis. Mol. Ecol. 7 915–917. [DOI] [PubMed] [Google Scholar]
- Isaac, R. E., C. Li, A. E. Leedale and A. D. Shirras, 2010. Drosophila male sex peptide inhibits siesta sleep and promotes locomotor activity in the post-mated female. Proc. Biol. Sci. 277 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher, E. S., and T. A. Markow, 2009. Duplication, selection and gene conversion in a Drosophila mojavensis female reproductive protein family. Genetics 181 1451–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., and E. Kubli, 2003. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100 9929–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw, L. A., A. C. Fiumera, M. Ramakrishnan, S. Madhavarapu, A. G. Clark et al., 2007. Larval rearing environment affects several post-copulatory traits in Drosophila melanogaster. Biol. Lett. 3 607–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, J. L., K. Ravi Ram, L. A. McGraw, M. C. Bloch Qazi, E. D. Siggia et al., 2005. Cross-species comparison of Drosophila male accessory gland protein genes. Genetics 171 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M., R. Nuttall, D. Naiman, G. Bouffard, J. Malley et al., 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, G. A., 1970. Sperm competition and its evolutionary consequences in the insects. Biol. Rev. 45 525–567. [Google Scholar]
- Prokupek, A., F. Hoffmann, S. I. Eyun, E. Moriyama, M. Zhou et al., 2008. An evolutionary expressed sequence tag analysis of Drosophila spermatheca genes. Evolution 62 2936–2947. [DOI] [PubMed] [Google Scholar]
- Prokupek, A. M., S. D. Kachman, I. Ladunga and L. G. Harshman, 2009. Transcriptional profiling of the sperm storage organs of Drosophila melanogaster. Insect Mol. Biol. 18 465–475. [DOI] [PubMed] [Google Scholar]
- Prokupek, A. M., S. I. Eyun, L. Ko, E. N. Moriyama and L. G. Harshman, 2010. Molecular evolutionary analysis of seminal receptacle sperm storage organ genes of Drosophila melanogaster. J. Evol. Biol. 23 1386–1398. [DOI] [PubMed] [Google Scholar]
- Ram, K. R., and M. F. Wolfner, 2009. A network of interactions among seminal proteins underlies the long-term postmating response in Drosophila. Proc. Natl. Acad. Sci. USA 106 15384–15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz, J. M., C. I. Castillo-Davis, C. D. Meiklejohn and D. L. Hartl, 2003. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 300 1742–1745. [DOI] [PubMed] [Google Scholar]
- Ravi Ram, K., and M. F. Wolfner, 2007. a Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr. Comp. Biol. 47 427–445. [DOI] [PubMed] [Google Scholar]
- Ravi Ram, K., and M. F. Wolfner, 2007. b Sustained post-mating response in Drosophila melanogaster requires multiple seminal fluid proteins. PLoS Genet. 3 e238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, W. R., 1984. Sex chromosome and the evolution of sexual dimorphism. Evolution 38 735–742. [DOI] [PubMed] [Google Scholar]
- Sackton, T. B., B. P. Lazzaro and A. G. Clark, 2010. Genotype and gene expression associations with immune function in Drosophila. PLoS Genet. 6 e1000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudan, P., K. Hauck, M. Soller, Y. Choffat, M. Ottiger et al., 2002. Ductus ejaculatorius peptide 99B (DUP99B), a novel Drosophila melanogaster sex-peptide pheromone. Eur. J. Biochem. 269 989–997. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., A. G. Clark, H. M. Waldrip-Dail, M. F. Wolfner and C. F. Aquadro, 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. USA 98 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W. J., A. Wong, M. F. Wolfner and C. F. Aquadro, 2004. Evolutionary EST analysis of Drosophila female reproductive tracts identifies several genes subjected to positive selection. Genetics 168 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaur, S. C., and C.-I. Wu, 1997. Positive selection and the molecular evolution of a gene of male reproduction, Acp26Aa of Drosophila. Mol. Biol. Evol. 14 544–549. [DOI] [PubMed] [Google Scholar]
- Tsaur, S. C., C. T. Ting and C.-I. Wu, 1998. Positive selection driving the evolution of a gene of male reproduction, Acp26Aa, of Drosophila: II. Divergence versus polymorphism. Mol. Biol. Evol. 15 1040–1046. [DOI] [PubMed] [Google Scholar]
- Venables, W. N., and B. D. Ripley, 2002. Modern Applied Statistics with S. Springer, New York.
- Wolfner, M. F., 2009. Battle and ballet: molecular interactions between the sexes in Drosophila. J. Hered. 100 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, A., S. N. Albright, J. D. Giebel, K. R. Ram, S. Ji et al., 2008. a A role for Acp29AB, a predicted seminal fluid lectin, in female sperm storage in Drosophila melanogaster. Genetics 180 921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, A., M. C. Turchin, M. F. Wolfner and C. F. Aquadro, 2008. b Evidence for positive selection on Drosophila melanogaster seminal fluid protease homologs. Mol. Biol. Evol. 25 497–506. [DOI] [PubMed] [Google Scholar]
- Yang, C. H., S. Rumpf, Y. Xiang, M. D. Gordon, W. Song et al., 2009. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron 61 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yapici, N., Y. J. Kim, C. Ribeiro and B. J. Dickson, 2008. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature 451 33–37. [DOI] [PubMed] [Google Scholar]