Abstract
The budding yeast Cdc14 phosphatase reverses Cdk1 phosphorylation to promote mitotic exit. Although Cdc14 activity is thought to be restricted to anaphase, we found that dephosphorylation of the Dsn1 kinetochore protein in metaphase requires Cdc14. These data suggest that there is a nonnucleolar pool of active Cdc14 prior to anaphase.
THE master regulator of cell cycle progression, Cdk1, drives the entry into S phase and mitosis by phosphorylating hundreds of substrates (Nurse 1990; Morgan 1997). To exit mitosis, Cdk1-dependent phosphorylation must be reversed and Cdk1 must be inactivated (Sullivan and Morgan 2007). The budding yeast Cdc14 phosphatase is essential for mitotic exit and is thus tightly controlled to ensure the accurate order of events (D'Amours and Amon 2004; Queralt and Uhlmann 2008; De Wulf et al. 2009; Mocciaro and Schiebel 2010). Cdc14 is sequestered in the nucleolus from G1 until the onset of anaphase when it is released into the nucleus and cytoplasm to dephosphorylate Cdk1 substrates and promote mitotic exit (Jaspersen et al. 1998; Visintin et al. 1998, 1999; Shou et al. 1999; Traverso et al. 2001; Lu and Cross 2010; Manzoni et al. 2010). Although it has been thought that Cdc14 is inhibited during metaphase, recent work suggests that there is a pool of active Cdc14 in the nucleolus prior to anaphase onset (Geil et al. 2008). Consistent with this, Cdc14 dephosphorylates one of its activators, Spo12, during metaphase in the nucleolus (Tomson et al. 2009). However, it is not known whether Cdc14 is active outside of nucleolus during metaphase or whether it dephosphorylates additional targets prior to anaphase. Dsn1 is an essential, conserved component of the Mis12 kinetochore subcomplex that is critical for kinetochore assembly, and the protein has not been detected in the nucleolus (Euskirchen 2002; Pinsky et al. 2003). Despite the importance of Dsn1 in kinetochore assembly and function (Cheeseman and Desai 2008), little is known about the regulation of the Dsn1 protein. Here, we show that a Cdk1 site on Dsn1 is dephosphorylated prior to anaphase in a Cdc14-dependent manner.
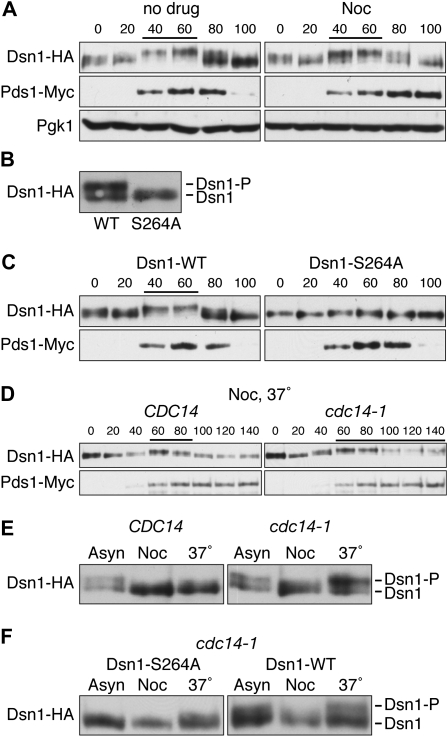
During the course of our studies on the Dsn1 kinetochore protein, we detected a cell cycle-dependent electrophoretic mobility shift. Cells containing Dsn1–HA released from a G1 arrest showed a dynamic migration pattern, high at 40–60 min when cells form buds (S phase) and low at 80–100 min (M phase) (Figure 1A). We confirmed the cell cycle stages by monitoring the levels of the anaphase inhibitor, Pds1 (Figure 1A). Dsn1 protein levels did not noticeably fluctuate during the cell cycle (Figure 1A). Dsn1 is known to be phosphorylated by multiple kinases including Cdk1 and Ipl1 ((Westermann et al. 2003; Gruhler et al. 2005; Albuquerque et al. 2008; B. Akiyoshi and S. Biggins, unpublished data), suggesting that the mobility shift may represent phosphorylation. We therefore repeated the experiment by releasing ipl1-321 (Aurora kinase), cdc5-1 (Polo kinase), or mps1-1 (Mps1 kinase) temperature-sensitive mutants from G1 to the restrictive temperature and found that the Dsn1 mobility shift still occurred, similar to wild-type (WT) cells (supporting information, Figure S1). We hypothesized that the potential phosphorylation shift could be due to Cdk1/Cdc28 activity, but we could not perform a similar experiment because cells do not progress through the cell cycle without Cdk1 activity (Reed 1980). We therefore tested whether mutation of Cdk1 consensus sites altered Dsn1 mobility. Although there are six potential Cdk1 consensus sites (S/T-P) in Dsn1, Ser264 is the only conserved site within the Saccharomyces lineage. When we mutated S264 to alanine, the Dsn1 mobility shift was reduced in both asynchronous (Figure 1B) and S phase cells (Figure 1C), suggesting that phosphorylation on S264 is largely responsible for the mobility shift on these gels. We confirmed that cell cycle progression was similar in WT and dsn1–S264A mutant cells (Figure 1C).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| SBY3 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 LYS2 can1-100 bar1Δ |
| SBY1841 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 LYS2 can1-100 bar1Δ cdc28-13 |
| SBY2225 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 lys2Δ can1-100 bar1Δ DSN1-GFP:HIS3 ndc80-1 |
| SBY5770 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 LYS2 can1-100 bar1Δ DSN1:HIS3:dsn1-S264A-3HA:TRP1 PDS1-myc18:LEU2 |
| SBY5866 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 LYS2 can1-100 bar1Δ DSN1-3HA:HIS3 PDS1-myc18:LEU2 |
| SBY5867 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 LYS2 can1-100 bar1Δ DSN1-3HA:HIS3 PDS1-myc18:LEU2 ipl1-321 |
| SBY6079 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 LYS2 can1-100 bar1Δ DSN1-3HA:HIS3 PDS1-myc18:LEU2 |
| SBY6085 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 LYS2 can1-100 bar1Δ DSN1-3HA:HIS3 PDS1-myc18:LEU2 cdc14-1 |
| SBY7223 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 LYS2 can1-100 bar1Δ DSN1-3HA:HIS3 cdc5-1 |
| SBY7837 | MATaura3-1:dsn1-6SA-3HA:URA3 leu2,3-112 his3-11 trp1-1 ade2-1 LYS2 can1-100 cdc28-13 dsn1ΔKAN |
| SBY7902 | MATaura3-1:DSN1-WT-3FLAG:URA3 leu2,3-112 his3-11 trp1-1 ade2-1 LYS2 can1-100 bar1Δ dsn1ΔKAN |
| SBY8008 | MATaura3-1 leu2,3-112 his3-11 trp1-1:pGAL-CDC20:TRP1 ade2-1 LYS2 can1-100 DSN1-3FLAG:KAN GLC7-3HA:HIS3 cdc20ΔLEU2 |
| SBY8042 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 lys2Δ can1-100 bar1Δ DSN1-3FLAG:KAN cdc28-as1 |
| SBY8123 | MATaura3-1:dsn1-S264A-3FLAG:URA3 leu2,3-112 his3-11 trp1-1 ade2-1 LYS2 can1-100 bar1Δ dsn1ΔKAN |
| SBY9139 | MATaura3-1:dsn1-6SA-3HA:URA3 leu2,3-112 his3-11 trp1-1 ade2-1 lys2Δ can1-100 bar1Δ ndc80-1 dsn1ΔKAN |
| SBY9149 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 LYS2 can1-100 bar1Δ DSN1-3HA:HIS3 mps1-1 |
| SBY9152 | MATaura3-1 leu2,3-112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:256lacO:TRP1 ade2-1 lys2Δ can1-100 bar1Δ mtw1-1 [2micron, LEU2] |
| SBY9153 | MATaura3-1 leu2,3-112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:256lacO:TRP1 ade2-1 lys2Δ can1-100 bar1Δ mtw1-1 [DSN1-WT-myc, 2micron, LEU2] |
| SBY9154 | MATaura3-1 leu2,3-112 his3-11:pCUP1-GFP12-lacI12:HIS3 trp1-1:256lacO:TRP1 ade2-1 lys2Δ can1-100 bar1Δ mtw1-1 [dsn1-S264A-myc 2micron, LEU2] |
| SBY9158 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 nsl1-5 [2micron, LEU2] |
| SBY9159 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 nsl1-5 [DSN1-WT-myc, 2micron, LEU2] |
| SBY9160 | MATaura3-1 leu2,3-112 his3-11 trp1-1 ade2-1 can1-100 nsl1-5 [dsn1-S264A-myc 2micron, LEU2] |
| SBY9187 | MATaura3-1:DSN1-WT-3HA:URA3 leu2,3-112 his3-11 trp1-1 ade2-1 lys2Δ can1-100 bar1Δ cdc14-1 dsn1ΔKAN |
| SBY9188 | MATaura3-1:dsn1-S264A-3HA:URA3 leu2,3-112 his3-11 trp1-1 ade2-1 lys2Δ can1-100 bar1Δ cdc14-1 dsn1ΔKAN |
All strains are isogenic with the W303 background. Plasmids are indicated in brackets. The cdc14-1 (Jaspersen et al. 1998), cdc28-as1 (Bishop et al. 2000), pGAL-CDC20 (Buvelot et al. 2003), cdc5-1 (Charles et al. 1998), mps1-1 (Winey et al. 1991), ipl1-321 (Biggins et al. 1999), cdc28-13 (Reed 1980), nsl1-5 (Nekrasov et al. 2003), ndc80-1 (Wigge et al. 1998), and mtw1-1 (Pinsky et al. 2003) alleles were crossed to make strains for this study. Deletions and epitope tags were made using a PCR-based integration system and confirmed by PCR (Longtine et al. 1998; Gelbart et al. 2001). Specific primers are available upon request.
Figure 1.—
Cdc14 dephosphorylates Dsn1 prior to anaphase. (A) Dsn1–HA shows a cell-cycle dependent mobility shift. Cells containing Dsn1–HA and Pds1–Myc (SBY6079) were released from G1 into complete media in the presence or absence of 15 μg/ml nocodazole at room temperature, lysates were prepared, and Dsn1–HA was monitored for mobility shift by immunoblotting as previously described (Biggins et al. 1999). Decreased mobility shifts are indicated by lines above immunoblots. Pgk1 was used as a loading control, and Pds1–Myc was used to monitor cell cycle progression. Yeast strains used in this study are listed in Table 1. (B and C) Ser264 phosphorylation is largely responsible for the mobility shift of Dsn1–HA. Lysates were prepared from asynchronously growing cells (B) or G1-released cells (C) expressing Pds1–Myc and either Dsn1–WT–HA (SBY5866) or Dsn1–S264A–HA (SBY5770) and immunoblotted with anti-HA and anti-Myc antibodies. (D) Cdc14 dephosphorylates Dsn1 prior to anaphase. The experiment in A was repeated using CDC14 (SBY6079) or cdc14-1 (SBY6085) cells containing Dsn1–HA and Pds1–Myc that were released to 37°. (E) Cdc14 is required to maintain the dephosphorylated status of Dsn1 during a preanaphase arrest. CDC14 (SBY6079) or cdc14-1 (SBY6085) cells were arrested with nocodazole for 3 hr and then shifted to 37° for 30 min. (F) The phosphorylation restored during a nocodazole arrest largely depends on Ser264 phosphorylation. The experiment in E was repeated using cdc14-1 cells containing Dsn1–S264A–HA (SBY9188) or Dsn1–WT–HA (SBY9187).
The time of Dsn1 dephosphorylation correlates with kinetochore biorientation (Goshima and Yanagida 2000 and data not shown), so we tested whether it depends on biorientation by treating cells with nocodazole, a microtubule depolymerizing drug. When cells containing Dsn1–HA were released from G1 into nocodazole, Dsn1 migrated similarly to untreated cells (Figure 1A). These data suggest that Dsn1 dephosphorylation occurs independently of kinetochore–microtubule attachment or biorientation. However, these results were surprising because the Cdc14 phosphatase that dephosphorylates Cdk1 sites is thought to be inhibited during a nocodazole arrest (e.g., Visintin et al. 1998). We therefore tested whether Cdc14 is required for the mobility shift by releasing WT and cdc14-1 cells into nocodazole at the restrictive temperature. Strikingly, Dsn1 was dephosphorylated in WT but not in cdc14-1 mutant cells, strongly suggesting that Cdc14 dephosphorylates Dsn1 prior to anaphase onset (Figure 1D). We confirmed that the cells stay arrested prior to anaphase because Pds1 was not degraded (Figure 1D). In addition, we obtained similar results when cells were released into the cell cycle in the absence of nocodazole (data not shown). To further validate that the dephosphorylation occurs prior to anaphase, we arrested WT and cdc14-1 cells with nocodazole for 3 hr and then shifted them to restrictive temperature for 30 min (Figure 1E). In cdc14-1 cells, Dsn1 phosphorylation was restored, suggesting that Cdc14 maintains the dephosphorylated status of Dsn1 during the preanaphase arrest. This mobility change largely depends on the phosphorylation of S264 because the Dsn1–S264A mutant showed only slight rephosphorylation (Figure 1F).
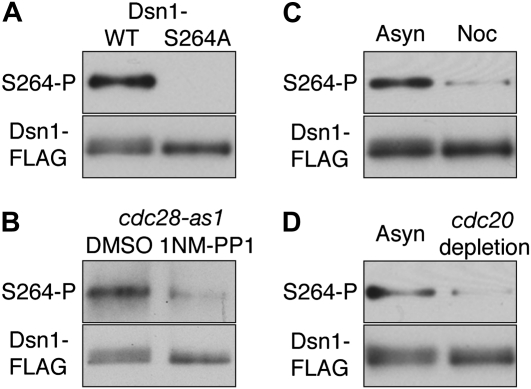
To ensure that the Dsn1 mobility shifts were indeed monitoring Cdk1-dependent phosphorylation of S264, we generated a phospho-specific antibody. Although the antibody could not be used on whole cell lysates due to a high background signal, it showed a specific signal against immunoprecipitated Dsn1 protein. We confirmed that the antibody specifically recognized phosphorylated Dsn1 because it reacted with wild-type Dsn1 that had been immunoprecipitated, but not with the Dsn1–S264A mutant (Figure 2A). The phosphorylation was reduced when Cdk1 was inhibited using an analog-sensitive allele (Figure 2B), suggesting that Ser264 is likely a direct target of Cdk1. Using the phospho-specific antibody, we next examined the phosphorylation status of Ser264 in preanaphase arrested cells where Cdc14 is sequestered in nucleolus (Visintin et al. 1999). Consistent with the mobility shifts, we found that S264 is hypophosphorylated in nocodazole-treated cells that arrest prior to anaphase (Figure 2C) as well as in cdc20-depleted cells that arrest in metaphase with bioriented kinetochores (Figure 2D). Taken together, these data support our finding that Cdc14 dephosphorylates Dsn1 prior to anaphase when Cdk1 activity is high (e.g., Surana et al. 1993; Jaspersen et al. 1998; Rahal and Amon 2008).
Figure 2.—
Dsn1–Ser264, a Cdk1 target site, is hypophosphorylated prior to anaphase. (A) A polyclonal phospho-specific antibody was custom-made by Pacific Immunology against phospho-Ser264 Dsn1 using peptide GGSTII(pS)PHKDIPEED. Dsn1–WT–FLAG (SBY7902) and Dsn1–S264A–FLAG (SBY8123) were immunoprecipitated using anti-FLAG antibodies as previously described (Akiyoshi et al. 2009), immunoblotted using the phospho-specific antibody at a 1:5000 dilution, and then reprobed with monoclonal FLAG antibodies. (B) Ser264 phosphorylation depends on Cdk1/Cdc28. Dsn1–FLAG proteins immunoprecipitated from cdc28–as1 mutant cells (SBY8042) (Bishop et al. 2000), treated with DMSO or 10 μm 1NM-PP1 for 30 min, were analyzed as in A. (C) Dsn1–FLAG proteins immunoprecipitated from asynchronous or preanaphase cells that were treated with 15 μg/ml nocodazole for 3 hr were analyzed as in A. (D) pGAL–CDC20 cells expressing Dsn1–FLAG (SBY8008) were asynchronously grown in galactose media and arrested in metaphase by adding glucose for 3 hr to deplete Cdc20. Dsn1–FLAG was immunoprecipitated and analyzed as in A.
In summary, we found that the Dsn1 kinetochore protein shows a cell cycle-dependent mobility shift that is largely due to Cdk1-dependent phosphorylation of S264. Unexpectedly, the Cdc14-dependent dephosphorylation of Dsn1 occurs prior to anaphase when Cdc14 is sequestered in nucleolus and the bulk of known Cdc14 targets are not dephosphorylated, including the Fin1 kinetochore protein that is readily dephosphorylated by Cdc14 at the onset of anaphase (e.g., Shou et al. 1999; Visintin et al. 1999; Jaspersen and Morgan 2000; Pereira and Schiebel 2003; Jin et al. 2008; Akiyoshi et al. 2009; Konig et al. 2010). To our knowledge, this is the first observation of a nonnucleolar protein that is dephosphorylated in a Cdc14-dependent manner prior to anaphase onset, suggesting that there is a pool of active Cdc14 outside of nucleolus. Interestingly, the Schizosaccharomyces pombe Cdc14 homolog, Clp1/Flp1, is released from the nucleolus after mitotic entry, yet it does not prematurely induce mitotic exit (Cueille et al. 2001; Trautmann et al. 2001; Wolfe et al. 2006). Clp1/Flp1 localizes to kinetochores and plays an important role in promoting biorientation during prometaphase (Trautmann et al. 2004). Kinetochore localization of budding yeast Cdc14 was also reported during anaphase (Pereira and Schiebel 2003; Stoepel et al. 2005). Although we do not know whether the Dsn1 dephosphorylation occurs on kinetochores or within the nucleus, these studies raise the possibility that Cdc14 could act on kinetochore-bound Dsn1 prior to anaphase. Regardless, these results suggest that in addition to control through nucleolar sequestration, Cdc14 activity may also be regulated by substrate specificity and processivity, enabling efficient dephosphorylation of specific targets when Cdk1 activity is high. In the future, it will be important to elucidate the mechanism that allows Dsn1 dephosphorylation by Cdc14 prior to anaphase. It is also important to understand how Cdk1-dependent phosphorylation and Cdc14-dependent dephosphorylation regulate the function of the Dsn1 kinetochore protein during cell cycle. Because kinetochores are assembled during S phase (Kitamura et al. 2007) when Dsn1 shows a dynamic phosphorylation pattern, Cdk1 may facilitate kinetochore assembly by targeting Dsn1. Although our preliminary experiments did not detect any obvious mitotic defects in dsn1–S264A or dsn1–S264D mutant cells (Figure 1C, Figure S2, and data not shown), it is possible that there are additional Cdk1 targets at kinetochores that function in parallel to Dsn1. Indeed, other kinetochore proteins have been reported to be phosphorylated by Cdk1 (Ubersax et al. 2003; Holt et al. 2009), and a slight biorientation defect was observed when cdc14 mutants were released from G1 into the restrictive temperature (D'Amours et al. 2004). It is therefore possible that Cdc14-dependent dephosphorylation of kinetochore targets contributes to accurate chromosome segregation, so it will be important to identify and characterize additional Cdk1 kinetochore substrates in the future.
Acknowledgments
We thank Christian Nelson for assistance and Sue Jaspersen and Frank Uhlmann for critical reading of the manuscript. This work was supported by National Institutes of Health grants (GM078069 and GM064386) (to S.B.). S.B. is a scholar of the Leukemia and Lymphoma Society.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.123653/DC1.
References
- Akiyoshi, B., C. R. Nelson, J. A. Ranish and S. Biggins, 2009. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 23 2887–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque, C. P., M. B. Smolka, S. H. Payne, V. Bafna, J. Eng et al., 2008. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics 7 1389–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., F. F. Severin, N. Bhalla, I. Sassoon, A. A. Hyman et al., 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, A. C., J. A. Ubersax, D. T. Petsch, D. P. Matheos, N. S. Gray et al., 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407 395–401. [DOI] [PubMed] [Google Scholar]
- Buvelot, S., S. Y. Tatsutani, D. Vermaak and S. Biggins, 2003. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles, J., S. L. Jaspersen, R. L. Tinker-Kulberg, L. Hwang, A. Szidon et al., 1998. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr. Biol. 9 497–507. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I. M., and A. Desai, 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9 33–46. [DOI] [PubMed] [Google Scholar]
- Cueille, N., E. Salimova, V. Esteban, M. Blanco, S. Moreno et al., 2001. Flp1, a fission yeast orthologue of the S. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J. Cell Sci. 114 2649–2664. [DOI] [PubMed] [Google Scholar]
- D'Amours, D., and A. Amon, 2004. At the interface between signaling and executing anaphase–Cdc14 and the FEAR network. Genes Dev. 18 2581–2595. [DOI] [PubMed] [Google Scholar]
- D'Amours, D., F. Stegmeier and A. Amon, 2004. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 117 455–469. [DOI] [PubMed] [Google Scholar]
- De Wulf, P., F. Montani and R. Visintin, 2009. Protein phosphatases take the mitotic stage. Curr. Opin. Cell Biol. 21 806–815. [DOI] [PubMed] [Google Scholar]
- Euskirchen, G. M., 2002. Nnf1p, Dsn1p, Mtw1p, and Nsl1p: a new group of proteins important for chromosome segregation in Saccharomyces cerevisiae. Eukaryot. Cell 1 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geil, C., M. Schwab and W. Seufert, 2008. A nucleolus-localized activator of Cdc14 phosphatase supports rDNA segregation in yeast mitosis. Curr. Biol. 18 1001–1005. [DOI] [PubMed] [Google Scholar]
- Gelbart, M. E., T. Rechsteiner, T. J. Richmond and T. Tsukiyama, 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21 2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and M. Yanagida, 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100 619–633. [DOI] [PubMed] [Google Scholar]
- Gruhler, A., J. V. Olsen, S. Mohammed, P. Mortensen, N. J. Faergeman et al., 2005. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4 310–327. [DOI] [PubMed] [Google Scholar]
- Holt, L. J., B. B. Tuch, J. Villen, A. D. Johnson, S. P. Gygi et al., 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325 1682–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen, S. L., and D. O. Morgan, 2000. Cdc14 activates cdc15 to promote mitotic exit in budding yeast. Curr. Biol. 10 615–618. [DOI] [PubMed] [Google Scholar]
- Jaspersen, S. L., J. F. Charles, R. L. Tinker-Kulberg and D. O. Morgan, 1998. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell 9 2803–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, F., H. Liu, F. Liang, R. Rizkallah, M. M. Hurt et al., 2008. Temporal control of the dephosphorylation of Cdk substrates by mitotic exit pathways in budding yeast. Proc. Natl. Acad. Sci. USA 105 16177–16182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura, E., K. Tanaka, Y. Kitamura and T. U. Tanaka, 2007. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 21 3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig, C., H. Maekawa and E. Schiebel, 2010. Mutual regulation of cyclin-dependent kinase and the mitotic exit network. J. Cell Biol. 188 351–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, 3rd, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Lu, Y., and F. R. Cross, 2010. Periodic cyclin-Cdk activity entrains an autonomous Cdc14 release oscillator. Cell 141 268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoni, R., F. Montani, C. Visintin, F. Caudron, A. Ciliberto et al., 2010. Oscillations in Cdc14 release and sequestration reveal a circuit underlying mitotic exit. J. Cell Biol. 190 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocciaro, A., and E. Schiebel, 2010. Cdc14: a highly conserved family of phophatases with non-conserved functions? J. Cell Sci. 123 2867–2876. [DOI] [PubMed] [Google Scholar]
- Morgan, D. O., 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13 261–291. [DOI] [PubMed] [Google Scholar]
- Nekrasov, V. S., M. A. Smith, S. Peak-Chew and J. V. Kilmartin, 2003. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14 4931–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse, P., 1990. Universal control mechanism regulating cell cycle timing of M-phase. Nature 344 503–508. [DOI] [PubMed] [Google Scholar]
- Pereira, G., and E. Schiebel, 2003. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science 302 2120–2124. [DOI] [PubMed] [Google Scholar]
- Pinsky, B. A., S. Y. Tatsutani, K. A. Collins and S. Biggins, 2003. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell 5 735–745. [DOI] [PubMed] [Google Scholar]
- Queralt, E., and F. Uhlmann, 2008. Cdk-counteracting phosphatases unlock mitotic exit. Curr. Opin. Cell Biol. 20 661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahal, R., and A. Amon, 2008. Mitotic CDKs control the metaphase-anaphase transition and trigger spindle elongation. Genes Dev. 22 1534–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, S. I., 1980. The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics 95 561–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed et al., 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97 233–244. [DOI] [PubMed] [Google Scholar]
- Stoepel, J., M. A. Ottey, C. Kurischko, P. Hieter and F. C. Luca, 2005. The mitotic exit network Mob1p-Dbf2p kinase complex localizes to the nucleus and regulates passenger protein localization. Mol. Biol. Cell 16 5465–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, M., and D. O. Morgan, 2007. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 8 894–903. [DOI] [PubMed] [Google Scholar]
- Surana, U., A. Amon, C. Dowzer, J. McGrew, B. Byers et al., 1993. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 12 1969–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomson, B. N., R. Rahal, V. Reiser, F. Monje-Casas, K. Mekhail et al., 2009. Regulation of Spo12 phosphorylation and its essential role in the FEAR network. Curr. Biol. 19 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann, S., S. Rajagopalan and D. McCollum, 2004. The S. pombe Cdc14-like phosphatase Clp1p regulates chromosome biorientation and interacts with Aurora kinase. Dev. Cell 7 755–762. [DOI] [PubMed] [Google Scholar]
- Trautmann, S., B. A. Wolfe, P. Jorgensen, M. Tyers, K. L. Gould et al., 2001. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol. 11 931–940. [DOI] [PubMed] [Google Scholar]
- Traverso, E. E., C. Baskerville, Y. Liu, W. Shou, P. James et al., 2001. Characterization of the Net1 cell cycle-dependent regulator of the Cdc14 phosphatase from budding yeast. J. Biol. Chem. 276 21924–21931. [DOI] [PubMed] [Google Scholar]
- Ubersax, J. A., E. L. Woodbury, P. N. Quang, M. Paraz, J. D. Blethrow et al., 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425 859–864. [DOI] [PubMed] [Google Scholar]
- Visintin, R., K. Craig, E. S. Hwang, S. Prinz, M. Tyers et al., 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2 709–718. [DOI] [PubMed] [Google Scholar]
- Visintin, R., E. S. Hwang and A. Amon, 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398 818–823. [DOI] [PubMed] [Google Scholar]
- Westermann, S., I. M. Cheeseman, S. Anderson, J. R. Yates, 3rd, D. G. Drubin et al., 2003. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 163 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge, P. A., O. N. Jensen, S. Holmes, S. Soues, M. Mann et al., 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., L. Goetsch, P. Baum and B. Byers, 1991. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 114 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, B. A., W. H. McDonald, J. R. Yates, 3rd and K. L. Gould, 2006. Phospho-regulation of the Cdc14/Clp1 phosphatase delays late mitotic events in S. pombe. Dev. Cell 11 423–430. [DOI] [PubMed] [Google Scholar]