Abstract
Left/right asymmetrically expressed genes permit an animal to perform distinct tasks with the right vs. left side of its brain. Once established during development, lateralized gene expression patterns need to be maintained during the life of the animal. We show here that a histone modifying complex, composed of the LSY-12 MYST-type histone acetyltransferase, the ING-family PHD domain protein LSY-13, and PHD/bromodomain protein LIN-49, is required to first initiate and then actively maintain lateralized gene expression in the gustatory system of the nematode Caenorhabditis elegans. Similar defects are observed upon postembryonic removal of two C2H2 zinc finger transcription factors, die-1 and che-1, demonstrating that a combination of transcription factors, which recognize DNA in a sequence-specific manner, and a histone modifying enzyme complex are responsible for inducing and maintaining neuronal laterality.
FEATURES of terminally differentiated cells not only need to be initiated through cell type-specific regulatory programs but also need to be continuously maintained throughout the life of the cell (Blau 1992). The mechanisms that maintain the identity of postmitotic, fully differentiated cells are, however, incompletely understood. We provide here some insights into these mechanisms, using a specific differentiation event in the nervous system of Caenorhabditis elegans. This neuronal differentiation event occurs differentially across the left/right axis of the animal and results in the left/right asymmetric expression of putative chemoreceptors (gcy genes) in the left vs. right ASE gustatory neuron (Figure 1A). This laterality, i.e., the left/right asymmetric expression of the gcy genes, is controlled by several gene regulatory factors that act in a bistable feedback loop (Figure 1, A and B) (Johnston and Hobert 2003; Johnston et al. 2005; Hobert 2006). Components of the bistable feedback loop are only transiently required around the time the ASE neurons are born in the embryo (Sarin et al. 2007). Postembryonic mechanisms that robustly and reproducibly maintain laterality throughout the life of the animal are unknown, yet must exist, considering the importance of ASE laterality for adult nervous system function (Suzuki et al. 2008). We describe here a locus, lsy-12, that executes such a maintenance function.
Figure 1.—
lsy-12, a MYST-type histone acetyltransferase, affects ASE laterality. (A) Genes known to be involved in controlling ASEL/R laterality (Hobert 2006; Didiano et al. 2010). (B) Schematic representation of phenotype of representative “2 ASEL” and “2 ASER” mutants. (C) Effects of lsy-12 on ASEL/R laterality markers. A subset of the defects were already reported, upon the initial identification of the lsy-12 locus (Sarin et al. 2007). Numbers below the panels indicate the penetrance of the phenotype, i.e., the fraction of animals that display the phenotype shown in the fluorescent image above. Data on other lsy-12 alleles were reported in Sarin et al. (2007, 2008, 2010). Animals that express the die-1 reporter fosmid also contain a ASEL/R-expressed red fluorescent reporter (che-1∷mCherry) for cell identification. A list of transgenes used in the study is provided in the File S1. Animals were scored as adults. (D) lsy-12 encodes a MYST-type histone acetyltransferase. Analysis of ESTs, expression tiling array clones, PCR specific rescue, and RT–PCR (see File S1 for more details) revealed that R07B5.8 and R07B5.9 are one genetic locus encoding at least two major splice isoforms. The 3′, polyadenylated end of yk82d06 and EX1785569 provide evidence for the existence of the lsy-12b, while other clones provide evidence of the lsy-12a isoform. RT–PCR data suggest that additional splice variants may be produced by the lsy-12 locus (some possibly even including the more upstream predicted gene T11A5.1), but we have not been able to conclusively identify the start and end of such alternative transcripts (data not shown; see also www.wormbase.org). The arrow indicates the predicted translational start site. lsy-12 expression constructs are shown in the lower part of the panel, with the bottom one being a negative control. Staggered red lines indicate that these constructs were generated by in vivo recombineering of co-injected, overlapping PCR fragments (Boulin et al. 2006), some of which were generated by an in vitro PCR fusion approach (Hobert 2002). The generation of constructs is described in File S1. Rescuing data is quantified in Table S1.
Laterality defects in animals lacking lsy-12, a histone acetyltransferase:
lsy-12 mutants were initially identified in a screen for ASEL/R laterality defects (Lsy phenotype) (Sarin et al. 2007). We characterized the lsy-12 defects in more detail by examining a panel of left/right asymmetrically expressed genes. We find that lsy-12 broadly affects lateralized gene expression; terminal markers (such as the gcy chemoreceptors), as well as upstream regulators, such as the miRNA lsy-6 or the die-1 zinc finger transcription factors, are affected (Figure 1C). All the defects sum up to a complete conversion of the ASEL state to the ASER state (“2-ASER” phenotype) (Figure 1B). Bilateral ASE fate is unaffected, as assessed by correct, bilateral expression of the Otx-type ceh-36 homeobox gene (Figure 1C).
lsy-12 was cloned in parallel by two independent strategies, one being classic fine mapping and ensuing transformation rescue (supporting information, Figure S1). The other strategy involved whole genome sequencing (Sarin et al. 2008). Both approaches showed that lsy-12 corresponds to the previously uncharacterized R07B5.9 locus, a notion confirmed by multiple alleles of lsy-12 each showing molecular lesions in R07B5.9, rescue experiments, and phenocopy of the lsy-12 defects by RNAi against R07B5.9 (Figure 1D, Table S1) (Sarin et al. 2008).
The R07B5.9 protein, as originally predicted in WormBase (WS215), codes for a 657-amino-acid protein with some limited homology to the C-terminal half of MYST-type histone acetyltransferases, yet it contains no predicted acetyltransferase domain or any other protein domain present in the databases. However, we noted that the gene predicted by WormBase to reside upstream of lsy-12 (R07B5.8) encodes a protein with a MYST-type histone acetyltransferase domain (previously called mys-3; Ceol and Horvitz 2004), which was not previously characterized with mutant alleles. Sequencing of available EST clones (kindly provided by Yuji Kohara) as well as our own RT–PCR analysis reveals that R07B5.9 and R07B5.8 were mispredicted as separate genes and that the downstream predicted gene R07B5.9 is always fused to the upstream R07B5.8 gene (we now refer to this gene as the “a” form of the lsy-12 locus) (Figure 1D). R07B5.8 in turn, is not always fused to R07B5.9 and can produce a smaller splice form with an alternative 3′-UTR (we refer to this variant now as the “b” form of the lsy-12 locus) (Figure 1D). The start site of lsy-12a and lsy-12b is confirmed by the presence of SL1 splice leader sequences. All six mutant alleles of lsy-12 locate to the long variant, lsy-12a (Figure 1D). None of the available lsy-12 alleles are unambiguous molecular nulls (Figure 1D). Attempts to retrieve such alleles by transposon mobilization have failed.
lsy-12 encodes one of four MYST-type histone acetyltransferase (HAT) proteins in the C. elegans genome (Ceol and Horvitz 2004). Other MYST family members have previously been implicated in vulval and ectodermal patterning (Ceol and Horvitz 2004; Shibata et al. 2010). On the basis of overall sequence homology, lsy-12/mys-3 (from here on referred to as lsy-12) and mys-4 are both members of the MOZ/MORF subfamily of MYSTs (Lee and Workman 2007). A deletion allele of mys-4 does not display a Lsy phenotype (data not shown).
Reporters of the lsy-12 locus that only contain upstream regulatory information showed restricted expression patterns and no expression in ASE (data not shown), likely because not all regulatory elements of this large locus are located in the 5′ upstream region. A fosmid-based gfp reporter rescues the lsy-12 mutant phenotype, and is expressed broadly throughout the animal; yet its expression was too weak to unambiguously assess expression in ASEL/R. To assess whether lsy-12 indeed acts in the ASE neurons, we expressed the lsy-12 coding region under control of the bilateral ASE promoter from the ceh-36 locus and found that this construct rescued the Lsy phenotype, demonstrating that lsy-12 acts cell autonomously within the ASE neuron class (Figure 1D; Table S1).
lsy-12 is continuously required to maintain ASE laterality:
Examining the onset of left/right asymmetric gene expression in the ASE neurons in the embryo, we find that in lsy-12 mutants the normally ASER-specific gcy-5 gene is expressed bilaterally from the onset of its expression in threefold embryos (Figure 2A). To address whether lsy-12 may not be involved only in the initial establishment of asymmetry but also in maintaining ASE asymmetry, we used an allele of lsy-12, ot563, which is strongly temperature sensitive. This allele was retrieved as a modifier of the lin-59 locus but was not yet characterized (Sarin et al. 2010). We find that animals continuously raised at 15° show a very lowly penetrant Lsy phenotype (∼10%), while animals continuously raised at 25° show an almost completely penetrant Lsy phenotype (Figure 2B). Animals shifted from the permissive temperature (15°) to the nonpermissive temperature (25°) at the postembryonic L4 stage or even the adult stage (i.e., long after ASE laterality has been established in the embryo) show an ∼50% penetrant ASEL-to-ASER conversion (Figure 2B). Similarly, animals grown at 25° until late larval or adult stages (at which animals would normally display an almost completely penetrant ASEL-to-ASER conversion), show a partial rescue of the mutant phenotype when shifted to 15° (Figure 2B). These findings underscore the plasticity and bipotentiality of the system in that it can revert from one state to the other even after the initial choice has been made.
Figure 2.—
lsy-12 is continually required in ASE but may also act early. (A) lsy-12 is required for the initial manifestation of asymmetry, as assessed by gcy-5∷gfp expression (otIs220 transgene). In wild-type animals, gcy-5 expression is first observed exclusively in ASER in threefold-stage embryos; in lsy-12(ot563) animals, expression of gcy-5∷gfp is bilateral from the onset. (B) Temperature-shift experiments indicate a sustained requirement for lsy-12 activity. lsy-12(ot563) animals that had been grown for several generations at either 15° or 25° were plated and temperature shifted up to 25° or down to 15° at the following timepoints: Embryo, 2-cell; pre-comma; two- to threefold; threefold (those embryos were collected from dissected adult) and postembryonic, L1; L2; L3; L4; and 2-day-old adult. All animals were then scored as 3-day-old adults.
Other known components of MOZ/MORF-type HATs also display Lsy phenotypes:
Work in other systems has revealed three proteins that functionally associate with MYST/LSY-12-type HATs namely the EAF6 protein, the BRPF1 bromodomain protein, and a PHD finger protein of the ING family (Doyon et al. 2006; Lee and Workman 2007; Ullah et al. 2008). There are C. elegans orthologs for each of the three proteins. Mutant alleles are available for the BRPF1 ortholog lin-49 and the ING-like genes T06A10.4 and ing-3. We had previously reported that mutant alleles in one of them, the bromodomain-encoding lin-49 locus, display Lsy defects (Chang et al. 2003). These defects are the same as in lsy-12 mutants: the fate of the ASEL neuron converts to that of the ASER neuron (Chang et al. 2003). We extended this previous observation by showing that not just terminal fate (as shown in Chang et al. 2003) is affected in lin-49 mutants, but that lin-49, like lsy-12, also affects the activity of the bistable feedback loop: ASEL-specific lsy-6 miRNA and die-1 expression is lost in lin-49 mutants (Figure S2A). Moreover, the laterality defects of a hypomorphic allele of lsy-12 allele are enhanced by lin-49 (Figure S2B; since stronger alleles of lsy-12 are completely penetrant, other interaction tests of this sort could not be performed).
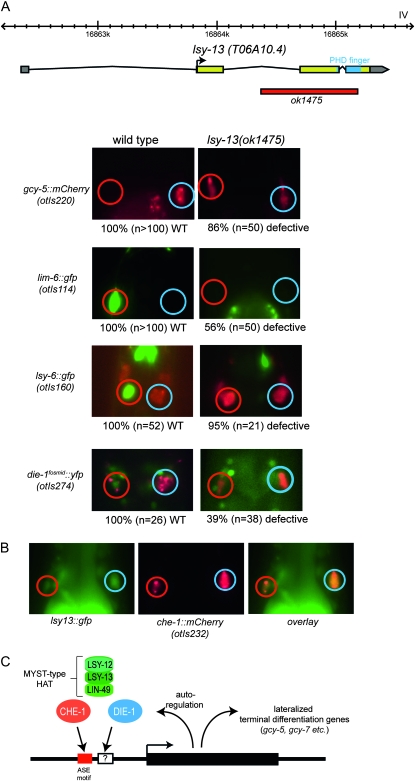
T06A10.4 and ing-3 encode PHD domain-containing proteins related to the ING subunit of MYST-type HAT complexes, with T06A10.4 being closer to the ING1/2/4/5 genes (http://www.treefam.org/cgi-bin/TFinfo.pl?ac=TF352014) and ing-3 being closer to the ING3 gene (http://www.treefam.org/cgi-bin/TFinfo.pl?ac=TF106497). The PHD domain is thought to recognize histone methylation marks and thereby recruit HAT activity to methylated histone substrates (Doyon et al. 2006; Ullah et al. 2008). Even though ING-like proteins have been shown to be involved in cell growth, apoptosis, and tumorgenesis (Coles and Jones 2009), their role in pattern formation and cell fate specification have been little explored. Deletion alleles of T06A10.4 and ing-3 were generated by the C. elegans knockout consortia. ing-3(tm2530) mutants display no Lsy phenotype (data not shown). In contrast, T06A10.4(ok1475) mutant animals display a “2 ASER” Lsy phenotype, such that expression of a terminal ASEL marker is lost while there is ectopic gain of a terminal ASER marker in ASEL (Figure 3A). This phenotype is indistinguishable from lsy-12/HAT and lin-49/BRPF mutants. Again similar to lsy-12 and lin-49 mutants, the expression of the ASEL inducers lsy-6 and die-1 is also lost in T06A10.4(ok1475) mutants, albeit at lower penetrance as observed in lsy-12 mutants. The Lsy phenotype of T06A10.4(ok1475) mutant animals can be rescued by a 4.4-kb PCR fragment spanning the entire locus [2.3 kb upstream of start codon to 0.7 kb downstream of stop; the 86% penetrant gcy-5 misexpression (n = 50) is rescued to 0% misexpression (n = 28)]. We therefore named the T06A10.4 locus lsy-13. A reporter construct for the lsy-13 locus displays a broad expression pattern throughout many tissue types, including expression in the two ASE neurons (Figure 3B), therefore resembling the broad expression of lin-49 (Chamberlin et al. 1999).
Figure 3.—
The MYST-complex component lsy-13 affects ASE laterality. (A) lsy-13 controls ASE laterality. The upper panel shows the structure of the lsy-13 locus and the lsy-13 null allele. The gene structure is confirmed by EST clones (www.wormbase.org). All other panels show the head regions of adult animals. Numbers below the panels indicate the penetrance of the phenotype, i.e. the fraction of animals that display the phenotype shown in the fluorescent image above. The die-1 and lsy-6 wild-type control images are the same as in other figures and shown for comparison only. The red fluorescent marker in the lsy-6 panel (ceh-36prom∷dsRed2) allows identification of the ASE neurons. Animals that express the die-1 reporter fosmid also contain a ASEL/R-expressed red fluorescent reporter (che-1∷mCherry). A list of transgenes used in the study is provided in File S1. We note that lsy-13 function can be maternally supplied (homozygous offspring of a heterozygous lsy-13 parent does not display a mutant phenotype). In contrast to the lin-49 null mutant animals, lsy-13(ok1475) null mutant animals are viable and display no obvious morphological abnormalities. (B) A reporter gene which contains 2.8 kb of 5′ sequences to the first exon of lsy-13, generated by PCR fusion (Hobert 2002), is broadly expressed, including in the two ASE neurons, marked with a red fluorescent reporter gene. Primer sequences for the construct are provided in File S1. (C) Model for lsy-12/lsy-13/lin-49 function, based on the phenotypic similarities between the genes shown here. che-1 directly regulates expression of terminal differentiation genes—both symmetrically and asymmetrically expressed ones—as well as regulators of the bistable feedback loop (Etchberger et al. 2007, 2009). Left/right asymmetrically expressed genes, contain cis-regulatory elements (indicated by “?”) in addition to the CHE-1 binding site (the ASE motif) that restrict CHE-1 activity to ASEL or ASER (Etchberger et al. 2009). Genetically, die-1 controls the activity of the factors that restrict che-1 activity, but it is not known whether die-1 fulfills this function directly (through binding to these additional motifs) or indirectly through the regulation of other factors. Since die-1 autoregulates its own transcription (L. Cochella and O. Hobert, unpublished data), the HAT complex also impinges on die-1 expression itself.
Taken together, the phenotypic similarity of lsy-12, lsy-13, and lin-49 in controlling terminal ASEL/R fate, together with the reported physical and functional interactions of their vertebrate homologs (Doyon et al. 2006; Ullah et al. 2008), suggest that LSY-12, LSY-13, and LIN-49 proteins act together in a complex to control ASEL/R lateralization. We note that lsy-13 is the only ING-like gene with a reported role in nervous system development and our studies provide the first phenotypic side-by-side comparisons of HAT and ING gene activities in a metazoan organism, thereby providing in vivo support for the biochemical studies that link these two proteins (Doyon et al. 2006; Ullah et al. 2008).
The die-1 Zn finger transcription factor is also continuously required to maintain ASE laterality and is a candidate recruiter of the MYST complex:
How could the maintenance function of the MYST complex be explained? Generally, the phenotypic specificity of histone-modifying enzymes must be conferred by transcription factors that recognize DNA in a sequence-specific manner (Struhl 1998). For example, the MYST-type HAT Tip60 is recruited to DNA via diverse transcription factors, such as nuclear hormone receptors or c-Myc (Sapountzi et al. 2006). In the context of ASE laterality control, the C2H2 die-1 Zn finger transcription factor may be such a recruiter. This is because, first, the lsy-12, lsy-13, and lin-49 phenotypes described here resemble those of the die-1 Zn finger transcription factor in that mutant alleles in all these loci display an ASEL-to-ASER fate conversion (“2 ASER” phenotype) (Chang et al. 2004). Second, both die-1 and lsy-12 are continuously required to maintain ASE laterality. In the case of lsy-12, this is demonstrated by the temperature-shift experiments described above; in the case of die-1, we uncovered such requirement through postdevelopmental treatment of animals with dsRNA directed against die-1. Using a nre-1lin-15b RNAi hypersensitive background (Schmitz et al. 2007) and the ASER-expressed gcy-5prom∷gfp transgene otIs186, we observed an ASEL-to-ASER conversion in the P0 generation of dsRNA treated animals [45% (n = 62) of animals showed such a conversion]. The maintained requirement of die-1 is also illustrated by the maintained expression of die-1 in ASEL throughout larval and adult stages (Figure 1C).
The che-1 C2H2 Zn finger transcription factor, a terminal selector for ASE fate, which acts through a cis-regulatory motif, the ASE motif, present in bilaterally and left/right asymmetrically expressed terminal differentiation genes, is, like die-1, also continuously required to maintain the differentiated state of the ASE neurons (Etchberger et al. 2007, 2009). In the case of asymmetrically expressed genes, CHE-1 cooperates with additional DNA-binding proteins—possibly DIE-1—to ensure left/right asymmetric expression (Etchberger et al. 2009). We therefore propose that continuously required CHE-1 and DIE-1 are the sequence-specific DNA binding proteins that recruit the LSY-12/LSY-13/LIN-49 MYST–HAT complex to maintain terminal differentiation features (Figure 3C).
A role in maintaining differentiated cellular states has also been recently reported for another MYST–HAT complex, composed of the C. elegans Bromodomain protein BET-1 (a paralog of LIN-49) and its associated MYST-type histone acetyltransferase mys-2 (a paralog of lsy-12) (Shibata et al. 2010). The vertebrate LIN-49 homolog BRPF1 and the histone acetyltransferase Myst3, as well as the fly MYST family member Chameau, are required to maintain HOX gene expression during development (Grienenberger et al. 2002; Laue et al. 2008), and the vertebrate MYST family member MOZ is required for the generation and maintenance of hematopoietic stem cells (Katsumoto et al. 2008). Together, these findings suggest that MYST function in maintenance of gene expression patterns has been broadly conserved during evolution and is employed in many different cell types.
Acknowledgments
We thank Baris Tursun for the die-1 fosmid reporter line, Sumeet Sarin for the ot563 allele, Richard J. Poole and Enkelejda Bashllari for sharing the first evidence of a die-1 maintenance role, Luisa Cochella for communicating unpublished results, the C. elegans Gene Knockout Consortia in Oklahoma and Tokyo for the ok1475 and tm2530 alleles, Yuji Kohara for EST clones, and Qi Chen for expert assistance in generating transgenic strains. We thank members of the Hobert lab for comments on the manuscript. This work was funded by the National Institutes of Health (R01NS039996-05; R01NS050266-03). O.H. is an investigator of the Howard Hughes Medical Institute.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.123661/DC1.
References
- Blau, H. M., 1992. Differentiation requires continuous active control. Annu. Rev. Biochem. 61 1213–1230. [DOI] [PubMed] [Google Scholar]
- Boulin, T., J. F. Etchberger and O. Hobert, 2006. Reporter gene fusions. WormBook, 1–23. [DOI] [PMC free article] [PubMed]
- Ceol, C. J., and H. R. Horvitz, 2004. A new class of C. elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev. Cell 6 563–576. [DOI] [PubMed] [Google Scholar]
- Chamberlin, H. M., K. B. Brown, P. W. Sternberg and J. H. Thomas, 1999. Characterization of seven genes affecting Caenorhabditis elegans hindgut development. Genetics 153 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S., R. J. Johnston, C. Frokjaer-Jensen, S. Lockery and O. Hobert, 2004. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 430 785–789. [DOI] [PubMed] [Google Scholar]
- Chang, S., R. J. Johnston, Jr. and O. Hobert, 2003. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 17 2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles, A. H., and S. N. Jones, 2009. The ING gene family in the regulation of cell growth and tumorigenesis. J. Cell Physiol. 218 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano, D., L. Cochella, B. Tursun and O. Hobert, 2010. Neuron-type specific regulation of a 3′UTR through redundant and combinatorially acting cis-regulatory elements. RNA 16 349–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon, Y., C. Cayrou, M. Ullah, A. J. Landry, V. Cote et al., 2006. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 21 51–64. [DOI] [PubMed] [Google Scholar]
- Etchberger, J. F., A. Lorch, M. C. Sleumer, R. Zapf, S. J. Jones et al., 2007. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 21 1653–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger, J. F., E. B. Flowers, R. J. Poole, E. Bashllari and O. Hobert, 2009. Cis-regulatory mechanisms of left/right asymmetric neuron-subtype specification in C. elegans. Development 136 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger, A., B. Miotto, T. Sagnier, G. Cavalli, V. Schramke et al., 2002. The MYST domain acetyltransferase Chameau functions in epigenetic mechanisms of transcriptional repression. Curr. Biol. 12 762–766. [DOI] [PubMed] [Google Scholar]
- Hobert, O., 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32 728–730. [DOI] [PubMed] [Google Scholar]
- Hobert, O., 2006. Architecture of a microRNA-controlled gene regulatory network that diversifies neuronal cell fates. Cold Spring Harb. Symp. Quant. Biol. 71 181–188. [DOI] [PubMed] [Google Scholar]
- Johnston, R. J., and O. Hobert, 2003. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426 845–849. [DOI] [PubMed] [Google Scholar]
- Johnston, Jr., R. J., S. Chang, J. F. Etchberger, C. O. Ortiz and O. Hobert, 2005. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. USA 102 12449–12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumoto, T., N. Yoshida and I. Kitabayashi, 2008. Roles of the histone acetyltransferase monocytic leukemia zinc finger protein in normal and malignant hematopoiesis. Cancer Sci. 99 1523–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue, K., S. Daujat, J. G. Crump, N. Plaster, H. H. Roehl et al., 2008. The multidomain protein Brpf1 binds histones and is required for Hox gene expression and segmental identity. Development 135 1935–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. K., and J. L. Workman, 2007. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8 284–295. [DOI] [PubMed] [Google Scholar]
- Sapountzi, V., I. R. Logan and C. N. Robson, 2006. Cellular functions of TIP60. Int. J. Biochem. Cell Biol. 38 1496–1509. [DOI] [PubMed] [Google Scholar]
- Sarin, S., M. O'Meara, E. B. Flowers, C. Antonio, R. J. Poole et al., 2007. Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics 176 2109–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin, S., S. Prabhu, M. M. O'Meara, I. Pe'er and O. Hobert, 2008. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat. Methods 5 865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin, S., V. Bertrand, H. Bigelow, A. Boyanov, M. Doitsidou et al., 2010. Analysis of multiple ethyl methanesulfonate-mutagenized Caenorhabditis elegans strains by whole-genome sequencing. Genetics 185 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, C., P. Kinge and H. Hutter, 2007. Axon guidance genes identified in a large-scale RNAi screen using the RNAi-hypersensitive Caenorhabditis elegans strain nre-1(hd20) lin-15b(hd126). Proc. Natl. Acad. Sci. USA 104 834–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, Y., H. Takeshita, N. Sasakawa and H. Sawa, 2010. Double bromodomain protein BET-1 and MYST HATs establish and maintain stable cell fates in C. elegans. Development 137 1045–1053. [DOI] [PubMed] [Google Scholar]
- Struhl, K., 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12 599–606. [DOI] [PubMed] [Google Scholar]
- Suzuki, H., T. R. Thiele, S. Faumont, M. Ezcurra, S. R. Lockery et al., 2008. Functional asymmetry in Caenorhabditis elegans taste neurons and its computational role in chemotaxis. Nature 454 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, M., N. Pelletier, L. Xiao, S. P. Zhao, K. Wang et al., 2008. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol. Cell. Biol. 28 6828–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]