A 1.35 Å resolution crystal structure of Cas2 from the bacterium Desulfovibrio vulgaris (DvuCas2) is reported.
Keywords: CRISPR-associated proteins, Cas2, Desulfovibrio vulgaris
Abstract
CRISPRs (clustered regularly interspaced short palindromic repeats) provide bacteria and archaea with RNA-guided acquired immunity to invasive DNAs. CRISPR-associated (Cas) proteins carry out the immune effector functions. Cas2 is a universal component of the CRISPR system. Here, a 1.35 Å resolution crystal structure of Cas2 from the bacterium Desulfovibrio vulgaris (DvuCas2) is reported. DvuCas2 is a homodimer, with each protomer consisting of an N-terminal βαββαβ ferredoxin fold (amino acids 1–78) to which is appended a C-terminal segment (amino acids 79–102) that includes a short 310-helix and a fifth β-strand. The β5 strands align with the β4 strands of the opposite protomers, resulting in two five-stranded antiparallel β-sheets that form a sandwich at the dimer interface. The DvuCas2 dimer is stabilized by a distinctive network of hydrophilic cross-protomer side-chain interactions.
1. Introduction
The prokaryal immune system known as CRISPR (clustered regularly interspaced short palindromic repeats) provides adaptive protection against foreign DNA elements (viruses and plasmids) as well as a heritable record of ‘past genetic aggressions’ (Barrangou et al., 2007 ▶; Mojica et al., 2005 ▶; Pourcel et al., 2005 ▶). CRISPRs are present in nearly all archaea and about 40% of bacteria for which genome sequences are available (Karginov & Hannon, 2010 ▶; Sorek et al., 2008 ▶; van der Oost et al., 2009 ▶). CRISPRs consist of an array of short direct repeat sequences (25–50 bp) separated by hypervariable spacer sequences (20–70 bp). The repeat array is flanked on one side by an AT-rich region called the leader. CRISPR loci are surrounded by a set of conserved protein-coding cas (CRISPR-associated) genes that vary in their orientation and order (Haft et al., 2005 ▶; Makarova et al., 2006 ▶). Cas proteins carry out the immune effector functions, entailing the incorporation of new spacer sequences into the CRISPR locus, the processing of CRISPR RNAs and interference with the nucleic acid targets. The CRISPR locus is transcribed into a single RNA transcript with the leader at the 5′ end followed by the tandem repeats. The primary transcript is processed by a subset of the Cas proteins to generate unit-sized CRISPR RNAs (crRNAs; Brouns et al., 2008 ▶), each consisting of one spacer flanked by 5′ and 3′ sequences derived from the direct repeats. The processed crRNAs can interfere with foreign genetic elements to which they display base complementarity (Horvath & Barrangou, 2010 ▶; Karginov & Hannon, 2010 ▶; Marraffini & Sontheimer, 2010 ▶; Sorek et al., 2008 ▶; van der Oost et al., 2009 ▶). The crRNA target can be either DNA or RNA, depending on the organism and the ensemble of Cas proteins (Brouns et al., 2008 ▶; Hale et al., 2009 ▶; Marraffini & Sontheimer, 2008 ▶).
The cas2 gene is always located near a CRISPR locus. Although the role of cas2 in CRISPR biology is not well defined, the available genetic studies indicate that cas2 is not required for the synthesis of the pre-CRISPR transcript or its processing to crRNAs (Brouns et al., 2008 ▶). Cas2 proteins are small (80–120 amino acids) and are conserved among CRISPR-bearing taxa. Crystal structures have been solved for four Cas2 proteins: the paralogs Sso1404 and Sso8090 from the crenarchaeon Sulfolobus solfataricus (PDB codes 2ivy, 2i8e and 3exc; Beloglazova et al., 2008 ▶), Pfu1117 from the euryarchaeon Pyrococcus furiosus (PDB code 2iox) and Tth1823 from the bacterium Thermus thermophilus (PDB code 1zpw). These Cas2s adopt a ferredoxin fold composed of a four-stranded antiparallel β-sheet plus two α-helices packed against one face of the sheet. Beloglazova et al. (2008 ▶) demonstrated metal-dependent endoribonuclease activity for the Sso1404 Cas2 protein. The Sso8090 paralog and the Cas2 homologs from Thermotoga maritima, Methanobacterium thermoautotrophicum, Archaeoglobus fulgidus and Nitrosomonas europea also had RNase activity. Mutational analysis identified six residues in Sso1404 as important for RNase activity; these were suggested to comprise the RNase active site. The potential roles of a Cas2 RNase in the CRISPR pathway might be to selectively degrade phage transcripts or to globally inhibit translation by mRNA cleavage (Beloglazova et al., 2008 ▶).
Here, we report the 1.35 Å resolution crystal structure of DvuCas2 from the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. The sequenced D. vulgaris genome consists of a 3.57 Mb chromosome and a 202 kb megaplasmid (Heidelberg et al., 2004 ▶). The D. vulgaris CRISPR and flanking cas2 gene are located on the megaplasmid. To our knowledge, there have been no reports to date concerning the structure or function of CRISPR-system components from this organism.
2. Materials and methods
2.1. Purification of DvuCas2
The D. vulgaris Hildenborough cas2 gene (DVUA0135; accession code YP_009175) was amplified by PCR from genomic DNA with sense and antisense primers that introduced a BamHI site over the start codon and a NotI site 3′ to the stop codon. The PCR product was digested with BamHI and NotI and inserted into pET-His10Smt3. The resulting pET-DvuCas2 plasmid encodes the 102-amino-acid DvuCas2 protein fused to an N-terminal His10Smt3 tag. Cultures (8 l) of Escherichia coli BL21 (DE3) pET-Cas2 were grown at 310 K in Terrific Broth containing 0.1 mg ml−1 kanamycin until A 600 reached ∼0.8. The cultures were adjusted to 0.5 mM isopropyl β-d-1-thiogalactopyranoside and incubation was continued for 18 h at 290 K with continuous shaking. Cells were harvested by centrifugation and stored at 193 K. All subsequent procedures were performed at 277 K. Thawed cells were resuspended in 100 ml buffer A (50 mM Tris–HCl pH 7.5, 1.2 M NaCl, 200 mM LiSO4, 15 mM imidazole, 10% glycerol) supplemented with 1% Triton X-100 and 1 mg ml−1 lysozyme to achieve cell lysis. The lysate was sonicated to reduce its viscosity and insoluble material was removed by centrifugation. His10Smt3-Cas2 was isolated from the soluble extract by adsorption to 15 ml Ni–NTA agarose (Qiagen) that had been equilibrated in buffer A. The bound protein was eluted stepwise with 50, 100, 200, 350 and 500 mM imidazole in buffer B (50 mM Tris–HCl pH 8.0, 50 mM NaCl, 10% glycerol). The elution profile was monitored by SDS–PAGE of the column fractions. The 200 and 350 mM imidazole eluate fractions containing His10Smt3-Cas2 were pooled (200 mg protein), supplemented with 300 µg Ulp1 (a Smt3-specific protease) and dialyzed overnight against buffer C (50 mM Tris pH 8.0, 250 mM NaCl, 5 mM DTT, 10% glycerol). The resulting mixture of cleaved His10Smt3 and tag-less Cas2 was applied onto a 10 ml Ni–NTA agarose column that had been equilibrated with buffer C. Cas2 was recovered in the flowthrough fractions, devoid of His10Smt3, which bound to the column. The Ni–agarose flowthrough material was applied onto a 10 ml column of DEAE-Sephacel that had been equilibrated with buffer C. Cas2 was recovered in the DEAE flowthrough fraction and was concentrated to a volume of 5 ml by centrifugal ultrafiltration. This material was applied onto a 120 ml HiPrep 16/60 Superdex-200 gel-filtration column equilibrated in 10 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT. The peak Cas2-containing fractions were concentrated by centrifugal ultrafiltration to 20 mg ml−1 in a 2 ml volume and the preparation was stored at 193 K.
2.2. Crystallization, diffraction data collection and structure determination
Crystallization was performed at room temperature by the hanging-drop vapor-diffusion method. Cas2 (1 µl) was mixed with an equal volume of reservoir solution consisting of 0.1 M trisodium citrate, 1 M lithium sulfate and 0.5 M ammonium sulfate. Plate-shaped crystals were harvested after 2–3 d and were transferred to a solution consting of 0.1 M trisodium citrate, 1 M lithium sulfate and 2 M ammonium sulfate prior to flash-freezing in liquid nitrogen. Alternatively, a Hg derivative was prepared by transferring crystals to the same solution containing 2 mM thimerosal for 30 min prior to flash-freezing in liquid nitrogen. Diffraction data for native crystals were collected to 1.35 Å resolution on APS beamline 24-ID-E at a wavelength of 0.9795 Å (200 consecutive 1° frames). Diffraction data for the thimerosal-soaked crystal were collected to 2.3 Å resolution using a Rigaku R-AXIS IV detector and a rotating-anode Cu Kα X-ray source at 1.5418 Å (208° of combined oscillation data). The data were processed using HKL-2000 and SCALEPACK (Otwinowski & Minor, 1997 ▶). The native and thimerosal-soaked crystals both belonged to space group C2 and had similar unit-cell parameters, but were nonisomorphous.
Single-wavelength anomalous diffraction (SAD) phasing calculations for the thimerosal-soaked crystal were implemented in PHENIX/Phaser (Adams et al., 2002 ▶). Anomalous difference Fourier maps revealed four peaks above 10σ. The initial phases were improved in RESOLVE with the aid of noncrystallographic symmetry determined from the heavy-atom sites (Terwilliger, 2000 ▶, 2003 ▶). The RESOLVE map revealed two Cas2 molecules in the asymmetric unit and allowed the automatic building of ∼75% of the amino acids in the Cas2 protomers. The two mercury sites per Cas2 protomer were located adjacent to cysteine side chains, as expected.
This initial model was used to derive phases for the native Cas2 crystal, which were improved in RESOLVE. The structural model was extended with the autobuild feature of PHENIX and was then edited manually in Coot (Emsley & Cowtan, 2004 ▶) and refined iteratively against experimental data in PHENIX using refinement of individual sites, occupancy refinement for alternate conformations and anisotropic atomic displacement parameters. The final refined model at 1.35 Å resolution (R and R free values of 0.128 and 0.177, respectively) included 99 and 102 amino acids from Cas2 protomers A and B with excellent geometry (Table 1 ▶).
Table 1. DvuCas2 crystallographic data and refinement statistics.
Standard definitions are used for all of the parameters. Values in parentheses are for the highest resolution bin. The data-collection statistics are from SCALEPACK. The refinement and geometric statistics are from PHENIX.
| Thimerosal-soaked crystal | Native crystal | |
|---|---|---|
| Space group | C2 | C2 |
| Unit-cell parameters at 130K (, ) | a = 107.30, b = 48.92, c = 41.61, = 98.68 | a = 107.88, b = 48.81, c = 41.32, = 99.20 |
| Radiation source | CuK | APS 24-ID-E |
| Wavelength () | 1.5418 | 0.9795 |
| Crystallographic data quality | ||
| Resolution () | 25.02.30 (2.342.30) | 50.01.35 (1.371.35) |
| R merge [for I> 3(I)] (%) | 9.3 (29.8) | 4.9 (43.0) |
| Unique reflections† | 18368 (909) | 45550 (2001) |
| Mean multiplicity | 3.35 (3.3) | 3.50 (2.7) |
| Completeness (%) | 99.2 (98.7) | 97.0 (85.7) |
| Mean I/(I) | 14.0 (3.8) | 29.1 (2.0) |
| Phasing statistics | ||
| Phasing method | Hg-SAD | |
| Resolution () | 25.03.0 | |
| Anomalous signal‡ | 0.08 | |
| Figure of merit§ | 0.539/0.78 | |
| Refinement statistics | ||
| Resolution () | 29.01.35 (1.401.35) | |
| R work/R free ¶ for F > 0 | 0.128/0.177 (0.174/0.249) | |
| Estimated coordinate error () | 0.13 | |
| Model statistics | ||
| R.m.s.d. bond lengths () | 0.006 | |
| R.m.s.d. bond angles () | 1.12 | |
| NCS deviations†† [C residues] () | 0.316 [96] | |
| Ramachandran outliers | 0 | |
| B factors (2) | ||
| Wilson | 12.1 | |
| Overall | 24.3 | |
| Protein | 22.1 | |
| Heteroatoms | 35.9 | |
| Model contents | ||
| Residues (chain A/chain B) | 99/102 | |
| Alternate conformations | 36 | |
| Heteroatoms | 5 sulfate, 3 citrate‡‡, 1 Tris, 1 sodium, 1 chloride | |
| Waters | 299 | |
| PDB code | 3oq2 | |
F + and F were treated as equivalent observations for native data sets, but as distinct in the data for the thimerosal-soaked crystal.
Anomalous signal as output by Phaser using the formula ([||F(+)| |F()||2]/{1/2[|F(+)|2 + |F()|2]})1/2.
Hg-SAD figures of merit are from Phaser/RESOLVE.
The R free sets for cross-validation consisted of 5% of data selected at random.
No NCS restraints were used during refinement.
Two sulfate positions are co-occupied by citrate.
3. Results and discussion
3.1. DvuCas2 structure and comparison with other Cas2 proteins
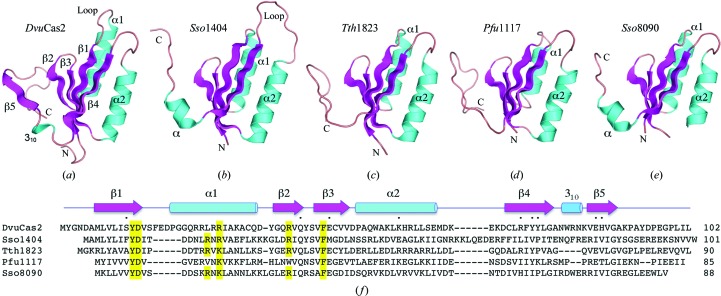
The A and B protomers of the DvuCas2 homodimer each consist of an N-terminal βαββαβ ferredoxin fold (amino acids 1–78) to which is appended a C-terminal segment (amino acids 79–102) that includes a short 310-helix and a fifth β-strand (Figs. 1 ▶ a and 1 ▶ f). The tertiary structures of the A and B protomers were similar overall (r.m.s.d. of 0.32 Å at 96 Cα positions), though punctuated by many local variations, e.g. in the main-chain conformation of the β1–α1 loop, in numerous side-chain orientations and the asymmetric association of sulfate ligands with one or the other subunit.
Figure 1.
Structure of DvuCas2 and comparison to other Cas2 homologs. The fold of the DvuCas2 protomer (PDB code 3oq2) is shown in (a) with cyan helices and magenta β-strands. (b)–(e) show the tertiary structures of the Cas2 homologs Sso1404 (PDB code 2i8e), Tth1823 (PDB code 1zpw), Pfu1117 (PDB code 2i0x) and Sso8090 (PDB code 3exc) superimposed on DvuCas2 and then offset horizontally. The N- and C-termini are labeled. (f) shows a structure-based alignment of the amino-acid sequences of the five Cas2 proteins. The secondary-structure elements of DvuCas2 are shown above the sequence, with β-strands depicted as arrows and helices as cylinders. Gaps in the alignment are indicated by dashes. The amino-acid side chains that comprise the DvuCas2 homodimer interface (Fig. 2 ▶) are denoted by dots above the sequence. Residues that are essential for the endoribonuclease activity of Sso1404 are highlighted in yellow.
A DALI search with DvuCas2 identified the four known Cas2 structures as the highest scoring hits, as follows: Tth1823 (Z score 11.6; r.m.s.d. of 1.9 Å at 82 Cα positions), Pfu1117 (Z score 10.4), Sso8090 (Z score 10.2) and Sso1404 (Z score 9.3). These four Cas2 structures are superimposed on DvuCas2 and offset laterally in Figs. 1 ▶(b)–1 ▶(e). A structure-based alignment of the amino-acid sequences of DvuCas2, Pfu1117, Tth1823, Sso8090 and Sso1404 is shown in Fig. 1 ▶(f). These comparisons reveal several distinctive features of DvuCas2. For example, DvuCas2 is unique in having a 310-helix as the connecting secondary-structure element to the C-terminal segment (Fig. 1 ▶ a); this contrasts with the α-helices found in the equivalent positions of Sso1404 (Fig. 1 ▶ b) and Sso8090 (Fig. 1 ▶ e) and the lack of any defined secondary structure at the corresponding sites in Tth1823 (Fig. 1 ▶ c) and Pfu1117 (Fig. 1 ▶ d). The β1–α1 loop is longer in DvuCas2 than in any of its homologs (Figs. 1 ▶ a and 1 ▶ f). [Note that Sso1404 is distinguished by its longer α2–β4 loop (Figs. 1 ▶ b and 1 ▶ f).] Finally, the paths of the C-termini distal to the equivalent of the DvuCas2 β5 strand vary among the Cas2 proteins. The C-terminus projects downwards in DvuCas2, Tth1823 and Pfu1117, but upwards in Sso1401 and Sso8090 (Figs. 1 ▶ a–1 ▶ e).
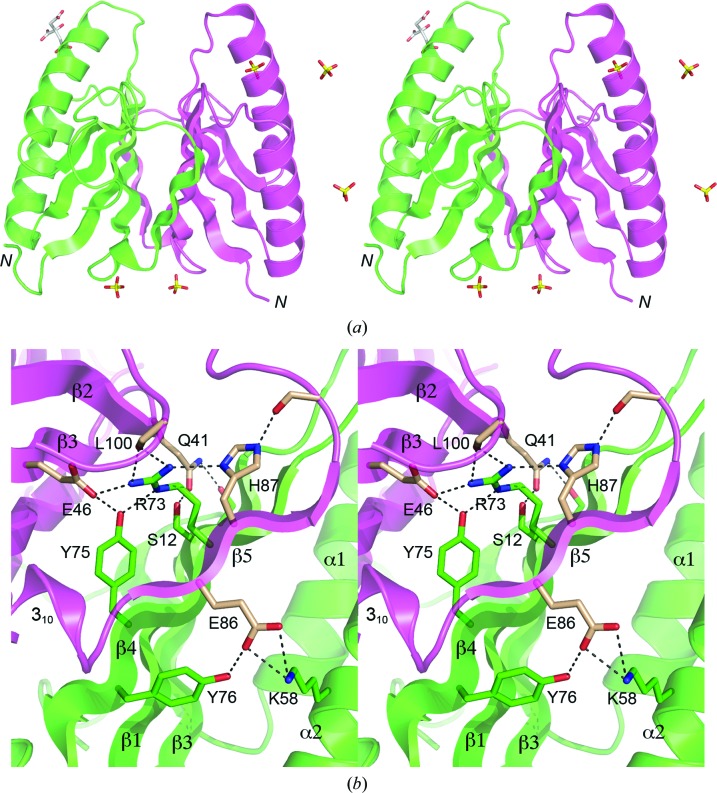
3.2. The DvuCas2 dimer interface
The β5 strands of the DvuCas2 A and B protomers project across a pseudo-twofold axis to align with the β4 strands of the opposite protomers, resulting in two five-stranded antiparallel β-sheets of topology β5*β4132 that form a β-sandwich at the dimer interface (Fig. 2 ▶ a). The dimer is stabilized by many hydrophilic cross-protomer side-chain interactions (Fig. 2 ▶ b). Glu86 tethers β5 to the trans protomer via a salt bridge to Lys58 (in α2) and a hydrogen bond to Tyr76 (β4). Arg73 and Tyr75 link β4 to the β3 and β5 strands and the C-terminal peptide of the trans protomer via multiple hydrogen bonds to the Glu46 carboxylate, His87 Nδ and the Leu100 main-chain carbonyl (Fig. 2 ▶ b). Gln41 tethers β2 to β1 and the β2–β3 loop of the trans protomer by hydrogen bonds to Ser12 Oγ and Ser43 Oγ, respectively (Fig. 2 ▶ b). The side-chain atomic contacts at the DvuCas2 dimer interface are substantially different from those seen in the Sso1404 and Sso8090 structures, which is consistent with the lack of side-chain conservation at most of the residues that comprise the interface in DvuCas2. Many (though not all) of the cross-protomer contacts seen in DvuCas2 are conserved in Tth1183.
Figure 2.
Stereoviews of the DvuCas2 dimer interface. (a) The folds of the A and B protomers are rendered in magenta and green, respectively. The N-termini are indicated. Five sulfates and a citrate located at various sites on the protein surface are shown as stick models. The two sulfate sites at the bottom of the image were refined as partially occupied by sulfate and partially by citrate; only the sulfates are shown for the sake of clarity. (b) This view highlights the hydrophilic cross-protomer side-chain interactions. The secondary-structure elements are labeled as in Fig. 1 ▶(f). Side chains of the A and B protomers that comprise the dimer interface are shown as stick models with beige and green C atoms, respectively. Ionic and hydrogen-bonding contacts of side-chain atoms at the dimer interface are indicated by dashed lines.
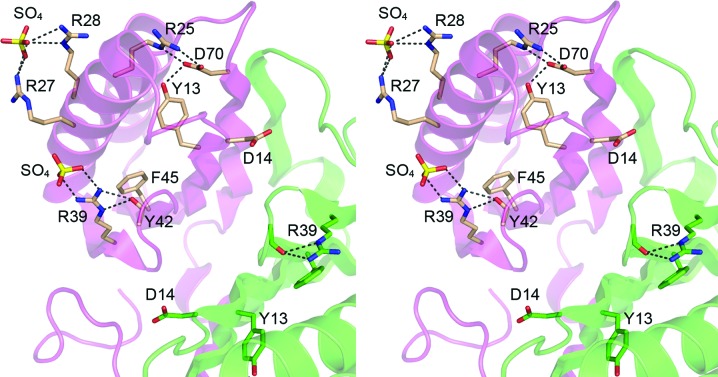
3.3. Does DvuCas2 have a phosphodiesterase active site?
Sso1404 was characterized as a metal-dependent endoribonuclease that incises single-stranded RNAs to yield 5′-phosphate and 3′-OH ends at the break site (Beloglazova et al., 2008 ▶). Alanine scanning identified six Sso1404 residues that are essential for RNase activity (Tyr9, Asp10, Arg17, Arg19, Arg31 and Phe37); these are highlighted in yellow in Fig. 1 ▶(f). Five of the six essential Sso1404 residues are conserved in DvuCas2; they correspond to DvuCas2 Tyr13, Asp14, Arg28, Arg39 and Phe45 (Fig. 3 ▶). Beloglazova et al. (2008 ▶) speculated that five of the essential side chains (Tyr9, Asp10, Arg17, Arg31 and Phe37) could form a phosphodiesterase active site. They specifically proposed that (i) Asp10 is critical for catalysis because it coordinates the divalent-cation cofactor and (ii) the two Asp10 carboxylates, which are located 6.5 Å from each other at the Sso1404 homodimer interface, cooperate to bind a single divalent cation. This detailed mechanistic hypothesis is most unlikely to apply to DvuCas2, insofar as the equivalent Asp14 side chains are separated by 15.4 Å (Fig. 3 ▶). It also strikes us as implausible that other homodimeric Cas2 homologs could form the proposed composite metal-binding site with contributions from both protomers because the equivalent aspartates are separated by 12.8 Å in Tth1823 and by 10.8 Å in Pfu1117. In order for these aspartates to bind a bridging metal, the Cas2 proteins would need to undergo a major reorientation of their homodimer interfaces. For DvuCas2, at least, this would entail the disruption and remodeling of a large number of cross-protomer contacts. Beloglazova et al. (2008 ▶) also suggested that the α2–β4 loop of Sso1404 might be responsible for RNA substrate recognition by the Cas2 endonuclease. The equivalent segment is missing from DvuCas2.
Figure 3.
Disposition of DvuCas2 residues corresponding to those implicated in the endoribonuclease activity of Cas2 homologs. The folds of the A and B protomers are rendered in magenta and green, respectively. The side chains of the A and B protomers that correspond to the putative Sso1404 phosphodiesterase active-site residues (Fig. 1 ▶ f) are shown as stick models with beige and green C atoms, respectively. Two sulfate anions docked on the surface of the DvuCas2 A protomer are also shown as stick models. Ionic and hydrogen-bonding interactions are indicated by dashed lines.
It is worth considering that endoribonuclease activity might not be shared by all members of the Cas2 family and that at least some of the residues found to be essential for RNase activity in Sso1404 might have a structural role. This scenario would be plausible in DvuCas2, where we see that Tyr13 (which is conserved in all Cas2 homologs) is buried in the protein core, where it donates a hydrogen bond to Asp70, which in turn makes a salt bridge to Arg25 (Fig. 3 ▶). Phe45, another invariant Cas2 residue, is buried in the hydrophobic core of the ferredoxin fold (Fig. 3 ▶). Neither Tyr13 nor Phe45 is disposed for a catalytic role. However, DvuCas2 Arg39 (equivalent to Arg31 in Sso1404) might be a candidate to interact with a nucleic acid because in one of the DvuCas2 protomers of the homodimer Arg39 coordinates a sulfate anion (a potential mimetic of a polynucleotide phosphate) via a bidentate salt bridge to the terminal guanidinium N atoms (Fig. 3 ▶). Note that Arg39 also contributes to the DvuCas2 fold in both protomers via a bifurcated hydrogen bond to the Tyr42 main-chain carbonyl (Fig. 3 ▶). Arg39 is conserved in Tth1823, Sso8090 and Sso1404, but is replaced by Trp in Pfu1117 (Fig. 1 ▶ f). Another sulfate anion in the DvuCas2 structure is coordinated by Arg27 and Arg28 (Fig. 3 ▶). Arg28 (equivalent to Arg19 in Sso1404) is conserved as a basic residue in other Cas2 homologs, but Arg27 is not (Fig. 1 ▶ f). We have been unable to convincingly demonstrate a metal-dependent endonuclease activity for purified DvuCas2 or to demonstrate its binding to ssRNA or ssDNA in a gel-shift assay format.
In summary, the 1.35 Å resolution crystal structure of DvuCas2 underscores a common fold for Cas2-family members, while revealing distinctive structural features of DvuCas2, especially at the homodimer interface. The DvuCas2 structure does not fit the current models of endonuclease catalysis proposed for Sso1404.
Supplementary Material
PDB reference: Cas2, 3oq2
Acknowledgments
We thank Dr Yehuda Goldgury for assistance with data collection. This research was supported by NIH grant GM63611. SS is an American Cancer Society Research Professor.
References
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., Romero, D. A. & Horvath, P. (2007). Science, 315, 1709–1712. [DOI] [PubMed]
- Beloglazova, N., Brown, G., Zimmerman, M. D., Proudfoot, M., Makarova, K. S., Kudritska, M., Kochinyan, S., Wang, S., Chruszcz, M., Minor, W., Koonin, E. V., Edwards, A. M., Savchenko, A. & Yakunin, A. F. (2008). J. Biol. Chem. 283, 20361–20371. [DOI] [PMC free article] [PubMed]
- Brouns, S. J., Jore, M. M., Lundgren, M., Westra, E. R., Slijkhuis, R. J., Snijders, A. P., Dickman, M. J., Makarova, K. S., Koonin, E. V. & van der Oost, J. (2008). Science, 321, 960–964. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Haft, D. H., Selengut, J., Mongodin, E. F. & Nelson, K. E. (2005). PLoS Comput. Biol. 1, e60. [DOI] [PMC free article] [PubMed]
- Hale, C. R., Zhao, P., Olson, S., Duff, M. O., Graveley, B. R., Wells, L., Terns, R. M. & Terns, M. P. (2009). Cell, 139, 945–956. [DOI] [PMC free article] [PubMed]
- Heidelberg, J. F. et al. (2004). Nature Biotechnol. 22, 554–559. [DOI] [PubMed]
- Horvath, P. & Barrangou, R. (2010). Science, 327, 167–170. [DOI] [PubMed]
- Karginov, F. V. & Hannon, G. J. (2010). Mol. Cell, 37, 7–19. [DOI] [PMC free article] [PubMed]
- Makarova, K. S., Grishin, N. V., Shabalina, S. A., Wolf, Y. I. & Koonin, E. V. (2006). Biol. Direct, 1, 7. [DOI] [PMC free article] [PubMed]
- Marraffini, L. A. & Sontheimer, E. J. (2008). Science, 322, 1843–1845. [DOI] [PMC free article] [PubMed]
- Marraffini, L. A. & Sontheimer, E. J. (2010). Nature Rev. Genet. 11, 181–190. [DOI] [PMC free article] [PubMed]
- Mojica, F. J., Diez-Villasenor, C., Garcia-Martinez, J. & Soria, E. (2005). J. Mol. Evol. 60, 174–182. [DOI] [PubMed]
- Oost, J. van der, Jore, M. M., Westra, E. R., Lundgren, M. & Brouns, S. J. J. (2009). Trends Biochem. Sci. 34, 401–407. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pourcel, C., Salvignol, G. & Vergnaud, G. (2005). Microbiology, 151, 653–663. [DOI] [PubMed]
- Sorek, R., Kunin, V. & Hugenholtz, P. (2008). Nature Rev. Microbiol. 6, 181–186. [DOI] [PubMed]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. (2003). Acta Cryst. D59, 38–44. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Cas2, 3oq2