Crystal structures of biosynthetic arginine decarboxylase (ADC, SpeA) from E. coli and C. jejuni are reported.
Keywords: biosynthetic arginine decarboxylases, SpeA, Campylobacter jejuni, Escherichia coli
Abstract
Biosynthetic arginine decarboxylase (ADC; also known as SpeA) plays an important role in the biosynthesis of polyamines from arginine in bacteria and plants. SpeA is a pyridoxal-5′-phosphate (PLP)-dependent enzyme and shares weak sequence homology with several other PLP-dependent decarboxylases. Here, the crystal structure of PLP-bound SpeA from Campylobacter jejuni is reported at 3.0 Å resolution and that of Escherichia coli SpeA in complex with a sulfate ion is reported at 3.1 Å resolution. The structure of the SpeA monomer contains two large domains, an N-terminal TIM-barrel domain followed by a β-sandwich domain, as well as two smaller helical domains. The TIM-barrel and β-sandwich domains share structural homology with several other PLP-dependent decarboxylases, even though the sequence conservation among these enzymes is less than 25%. A similar tetramer is observed for both C. jejuni and E. coli SpeA, composed of two dimers of tightly associated monomers. The active site of SpeA is located at the interface of this dimer and is formed by residues from the TIM-barrel domain of one monomer and a highly conserved loop in the β-sandwich domain of the other monomer. The PLP cofactor is recognized by hydrogen-bonding, π-stacking and van der Waals interactions.
1. Introduction
Polyamines (putrescine, spermidine and spermine) are involved in a variety of important cellular processes in all living cells, including cell growth and differentiation, DNA replication and repair, RNA synthesis and processing, and protein synthesis and metabolism (Wallace, 2009 ▶; Wallace et al., 2003 ▶; Tabor & Tabor, 1985 ▶). In animals, arginine serves as the primary precursor for the biosynthesis of polyamines through its hydrolysis to ornithine by arginases (Morris, 2004 ▶, 2009 ▶). Ornithine is then converted to putrescine by ornithine decarboxylase (ODC), which is a proven target for drug action (Wallace, 2009 ▶). In bacteria and plants, the biosynthesis of polyamines from arginine proceeds through a different pathway. Arginine is first converted to agmatine by a biosynthetic arginine decarboxylase (ADC; also known as SpeA; Wu & Morris, 1973a ▶; Moore & Boyle, 1990 ▶), which is then hydrolyzed to generate putrescine and urea by agmatinase (Morris, 2004 ▶). These organisms can also produce polyamines through ODC activity.
SpeA is a pyridoxal-5′-phosphate (PLP)-dependent enzyme that is widely distributed in plants and bacteria. It shares weak sequence homology to several other PLP-dependent decarboxylases, especially ODC. Previous biochemical studies suggested that the holoenzyme of Escherichia coli SpeA is a tetramer, in which one molecule of PLP is bound to each subunit, with a molecular weight of 70 kDa (Wu & Morris, 1973a ▶,b ▶). This tetrameric form of the enzyme depends on the presence of an Mg2+ ion. In its absence, SpeA exists in an equilibrium between monomeric and dimeric forms, in which the ratio of monomer to dimer population depends on the pH (Wu & Morris, 1973b ▶).
E. coli also contains an acid-induced arginine decarboxylase (AdiA) which plays a role in pH homeostasis (Stim & Bennett, 1993 ▶). The two ADCs share only about 10% amino-acid sequence identity and AdiA does not have the same backbone fold as SpeA (Andrell et al., 2009 ▶).
Crystallization of E. coli (Rodrigez et al., 1994 ▶), Yersinia pestis (Patel et al., 2004 ▶) and Bacillus subtilis (Liu et al., 2009 ▶) SpeA has been reported, although a crystal structure of SpeA was not available when we initiated this project. In this paper, we describe the crystal structures of the PLP-bound form of SpeA from Campylobacter jejuni at 3.0 Å resolution and of E. coli SpeA in complex with a sulfate ion at 3.1 Å resolution.
2. Materials and methods
The production of the SpeA proteins from C. jejuni and E. coli was carried out as part of the high-throughput protein-production process of the Northeast Structural Genomics Consortium (NESG; Acton et al., 2005 ▶). These SpeA proteins correspond to NESG targets BR53 and ER600. The full-length speA genes from C. jejuni (strain NCTC 11168) and E. coli (strain K-12) were cloned into a pET21d derivative (Novagen), generating plasmids pBR53-21.11 and pER600-21.16, respectively. The resulting recombinant proteins contain eight non-native residues (LEHHHHHH) at the C-terminus. E. coli BL21 (DE3) pMGK cells, a rare-codon-enhanced strain, were transformed with pBR53-21.11 and pER600-21.16. A single isolate was cultured in MJ9 minimal medium (Jansson et al., 1996 ▶) supplemented with selenomethionine, lysine, phenylalanine, threonine, isoleucine, leucine and valine for the production of selenomethionine-labeled SpeA (Doublié et al., 1996 ▶). Initial growth was carried out at 310 K until the OD600 of the culture reached 0.6–0.8. The incubation temperature was then decreased to 290 K and protein expression was induced by the addition of IPTG (isopropyl β-d-1-thiogalactopyranoside) to a final concentration of 1 mM. Following overnight incubation, the cells were harvested by centrifugation.
Selenomethionyl SpeA was purified using standard methods. Cell pellets were resuspended in lysis buffer (50 mM Tris pH 7.5, 500 mM NaCl, 40 mM imidazole, 1 mM TCEP and 0.02% NaN3) and disrupted by sonication. The resulting lysate was clarified by centrifugation at 26 000g for 45 min at 277 K. The supernatant was loaded onto an ÄKTAxpress system (GE Healthcare) with a two-step protocol consisting of IMAC (HisTrap HP) and gel-filtration (HiLoad 26/60 Superdex 75) chromatography. The purified SpeA proteins were concentrated to 8–10 mg ml−1, flash-frozen in aliquots and used for crystallization screening. The sample purity (>95%) and molecular weight were verified by SDS–PAGE and MALDI–TOF mass spectrometry, respectively. The yields of the purified SpeA proteins from C. jejuni and E. coli were approximately 22 and 64 mg l−1, respectively. The purified SpeA from C. jejuni was yellow in color, whereas that from E. coli was colorless, suggesting that the C. jejuni SpeA possibly contained the PLP cofactor.
C. jejuni SpeA was crystallized by the microbatch method at 277 K. 2 µl protein solution consisting of C. jejuni SpeA (7.9 mg ml−1), 5 mM Tris pH 7.5, 100 mM NaCl, 5 mM DTT and 0.02% NaN3 was mixed with 2 µl precipitating solution consisting of 100 mM bis-tris propane pH 7 and 1.88 M sodium sulfate. Yellow hexagonal crystals of SpeA appeared overnight and grew to full size after several days; they were subsequently cryoprotected using 25%(v/v) glycerol and flash-frozen in liquid propane for data collection at 100 K. The crystallization of E. coli SpeA was carried out using a similar method and using a similar protein buffer except that 100 mM HEPES pH 8.5 and 1.6 M ammonium sulfate were used as precipitants.
The crystals of C. jejuni SpeA and E. coli SpeA belonged to space group P3121 with unit-cell parameters a = 203.1, c = 101.8 Å and space group P41212 with unit-cell parameters a = 192.5, c = 119.9 Å, respectively. There are two monomers of SpeA in the crystallographic asymmetric unit of both enzymes. For C. jejuni SpeA, a single-wavelength anomalous diffraction (SAD) data set was collected to 3.0 Å resolution at the peak absorption wavelength of selenium on the X6A beamline of the National Synchrotron Light Source (NSLS). Diffraction images were processed with the HKL package (Otwinowski & Minor, 1997 ▶) and selenium sites were located with the program SHELX (Sheldrick, 2008 ▶). SOLVE/RESOLVE (Terwilliger, 2003 ▶) was used for phasing the reflections and for automated model building, which correctly placed 20% of the residues with side chains in each protomer of the asymmetric unit. The majority of the model building was rapidly built manually as the crystal has a large solvent content (72%). Each subunit, including the initiating (seleno)methionine, was built with the program XtalView (McRee, 1999 ▶) and refined using CNS (Brünger et al., 1998 ▶). Noncrystallographic symmetry restraints were applied in most stages of the refinement of the structure. A similar method was used for structure determination of E. coli SpeA. The process of model building was facilitated using the refined model of the C. jejuni SpeA structure. Data-processing and refinement statistics are summarized in Table 1 ▶.
Table 1. Summary of crystallographic information.
Values in parentheses are for the highest resolution shell.
| C. jejuni SpeA (PLP complex) | E. coli SpeA (sulfate complex) | |
|---|---|---|
| Maximum resolution (Å) | 3.0 | 3.1 |
| No. of observations | 460394 | 511680 |
| Rmerge (%) | 9.8 (48.8) | 17.2 (76.9) |
| No. of reflections | 93958 | 41600 |
| Resolution range used in refinement | 20–3.0 | 20–3.1 |
| Completeness (%) | 100 (100) | 100 (97) |
| R factor (%) | 18.0 (25.5) | 20.0 (37.4) |
| Free R factor† (%) | 21.9 (29.4) | 23.9 (39.2) |
| R.m.s.d. in bond lengths (Å) | 0.009 | 0.009 |
| R.m.s.d. in bond angles (°) | 1.1 | 1.2 |
| PDB code | 3nzp | 3nzq |
5% of the reflections were selected for free R calculation.
3. Results and discussion
The crystal structures of C. jejuni and E. coli SpeA were determined using the selenomethionyl single-wavelength anomalous diffraction (SAD) method (Hendrickson, 1991 ▶). The crystals generally diffracted X-rays poorly, possibly because of their high solvent content (70%), and the best diffraction data sets were collected after screening many crystals. On the other hand, the quality of the electron-density map after solvent flattening was rather good because of the high solvent content. This is also supported by the low R and free R values and the excellent agreement with expected geometric parameters for the refined models (Table 1 ▶). Residues 190–202 and 422–434 are not included in the C. jejuni SpeA structure owing to a lack of electron density. None of the residues of the protein are located in the disallowed region, while 85% of the residues are located in the most favored region of the Ramachandran plot (data not shown). The overall structures of E. coli and C. jejuni SpeA are similar to each other, with an r.m.s. distance of 1.9 Å among their structurally equivalent Cα atoms, consistent with the fact that the two enzymes share 33% amino-acid sequence identity.
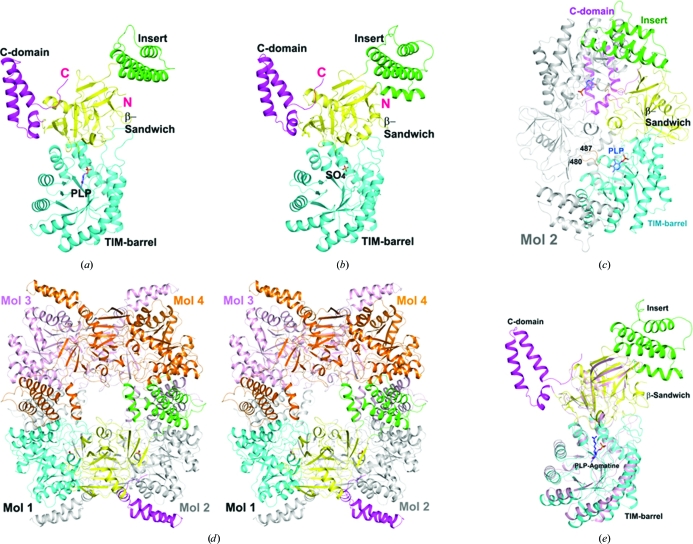
The structure of the SpeA monomer contains two large domains, an N-terminal TIM-barrel domain followed by a β-sandwich domain, as well as two smaller helical domains (Figs. 1 ▶ a and 1 ▶ b). The first of these helical domains is an insert into the β-sandwich domain, while the second is formed by residues at the C-terminus.
Figure 1.
Crystal structures of bacterial biosynthetic arginine decarboxylases (ADC, SpeA). (a) Structure of the C. jejuni SpeA monomer in complex with PLP. The four domains, TIM-barrel, β-sandwich, insert and C-terminal domain, are colored cyan, yellow, green and magenta, respectively. The PLP cofactor (light blue C atoms) is shown as a ball-and-stick model. (b) Structure of the E. coli SpeA monomer. A sulfate ion is located in the phosphate-binding site of PLP. (c) Structure of the C. jejuni SpeA dimer. The second monomer is colored gray, except for the loop that reaches into the active site of the first monomer, which is colored orange and its residue range is indicated. (d) Stereo drawing of the structure of the E. coli SpeA tetramer. (e) Superposition of the structure of the E. coli SpeA monomer (colored by domains) and ADC from P. bursaria Chlorella virus (pink) in complex with PLP–agmatine (Shah et al., 2007 ▶). All structural figures were created with PyMOL (http://www.pymol.org).
The asymmetric unit of the crystals contains a tightly associated SpeA dimer, in which the TIM-barrel domain of one subunit is surrounded on three sides by the β-sandwich, insert and C-terminal domains of the other subunit (Fig. 1 ▶ c). A total of 3000 Å2 of the surface area of each subunit is buried in this dimer interface, indicating that the dimer is likely to be stable, which is supported by our solution light-scattering studies on E. coli SpeA (data not shown). The C-terminal, β-sandwich and insert domains provide roughly 1300, 1000 and 700 Å2 surface-area burial in the dimer interface, respectively.
A tetramer of SpeA can be constructed using crystallographic symmetry in the crystals of both E. coli and C. jejuni SpeA, with generally similar quaternary organizations (Fig. 1 ▶ d). The insert domain is located in the tetramer interface, burying roughly 1800 Å2 of its surface area for E. coli SpeA. There is a large cavity in the center of both tetramers and the tetramer interface is not tightly packed, presumably owing to the absence of PLP, arginine and magnesium ions. Nonetheless, the fact that a similar tetramer was observed for both enzymes in different crystal forms suggests that this is likely to be the structure of the SpeA holoenzyme tetramer (Wu & Morris, 1973a ▶,b ▶).
The TIM-barrel and β-sandwich domains of SpeA have many structural homologs among various PLP-dependent decarboxylases based on a search of the Protein Data Bank with the DaliLite program (Holm et al., 2008 ▶). The closest structural homologs include diaminopimelate decarboxylase (DAPDC; Z score of 35.2, an r.m.s.d. of 2.9 Å for 414 aligned residues and 25% sequence identity with SpeA; Ray et al., 2002 ▶), lysine/ornithine decarboxylase (L/ODC; Z score of 31.6, an r.m.s.d. of 3.1 Å for 356 aligned residues and 19% sequence identity; Lee et al., 2007 ▶), ODC (Z score of 30.4, an r.m.s.d. of 3.0 Å for 340 aligned residues and 21% sequence identity; Jackson et al., 2004 ▶) and the Paramecium bursaria Chlorella virus ADC (Fig. 1 ▶ e; Z score of 29.7, an r.m.s.d. of 3.1 Å for 351 aligned residues and 18% sequence identity; Shah et al., 2007 ▶). The strong structural and functional homology of SpeA to these other enzymes suggests that they have evolved from a common ancestor, despite their low degree of sequence conservation. On the other hand, the insert and the C-terminal domains appear to be unique to SpeA (Fig. 1 ▶ e) and mediate the dimerization and tetramerization of SpeA.
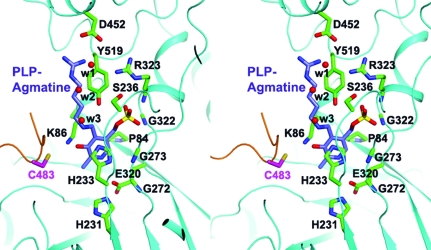
The active site of SpeA is located in the TIM-barrel domain, which is also supported by the observation of a bound PLP cofactor in our structure of the C. jejuni enzyme (Fig. 2 ▶). In addition, the other monomer of the dimer makes crucial contributions to the active site. Specifically, residues 480–487 in the β-sandwich domain of the other monomer are located in the active site (Fig. 1 ▶ c). This loop contains highly conserved residues among SpeA homologs, especially Cys483, which is equivalent to a Cys residue that has been proposed to be the catalytic residue in close structural homologs of SpeA (Shah et al., 2007 ▶; Buschiazzo et al., 2006 ▶). Therefore, monomers of SpeA are unlikely to be catalytically active.
Figure 2.
The active site of SpeA. Stereo drawing of the interactions between the PLP cofactor and C. jejuni SpeA. A model of PLP–agmatine is shown based on a structural overlay with P. bursaria Chlorella virus ADC (Shah et al., 2007 ▶). The binding mode of PLP is essentially the same in SpeA and this enzyme. Three water molecules in our structure occupying the expected binding site of the Arg substrate are shown as red spheres.
The phosphate group of PLP is held in place by five hydrogen bonds to the side chains of the conserved residues Ser236 and Tyr519 (from the β-sandwich domain) and to the backbone amides of the invariant residues Gly274, Gly322 and Arg323 (Fig. 2 ▶). The pyridoxal group of PLP forms a π-stacking interaction with the side chain of His233 on one face and is positioned against Pro84 on the other face. The N1 atom of PLP makes a hydrogen bond to the side chain of the strictly conserved Glu320. Most of these residues have equivalents in the close structural homologs of SpeA, suggesting that these enzymes share a common mechanism for recognizing the PLP cofactor. In fact, the bound position of PLP is essentially the same in SpeA and the structure of P. bursaria Chlorella virus ADC in complex with PLP–agmatine (compare Figs. 1 ▶ a and 1 ▶ e; Shah et al., 2007 ▶). Based on this structure, we can also predict the binding site for the Arg substrate, which is occupied by three water molecules in our structure (Fig. 2 ▶). The guanidinium group of Arg may interact with the side chain of Asp452.
While this manuscript was in preparation, the crystal structure of Vibrio vulnificus ADC was reported (Deng et al., 2010 ▶). The overall structure of this ADC is similar to those of C. jejuni and E. coli SpeA, including the binding mode of the PLP cofactor and the presence of the two helical domains that help to stabilize the tetramer.
Supplementary Material
PDB reference: C. jejuni SpeA, 3nzp
PDB reference: E. coli SpeA, 3nzq
Acknowledgments
We thank Angela Lauricella and George DeTitta for setting up initial crystallization screenings, Rachel Belote, Colleen Ciccosanti and Seema Sahdev for technical assistance, Randy Abramowitz and John Schwanof for setting up the X4A beamline and Jean Jakoncic for setting up the X6A beamline. This research was supported by the Protein Structure Initiative of the National Institutes of Health grants P50 GM062413 and U54 GM074958.
References
- Acton, T. B. et al. (2005). Methods Enzymol.394, 210–243. [DOI] [PubMed]
- Andrell, J., Hicks, M. G., Palmer, T., Carpenter, E. P., Iwata, S. & Maher, M. J. (2009). Biochemistry, 48, 3915–3927. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Buschiazzo, A., Goytia, M., Schaeffer, F., Degrave, W., Shepard, W., Gregoire, C., Chamond, N., Cosson, A., Berneman, A., Coatnoan, N., Alzari, P. M. & Minoprio, P. (2006). Proc. Natl Acad. Sci. USA, 103, 1705–1710. [DOI] [PMC free article] [PubMed]
- Deng, X., Lee, J., Michael, A. J., Tomchick, D. R., Goldsmith, E. J. & Phillips, M. A. (2010). J. Biol. Chem.285, 25708–25719. [DOI] [PMC free article] [PubMed]
- Doublié, S., Kapp, U., Aberg, A., Brown, K., Strub, K. & Cusack, S. (1996). FEBS Lett.384, 219–221. [DOI] [PubMed]
- Hendrickson, W. A. (1991). Science, 254, 51–58. [DOI] [PubMed]
- Holm, L., Kaariainen, S., Rosenstrom, P. & Schenkel, A. (2008). Bioinformatics, 24, 2780–2781. [DOI] [PMC free article] [PubMed]
- Jackson, L. K., Baldwin, J., Akella, R., Goldsmith, E. J. & Phillips, M. A. (2004). Biochemistry, 43, 12990–12999. [DOI] [PubMed]
- Jansson, M., Li, Y.-C., Jendeberg, L., Anderson, S., Montelione, G. T. & Nilsson, B. (1996). J. Biomol. NMR, 7, 131–141. [DOI] [PubMed]
- Lee, J., Michael, A. J., Martynowski, D., Goldsmith, E. J. & Phillips, M. A. (2007). J. Biol. Chem.282, 27115–27125. [DOI] [PubMed]
- Liu, X.-Y., Lei, J., Liu, X., Su, X.-D. & Li, L. (2009). Acta Cryst. F65, 282–284. [DOI] [PMC free article] [PubMed]
- McRee, D. E. (1999). J. Struct. Biol.125, 156–165. [DOI] [PubMed]
- Moore, R. C. & Boyle, S. M. (1990). J. Bacteriol.172, 4631–4640. [DOI] [PMC free article] [PubMed]
- Morris, S. M. Jr (2004). Curr. Opin. Clin. Nutr. Metab. Care, 7, 45–51. [DOI] [PubMed]
- Morris, S. M. Jr (2009). Br. J. Pharmacol.157, 922–930. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Patel, C. N., Adcock, R. S., Sell, K. G. & Oliveira, M. A. (2004). Acta Cryst. D60, 2396–2398. [DOI] [PubMed]
- Ray, S. S., Bonanno, J. B., Rajashankar, K. R., Pinho, M. G., He, G., de Lencastre, H., Tomasz, A. & Burley, S. K. (2002). Structure, 10, 1499–1508. [DOI] [PubMed]
- Rodrigez, B. R., Carroll, D. W., Mitchell, D., Momany, C. & Hackert, M. L. (1994). Acta Cryst. D50, 175–177. [DOI] [PubMed]
- Shah, R., Akella, R., Goldsmith, E. J. & Phillips, M. A. (2007). Biochemistry, 46, 2831–2841. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Stim, K. P. & Bennett, G. N. (1993). J. Bacteriol.175, 1221–1234. [DOI] [PMC free article] [PubMed]
- Tabor, C. W. & Tabor, H. (1985). Microbiol. Rev.49, 81–99. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. (2003). Methods Enzymol.374, 22–37. [DOI] [PubMed]
- Wallace, H. M. (2009). Essays Biochem.46, 1–9. [DOI] [PubMed]
- Wallace, H. M., Fraser, A. V. & Hughes, A. (2003). Biochem. J.376, 1–14. [DOI] [PMC free article] [PubMed]
- Wu, W. H. & Morris, D. R. (1973a). J. Biol. Chem.248, 1687–1695. [PubMed]
- Wu, W. H. & Morris, D. R. (1973b). J. Biol. Chem.248, 1696–1699. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: C. jejuni SpeA, 3nzp
PDB reference: E. coli SpeA, 3nzq