The cloning, purification and crystallization of the E. coli lipoproteins BamC, BamD and BamE is reported. X-ray diffraction data at high resolution were obtained for each of the proteins or protein domains.
Keywords: lipoproteins, Escherichia coli, BamC, BamD, BamE
Abstract
In Escherichia coli, the β-barrel assembly machinery (or BAM complex) mediates the recognition, insertion and assembly of outer membrane proteins. The complex consists of the integral membrane protein BamA (an Omp85-family member) and the lipoproteins BamB, BamC, BamD and BamE. The purification and crystallization of BamC, BamD and BamE, each lacking the N-terminal membrane anchor, is described. While the smallest protein BamE yielded crystals under conventional conditions, BamD only crystallized after stabilization with urea. Full-length BamC did not crystallize, but was cleaved by subtilisin into two domains which were subsequently crystallized independently. High-resolution data were acquired from all proteins.
1. Introduction
Gram-negative bacteria are covered by an inner membrane and an outer membrane (OM). Proteins destined for insertion into the outer membrane (OMPs) are synthesized in the cytosol with an ∼20-amino-acid presequence and are translocated into the periplasm by the Sec machinery. After entry into the periplasm, the presequence is cleaved and the OMP is delivered to the outer membrane by the periplasmic chaperones Skp, SurA and DegP (Knowles, Scott-Tucker et al., 2009 ▶; Jacob-Dubuisson et al., 2009 ▶). These chaperones prevent unfolded OMPs from early precipitation and guide them to the BAM complex located in the outer membrane. This complex is responsible for the correct recognition, insertion and assembly of OMPs into the outer membrane (Bos et al., 2007 ▶). In Escherichia coli, this complex is composed of the integral membrane protein BamA and the four lipoproteins BamB, BamC, BamD and BamE (previously Omp85/YaeT and YfgL, NlpB, YfiO and SmpA, respectively; Walther et al., 2009 ▶; Knowles, Scott-Tucker et al., 2009 ▶). BamA consists of a large ‘periplasmic polypeptide-transport associated’ (POTRA) N-terminal domain and an outer membrane-embedded C-terminal β-barrel. The POTRA domain, which can be divided into five distinct subdomains of nearly equal structure, is involved in the recognition of substrates and the binding of other components of the BAM complex. From biochemical and mutational studies, it has been shown that the fifth subdomain, which is near to the β-barrel part of BamA, recognizes unfolded OMPs. On the other hand, all subdomains except for the first subdomain appear to be essential for binding of the four lipoproteins to BamA (Kim et al., 2007 ▶). Direct interactions between BamA and BamB as well as between BamA and BamD have been shown (Malinverni et al., 2006 ▶), while BamC (Malinverni et al., 2006 ▶) and BamE (Sklar et al., 2007 ▶) may bind to BamA indirectly via a BamD link. Binding of BamC to the complex requires the C-terminus of BamD (Malinverni et al., 2006 ▶). From studies of BamB mutants, residues in BamB have been identified that are critical for the binding to BamA (Vuong et al., 2008 ▶). Based on these physiological data, a concept of the complex architecture and of the mechanism of OMP recognition and insertion has started to emerge, but a variety of structural and mechanistic details remain unknown. While the POTRA domain of BamA has been structurally characterized by X-ray crystallography (Kim et al., 2007 ▶; Gatzeva-Topalova et al., 2008 ▶) and NMR (Knowles et al., 2007 ▶, 2008 ▶), the structures of other components of the E. coli BAM complex have not yet been reported, although a chemical shift assignment of BamC on the basis of NMR has been published (Knowles, McClelland et al., 2009 ▶).
Here, we report the expression, purification and crystallization of BamC as two domains, of BamD and of BamE. In all cases we collected high-resolution data which will allow de novo determination of the individual structures.
2. Materials and methods
2.1. Cloning
The DNA sequences corresponding to the open reading frames of BamC (residues 65–320), BamD (14–226) and BamE (21–94) were amplified from genomic E. coli DNA by PCR using primers that included BamHI and EcoRI restriction sites into the PCR products. The purified PCR products were digested with BamHI and EcoRI and ligated to the respective cloning sites of the vector pET24d, resulting in constructs coding for the expression of protein with six additional His residues at the C-terminus. Two additional amino acids, Leu and Glu, derived from the cloning strategy are located between the His tag and the protein sequence. Positive clones were verified by DNA sequencing.
2.2. Expression and purification
Plasmids were transformed into BL21 (DE3) RIL expression cells (Stratagene) by electroporation. Precultures were induced by inoculating LB broth supplemented with kanamycin (Kan) and chloramphenicol (CM) (25 µg ml−1 each) with positive clones. Precultures were grown overnight at 303 K. A 40 ml volume taken from these cultures was added to 800 ml LB/Kan/CM and the suspension was vigorously aerated at 310 K. When the optical density reached 0.9, isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 1 mM. After induction, the cells were grown for an additional 4 h. The cells were harvested by centrifugation and frozen until further use. For lysis, the cells were resuspended in buffer A (20 mM HEPES, 300 mM NaCl, 10 mM imidazole pH 7.8) supplemented with DNaseI, lysozyme and phenylmethylsulfonyl fluoride (PMSF). The cells were lysed with a French press. The lysate was centrifuged at 160 000g for 1 h. The supernatant was filtered through a 0.22 µm filter and loaded onto a 10 ml nickel–nitrilotriacetic acid (Ni–NTA) column (Qiagen). After loading the sample, the column was extensively washed with buffer A. Further washing steps with imidazole concentrations of 34 and 58 mM were conducted before the His-tagged protein was eluted using buffer B (buffer A plus 250 mM imidazole).
BamE and BamC were further purified by size-exclusion chromatography (SEC) on Superdex 75 and Superdex 200 columns (GE Healthcare), respectively, using 20 mM Tris, 150 mM NaCl pH 7.8 (for BamE) or 20 mM HEPES, 150 mM NaCl, 5 mM MgCl2 pH 7.5 (for BamC). In the case of BamE two peaks were obtained, which were concentrated separately using a 10 kDa membrane, and used for crystallization. In the case of BamC only a single peak was observed. A selenomethionine-labelled derivative of BamE was expressed using the same cells as used for wild-type expression but working in M9 minimal medium supplemented with the six amino acids Leu, Ile, Phe, Ser, Thr and Val (see, for example, Hanoulle et al., 2004 ▶). Expression and purification were conducted using essentially the same protocol as used for the native proteins.
BamD started to precipitate from the elution fraction of the Ni–NTA column soon after elution had finished. The precipitate formed overnight was partially redissolved in 50 mM Tris, 1.5 M urea pH 8. This solution was concentrated using a 30 kDa membrane to an OD280 value of 63. A 1:2 dilution with Tris buffer gave a clear solution with an OD280 value of 45, which was further used in crystallization attempts.
2.3. Subtilisin treatment of BamC
The BamC protein was treated with subtilisin on ice at a mass ratio of 1:70 (subtilisin:BamC). After 1 h, the reaction was stopped by the addition of PMSF. After centrifugation, the supernatant was treated with concentrated NaCl and imidazole at final concentrations of 0.3 M and 20 mM, respectively, filtered through a 0.22 µm filter and passed through an Ni–NTA column pre-equilibrated with 10 mM HEPES, 300 mM NaCl, 5 mM MgCl2, 10 mM imidazole (buffer C). The flowthrough was collected and protein bound to the column was eluted with buffer C containing 250 mM imidazole. The flowthrough was concentrated and desalted using a 10 kDa membrane and was immediately subjected to crystallization trials. The protein eluted from the column was further purified by size-exclusion chromatography on Superdex 75 using 10 mM HEPES, 85 mM NaCl, 20 mM KCl, 3 mM MgCl2, 1 mM β-mercaptoethanol (β-ME) pH 7.5. Elution fractions were concentrated to ∼45 mg ml−1 and subjected to crystallization trials.
2.4. CD and ES mass spectrometry
Circular-dichroism (CD) spectra were measured between 205 and 240 nm on a Jasco J-810 spectropolarimeter using 1 mm cuvettes at 0.2 nm resolution and 1 nm bandwidth with a 1 s time constant and a sensitivity of 100 mdeg. BamD dissolved in 50 mM Tris, 1 M urea was diluted to 0.2 mg ml−1 (5 µM) with 10 mM HEPES pH 7.5, 15 mM NaCl buffer. Electrospray mass spectra were recorded on a SCIEX API 165 single-quadrupole mass spectrometer (Perkin–Elmer) equipped with a Nucleosil C8-HD reverse-phase column (Macherey & Nagel). Elution was performed according to standard conditions using an acetonitrile gradient.
2.5. Crystallization
Sitting-drop crystallization trials were performed at 293 K by mixing 0.4 µl protein solution with 0.4 µl precipitant solution (0.6 + 0.35 µl for BamC-NT; see below) in 96-3 Intelli-Plates (Art Robbins) using a Honeybee 961 crystallization robot (Genomic Solutions). Up to 14 different screens from Hampton Research, Qiagen and Emerald BioSystems were applied to each protein. The drops were analyzed using a Rock Imager 54 imaging system (Formulatrix, Waltham, Massachusetts, USA) and images were inspected for the formation of crystals. The crystallization conditions yielding diffracting crystals are listed in Table 1 ▶. In all cases the initial crystals obtained in the 96-well plates were used for data collection. Crystals were picked up in small nylon loops (Hampton), briefly immersed in cryoprotectant solution if necessary and immediately flash-frozen in liquid nitrogen.
Table 1. Crystallization conditions of the proteins.
| Protein | Protein solution | Reservoir solution |
|---|---|---|
| BamE (SeMet derivative) | 12.8 mg ml−1 in 10 mM HEPES pH 7.5, 100 mM NaCl, 3 mM MgCl2, 1 mM β-ME | 0.1 M HEPES pH 7.5, 10% 2-propanol, 20% PEG 4000 |
| BamD | 30 mg ml−1 in 50 mM Tris pH 8, 1 M urea | 0.1 M HEPES pH 7.5, 20% PEG 6000 |
| BamC-NT | 6.5 mg ml−1 in 10 mM HEPES pH 7.5, 100 mM NaCl, 20 mM KCl, 3 mM MgCl2, 1 mM β-ME, 5% glycerol | 0.1 M citric acid pH 3.5, 2 M (NH4)2SO4 |
| BamC-CT | 45 mg ml−1 in 20 mM HEPES pH 7.5, 50 mM NaCl, 1 mM β-ME | 0.1 M MES pH 6.5, 25% PEG 1000 |
2.6. Data collection and processing
X-ray data were collected on beamline PXII at the Swiss Light Source (SLS, Villigen, Switzerland). Diffraction experiments were conducted at 100 K and images were recorded on a 225 mm MAR CCD camera (MAR Research, Norderstedt, Germany). Data were indexed, integrated and scaled using the programs XDS and XSCALE (Kabsch, 2010 ▶).
3. Results
Genes encoding lipoproteins in E. coli typically include an N-terminal signal sequence of 19–24 amino acids in length for their export into the periplasm. The mature lipoproteins start with a cysteine residue which is chemically linked to the lipid molecule to subsequently allow targeting and to form the membrane anchor (Tokuda & Matsuyama, 2004 ▶). The lipoprotein sequences of BamC, BamD and BamE of the BAM complex were designed on the basis of secondary-structure predictions and N-terminal parts that were predicted to be disordered were omitted. The proteins BamC, BamD and BamE, which were all derived from E. coli, were subsequently cloned using a C-terminal His tag (see Fig. 1 ▶).
Figure 1.
Scheme showing the size of the mature lipoproteins and the fragments cloned on the basis of secondary-structure predictions. For BamC, the fragments obtained from subtilisin cleavage are also shown.
Not unexpectedly, given the homologous expression, all of the proteins were strongly overproduced. BamC and BamE were purified via two-step procedures using Ni–NTA affinity and size-exclusion chromatography. For BamC this purification resulted in a homogeneous protein sample. However, SEC showed BamE to be present as a mixture of two oligomers in an estimated ratio of ∼3:1. From the retention volumes, we assumed these oligomers to be dimers and tetramers (data not shown). Upon separate handling of these fractions it became obvious that the equilibrium between the higher and lower molecular-weight particles was not stable and the smaller oligomer reassembled to form the larger oligomer over time.
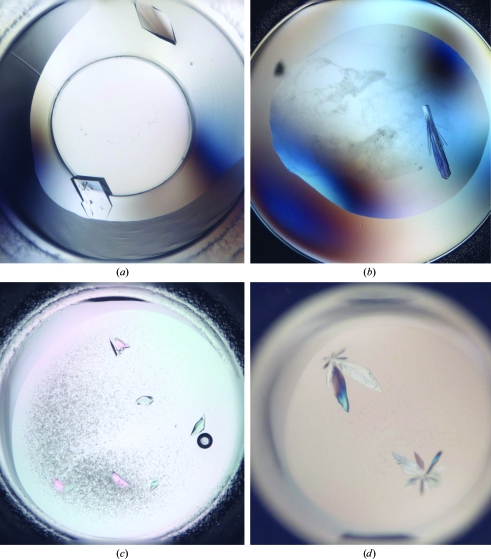
Initially, wild-type BamE yielded crystals that diffracted to a resolution of 3.5 Å. Despite many attempts to reproduce the crystallization condition and to improve the crystal quality, we could not reproduce these crystals from the protein using either the sitting-drop or hanging-drop methods. We then decided to directly reproduce the crystallization conditions using selenomethionine-labelled protein. This derivative formed crystals of 250 × 150 µm in size after 35 d (Fig. 3a) and these crystals diffracted to a resolution of 1.8 Å.
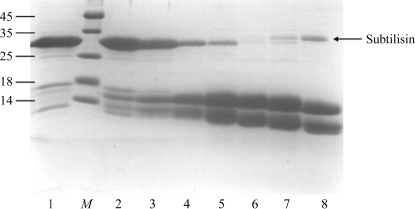
In the case of BamC we did not obtain any crystals of the full-length protein, although the domain being cloned appeared to be reasonably well folded according to secondary-structure prediction. However, we noticed slow degradation of the protein upon long-term storage of the protein solution. To systematically study this effect, we subjected the protein to degradation using the nonspecific protease subtilisin. Surprisingly, we found that subtilisin treatment resulted in the quantitative cleavage of BamC into only two stable fragments, both of which had a molecular size of around 14 kDa based on SDS gel electrophoresis (see Fig. 2 ▶). We subjected the reaction mixture to electrospray mass spectrometry, which returned fragments of between 12.2 and 14.5 kDa; the majority of these peaks could be assigned to fragments originating from the region 75–193 as well as 196–328 (data not shown), keeping the C-terminal His tag intact for separation of the two domains. In fact, Ni–NTA chromatography of the reaction mixture showed the N-terminal domain (BamC-NT) in the flowthrough and another slightly larger domain (the C-terminal domain; BamC-CT) that only eluted at a high imidazole concentration. We used the concentrated solutions in crystallization screens and observed crystals for both fragments. The crystals of BamC-NT (Fig. 3 ▶ c) diffracted to 1.6 Å resolution and the crystals of BamC-CT (Fig. 3 ▶ d) diffracted to 1.3 Å resolution (for further information, see Table 2 ▶).
Figure 2.
17% SDS–PAGE gel showing the subtilisin treatment of BamC. Lane 1, pure BamC isolated after Ni–NTA affinity chromatography. Lanes 2–8, subtilisin treatment of BamC with increasing (from left to right) subtilisin:BamC ratios under standard conditions (277 K for 1 h followed by addition of PMSF). The subtilisin:BamC ratios were 1:10 000 (lane 2), 1:3000 (lane 3), 1:1000 (lane 4), 1:300 (lane 5), 1:100 (lane 6), 1:30 (lane 7) and 1:10 (lane 8). Lane M contains molecular-weight markers (kDa).
Figure 3.
Crystals of SeMet-labelled BamE (a), BamD (b), BamC-NT (c) and BamC-CT (d).
Table 2. Crystal data and statistics of measurements.
Values in parentheses are for the outer shell.
| SeMet BamE | BamD | BamC-NT | BamC-CT | |
|---|---|---|---|---|
| Crystal dimensions (µm) | 270 × 180 | 180 × 40 | 80 × 120 | 370 × 120 |
| Space group | C2 | P21 | P1 | P21 |
| Unit-cell parameters | ||||
| a (Å) | 69.66 | 53.13 | 46.57 | 29.74 |
| b (Å) | 96.52 | 33.44 | 46.63 | 59.12 |
| c (Å) | 50.49 | 57.80 | 60.20 | 31.06 |
| α (°) | 90.00 | 90.00 | 102.70 | 90.00 |
| β (°) | 134.06 | 111.44 | 92.86 | 116.37 |
| γ (°) | 90.00 | 90.00 | 118.20 | 90.00 |
| Resolution limits (Å) | 50–1.8 (1.88–1.80) | 50–1.8 (1.90–1.80) | 50–1.55 (1.65–1.55) | 50–1.28 (1.33–1.28) |
| Wavelength (Å) | 0.9789 | 1.0 | 1.06 | 1.07 |
| Oscillation angle (°) | 1 | 1 | 1 | 1 |
| Observed reflections | 120092 | 111534 | 191281 | 126101 |
| Unique reflections | 21093 | 17881 | 56570 | 25297 |
| Completeness (%) | 100 (99.8) | 99.4 (97.8) | 95.1 (93.8) | 98.4 (95.6) |
| Multiplicity | 5.9 (5.7) | 6 (5.9) | 3.3 (3.1) | 2.5 (1.9) |
| 〈I〉/〈σ(I)〉 | 25.7 (2.8) | 20.3 (3.9) | 11.15 (2.76) | 10.3 (1.6) |
| Rmerge† | 0.07 (0.472) | 0.044 (0.513) | 0.059 (0.443) | 0.065 (0.572) |
R
merge = 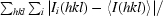
 .
.
BamD appeared to be an unstable protein with a tendency to slowly aggregate and precipitate. While searching for small molecules that might help to prevent these solubility problems, we used 1.5 M urea to stabilize the protein and followed a protocol described elsewhere (Dines et al., 2007 ▶). To ensure that the protein was still folded in the presence of urea, we performed CD spectroscopy. The spectrum indicated a strong α-helical fold of the protein (data not shown). We could subsequently concentrate the protein to ∼50 mg ml−1 without any precipitation, while in the absence of urea the protein started precipitating at a concentration of ∼4 mg ml−1. Using this highly concentrated solution we obtained crystals (Fig. 3 ▶ b) that diffracted to 1.8 Å resolution.
4. Discussion
The precise mechanism of outer membrane protein recognition and insertion, the role of the lipoproteins and their mutual interactions have still to be elucidated. In an effort to shed more light on these points, we set out to determine the three-dimensional structures of these isolated lipoproteins. As a first step, we describe the purification and successful crystallization of the lipoproteins BamC, BamD and BamE. The crystallization of BamE was straightforward using SeMet-labelled protein and a data set could be collected for the SeMet-labelled protein. BamC initially did not crystallize, but could be cleaved into an N-terminal domain and a C-terminal domain that could be separated from each other and crystallized independently. Purified BamD readily started to precipitate from solution and could not be concentrated, but the precipitate could be dissolved in a buffer containing urea, which increased the protein solubility and resulted in a highly concentrated solution from which BamD could finally be crystallized. The addition of urea to a solution of sparingly soluble proteins seems to be an interesting method of increasing their solubility and stability (Dines et al., 2007 ▶).
High-resolution X-ray data could be collected from all four lipoproteins. While determination of the BamE structure is currently under way, heavy-atom-soaked crystals or crystals of SeMet derivatives will be needed in order to be able to solve the other structures. These experiments are currently under investigation. In combination with additional methods such as small-angle X-ray scattering, we plan to model the entire complex using the individual domain structures of the lipoproteins.
Acknowledgments
We thank Dr Reinhold Horlacher (Trenzyme GmbH, Konstanz) for cloning support and the staff of the PXII beamline for excellent support. We also would like to thank Elisabeth Weyher-Stingl (Max Planck Institute for Biochemistry, Martinsried) for recording the mass spectra. This project was supported by institutional funds of the Max Planck Society.
References
- Bos, M. P., Robert, V. & Tommassen, J. (2007). Annu. Rev. Microbiol.61, 191–214. [DOI] [PubMed]
- Dines, M., Sendersky, E., Schwarz, R. & Adir, N. (2007). J. Struct. Biol.158, 116–121. [DOI] [PubMed]
- Gatzeva-Topalova, P. Z., Walton, T. A. & Sousa, M. C. (2008). Structure, 16, 1873–1881. [DOI] [PMC free article] [PubMed]
- Hanoulle, X., Rollet, E., Clantin, B., Landrieu, I., Ödberg-Ferragut, C., Lippens, G., Bohin, J.-P. & Villeret, V. (2004). J. Mol. Biol.34, 195–205. [DOI] [PubMed]
- Jacob-Dubuisson, F., Villeret, V., Clantin, B., Delattre, A. S. & Saint, N. (2009). Biol. Chem.390, 675–684. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kim, S., Malinverni, J. C., Sliz, P., Silhavy, T. J., Harrison, S. C. & Kahne, D. (2007). Science, 317, 961–964. [DOI] [PubMed]
- Knowles, T. J., Bobat, S., Jeeves, M., Henderson, I. R. & Overduin, M. (2007). Biomol. NMR Assign.1, 113–115. [DOI] [PubMed]
- Knowles, T. J., Jeeves, M., Bobat, S., Dancea, F., McClelland, D., Palmer, T., Overduin, M. & Henderson, I. R. (2008). Mol. Microbiol.68, 1216–1227. [DOI] [PubMed]
- Knowles, T. J., McClelland, D. M., Rajesh, S., Henderson, I. R. & Overduin, M. (2009). Biomol. NMR Assign.3, 203–206. [DOI] [PubMed]
- Knowles, T. J., Scott-Tucker, A., Overduin, M. & Henderson, I. R. (2009). Nature Rev. Microbiol.7, 206–214. [DOI] [PubMed]
- Malinverni, J. C., Werner, J., Kim, S., Sklar, J. G., Kahne, D., Misra, R. & Silhavy, T. J. (2006). Mol. Microbiol.61, 151–164. [DOI] [PubMed]
- Sklar, J. G., Wu, T., Gronenberg, L. S., Malinverni, J. C., Kahne, D. & Silhavy, T. J. (2007). Proc. Natl Acad. Sci. USA, 104, 6400–6405. [DOI] [PMC free article] [PubMed]
- Tokuda, H. & Matsuyama, S. (2004). Biochim. Biophys. Acta, 1694, IN1–IN9. [PubMed]
- Vuong, P., Bennion, D., Mantei, J., Frost, D. & Misra, R. (2008). J. Bacteriol.190, 1507–1517. [DOI] [PMC free article] [PubMed]
- Walther, D. M., Rapaport, D. & Tommassen, J. (2009). Cell. Mol. Life Sci.66, 2789–2804. [DOI] [PMC free article] [PubMed]