Recombinant thiol peroxidase from Y. pseudotuberculosis has been purified and crystallized in three crystal forms.
Keywords: Yersinia pseudotuberculosis, thiol peroxidases, Tpx, peroxiredoxins
Abstract
Thiol peroxidase is an atypical 2-Cys peroxiredoxin that reduces alkyl hydroperoxides. Wild-type and C61S mutant protein have been recombinantly expressed in Escherichia coli and purified using nickel-affinity chromatography. Initial crystallization trials yielded three crystal forms in three different space groups (P21, P64 and P212121) both in the presence and the absence of DTT.
1. Introduction
Yersinia pseudotuberculosis is a Gram-negative bacterium that is capable of causing a tuberculosis-like disease in humans and animals, as well as being a model organism for Y. pestis (Robins-Browne & Hartland, 2003 ▶). Thiol peroxidase (Tpx; p20; UniProt accession No. Q66A71) is an atypical 2-Cys peroxiredoxin that uses the redox potential from thioredoxin reductase and thioredoxin I to reduce alkyl hydroperoxides (Baker & Poole, 2003 ▶). Tpx was initially suggested to be localized to the periplasm and to be involved in the removal of lipid hydroxyperoxides produced by oxidative stress (Cha et al., 1995 ▶). However, recent studies of fractionated cells show that Tpx is cytoplasmic and is released into the periplasm as a response to stress (Tao, 2008 ▶).
Tpx contains three cysteines, two of which (Cys61 and Cys95, with Cys95 being the resolving cysteine) form the redox-active pair (Baker & Poole, 2003 ▶). Despite the presence of a third cysteine, there is no covalent dimerization in the oxidized state as is observed for most 2-Cys peroxiredoxins (Baker & Poole, 2003 ▶). The structure of Tpx from Escherichia coli has been elucidated in the oxidized form and the mutated C61S form (Hall et al., 2009 ▶; Choi et al., 2003 ▶) and shows a sequence identity of approximately 80% to Y. pseudotuberculosis Tpx (ypTpx). Here, we describe the crystallization of ypTpx in three different crystal forms grown under different conditions, as well as the crystallization of the catalytically inactive (Baker & Poole, 2003 ▶) mutant ypTpxC61S.
2. Material and methods
2.1. Cloning
The whole gene encoding ypTpx was amplified from Y. pseudotuberculosis YPIII pIB102 genomic DNA using proofreading DNA polymerase (Accuprime Pfx Supermix, Invitrogen) in conjunction with the primer pair ypTpx5 and ypTpx3 (CACCATGACACAGACCGTACATTTC and TTATTTCAGTGCAGCCAGCG, respectively). The amplified product was cloned into the TOPO pET-151 (Invitrogen) expression vector, thereby fusing a hexahistidine tag and TEV cleavage site to the N-terminus of ypTpx.
The mutant ypTpxC61S was obtained by site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit, Stratagene) using the primer pair mut5 and mut3 (GATACCGGCGTTTCGGCCGCCTCGGTACG and CGTACCGAGGCGGCCGAAACGCCGGTATC, respectively). The DNA sequences of the resultant expression vectors (pDW121 for the wild type and pDW121-C61S for the mutant) were confirmed using DNA sequencing.
2.2. Expression and purification of ypTpx
pDW121 or pDW121-C61S was transformed into E. coli BL21 (λDE3) cells and grown in 400 ml LB medium containing ampicillin (100 mg l−1). The cells were grown at 310 K until an optical density (OD600) of approximately 0.6 was reached, upon which expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM and the culture was left to grow overnight at 298 K. The cells were harvested and stored at 253 K until required. The cells were suspended in 20 mM Tris pH 7.5, 500 mM NaCl (buffer A) with 10 mg DNAse (Sigma) and lysed in a French press. The lysate was cleared by low-speed centrifugation at 8000g and loaded onto a 5 ml Ni2+ HisTrap column (GE Healthcare) pre-equilibrated in buffer A, washed with five column volumes (CV) of buffer A containing 20 mM imidazole followed by 3 CV buffer A containing 100 mM imidazole and finally eluted with 2 CV buffer A containing 300 mM imidazole. The ypTpx-containing fractions were identified on an SDS–PAGE gel and pooled. The purity based on the SDS–PAGE was deemed to be greater than 95%.
2.3. Crystallization
Purified protein was exchanged into 20 mM Tris pH 7.5, 50 mM NaCl (buffer B) with or without dithiothreitol (DTT) using a PD-10 desalting column (GE Healthcare) before being concentrated to approximately 8 mg ml−1 based on the absorbance at 280 nm (using an absorption coefficient of 0.261 M −1 cm−1). Initial crystallization screens were set up using sitting-drop vapour diffusion, with drops consisting of 500 nl protein solution and 500 nl reservoir solution. All trays were kept at 293 K. Three crystal forms were observed of wild-type ypTpx (crystal forms 1, 2 and 3) and one of ypTpxC61S (crystal form M).
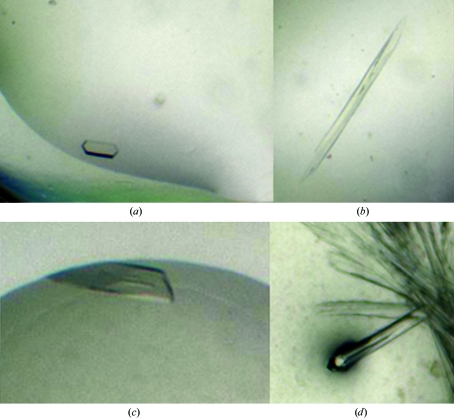
Crystal form 1 appeared in condition No. 46 of the PEG/Ion screen [Hampton Research; 20% polyethylene glycol (PEG) 3350, 0.2 M sodium citrate tribasic]. This hexagonal rod grew to dimensions of approximately 150 × 50 × 20 µm within 5 d (Fig. 1 ▶ a). The protein was kept in buffer B with the addition of 2 mM DTT.
Figure 1.
Crystals of Y. pseudotuberculosis Tpx obtained using different crystallization conditions. (a) Crystal form 1 obtained using 20% polyethylene glycol (PEG) 3350, 0.2 M sodium citrate tribasic, (b) crystal form 2 obtained using 0.1 M sodium malonate pH 5, 12% PEG 3350, (c) crystal form 3 obtained using 0.2 M tripotassium citrate, 20% PEG 3350 and (d) crystals of the mutant protein in crystal form 2 obtained using 1% tryptone, 0.05 M Na HEPES pH 7.0, 12% PEG 3350.
Crystal form 2 appeared in condition No. 3 of the PEG/Ion 2 screen (Hampton Research; 0.1 M sodium malonate pH 5, 12% PEG 3350). The protein sample was kept in buffer B with 2 mM DTT. A single crystal, which appeared within 24 h, with dimensions of 900 × 70 × 50 µm (Fig. 1 ▶ b) was broken into smaller pieces for data collection.
Crystal form 3 appeared in condition No. 24 of the JCSG+ Screen (Molecular Dimensions; 0.2 M tripotassium citrate, 20% PEG 3350). The protein was kept in buffer B with no additives in the hope of obtaining crystals of the oxidized protein sample. The crystals measured 300 × 80 × 20 µm and appeared within 7 d (Fig. 1 ▶ c).
Crystals of ypTpxC61S appeared in condition No. 47 of the PEG/Ion 2 screen (Hampton Research; 12% PEG 3350, 0.05 M Na HEPES pH 7.0, 1% tryptone) and belonged to crystal form 2. The protein sample was kept in buffer B with no additives. Crystals appeared within 24 h with dimensions of 2000 × 70 × 60 µm and a small fragment was broken off for data collection (Fig. 1 ▶ d).
2.4. Data collection and processing
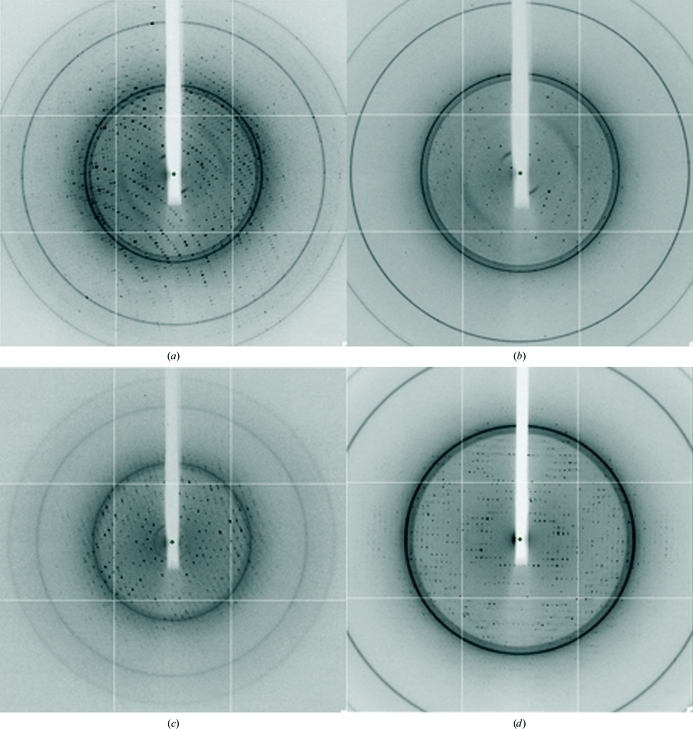
Crystals were plunged into a stream of cooled nitrogen gas (110 K; Oxford Cryosystems) with no further cryoprotection. Crystal forms 1–3 and M were transported to the Diamond Light Source for diffraction studies, in which data were collected on an ADSC Q315 CCD detector. Data were collected with 1° oscillations for a total of between 150 and 360 images at a wavelength of 0.9763 Å (Fig. 2 ▶).
Figure 2.
X-ray diffraction of the three crystal forms. (a) Crystal form 1, (b) crystal form 2, (c) crystal form 3 and (d) crystals of ypTpxC61S.
Data were processed using MOSFLM (Leslie, 1992 ▶) and scaled and merged using SCALA (Evans, 1993 ▶) from the CCP4 suite of programs (Collaborative Computational Project, Number 4, 1994 ▶) or d*TREK (Pflugrath, 1999 ▶). The space groups were confirmed using POINTLESS (Evans, 2006 ▶).
3. Results
The crystals were found to grow in three different space groups. Crystal form 1 crystallized in space group P21 (unit-cell parameters a = 64.86, b = 92.07, c = 85.60 Å, β = 91.41°), crystal form 2 in space group P62 or P64 (unit-cell parameters a = 65.01, c = 86.28 Å) and crystal form 3 in space group P212121 (unit-cell parameters a = 56.37, b = 63.44, c = 91.39 Å). The data from the best diffracting crystal, crystal form 1, were scaled to 2.00 Å. The data sets from crystal forms 2 and 3 were scaled to 2.35 and 2.5 Å, respectively. The data from the mutant crystal were scaled to 2.55 Å. All relevant diffraction statistics are presented in Table 1 ▶.
Table 1. Data-collection and reduction statistics.
Values in parentheses are for the highest resolution bin.
| Crystal form 1 | Crystal form 2 | Crystal form 3 | ypTpxC61S | |
|---|---|---|---|---|
| Space group | P21 | P64 | P212121 | P64 |
| Unit-cell parameters (Å, °) | a = 64.86, b = 92.07, c = 85.60, β = 91.41 | a = 65.01, c = 86.28 | a = 56.37, b = 63.44, c = 91.39 | a = 65.03, c = 85.60 |
| Resolution (Å) | 44.67–2.00 (2.11–2.00) | 47.15–2.35 (2.48–2.35) | 42.14–2.50 (2.59–2.50) | 56.32–2.55 (2.69–2.55) |
| Observed reflections | 182651 | 62950 | 160225 | 59770 |
| Unique reflections | 64433 | 8685 | 11830 | 6124 |
| Multiplicity | 2.8 (2.8) | 7.2 (7.4) | 13.54 (14.25) | 9.8 (9.6) |
| Completeness (%) | 94.9 (91.5) | 99.9 (100) | 99.9 (100) | 90.5 (72.6) |
| Matthews coefficient (Å3 Da−1) | 2.09 | 2.58 | 2.01 | 2.56 |
| Solvent content (%) | 41 | 52 | 38 | 52 |
| Monomers in asymmetric unit | 6 | 1 | 2 | 1 |
| Rmeas (%) | 15.4 (79.1) | 7.7 (78.9) | 13.6 (46.6) | 10.6 (60.6) |
| Rp.i.m. (%) | 8.8 (45.7) | 3.0 (28.7) | — | 3.5 (18.9) |
| 〈I/σ(I)〉 | 5.3 (1.7) | 14.6 (2.2) | 8.6 (4.0) | 11.6 (3.1) |
| Wilson B factor (Å2) | 20.5 | 54.8 | 45.4 | 55.8 |
The molecular weight of ypTpx is 17 592 Da, comprising 167 residues; as the crystallized construct also included the tag and the TEV cleavage site (MHHHHHHGKPIPNPLLGLDSTENLYFQGIDPFT), its full weight and length are 20 375 Da and 200 residues, respectively. Calculated Matthews coefficients (Matthews, 1968 ▶) for the data sets (Table 1 ▶) suggest that there are six monomers in the asymmetric unit for crystal form 1, one monomer for crystal form 2 and two monomers for crystal form 3, which are in agreement with the molecular-replacement results (see below). As Tpx from E. coli has been shown to be a dimer in both solution and crystal structure (Baker & Poole, 2003 ▶), it is likely that ypTpx crystallizes with dimers or multiples of dimers in the various crystal forms. The substrate-binding site of the oxidized form of Tpx is made up by both parts of the dimer (Choi et al., 2003 ▶).
We have crystallized ypTpx, an atypical 2-Cys peroxiredoxin from Y. pseudotuberculosis, and its mutant ypTpxC61S in three different space groups; all of the crystal froms diffracted to better than 2.5 Å resolution. As the crystals were obtained with and without a reducing agent present, it is hoped that the protein will have crystallized in both the oxidized and the reduced state. Initial solutions have been found by positioning the known structure of E. coli Tpx [PDB codes 1qxh (Choi et al., 2003 ▶) and 3hvv (Hall et al., 2009 ▶)] by molecular replacement (Phaser; McCoy et al., 2007 ▶). The solutions had final Z scores of 32.4, 27.3, 21.4 and 26.6 for crystal forms 1, 2, 3 and M, respectively. Phaser also identified P64 as the correct space group for crystal form 2. Work is now being carried out to refine the crystal structures.
Acknowledgments
This work was carried out with the support of the Diamond Light Source. The work was supported by a grant from the Biotechnology and Biological Sciences Research Council to AJR and MG (BB/G011389/1) and a Medical Research Scotland grant (reference 223 ORG G 0709) to AJR and DW. KSHB is supported by a Wellcome Trust studentship.
References
- Baker, L. M. & Poole, L. B. (2003). J. Biol. Chem.278, 9203–9211. [DOI] [PMC free article] [PubMed]
- Cha, M.-K., Kim, H.-K. & Kim, I.-H. (1995). J. Biol. Chem.270, 28635–28641. [DOI] [PubMed]
- Choi, J., Choi, S., Cha, M.-K., Kim, I.-H. & Shin, W. (2003). J. Biol. Chem.278, 49478–49486. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Evans, P. R. (1993). Proceedings of the CCP4 Study Weekend. Data Collection and Processing, edited by L. Sawyer, N. Isaacs & S. Bailey, pp. 114–122. Warrington: Daresbury Laboratory.
- Hall, A., Sankaran, B., Poole, L. B. & Karplus, P. A. (2009). J. Mol. Biol.393, 867–881. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed]
- Robins-Browne, R. & Hartland, E. (2003). International Handbook of Foodborne Pathogens, edited by M. D. Miliotis & J. W. Bier, pp. 323–356. New York: Marcel Dekker.
- Tao, K. (2008). FEMS Microbiol. Lett.289, 41–45. [DOI] [PubMed]