Zebrafish Drgal1-L2 was expressed in E. coli, purified and crystallized in the presence of lactose. X-ray diffraction data have been collected to a resolution of 1.5 Å.
Keywords: Drgal1-L2, zebrafish, galectin
Abstract
Zebrafish (Danio rerio) are an important developmental and embryological model given the optical clarity of the embryos and larvae, which permits real-time viewing of developing pathologies. More recently, a broader scope for these vertebrates to model a range of human diseases, including some cancers, has been indicated. Zebrafish Drgal1-L2 has been identified as an orthologue of mammalian galectin-1, which is is a carbohydrate-binding protein that exhibits β-galactoside-binding specificity and which is overexpressed by many aggressive human cancers. This study describes the cloning, expression in Escherichia coli, purification and crystallization of recombinant Drgal1-L2 protein in the presence of lactose (ligand). X-ray diffraction data from these novel crystals of zebrafish Drgal1-L2 were collected to a resolution of 1.5 Å using a synchrotron-radiation source, enabling their characterization.
1. Introduction
Owing to their highly conserved molecular pathways compared with those of humans, lower organisms such as fruit flies (Drosophila melanogaster), nematodes (Caenorhabditis elegans) and zebrafish (Danio rerio) are often used as alternatives for modelling human development and disease (Ma, 2004 ▶). Being a vertebrate species, the zebrafish has advantages as it is more closely related to humans, having most of the same organs and also homologues of the majority of human genes (Shin & Fishman, 2002 ▶). A number of zebrafish proteins have demonstrated similar functions to their human homologues, which validates the use of zebrafish to model human development and disease (Lassen et al., 2005 ▶; Chang et al., 2006 ▶; Zhao et al., 2010 ▶). Zebrafish are a particularly useful animal model for human embryogenesis and developmental studies because fertilization is external and the embryos are transparent and grow rapidly in vitro, allowing rapid phenotypic assessment of genetic mutations in the developing embryo (Ahmed et al., 2004 ▶). These properties circumvent some of the difficulties associated with the investigation of human embryogenesis and developmental processes using mammalian models, such as the surgical challenges and invasive monitoring techniques associated with ex utero analysis of developing mammalian embryos (Ahmed et al., 2004 ▶; Ma, 2004 ▶). Furthermore, zebrafish mutants that mimic human disease are used to assess the toxicity and efficacy of small-molecule drugs designed to combat disease in high-throughput in vivo drug screens. The relatively small size of zebrafish and the simple diffusion of drugs into developing embryos from fish water enable such screens to be performed with relative ease and cost-effectiveness (Lieschke & Currie, 2007 ▶). The zebrafish disease model has been extended in recent years to include xenograft tumour zebrafish models and transgenic zebrafish models which overexpress cancer-associated genes (Lieschke & Currie, 2007 ▶; Feitsma & Cuppen, 2008 ▶; Flores et al., 2010 ▶). It has been found that zebrafish are responsive to carcinogens and develop histologically similar neoplasms to those in human cancers (reviewed in Stern & Zon, 2003 ▶).
Galectins are a family of eukaryotic carbohydrate-binding proteins (lectins) that recognize β-galactoside glycoconjugates and have been identified in phylogenetically distinct species ranging from nematodes to mammals (Barondes et al., 1994 ▶). Recently, galectin orthologues have been identified in zebrafish. Specifically, three zebrafish galectin prototype proteins (Drgal1-L1, Drgal1-L2 and Drgal1-L3), a chimera type (Drgal3) and a tandem-repeat galectin (Drgal9-L1) have been characterized (Ahmed et al., 2004 ▶). Here, we investigate zebrafish Drgal1-L2 which, akin to human galectin-1 (galectin-1 is the prototype galectin), is expressed in the adult (brain, spleen, liver, intestine and muscle of zebrafish) and exhibits a binding specificity preference for N-acetyllactosamine and thiodigalactoside over lactose. In addition, the Drgal1-L2 gene contains four exons and the exon–intron boundaries are highly conserved between Drgal1-L2 and mammalian galectin-1 genes (Ahmed et al., 2004 ▶).
Human galectin-1 is expressed within different tissues during human embryogenesis (Camby et al., 2006 ▶) and is overexpressed by most cancers (Horiguchi et al., 2003 ▶; Hsieh et al., 2008 ▶; Scott & Weinberg, 2002 ▶; van der Brule et al., 2004 ▶). Recent studies have also shown that galectin-1 plays a role in skeletal muscle development and regeneration (Georgiadis et al., 2007 ▶). Interestingly, knockdown experiments demonstrated the role of Drgal1-L2 in development, with its absence giving a phenotype with a bent tail and disorganized muscle fibres (Ahmed et al., 2009 ▶). Within the developing zebrafish embryo, Drgal1-L2 is strongly expressed in the notochord (Ahmed et al., 2004 ▶). As the notochord is used as the primary source of signalling molecules and mechanical function essential for proper patterning of adjacent tissues, such as somites and heart, this suggests that galectin-like proteins produced by the notochord play a key role in somatic cell differentiation and development. Furthermore, the notochord plays a very important role in the specification and differentiation of skeletal muscles. The knockdown of Drgal1-L2 expression appears to indirectly cause defects in zebrafish skeletal muscle development by influencing notochord formation (Ahmed et al., 2009 ▶).
The bovine, murine and human galectin-1 proteins share 37–40% amino-acid sequence identity to Drgal1-L2, and Drgal1-L2 contains the same eight residues (His44, Asn46, Arg48, Val59, Asn61, Trp68, Glu71, Arg73; human galectin-1 residue numbering) that interact with the ligand at the carbohydrate-binding sites of toad galectin-1 (PDB code 1gan; Bianchet et al., 2000 ▶), bovine galectin-1 (PDB code 1slt; Liao et al., 1994 ▶) and human galectin-1 (PDB code 1gzw; López-Lucendo et al., 2004 ▶) as shown by structural analysis (Camby et al., 2006 ▶; Ahmed et al., 2009 ▶). The strict conservation of such crucial residues suggests that zebrafish Drgal1-L2 could be an alternative target to mammalian galectin-1 for in vivo investigation of knockdown effects and/or the physiological and pathological effects of human galectin-1-specific inhibitors using zebrafish. Within a reducing environment, human galectin-1 exists as a noncovalently bound homodimer comprising two 14.5 kDa subunits, with one carbohydrate binding site per subunit located at each end of the homodimer (López-Lucendo et al., 2004 ▶). When oxidized, galectin-1 shows extracellular lectin-independent biological activity and its protein structure undergoes significant rearrangement, enabling the formation of intramolecular disulfide bonds, the exact atomic details of which are yet to be elucidated. In order to assess the structural similarities and differences between Drgal1-L2 and reduced human galectin-1, in particular in regard to the carbohydrate-binding site and adjacent regions, we are pursuing the elucidation of the X-ray crystallographic structure of Drgal1-L2. Here, we present the first report of the cloning and expression in Escherichia coli and the purification and crystallization of recombinant Drgal1-L2 protein in the presence of lactose and under reducing conditions. X-ray diffraction data were obtained that have enabled the assignment of unit-cell parameters, crystal system and space group for these novel crystals of zebrafish Drgal1-L2.
2. Experimental procedures and results
2.1. Construction of the Drgal1-L2 expression vector
The zebrafish galectin-1-like (Drgal1-L2) gene sequence was amplified by PCR using zebrafish cDNA (prepared from RNA extracted from embryos 22–24 h post-fertilization) as template and primers 5′-CATATGGCCGGTGTGCTTATACAG-3′ and 5′-GCTCAGCCTATTTAATTTCAACCCC-3′ derived from the published sequence (Ahmed et al., 2004 ▶). The PCR product was blunt-end ligated into the pCR-Blunt vector (Invitrogen) to generate the pCR-Blunt-Drgal1-L2 plasmid and the sequence was verified. The cloned Drgal1-L2 gene varies at some nucleotide positions when compared with the two zebrafish sequences available in GenBank (AY421704.1 and BC071424.1). Our Drgal1-L2 clone contains cytosine-to-thymine nucleotide changes at positions 66 and 350 of the BC071424.1 coding sequence. These are silent mutations and the amino-acid residue sequence of our Drgal1-L2 clone is identical to that derived from the BC071424.1 gene sequence (Protein ID AAH71424.1 in GenPept). The Drgal1-L2 gene was subcloned into the pET-3a vector (Novagen) using NdeI and BlpI restriction enzymes to produce the expression plasmid pET-3a-Drgal1-L2.
2.2. Expression and purification of recombinant Drgal1-L2
pET-3a-Drgal1-L2 was propagated in BL21 (DE3) E. coli. In the first instance, cultures of recombinant Drgal1-L2 BL21 (DE3) E. coli were expressed following a previously described protocol (Scott et al., 2007 ▶). Optimization by performing expression at 310 K and with an induction period of 5 h using IPTG (final concentration of 1 mM) significantly increased protein expression from approximately 2 to 4 mg Drgal1-L2 per litre of culture. Drgal1-L2 exhibits specificity for β-galactosides, as does the noncovalently bound homodimeric form of mammalian galectin-1 (Bourne et al., 1994 ▶; López-Lucendo et al., 2004 ▶). Consequently, Drgal1-L2 protein was purified from the lysate by affinity chromatography using lactosyl Sepharose resin following the protocol within our previous report on the purification of human galectin-1 (Scott et al., 2007 ▶). The eluant fraction was then extensively dialysed against phosphate-buffered saline supplemented with 4 mM β-mercaptoethanol and 5 mM lactose (‘crystallization buffer’) over 1–2 d using dialysis tubing (3.5 kDa molecular-weight cutoff).
2.3. Assessment of the oligomeric state of Drgal1-L2 and protein-sample purity
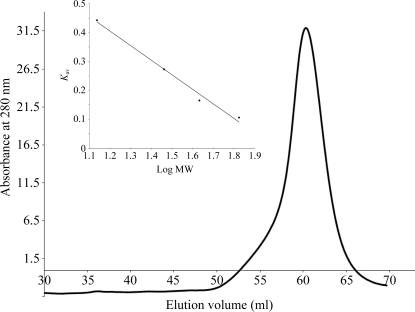
The calculated molecular weight of Drgal1-L2 (134 amino acids) is 15.26 kDa and that of the expressed protein (from which the N-terminal methionine would be expected to have been removed by E. coli methionine aminopeptidase during expression) is 15.13 kDa. To estimate the molecular weight of the purified Drgal1-L2 protein, a calibrated size-exclusion chromatography (SEC) column was used. Specifically, a HiPrep 16/60 Sephacryl S-100 High Resolution column (GE Healthcare) was calibrated with 2.5 ml samples of blue dextran (2000 kDa) and standard proteins [bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa) and ribonuclease A (13.7 kDa)] eluted at 0.3 ml min−1. K av was plotted versus log(molecular weight of standards) to obtain a standard curve (Fig. 1 ▶, inset). Following calibration, 2.5 ml Drgal1-L2 protein was loaded onto the HiPrep 16/60 Sephacryl S-100 High Resolution column equilibrated with crystallization buffer and eluted at 0.3 ml min−1. The molecular weight of recombinant Drgal1-L2 in crystallization buffer was estimated by SEC to be 33 kDa (Fig. 1 ▶) and is consistent with that of a homodimeric protein structure.
Figure 1.
Elution profile of Drgal1-L2 from a HiPrep 16/60 Sephacryl S-100 High Resolution column (GE Healthcare). Inset: the standard curve was obtained using the proteins bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa) and ribonuclease A (13.7 kDa) and estimated the molecular weight of Drgal1-L2 to be 33 kDa (the molecular weight is consistent with that of a dimeric protein structure). K av = (V e − V o)/(V t − V o), where V e is the elution volume of the protein, V o is the void volume and V t is the bed volume.
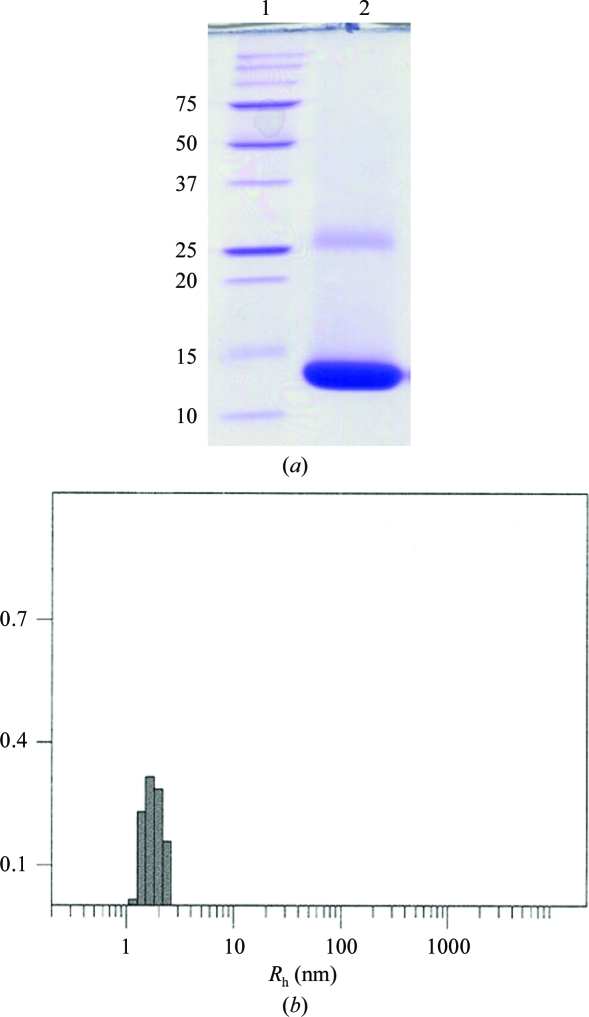
SEC analysis revealed a single protein peak in the elution profile, which indicates a highly pure protein sample of dimeric Drgal1-L2. The high biochemical purity and structural homogeneity was corroborated by subsequent SDS–PAGE (15% acrylamide) and dynamic light-scattering (DLS) analyses. Drgal1-L2 protein was heated at 368 K for 10 min in reducing loading buffer before being loaded onto an SDS–PAGE gel. Previously, we have shown that the noncovalently bound subunits of human galectin-1 dissociate under SDS–PAGE analysis, yielding a band (∼14.5 kDa) correlating with the subunit molecular weight (Scott et al., 2007 ▶). SDS–PAGE analysis of the Drgal1-L2 protein gave a band (Fig. 2 ▶ a, lane 2) that constitutes >95% of the protein sample which is notably smaller than the predicted subunit mass of the expressed protein of 15.13 kDa. The low apparent molecular weight appears to a consequence of the inherent nature of the Drgal1-L2 amino-acid sequence since both the SEC molecular-weight estimation and the elucidated crystal structure revealed that the protein was not truncated. The band larger than 25 kDa in size is representative of homodimeric remnants (Fig. 2 ▶ a, lane 2). The dimeric association is quite strong and depending on protein concentration dimers can persist despite SDS–PAGE sample preparation and migration through the acrylamide gel. Prior to dynamic light-scattering (DLS) analysis, samples of the homodimeric Drgal1-L2 protein in crystallization buffer were concentrated to either 10 or 20 mg ml−1 using an Amicon Ultracel 3k concentrator [Millipore; Bradford reagent (Sigma) was used to estimate concentration]. The hydrodynamic radius (R h) of Drgal1-L2 in crystallization buffer was measured by DLS using a CoolBatch 90T DLS instrument (Precision Detectors). Just prior to DLS analysis, 30 µl Drgal1-L2 protein (2 mg ml−1) was centrifuged at 13 000 rev min−1 for 10 min to remove large insoluble aggregates and dust. PrecisionDeconvolve32TM software parameters were set as follows: intensity 106; temperature 293 K; viscosity 0.01002 and refraction index 1.333 (based on water at 293 K); run time 1 s; accumulate 60 and smoothness 10 before each experiment. Sample-time parameters were adjusted as required to optimize DLS experiments. A reproducible protein peak was observed across 20 experimental repeats (Fig. 2 ▶ b). The average R h of this peak is 1.83 nm with an average polydispersity (peak spread %) of 23.8%. The R h value is consistent with that which we elucidated for dimeric human galectin-1 (1.88 nm; Scott et al., 2007 ▶) and the low polydispersity (<30%) is indicative of a homogeneous Drgal1-L2 protein sample.
Figure 2.
(a) SDS–PAGE (15% acrylamide) analysis of Drgal1-L2 after affinity chromatography. Lane 1, molecular-weight ladder (labelled in kDa); lane 2, the dissociated subunits of Drgal1-L2 run to an apparent molecular weight of approximately 14.5 kDa. The gel was stained with Coomassie Blue. (b) Dynamic light-scattering analysis of Drgal1-L2 (2 mg ml−1) undertaken in the crystallization buffer and thus under reducing conditions. Depicted is the output from one experimental repeat. The protein peak is consistently produced over 20 experimental repeats. The average of 20 repeats reveals a homogeneous protein sample of Drgal1-L2 that has a hydrodynamic radius R h of 1.83 nm indicative of a homodimeric structure.
2.4. Crystallization of Drgal1-L2
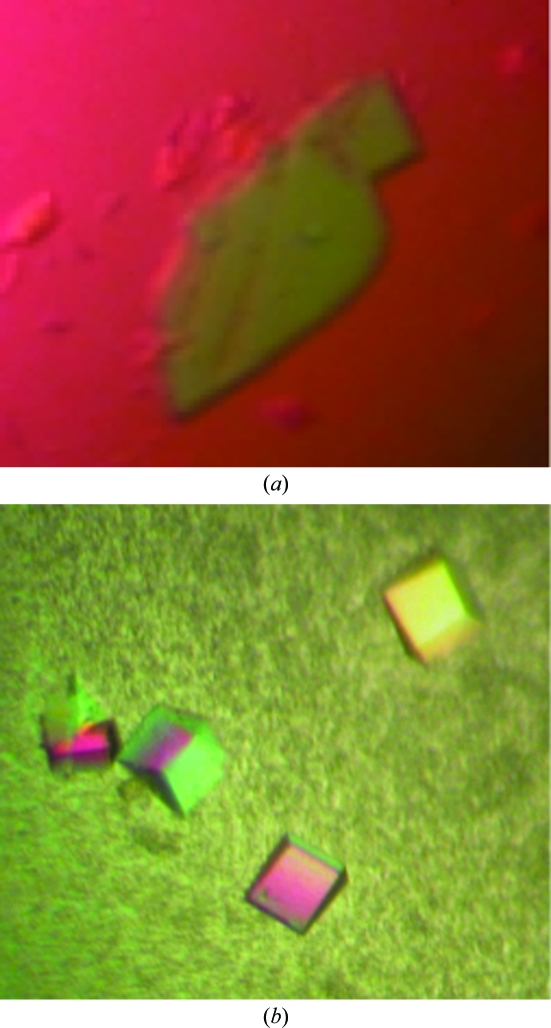
Hanging-drop vapour-diffusion crystallization trials (using Crystal Screen kits from Hampton Research) were undertaken at 293 K using 24-well plates containing 0.5 ml reservoir solution supplemented with β-mercaptoethanol (to a final concentration of 1%). Drgal1-L2 protein in the presence of lactose was initially concentrated to 10 mg ml−1 in crystallization buffer and drops comprising 1 µl Drgal1-L2 solution and 1 µl reservoir solution were set up. Small thin crystals were obtained within days from a PEG 4000-containing condition. Optimization via variation of protein concentration (to a maximum of 20 mg ml−1), precipitant concentration and pH as well as macroseeding successfully reduced nucleation and led to larger crystals (Fig. 3 ▶ a). Ultimately, the macroseeding of small crystals that were grown when the reservoir solution comprised 40% PEG 4000, 0.1 M Tris–HCl pH 8.0, 0.2 M sodium acetate and 1% β-mercaptoethanol into newly equilibrated drops formed using an identical reservoir solution except with a lower PEG 4000 content (30%) proved to be most effective for generating large crystals. Although the majority of the crystals generated were multiple, a few good-quality single crystals were obtained (Fig. 3 ▶ b).
Figure 3.
Drgal1-L2 co-crystallized with lactose in the presence of a reducing environment. Crystals A and B are shown (with different magnification scales). The dimensions of crystal A are 0.5 × 0.25 × 0.025 mm and those of crystal B are 0.125 × 0.1 × 0.075 mm.
2.5. X-ray diffraction analysis
X-ray diffraction analysis was performed on the MX1 beamline at the Australian Synchrotron (wavelength 0.9537 Å at 100 K; equipped with an ADSC Quantum detector). Cryoprotectant solutions consisted of reservoir solution supplemented with either 15% glycerol (for crystal A) or 10% PEG 400 (for crystal B). The plate-shaped crystal A diffracted to ∼2 Å resolution and showed that the crystals belonged to an orthorhombic crystal system. Data were collected using the Blu-Ice software (McPhillips et al., 2002 ▶; crystal-to-detector distance 200 mm; frames with 1° ϕ oscillation and with 2 s exposure); however, these data showed high mosaicity in some sections of reciprocal space and evidence of diffraction from a second lattice. Crystal B diffracted to a resolution of 1.5 Å and a complete data set was collected (crystal-to-detector distance 150 mm; 360 frames with 1° ϕ oscillation and 1 s exposure time; Table 1 ▶). The crystal exhibits an orthorhombic crystal system, with unit-cell parameters a = 60.34, b = 66.57, c = 70.87 Å. The data were processed using iMOSFLM (Leslie, 1992 ▶) and scaled using SCALA (Evans, 2006 ▶) as implemented in the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). A Matthews coefficient (V M) of 2.36 Å3 Da−1 and an associated solvent content of 47.9% were calculated with one Drgal1-L2 homodimer in the asymmetric unit. A homology model of zebrafish Drgal1-L2 (predicted to be a homodimer, each monomer comprising 134 amino acids) was built using Coot (Emsley & Cowtan, 2004 ▶) based on the homodimeric human galectin-1 structure (PDB code 1gzw; López-Lucendo et al., 2004 ▶), with which Drgal1-L2 shares 40% amino-acid sequence identity. Using this as a search model, molecular replacement was performed using AMoRe (Navaza, 1994 ▶). Rotation and translation functions were calculated at 15.0–3.0 Å and a solution in space group P212121 was determined for one homodimer in the asymmetric unit. Rigid-body refinement was applied followed by an initial ten cycles of refinement (REFMAC5; Murshudov et al., 1997 ▶) of the model, giving an R factor of 40.4% and an R free of 43.5%. The electron density unambiguously revealed the complete Drgal1-L2 amino-acid chain encompassing residues Ala2–Lys134 and also revealed that lactose was clearly defined bound at each of the two carbohydrate-binding sites within the homodimer. Initial model improvement reduced the R factor to 37.2% and R free to 39.9%. Model building and refinement is in progress. Ultimately, our zebrafish Drgal1-L2 crystal structure will be used for comparative structural analysis with human galectin-1 crystal structure(s) to assess structural conservation and to provide insight into the details of the interactions with carbohydrates. The structural resemblance between human and zebrafish galectin-1 will provide evidence supporting the use of zebrafish Drgal1-L2 as a potential alternative target for galectin-1 inhibitor design and development.
Table 1. Diffraction data statistics for crystal B (Fig. 3 ▶ b).
Data collection was undertaken with a synchrotron-radiation source. Values in parentheses are for the highest resolution shell.
| Resolution (Å) | 1.50 (1.58–1.50) |
| Total no. of observations | 630815 (64480) |
| Total no. of unique observations | 45777 (6088) |
| Multiplicity | 13.8 (10.6) |
| Completeness (%) | 98.6 (91.4) |
| 〈I/σ(I)〉 | 5.6 (1.8) |
| Rmerge† (%) | 6.8 (21.4) |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 60.34, b = 66.57, c = 70.87 |
R
merge = 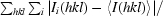
 .
.
Acknowledgments
HB gratefully acknowledges the financial support of the Cancer Council Queensland. MR and KS thank the University of Auckland Staff Research Fund for support. This research was undertaken on the MX1 beamline at the Australian Synchrotron, Victoria, Australia.
References
- Ahmed, H., Du, S.-J., O’Leary, N. & Vasta, G. R. (2004). Glycobiology, 14, 219–232. [DOI] [PubMed]
- Ahmed, H., Du, S.-J. & Vasta, G. R. (2009). Glycoconj. J.26, 277–283. [DOI] [PMC free article] [PubMed]
- Barondes, S. H., Cooper, D. N., Gitt, M. A. & Leffler, H. (1994). J. Biol. Chem.269, 20807–20810. [PubMed]
- Bianchet, M. A., Ahmed, H., Vasta, G. R. & Amzel, L. M. (2000). Proteins, 40, 378–388. [DOI] [PubMed]
- Bourne, Y., Bolgiano, B. & Liao, D. (1994). Nature Struct. Biol.1, 863–870. [DOI] [PubMed]
- Brule, F. van der, Califice, S. & Castronovo, V. (2004). Glycoconj. J.19, 537–542. [DOI] [PubMed]
- Camby, I., Le Mercier, M., Lefranc, F. & Kiss, R. (2006). Glycobiology, 16, 137R–157R. [DOI] [PubMed]
- Chang, W.-N., Tsai, J.-N., Chen, B.-H. & Fu, T.-F. (2006). Protein Expr. Purif.46, 212–220. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Feitsma, H. & Cuppen, E. (2008). Mol. Cancer Res. 6, 685–694. [DOI] [PubMed]
- Flores, M. V., Hall, C. J., Crosier, K. E. & Crosier, P. S. (2010). Dev. Dyn.239, 2128–2135. [DOI] [PubMed]
- Georgiadis, V., Stewart, H. J., Pollard, H. J., Tavsanoglu, Y., Prasad, R., Horwood, J., Deltour, L., Goldring, K., Poirier, F. & Lawrence-Watt, D. J. (2007). Dev. Dyn.236, 1014–1024. [DOI] [PubMed]
- Horiguchi, N., Arimoto, K., Mizutani, A., Endo-Ichikawa, Y., Nakada, H. & Taketani, S. (2003). J. Biochem.134, 869–874. [DOI] [PubMed]
- Hsieh, S. H., Ying, N. W., Wu, M. H., Chiang, W. F., Hsu, C. L., Wong, T. Y., Jin, Y. T., Hong, T. M. & Chen, Y. L. (2008). Oncogene, 27, 3746–3753. [DOI] [PubMed]
- Lassen, N., Estey, T., Tanguay, R. L., Pappa, A., Reimers, M. J. & Vasiliou, V. (2005). Drug Metab. Dispos.33, 649–656. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Liao, D. I., Kapadia, G., Ahmed, H., Vasta, G. R. & Herzberg, O. (1994). Proc. Natl Acad. Sci. USA, 91, 1428–1432. [DOI] [PMC free article] [PubMed]
- Lieschke, G. J. & Currie, P. D. (2007). Nature Rev. Genet.8, 353–367. [DOI] [PubMed]
- López-Lucendo, M. F., Solis, D., Andre, S., Hirabayashi, J., Kasai, K., Kaltner, H., Gabius, H. & Romero, A. (2004). J. Mol. Biol.343, 957–970. [DOI] [PubMed]
- Ma, C. (2004). Mod. Drug Discov.7, 30–36.
- McPhillips, T. M., McPhillips, S. E., Chiu, H.-J., Cohen, A. E., Deacon, A. M., Ellis, P. J., Garman, E., Gonzalez, A., Sauter, N. K., Phizackerley, R. P., Soltis, S. M. & Kuhn, P. (2002). J. Synchrotron Rad.9, 401–406. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Scott, K. & Weinberg, C. (2002). Glycoconj. J.19, 467–477. [DOI] [PubMed]
- Scott, S. A., Scott, K. & Blanchard, H. (2007). Acta Cryst. F63, 967–971. [DOI] [PMC free article] [PubMed]
- Shin, J. T. & Fishman, M. C. (2002). Annu. Rev. Genomics Hum. Genet.3, 311–340. [DOI] [PubMed]
- Stern, H. M. & Zon, L. I. (2003). Nature Rev. Cancer, 3, 533–539. [DOI] [PubMed]
- Zhao, X., Liu, M., Wu, N., Ding, L., Liu, H. & Lin, X. (2010). Protein Expr. Purif.72, 262–266. [DOI] [PubMed]