The ferritin homolog, BfrB (Rv3841), from Mycobacterium tuberculosis has been purified, crystallized, and diffraction data were collected to 2.5 Å resolution. Here, preliminary crystallographic characterization and SAXS analyses are reported.
Keywords: BfrB, Mycobacterium tuberculosis, ferritins
Abstract
Mycobacterium tuberculosis (Mtb) is the causative agent of the deadly disease tuberculosis. Iron acquisition, regulation and storage are critical for the survival of this pathogen within a host. Thus, understanding the mechanisms of iron metabolism in Mtb will shed light on its pathogenic nature, as iron is important for infection. Ferritins are a superfamily of protein nanocages that function in both iron detoxification and storage, and Mtb contains both a predicted ferritin and a bacterioferritin. Here, the cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of the ferritin homolog (Mtb BfrB, Rv3841) is reported. An Mtb BfrB crystal grown at pH 6.5 using the hanging-drop vapor-diffusion technique diffracted to 2.50 Å resolution and belonged to space group C2, with unit-cell parameters a = 226.2, b = 226.8, c = 113.7 Å, β = 94.7° and with 24 subunits per asymmetric unit. Furthermore, modeling the crystal structure of a homologous ferritin into a low-resolution small-angle X-ray scattering (SAXS) electron-density envelope is consistent with the presence of 24 subunits in the BfrB protein cage quaternary structure.
1. Introduction
Iron is an essential element for virtually all forms of life and most bacterial pathogens must import iron directly from a host for survival. Successful iron acquisition is a critical virulence determinant for infection and understanding the process could potentially unveil new drug targets. However, iron homeostasis poses a serious problem. Despite its abundance in nature, iron is poorly soluble under physiological conditions, thus requiring a highly regulated iron metabolism in biological systems. Ferritins are a superfamily of proteins that are ubiquitous in nature, with their role in iron storage being their most often characterized function. Ferritins are large protein nanocages that catalyze the reversible oxidation of ferrous iron into a ferric oxide core within their protein shell and thus protect the cell against Fe2+-induced oxidative damage (Pierre & Fontecave, 1999 ▶; Arosio et al., 2009 ▶).
Structures of ferritins have been described from all three domains of life: eukaryotes, bacteria and archaea. Nearly all known ferritins spontaneously assemble into highly conserved 24-subunit oligomers folded into four-helix bundles that form a hollow spherical nanocage complex related by fourfold, threefold and twofold symmetry axes (Carrondo, 2003 ▶). Bacterial ferritins are subdivided into two types of ferritins: ferritins (Ftns) and bacterioferritins (Bfrs). They both share essentially the same architecture; however, Bfr also contains noncovalently bound heme in a pocket formed by the interface between a pair of symmetry-related subunits and thus Bfr contains 12 heme molecules per 24-subunit cage.
The causative agent of tuberculosis, Mycobacterium tuberculosis (Mtb), infects approximately 11.1 million people per year and results in over 1.3 million deaths worldwide (World Health Organization, 2009 ▶). Iron acquisition is necessary for Mtb pathogenicity and ferritins have been shown to be important in mycobacterial iron metabolism. Mtb BfrB is up-regulated in response to hypoxic conditions (Sherman et al., 2001 ▶) and during adaptation to the stationary phase (Voskuil et al., 2004 ▶), which could potentially be important in latent Mtb infections. Additionally, Mtb BfrB can be secreted (Rosenkrands et al., 2002 ▶), which is suggestive of an as yet uncharacterized function for Mtb BfrB.
Interestingly, Mtb has two homologs of ferritin, one of which has high sequence similarity to bacterioferritin (Mtb BfrA, Rv1876) and the other to ferritin (Mtb BfrB, Rv3841). While Mtb BfrA and Mtb BfrB may perform similar functions in iron metabolism, their specific roles and regulatory pathways are still unclear. Recently, the crystal structure of Mtb BfrA was reported (Gupta et al., 2009 ▶), but its role in iron metabolism has not yet been characterized. Less is known about Mtb BfrB. In an effort to gain a better understanding of these important players in Mtb iron metabolism, we report here the crystallization and preliminary structural characterization of Mtb BfrB.
2. Experimental methods
2.1. Construction of the expression vectors for Mtb BfrB
The Mtb gene Rv3841 encoding the protein BfrB was PCR-amplified from Mtb H37Rv genomic DNA using the KOD HotStart Polymerase Kit (Novagen) with forward (5′-CCATATGACAGAATACGAAGGGCCTAAG) and reverse (5′-GGCTCGAGGAGGCGGCCCCCGGCAGCG) primers containing NdeI and XhoI restriction-enzyme sites, respectively. The primers were designed based on the sequence available from the TB Structural Genomics Consortium and in such a way as to add a C-terminal polyhistidine tag to facilitate purification. Double digestion with NdeI and XhoI was performed on the PCR-amplified Rv3841 as well as the plasmid pET-28b+ (Novagen). Excised Rv3841 and pET-28b+ were ligated and transformed into Escherichia coli BL21-Gold (DE3) cells (Novagen). The gene sequence was confirmed by nucleotide sequencing using standard primers for the T7 promoter and terminator.
2.2. Protein expression and purification
E. coli BL21-Gold (DE3) cells harboring pET-28b-Rv3841 were grown aerobically at 310 K at 250 rev min−1 in LB medium containing 30 µg ml−1 kanamycin. Protein expression was induced by adding isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mM at an OD600nm of ∼0.8 and the cells were harvested after 4 h by centrifugation at 5500g for 20 min. After the addition of resuspension buffer (50 mM Tris–HCl pH 7.4, 350 mM NaCl, 10% glycerol) containing phenylmethylsulfonyl fluoride and hen egg-white lysozyme, the cells were disrupted by sonication, clarified by centrifugation at 18 000g for 30 min and syringe-filtered (1 µm pore size) to remove cell debris. The clarified cell lysate was then loaded onto a 5 ml Ni2+-charged HisTrap column (GE Healthcare) and washed with 50 mM HEPES pH 7.8, 350 mM NaCl, 10 mM imidazole and 10% glycerol. The protein was eluted with a linear gradient of 10–500 mM imidazole (100 ml); the purified protein eluted between 75 and 200 mM imidazole. The fractions were collected and concentrated to 1 ml in a 100 kDa molecular-weight cutoff concentrator (Millipore). After the addition of glycerol to a final concentration of 25%(v/v), protein samples in the elution–filtration buffer were flash-frozen in liquid nitrogen for storage at 193 K until use. Protein samples were thawed and further purified on a Superdex 200 HiLoad 16/60 gel-filtration chromatography column (GE) equilibrated with 50 mM Tris–HCl pH 7.4, 350 mM NaCl and 10% glycerol prior to use. In addition to its native amino-acid sequence, the recombinant protein has eight extra residues at the C-terminus (GSHHHHHH); the last six residues constitute the polyhistidine tag. The predicted molecular weight of the purified recombinant protein monomer is 21 409 Da. Protein purity was analyzed by SDS–PAGE. Protein concentration was determined by UV spectroscopy, assuming a molar extinction coefficient of 10 810 M −1 cm−1 at 280 nm as determined by the program Protein Calculator (Putnam, 2006 ▶).
The selenomethionine derivative of Mtb BfrB (SeMet-Mtb BfrB) was prepared by the metabolic inhibition method using the Overnight Express Autoinduction System 2 (EMD Biosciences; Grabski et al., 2005 ▶). Briefly, cells harboring pET-28b-Rv3841 were initially grown aerobically at 310 K at 250 rev min−1 in LB medium containing 30 µg ml−1 kanamycin. At an OD600nm of ∼0.8, cells were harvested by centrifugation, washed and transferred to Overnight Express System medium supplemented with l-selenomethionine to a final concentration of 125 mg l−1. Cultures were incubated for a further 16 h at 300 rev min−1, harvested by centrifugation and purified under identical conditions to those used for the native protein. The incorporation of selenomethionine was confirmed by MALDI–TOF mass spectrometry; the predicted molecular weight was 21 690 Da (six methionine residues per subunit; 144 per nanocage). The purity and concentration were determined as described above.
2.3. Small-angle X-ray scattering (SAXS) data collection and processing
Small-angle X-ray scattering (SAXS) data were collected on ALS beamline 12.3.1. Mtb BfrB was diluted in native buffer (50 mM Tris–HCl pH 7.4, 350 mM NaCl, 10% glycerol and 10 mM β-mercaptoethanol) to give 20 µl samples at final concentrations of 1.1 and 0.55 mg ml−1. For each protein sample and a buffer blank, SAXS data were collected using a 0.5 s exposure, a 5 s exposure and a second 0.5 s exposure. Buffer-subtracted files were analyzed using PRIMUS (Konarev et al., 2003 ▶) and the GNOM (Svergun, 1992 ▶) P(r) output file with d max = 143 was used to calculate electron-density envelopes. Each envelope is the product of 16 GASBOR (Svergun et al., 2001 ▶) runs (P1 symmetry and 4000 residues) averaged with DAMMAVER (Volkov & Svergun, 2003 ▶).
2.4. Crystallization
Purified native and SeMet-derivatized Mtb BfrB were concentrated to 10 mg ml−1 in 50 mM Tris pH 7.4, 350 mM NaCl, 10% glycerol and supplemented with 10 mM β-mercaptoethanol for crystallization trials. Initially, high-throughput sparse-matrix crystallization screening was carried out using a Mosquito nanolitre-dispensing robot (TTP LabTech). Conditions from five crystallization kits (Crystal Screen HT, Index HT and PEG/Ion HT from Hampton Research and JCSG+ Suite and PACT Suite from Qiagen) were screened using the hanging-drop vapor-diffusion technique under aerobic conditions at 277 K with drops consisting of 0.2 µl protein solution mixed with 0.2 µl reservoir solution and equilibrated against a reservoir volume of 100 µl. Positive hits were optimized to obtain diffraction-quality crystals using siliconized glass slides (Hampton Research) and 24-well VDX plates (Hampton Research) with drops consisting of 1 µl protein solution mixed with 1 µl reservoir solution and equilibrated against a reservoir volume of 1 ml. Cryoprotection was performed in reservoir solution enriched with 30%(v/v) ethylene glycol and crystals were immediately flash-cooled in liquid nitrogen.
2.5. Data collection and processing
Preliminary screening was performed in-house using a Rigaku MicroMax-007 HF high-intensity microfocus rotating-anode X-ray generator with a rotating copper anode (operating at 40 kV and 20 mA) equipped with VariMax HF optics and a Saturn 944+ detector (Rigaku). Promising crystals were then sent for diffraction analysis on beamline 5.0.2 by the Collaborative Crystallography (CC) group at the Advanced Light Source (ALS). A complete native data set was collected from a single crystal at ∼100 K under a nitrogen-gas stream using an ADSC Quantum 315 area detector. The crystal-to-detector distance was set to 250 mm and a total of 440 images were recorded with an oscillation angle of 1° and an exposure time of 5 s per image at a wavelength of 1 Å. A complete SeMet-derivative data set was collected from a single crystal; the crystal-to-detector distance was set to 250 mm and a total of 220 images were recorded with an oscillation angle of 1° and an exposure time of 3 s per image at wavelengths of 0.9797 and 1 Å. Images were indexed, integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
3.1. Mtb BfrB crystallization and X-ray diffraction analysis
Diffraction-quality crystals of native Mtb BfrB were grown in 11.9%(w/v) PEG MME 5000, 100 mM Tris maleate pH 6.5, 0.54%(w/v) 1,6-hexanediol and 0.44%(w/v) lauryldimethylamine oxide. Crystals grew to dimensions of approximately 0.1 × 0.1 × 0.05 mm and the Mtb BfrB crystals were colorless (Fig. 1 ▶ a). In contrast, Mtb BfrA crystals were brown–orange in color owing to heme binding (Gupta et al., 2008 ▶). The lack of color of the Mtb BfrB crystals indicate that heme is not present, suggesting that Mtb BfrB is more similar to the ferritins than to the heme-containing bacterioferritins and verifying its high sequence identity to ferritin. The native crystal diffracted to 2.50 Å resolution and belonged to space group C2, with unit-cell parameters a = 226.2, b = 226.8, c = 113.7 Å, β = 94.7° and with 24 subunits per asymmetric unit (Table 1 ▶). The native crystal had a solvent content of 56.5% and a Matthews coefficient of 2.83 Å3 Da−1 (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶), which is consistent with 24 molecules packing in the asymmetric unit. The overall R merge of 11.6% for the native data set was slightly high and suggested that the space group may be incorrect. However, processing the data in a variety of space groups resulted in considerably higher R merge values, verifying that the space group was indeed C2. It should be noted that the data are anisotropic, which may account for the overall high R merge. Mtb SeMet-BfrB was crystallized with the intention of solving the crystal structure using the multiple-wavelength anomalous diffraction (MAD) method; however, molecular replacement (MR) will be attempted first. SeMet-Mtb BfrB crystals were grown in 14%(w/v) PEG 3350, 5%(w/v) Tacsimate pH 6.4 and 100 mM glycine for two weeks at 277 K (Fig. 1 ▶ b). The SeMet-Mtb BfrB diffraction data were indexed in space group C2 and are reported to 3.20 Å resolution for both the peak and remote data sets. The data-collection and processing statistics are given in Table 1 ▶ for the native and SeMet-derivative data sets.
Figure 1.
Crystals of Mtb BfrB. Crystals grew to dimensions of about 0.1 × 0.1 × 0.05 mm using the hanging-drop vapor-diffusion technique at 277 K. (a) Native crystals, (b) SeMet-derivative crystals.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| SeMet derivative | |||
|---|---|---|---|
| Native | Peak | Remote | |
| No. of crystals | 1 | 1 | 1 |
| Beamline | Beamline 5.0.2, ALS, Berkeley | Beamline 5.0.2, ALS, Berkeley | Beamline 5.0.2, ALS, Berkeley |
| Wavelength (Å) | 1.0000 | 0.9797 | 1.0000 |
| Detector | ADSC Quantum 315 area detector | ADSC Quantum 315 area detector | ADSC Quantum 315 area detector |
| Crystal-to-detector distance (mm) | 250 | 250 | 250 |
| Rotation range per image (°) | 1 | 1 | 1 |
| Total rotation range (°) | 440 | 220 | 220 |
| Exposure time per image (s) | 5 | 3 | 3 |
| Resolution range (Å) | 50.00–2.50 (2.59–2.50) | 50.00–3.20 (3.31–3.20) | 50.00–3.20 (3.31–3.20) |
| Space group | C2 | C2 | C2 |
| Unit-cell parameters (Å, °) | a = 226.2, b = 226.8, c = 113.7, β = 94.7 | a = 232.1, b = 233.1, c = 114.5, β = 94.7 | a = 231.8, b = 232.0, c = 114.3, β = 94.7 |
| Solvent content (%) | 56.5 | 59.1 | 58.7 |
| Matthews coefficient (Å3 Da−1) | 2.83 | 3.01 | 2.98 |
| Mosaicity (°) | 0.65 | 0.76 | 0.69 |
| Total no. of measured intensities | 853274 | 467420 | 453221 |
| Unique reflections | 194323 (19297) | 99366 (9865) | 98514 (9788) |
| Multiplicity | 4.4 (4.1) | 4.7 (4.7) | 4.6 (4.6) |
| Mean I/σ(I) | 13.0 (3.6) | 8.5 (2.8) | 11.9 (6.6) |
| Completeness (%) | 99.9 (99.7) | 99.8 (99.6) | 99.8 (99.7) |
| Rmerge† (%) | 11.6 (30.7) | 18.3 (47.9) | 12.3 (23.3) |
| Overall B factor from Wilson plot‡ (Å2) | 26.6 | 38.5 | 30.8 |
R
merge = 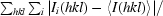
 , where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
, where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
Overall B factors from the Wilson plot were calculated using phenix.xtriage from the PHENIX suite (Adams et al., 2010 ▶).
3.2. Self-rotation Patterson plot
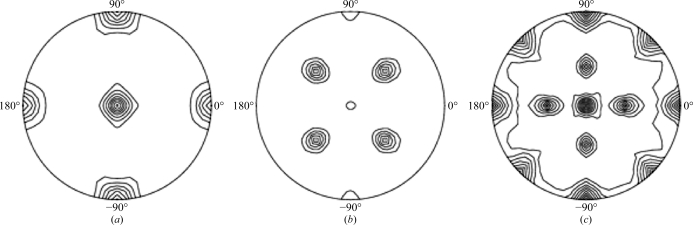
Ferritins are composed of monomeric subunits that fold into four-α-helix bundles with a short C-terminal α-helix almost perpendicular to the bundle. They assemble into 24-mer complexes with fourfold, threefold and twofold symmetry axes. A self-rotation function analysis of the data at κ = 90°, κ = 120° and κ = 180° in the resolution range 9–4 Å with an integration radius of 25 Å was carried out using the Self Rotation Function Server (http://www.doe-mbi.ucla.edu/~sawaya/selfrot/) and showed peaks consistent with 432 point-group symmetry for Mtb BfrB (Fig. 2 ▶).
Figure 2.
Self-rotation Patterson plots for Mtb BfrB. The peaks in the (a) κ = 90°, (b) κ = 120° and (c) κ = 180° sections confirm the fourfold, threefold and twofold molecular symmetry, respectively. The self-rotation function was calculated using the Self Rotation Function Server (http://www.doe-mbi.ucla.edu/~sawaya/selfrot/) using default parameters.
3.3. Best PDB homologs for MR
Ferritins have very high structural conservation, in which each monomer folds into a four-helix bundle, although they vary in amino-acid sequence by as much as 80% or more (Theil et al., 2006 ▶). We are currently in the process of determining the structure by MR using Phaser (McCoy et al., 2007 ▶). The homologous ferritins from Archaeoglobus fulgidus (27.9% sequence identity; PDB code 1sq3; Johnson et al., 2005 ▶), Pyrococcus furiosus (25.9% sequence identity; PDB code 2x17; Kasyutich et al., 2010 ▶), Thermatoga maritima (25.2% sequence identity; PDB code 1vlg; Joint Center for Structural Genomics, unpublished work) and bullfrog (23.9% sequence identity; PDB code 1mfr; Ha et al., 1999 ▶) will be used as initial search models.
3.4. Small-angle X-ray scattering (SAXS)
Ferritins generally assemble into a 24-mer spherical complex. To confirm this, a SAXS experiment was performed with Mtb BfrB protein in solution. Fig. 3 ▶ shows the electron-density envelope of the average result of 16 individual calculations visualized using CHIMERA (Pettersen et al., 2004 ▶) overlaid onto the crystal structure of the ferritin from T. maritima (PDB entry 1vlg). The modeling of T. maritima BfrB into a low-resolution SAXS electron-density envelope of Mtb BfrB is consistent with 24 subunits of a ferritin protein cage quaternary structure. Furthermore, the electron-density envelope is hollow, confirming that Mtb BfrB does not contain iron in this instance.
Figure 3.
SAXS. A fit of the crystal structure of T. maritima ferritin (PDB entry 1vlg) into the surface envelope of Mtb BfrB confirms the conserved hollow spherical topology of a ferritin. The overlay with the electron-density envelope was created using CHIMERA. The envelope is calculated from an average of 16 GASBOR runs with P1 symmetry and 4000 residues.
4. Conclusions
We report here the crystallization conditions and preliminary X-ray crystallographic analyses of native and SeMet-derivative crystals of the Mtb ferritin homolog Mtb BfrB. Elucidation of the three-dimensional structure of Mtb BfrB will provide insight into the mechanism of putative iron storage and detoxification, and will lead to a better understanding of its role in the iron metabolism of this deadly pathogen.
Acknowledgments
This work has been supported by National Institutes of Health grant AI068135 (subcontract to CWG). The authors wish to thank Dr John T. Belisle, Colorado State University (NIH NIAID Contract NO1 AI-75320) for the generous supply of M. tuberculosis H37Rv genomic DNA. We would like to thank Lana Cong, Jared Wing Lee and Vincent Ngo and give special thanks to the TB Structural Genomics Consortium for their assistance on this project.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Arosio, P., Ingrassia, R. & Cavadini, P. (2009). Biochim. Biophys. Acta, 1790, 589–599. [DOI] [PubMed]
- Carrondo, M. A. (2003). EMBO J.22, 1959–1968. [DOI] [PMC free article] [PubMed]
- Grabski, A., Mehler, M. & Drott, D. (2005). Nature Methods, 2, 233–235.
- Gupta, V., Gupta, R. K., Khare, G., Salunke, D. M. & Tyagi, A. K. (2008). Acta Cryst. F64, 398–401. [DOI] [PMC free article] [PubMed]
- Gupta, V., Gupta, R. K., Khare, G., Salunke, D. M. & Tyagi, A. K. (2009). PLoS ONE, 4, e8028. [DOI] [PMC free article] [PubMed]
- Ha, Y., Shi, D., Small, G. W., Theil, E. C. & Allewell, N. M. (1999). J. Biol. Inorg. Chem.4, 243–256. [DOI] [PubMed]
- Johnson, E., Cascio, D., Sawaya, M. R., Gingery, M. & Schroder, I. (2005). Structure, 13, 637–648. [DOI] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci.12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Kasyutich, O., Ilari, A., Fiorillo, A., Tatchev, D., Hoell, A. & Ceci, P. (2010). J. Am. Chem. Soc.132, 3621–3627. [DOI] [PubMed]
- Konarev, P. V., Volkov, V. V., Sokolova, A. V., Koch, M. H. J. & Svergun, D. I. (2003). J. Appl. Cryst.36, 1277–1282.
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. & Ferrin, T. E. (2004). J. Comput. Chem.25, 1605–1612. [DOI] [PubMed]
- Pierre, J. L. & Fontecave, M. (1999). Biometals, 12, 195–199. [DOI] [PubMed]
- Putnam, C. D. (2006). Protein Calculator. http://www.scripps.edu/~cdputnam/protcalc.html.
- Rosenkrands, I., Slayden, R. A., Crawford, J., Aagaard, C., Barry, C. E. & Andersen, P. (2002). J. Bacteriol.184, 3485–3491. [DOI] [PMC free article] [PubMed]
- Sherman, D. R., Voskuil, M., Schnappinger, D., Liao, R., Harrell, M. I. & Schoolnik, G. K. (2001). Proc. Natl Acad. Sci. USA, 98, 7534–7539. [DOI] [PMC free article] [PubMed]
- Svergun, D. I. (1992). J. Appl. Cryst.25, 495–503.
- Svergun, D. I., Petoukhov, M. V. & Koch, M. H. (2001). Biophys. J.80, 2946–2953. [DOI] [PMC free article] [PubMed]
- Theil, E. C., Matzapetakis, M. & Liu, X. (2006). J. Biol. Inorg. Chem.11, 803–810. [DOI] [PubMed]
- Volkov, V. V. & Svergun, D. I. (2003). J. Appl. Cryst.36, 860–864.
- Voskuil, M. I., Visconti, K. C. & Schoolnik, G. K. (2004). Tuberculosis, 84, 218–227. [DOI] [PubMed]
- World Health Organization (2009). Global Tuberculosis Control: A Short Update to the 2009 Report. http://www.who.int/tb/publications/global_report/2009/update/tbu_9.pdf. Geneva: World Health Organization.