Introduction
Studying biomolecules on surfaces is of great importance in the design of biosensors, advanced materials, and bioassays. Single molecule techniques, in particular, enable the understanding of properties and dynamic behavior that are usually obscured and inaccessible through ensemble (bulk) experiments. The nanomechanics of proteins has been studied via single molecule force spectroscopy on surfaces to identify energy landscapes and to reveal unique structural or temporal states of molecules under stress.(1-5) Atomic force microscopy (AFM)(6) is an ideal tool for imaging single molecules(7, 8) as well as studying molecular interactions and nanomechanics. Application of force spectroscopy(9, 10) to molecules such as titin,(1) myosin,(11, 12) and signaling proteins(13-17) has successfully demonstrated molecular rupture forces,(18) protein domain unfolding,(19-24) and receptor–ligand interactions.(5, 25-27)
Force spectroscopy of biopolymers requires the attachment of polymers of sufficient length between the tip and surface as well as the stretching of a single or a few polymers at a defined loading rate. Protein attachment has been the subject of intensive investigation. The commonly used physisorption on substrate allows polymers to be adsorbed and tethered by AFM cantilever but only at random, unspecified locations and yields hard-to-interpret force curves. The use of site-specific antibodies directed to protein sequences allows for tethering and stretching the protein between known sites.(28-30) Force curves of definitive single molecule events are even more challenging to achieve consistently.(31) Single molecule events are enhanced by reducing the surface density of attachment with either dilute solutions for physisorption or with limiting number of chemically reactive sites.(32) Tip–surface or adhesive interactions can obscure the first portion of the force spectrum,(32, 33) thus allowing only long polymers to yield any interpretable data. Hence, some of the first force spectra of single molecules were from long biopolymers such as polysaccharides,(34) DNA,(35) and titin(1) (the longest known protein). Refinement of protein immobilization techniques and surfaces have greatly reduced the obscuring effects of tip–surface adhesion, thereby allowing smaller proteins to be studied.(31, 32) For large modular proteins where there are significant conformational data on the fundamental domains, each of these domains can be studied by the use of engineered polyproteins of 8–12 identical domains.(36) Polymers of tandem repeats of identical domains render regularly spaced force events (e.g., the sawtooth pattern) and can be interpreted in fine detail by the unfolding of a single domain, e.g., from titin Ig/Fn3.(1, 37)
While these engineered polyproteins have been powerful in revealing the mechanism of mechanical unfolding of protein domains, many cytoskeletal proteins consist of numerous distinct modules that may interact intimately and fulfill collectively complicated mechanical roles in cells. Therefore, direct nanomechanical measurements of the full length, intact protein are required to understand biomechanics at the cellular and molecular levels. Such force curves are composites of many overlapping force events and are too complex to interpret with any degree of confidence. Dynamics stiffness measurement, taken simultaneously with the force curves, is a complementary technique that holds promise of resolving some of the complexity.(20, 38)
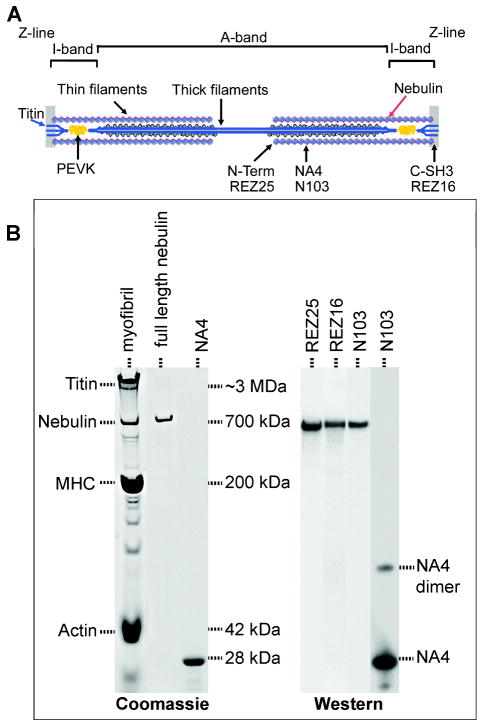
The giant modular protein nebulin (MW 700–800 kDa) plays a critical role in the structure and function of thin filaments and active contraction of skeletal muscles (Figure 1A).(39) Mutations in the nebulin gene have been linked to the muscle disease nemaline myopathy.(40) Nebulin associates with actin to form a 1 μm long composite thin filament in skeletal muscles and is thought to act as a protein ruler to regulate thin filament length,(41, 42) and perhaps as a Ca2+/calmodulin or S100-mediated regulatory protein.(43, 44) The protein (e.g., 6669 residues in the human adult nebulin isoform(45)) comprises highly homologous repeats of 35 amino acids that correlate with the number of actin monomers in the thin filament.(45, 46) Biochemical, structural, and developmental studies support the notion that nebulin, in conjunction with tropomodulin and other actin capping proteins, regulates thin filament length, but the architecture of nebulin-containing composite thin filaments remains speculative.(47, 48)
Figure 1.
Nebulin is a major sarcomeric protein localized to the thin filaments. (A) Schematic of a striated muscle sarcomere showing actin thin filaments with nebulin as well as myosin thick filaments with the associated titin and its elastic PEVK segment. One nebulin molecule spans the entire 1 μm thin filament length. The locations of the binding sites of the antibodies used in this study are also shown. (B) Native nebulin and site specific antibodies. Native nebulin from the rabbit longissimus dorsi muscle showed a single band on 3–8% gradient Tris-acetate–SDS gels and was recognized specifically by antibodies to the N-terminus (REZ25), C-terminus (REZ16), and NA4 super-repeat (N103) of nebulin. Rabbit myofibrils and bacterially expressed NA4 were blotted for comparison. Trace of NA4 dimer was detected.
Nebulin is an attractive candidate for study by force spectroscopy because it plays a direct role in the force generating machinery of muscle and is likely to play a mechanical role in the stability and function of the muscle sarcomere. Force spectroscopy would help identify those structural features and transitions that are important in these mechanical roles. Because of its intimate association with actin, myosin, and its anchorage into the Z-bands of the muscle sarcomere, the isolation of native nebulin has remained elusive, and it is still much too large for recombinant expression in toto. Through the use of the mild detergent deoxycholate, we have successfully extracted and purified full-length, native nebulin from rabbit muscle for the first time. In the course of our study on the nanomechanical properties of nebulin, we have approached and resolved several key technical obstacles for investigating giant, modular proteins in general: (1) the use of pairs of antibodies to either termini or internal site to tether selected regions of the full-length nebulin; (2) the use of self-assembled monolayer surfaces (OEG-SAM) to isolate and covalently tether single proteins while avoiding tip–surface interactions and adsorption of protein to the surface;(32) (3) the analysis of the entire set of force curves without bias to establish empirical criteria for identifying single molecule events versus multiple molecule events; (4) the use of the adaptive, model independent HHT(49-51) method to search for periodicity of key mechanical events in the force spectra.
Experimental Details
Materials
Oligoethylene glycol thiol (OEG), HS-C11-(EG)6OH, and NHS-terminated thiol, HS-C15COO-NHS, were purchased from Prochimia (Gdansk, Poland). Ethanol (200-proof) was purchased from the Warner-Graham Company (Cockeysville, MD). To minimize contamination, solutions were prepared in Teflon beakers and containers (Savillex Corp., Minnetonka, MN). Epotek 377 glue was purchased from Epoxy Technology (Billerica, MA). Tetrahydrofuran (THF), acetone, glycidyl silane, ethyl acetate, trifluoromethylsulfonyl chloride, and dimethyl amino pyridine (DMAP) were purchased from Sigma Aldrich (St. Louis, MO) and used as received. Phosphate buffered saline (1× PBS, 1.06 mM KH2PO4, 155.17 mM NaCl, and 2.97 mM Na2HPO4, pH 7.4) was obtained from Gibco and sterile filtered using a Millex-GS 0.45 μm filter from Millipore (Billerica, MA) prior to use. Silicon nitride (Si3N4) microlevers from Veeco (Santa Barbara, CA) and ultrasharp type I MAC cantilevers from Molecular Imaging (Agilent) were used for imaging and force measurements.
Protein Preparation
Full-length nebulin was prepared from the rabbit longissimus dorsi muscle, and details will be published elsewhere (Gutierrez, G., Tsai, W., and Wang, K., unpublished work). Briefly, muscle was extracted with 0.45% (w/v) sodium deoxycholate (DOC), 20 mM Tris-HCl, and 1 mM DTT at pH 6.5 and centrifuged at 27,000g for 20 min at 4 °C. The extract was applied to a Q-type exchanger cartridge (Sartorius # Q15F) previously equilibrated with 0.45% DOC, 20 mM Tris-HCl, and 0.5 mM DTT at pH 6.5. Nebulin was eluted with a linear gradient of 0–1.0 M NaCl. SDS gel electrophoresis and Western blotting with custom rabbit polyclonal antibodies toward the C-terminus peptide (REZ16) (nebulin SH3 primary sequence VQRTGRTGMLPANYVE), N-terminus peptides (REZ25) (primary sequence MADDEDYEEVVE), and monoclonal anti-NA4 antibody (N103)(41) were done on 3–8% Tris-acetate gels (Invitrogen, Carlsbad CA) (Figure 1B). Nebulin was freshly gel filtered at room temperature (Sephacryl S1000 column, 20 mM Tris-Cl, 0.45% DOC, and 0.5 mM DTT at pH 8.5), and the monodisperse fractions were selected immediately for AFM experiments.
Mixed Function Self-Assembled Monolayers
Functionalized SAMs were prepared as described.(32) Briefly, freshly prepared ultraflat gold surfaces were incubated with a mixture of OEG thiols ((1-mercaptoundec-11-yl) hexa(ethylene glycol)) and NHS thiols ((1-mercaptohexadecanoic acid)-N-succinimidyl ester)) in absolute ethanol for 16–24 h. After incubation, the gold surfaces were rinsed with ethanol and dried in a stream of purified nitrogen. The molar ratio of the OEG thiol to the NHS thiol was empirically determined to obtain well-separated attachment sites on gold. The functionalized surfaces were incubated with affinity-purified anti-SH3 antibody REZ16 (1 μg/mL in PBS at pH 7.4) for 1 h. The unreacted NHS groups were then quenched for 1 h in 0.1 M ethanolamine solution at pH 7.4. Nebulin (1 μg/mL in 20 mM Tris-Cl, 0.45% DOC, and 0.5 mM DTT at pH 8.5) was subsequently incubated with the conjugated surface by injecting underneath a drop of PBS buffer at a 1:100 volume ratio. The surfaces were washed with PBS and imaged by AFM immediately in noncontact mode (MAC mode) to obtain topography, phase, and amplitude images. All experiments were performed at room temperature unless otherwise specified.
AFM Cantilever Functionalization
The cantilever tips were cleaned by exposure to ozone generated in the presence of high-intensity UV light for 15 min to remove any contamination by organics on the surface. Cleaned AFM cantilevers were exposed to glycidyl silane vapor in a jar at 80 °C. The tips were then rinsed in ethyl acetate followed by water and cured at 80 °C for 30 min. This was followed by hydrolysis in 10 mM HCl for 30 min at 80 °C. The tips were then rinsed in water and dried. Tips were activated by exposure to 5 mL of acetone containing 10 μL of trifluoromethylsulfonyl chloride and 5 mg of dimethylamino pyridine. Finally, the tips were washed in acetone and dried at 80 °C and stored in vacuum prior to use. Reactive tips were functionalized with N103 or REZ-25 antibody by dipping into 10 μL of a 0.5 μg/mL antibody solution in 1× PBS (pH 7.4) for 1 h in a Teflon well, followed by washing in PBS and quenching of the unreacted groups in 0.1 M ethanolamine-HC1 (pH 7.4). The tips were immediately mounted and used for stretching nebulin tethered to the SAM/gold surface.
Atomic Force Microscopy
A PicoSPM AFM instrument (Molecular Imaging, Phoenix, AZ) was used for force spectroscopy and imaging measurements in both contact and noncontact (MAC) modes. Soft cantilevers (nominal force constant 0.01 N/m; resonance frequency 7 kHz) were used for the force–distance analysis. Stiffer cantilevers (nominal force constant 0.6–0.95 N/m; resonance frequency 75–105 kHz) were used for imaging in MAC mode. For imaging of nebulin morphology, experiments were also conducted on a mica surface by placing a drop of nebulin solution and then rinsed off by flowing PBS buffer across the surface. Images were analyzed using the Scanning Probe Image Processor (SPIP) software (Image Metrology, Denmark).
Force-Extension Curves of Single Nebulin Molecules Tethered by Site-Specific Antibodies
Cantilevers with and without attached protein were calibrated and their force constants measured using the thermal fluctuation method.(52) The tips used for covalent attachment of antibodies and subsequent force measurements had a force constant k ≈ 0.012 N/m. Force-extension curves were obtained by moving the tip into contact with the surface whereupon the antibody and nebulin bind, then retracting until detachment occurred. Generally, detachment of a tether between the tip and surface is expected to occur at the weakest link, in this case, the noncovalent antibody–antigen interaction. The force required to break a covalent bond has been shown to be on the order of several nanonewtons (e.g., the sulfur–gold bond ruptures at 1.4 ± 0.3 nN(18)) and thus represents an operational upper limit for meaningful analysis.
Several hundred curves were obtained for each experiment by moving the tip to different points on each surface. The extension sweep speed was 1 μm/s, well below where viscous drag exerts an effect on the cantilever and fast enough to minimize the instrument drift during a prolonged stretch. The nominal loading rate was 10,000 pN/s. The force of contact was < 1 nN in order to avoid damaging the surface and immobilized proteins. Each force curve had 2048 points, the maximum allowed by the control software. Detachment forces and distances were tabulated and binned at 25–200 pN and 25 –200 nm to obtain a 2D histogram. Since the force-extension curves consist of irregularly spaced peaks superimposed on a rapidly rising base, these craggy mountain range profiles were analyzed with the worm-like chain (WLC) model of polymer elasticity. The ascending limb of the peaks were fitted with the following equation:(53)
| (1) |
where f(x) is the force at a distance of x, p is the persistence length, kB is the Boltzmann's constant, T is the absolute temperature, and L is the molecular contour length of the polymer being stretched. Two parameter fits (L and p) of the available points were performed, subject to the constraints p, L > 0, and L > p. The extension stiffness (elastic modulus) is estimated as δF/δx along the WLC curves.
Periodicity Analysis of Peak-to-Peak Spacing
Since nebulin consists of tandem repeats of nebulin modules, the possible presence of periodicity in the unfolding events, manifested as periodic peak-to-peak in the force curves, was then investigated. All peaks were identified from the force (F) versus distance (x) curves by a nearest neighbor approach. The force at point xn was compared to those at positions xn−2, xn−1, xn+1 and xn+2. If the value of the force Fn at xn is greater than the forces at the neighbors, then it is considered a peak. The numerical derivatives at the peak positions were used to further confirm the extrema. Finally, visual examination was used to check the results. Full-length nebulin force curves each showed 6–20 major peaks as identified by this method. All peaks, large and small, were analyzed using a pairwise distribution function. The data were initially verified to be nonrandom via an autocorrelation test. The presence of periodicity in the peaks or large spikes in the force–distance curves was determined by computing a pairwise distance distribution function (PDF).(54) The PDF was first obtained by binning the distance differences between two neighboring peaks (Lpp = xn − xn−1) in histograms with bin widths of 2, 5, 7, and 10 nm. The PDF was smoothed using a binomial smoothing filter,(55) and periodicity was then determined from the power spectrum of the PDF. Computation was performed in open-source GNU-Octave. Optimal results showing clear periodicities were obtained with a bin of 5, which was the predicted optimal bin width using the method of Scott.(56) Results from a bin width of 5 nm were used in further analysis.
Periodicity in the peak-to-peak spacing, Lpp, was determined by the application of Hilbert–Huang transform (HHT). HHT is an adaptive empirical (i.e., model independent) method for data analysis that has been particularly useful for nonstationary and nonlinear data (typically data from physical and natural processes). The first step of the transform involves empirical mode decomposition (EMD) of the data into intrinsic oscillation modes, the superposition of which results in the existing data set. This intrinsic mode function contains information with regard to any existing period, and a subsequent Fourier or Hilbert transform can then be used for feature extraction from this frequency–energy space.(49, 50) The empirical mode decomposition was implemented using available MATLAB code.(57, 58)
Results
Following the integrated strategy outlined in the introduction, we stretched native nebulin via site-specific antibodies over a protein-resistant self-assembled monolayer (SAM) of oligoethylene glycols (OEG) on an ultraflat gold surface(32) (Figure 2). Such nonsticky surfaces facilitated single molecule imaging as well as the detection of force signatures over the entire force range using an atomic force microscope (AFM).(10) One pair of antibodies specific to the N- and C-termini was used to tether the full-length molecule (REZ25 on tip and REZ16 on SAM), and another pair specific to the NA4 and C-terminus (N103 on tip and REZ16 on SAM) was used to bracket a large segment of the intact nebulin. The NA4 super-repeat is located as a single band 0.85 μm from the Z-line in rabbit muscle(46, 59) and spans residues 1488–1729 from the C-terminus of the 6669 residue human nebulin. The site specificity of attachment of these antibodies is supported by the Western blots (Figure 1B).
Figure 2.
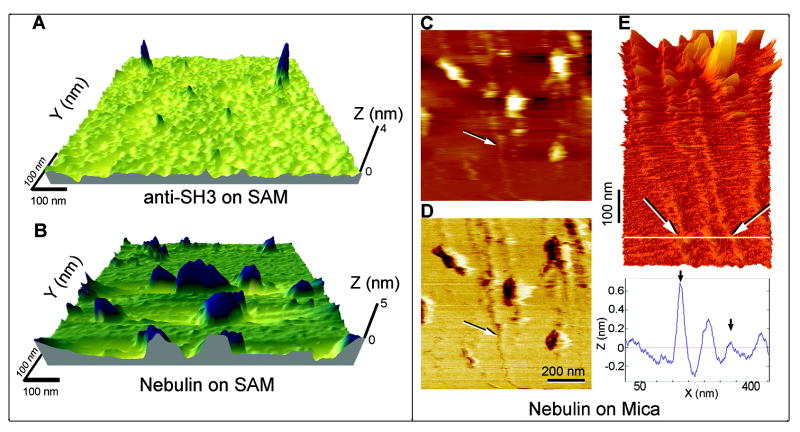
AFM images of single nebulin molecules on self-assembled monolayer and mica. (A) Sparsely populated anti-SH3 antibodies conjugated to the OEG-SAM. (B) Nebulin tethered onto anti-SH3 on the OEG-SAM. Average feature height of nebulin is 9.1 nm, with a monolayer surface roughness of 0.5 nm. Note that the Z dimension is greatly exaggerated relative to X and Y. (C–E) Nebulin on mica. Some of the nebulin molecules were stretched long and thin on the adhesive mica surface, without the antibody tethers. The arrows show filaments >650 nm in length and 3–7 Å in height. (C) The topographic image and (D) the phase image of the same field as in panel A show the heterogeneity of the flexible molecules. (E) Three-dimensional rendering of nebulin stretched on mica, with the height of the two protein strands at the white line (arrows) indicated on the height profile below.
Anti-SH3 antibodies (C-terminus) covalently attached onto the monolayer were visible via AFM imaging in noncontact (MAC) mode, 3–5 nm in height (Figure 2A). Full-length nebulin molecules tethered at its C-terminus by this antibody appeared as tumuli or mushrooms, 5–15 nm in height (Figure 2B). Since the separation of the mushrooms is much greater than the Stokes radius of 6 nm (unpublished data), the tethered polypeptides are not expected to intersect with one another on the surface.(60) In contrast, nebulin's filamentous nature was revealed when it was adsorbed on an adhesive mica surface without the antibody tether (Figures 2C–E). Thin strands up to 1 μm long were prevalent among compact mushrooms (5–15 nm in height). Filament height (0.3–0.7 nm) is comparable to a flattened single α-helix (0.8 nm) or a β-strand (0.4–1.1 nm). Thus, flexible, convoluted nebulin filaments may have been stretched by surface tension at the receding meniscus on the mica surface (molecular combing).(61)
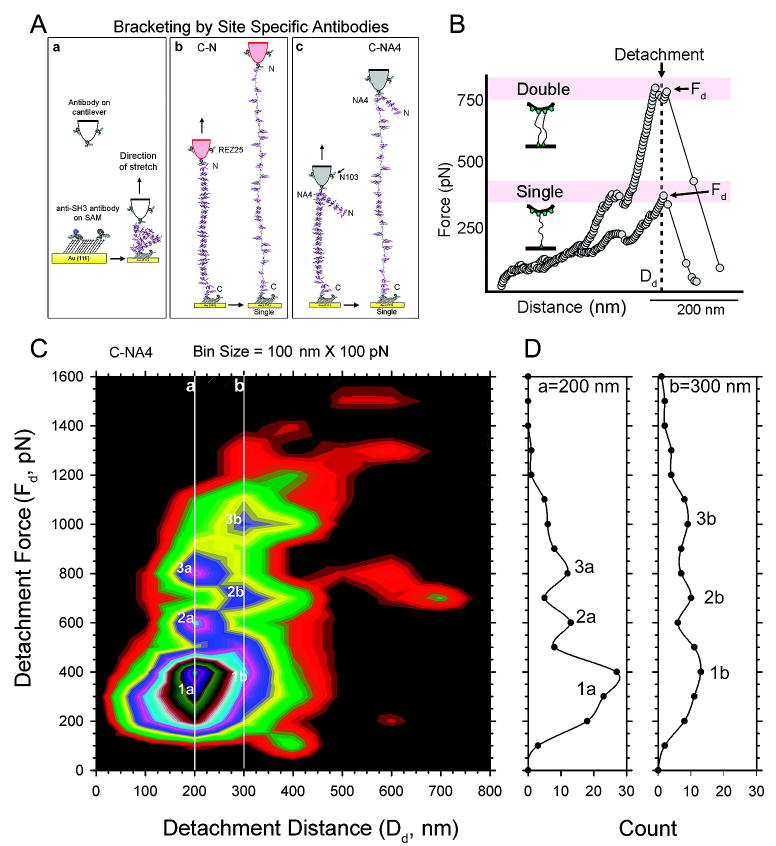
Stretching nebulin molecules tethered by anti-SH3 antibodies on the SAM (Figure 3A(a)) with the AFM tip conjugated with anti-N terminus (REZ25) (Figure 3A(b)) or anti-NA4 (N103) (Figure 3A(c)) antibodies gave rise to force-extension curves consisting of irregularly spaced peaks superimposed on a rapidly rising base. These craggy mountain range profiles (Figures 3B and 4) are distinct from the regularly spaced sawtooth patterns arising from the sequential unfolding of Ig or Fn3 domains of titin or polyprotein constructs.(1, 62) The analysis of force-extension curves first took into consideration the unbinding force of antibodies, its site-specificity, as well as the number of molecules being stretched.
Figure 3.
Stretching nebulin molecules between site-specific antibodies on a SAM. (A) Stretching single nebulin molecules tethered between two pairs of site-specific antibodies. Anti-SH3 antibodies (REZ16) are attached sparsely to a functionalized OEG-SAM on an ultraflat gold surface. Antibodies to either the N-terminus (REZ25) or the NA4 epitope (N103) are conjugated to a functionalized AFM cantilever (a). These antibody pairs thus tether and stretch nebulin molecules and measure the elasticity of two segments C-N (b) or C-NA4 (c) of the same protein. The nonbracketed region (N-NA4 in the C-NA4 bracket) is not stretched. (B) Representative mountain range force spectra of antibody tethered nebulin. The molecule detaches (arrows) when the detachment force, Fd, reaches the lowest unbinding force of the antibodies (300–400 pN). Stretching two molecules in parallel gives rise to a proportionally higher detachment force Fd. (C) Two-dimensional histogram of 500 detachment points. Detachment forces (Fd) with a bin size of 100 pN vs detachment distance (Dd) with a bin size of 100 nm. Note the peaks in the histogram are related to each other by the proportionally higher force values for both the 200 nm (line a) and 300 nm (line b) detachment distance. (D) Traces through Dd = 200 nm (line a) and Dd = 300 nm (line b) showing multiple peaks of Fd for these two Dd values.
Figure 4.
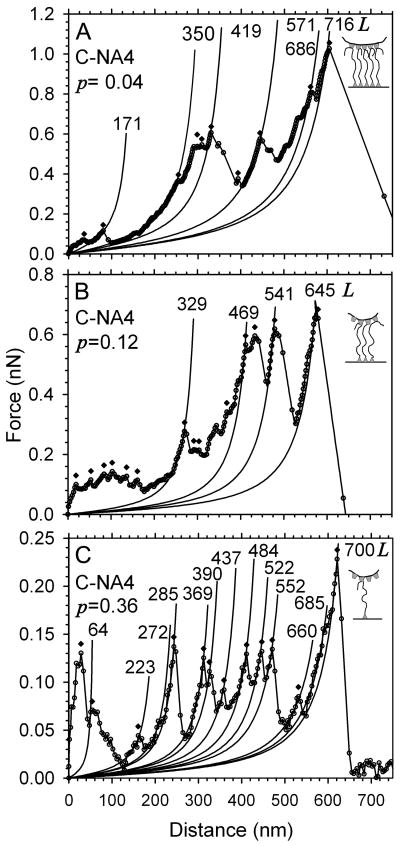
Force signature of stretched nebulin and force peak analysis. Examples of the mountain-range types of force spectra of C-NA4 bracketed nebulin with similar detachment lengths. The major peaks were subjected to WLC analysis to determine the persistence length and contour length for each. (A) Detachment force at 1000 pN and p ≈ 0.04 nm. (B) Detachment force at 660 pN and p ≈ 0.12 nm. (C) Detachment force at 220 pN and p ≈ 0.36 nm. Note that all major peaks have the same persistence length (p) but increasing contour length (L) by 75–200 nm as nebulin is stretched. Major peaks are identified () for periodicity analysis. The spacing between nearest peaks is Lpp. This set of curves thus arises from stretching 5, 3, and 1 nebulin molecule(s)
In our experimental design, stretched nebulin molecules would detach when the force at a given extension exceeded the unbinding forces between the antibodies and corresponding epitopes. Since the same fragment is bracketed and stretched for each antibody pair, we expect that when multiple molecules are stretched, identical segments will extended independently and in parallel, resulting in a proportionally higher detachment force at a given detachment distance (Figure 3B). We pursued this idea by examining the 2D histogram (Figure 3C) of a total of 488 curves for the C-NA4 bracket where the detachment occurred over a broad range of extensions (Dd) from 100 nm to 1 μm and forces (Fd) from 150 pN to 1 nN. The contour plot (Fd vs Dd) revealed a striking pattern that Fd's are vertically aligned at Dd at 200 nm (line a in Figure 3C) and 300 nm (line b in Figure 3C) and, moreover, are integral multiples of 300 pN and 350 pN. For example, one major and broad peak centered at 300 pN/200 nm (spot1a); four minor peaks at 600 pN/200 nm (spot 2a), 800 pN/200 nm (spot 3a), 1000 pN/300 nm (spot 3b), and 700 pN/300 nm (spot 2b) as well as a shoulder at 350 pN/300 nm (spot 1b). That these peaks are clustered around multiple Fd values at Dd of 200 nm (line a) and 300 nm (line b) is also clearly seen in the line profiles shown in Figure 3D.
This pattern indicates that the majority of the force curves resulted for stretching one, two, and three parallel nebulin molecules and that the C-NA4 bracket detached with an unbinding force that centered broadly at 300 pN and 350 pN. Similar analysis for the full-length nebulin C–N bracket was less successful and led to a tentative identification of single molecule events at 300pN/350 nm. Taken together, our data indicate that the unbinding force distribution for the anti-C is centered at 300 pN, anti-NA4 at 350 pN, and anti-N perhaps at 300 pN. On the basis of this analysis, the total single molecule events account for 38% of the total curves for the C-NA4 bracket. These unbinding forces for these polyclonal antinebulin antibodies are at the high end of the range reported for a single antigen–antibody pair from 50 to 300 pN.(5, 63-65) We cannot yet rule out that the unbinding force reflects the sum of two antibody binding sites per immunoglobulin G.
Closer examination of curves at the same detachment force and distance revealed significant variation in number, position, and magnitude of the intervening peaks (Figure 4). For a giant, modular protein such as nebulin, variability is expected and may reflect stochastic unfolding of the tandem repeats of similar but nonidentical nebulin modules.(46, 66) In studies of full-length titin (3 MDa), the force spectra also varied widely in the number and spacing of peaks.(1, 16, 20) Most nebulin force curves have a craggy mountain range profile and contained 6–20 major peaks each (Figure 4). Further analysis was done by first identifying all major peaks for all curves and then fitting the ascending limbs with a series of worm-like chain curves with small changes in the contour length (Figure 4). Analysis of the detachment force and distance, as well as the persistent length, of major peaks was facilitated by applying the following considerations: first, forces curves resulting from multiple nebulin molecules were expected to have proportionally higher detachment force for a give detachment distance (Figure 3B); second, if one or more tethers broke during the stretch, the persistent length of subsequent peaks would increase since theoretically the persistence length is inversely proportional to the number of remaining molecules, under identical conditions. The data in Figures 3B and 4 illustrate how these criteria are used to identify single versus multiple molecule curves.
The curves in Figure 4 have a similar detachment distance of 600 nm but a range of detachment forces from 1000 pN (Figure 4A), 650 pN (Figure 4B), and 220 pN (Figure 4C). Since all major peaks for each curve were fit with the same persistence length, there is no evidence of tether breakage during the stretch that increased the contour length due to domain unfolding. On the basis of the ratio of detachment force, there are likely to result from stretching five, three, and one nebulin molecules for Figures 4A–C, respectively. Moreover, this interpretation is also consistent with the increasing persistence length resulting from decreasing number of molecules. The changes in contour length (L) from the successive peaks also provide an intriguing clue on the structural basis of the major force generating events.
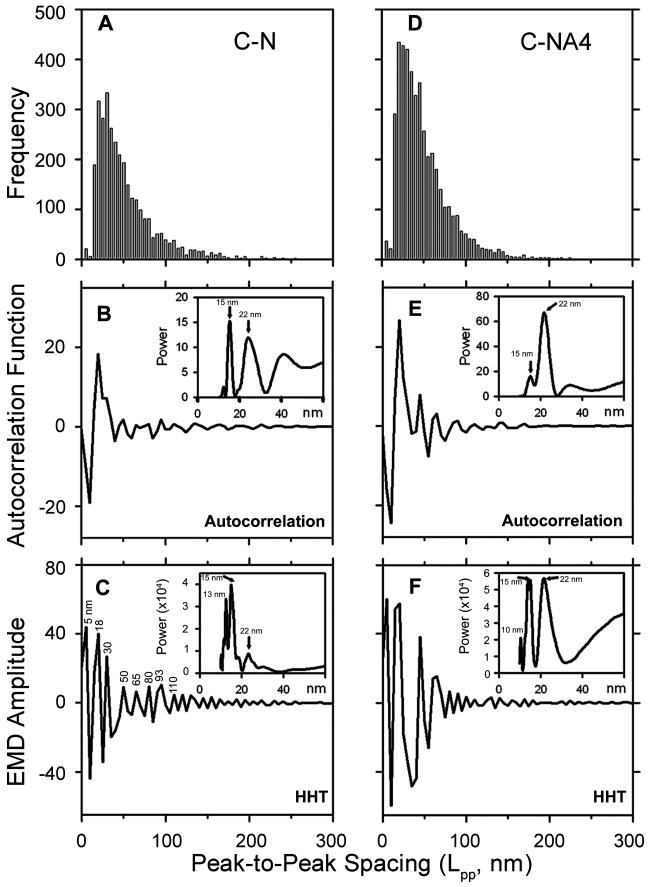
Besides the major peaks that are suitable for WLC analysis, there are also many small peaks or shoulders that are not as amendable. As a second approach to estimated changes in contour length for both major and minor peaks, we analyzed the peak to peak spacing of nearest neighbors, Lpp, from a total of 7000 peaks from both C–N and C-NA4 brackets curves using a pairwise distribution function (PDF).(54) The periodicity of the PDF (Figure 5A and D) was analyzed by two independent methods: autocorrelation spectra(55, 67, 68) (Figures 5 B and E) and the adaptive, empirical mode decomposition (EMD) method of the Hilbert–Huang Transform (HHT)(50) (Figures 5C and F). For the autocorrelation analysis, the Lpp distribution was first smoothed using a binomial smoothing filter(55) followed by the application of an autocorrelation function. Two major periodicities were seen in both brackets: a peak at 15 nm and a broad peak at 22 nm. Smaller amplitude peaks represent minor contributions to the period, as well as (sub)harmonics of the primary peaks. For the C–N bracket, the major peak was 15 nm, and the 22 nm peak was less than 50% in power. For the C-NA4, the two peaks were of similar amplitude. For the HHT analysis, the empirical mode decomposition of the distribution of Lpp (Figures 5A and D) yielded the set of intrinsic mode functions. One mode, IMF1, is shown in Figures 5C and F. (The complete set of IMFs is shown in Supporting Information, Figure S1). A Fourier Transform (FT) of the decomposed data resulted in power spectra that revealed periodicities in the histograms of Lpp (insets in Figures 5C and F). Two major periodicities were seen in both brackets: a doublet near 13/15 nm and one peak at 22 nm. Again, the relative powers of the 13/15 nm and 22 nm peaks were different for the two different brackets, albeit opposite of those determined using the autocorrelation method. It was noted that periodicity inherent in the full-length nebulin force curves was more clearly revealed by the HHT method. While similar periodicities were obtained from FT, the power of the peaks in the FT from the autocorrelation method (insets in Figures 5B and E) was 800- to 3000-fold lower than those in the FT from the IMF (insets in Figures 5C and F). This comparison demonstrates that the simpler, adaptive HHT method is a more powerful filtering/autocorrelation method for determining periodicities in histograms.
Figure 5.
Analysis of peak spacing by two methods. (A, D) The periodicity of force peak spacing Lpp was revealed by analysis of the pairwise distribution function (PDF) histograms binned at 5 nm. (B, E) Periodicity in the PDFs of Lpp for C-N (A) and C-NA4 (D) brackets was estimated by binomial smoothing and autocorrelation analysis. Insets show the power spectrum of the autocorrelation function obtained by Fourier transform. (C, F) Periodicities were analyzed by empirical mode decomposition(50) (EMD) of the PDFs followed by a Fourier transform to obtain the power spectra (insets). The dominant peaks in the power spectra were at 15 nm and 22 nm.
For proteins with multiple modules, force peak spacing is generally accountable by the extension of one or more modules that unfold between successive peaks. Single nebulin modules from many species range in size from 31 to 39 residues,(46, 66) corresponding to 11.8 to 14.8 nm of completely unfolded single modules, assuming a 0.38 nm span per residue.(69) Similarly, two modules, totaling 62–80 residues, would give 23.6 to 29.6 nm maximal extension. Since Lpp ranged from 13/15 and 22 nm (with 10–15% experimental error), the data suggest that one or more modules unfolded and extended roughly 70% to 100% of the maximal extension between the force peaks.
The extension stiffness of nebulin was estimated in two ways. First, as the instantaneous slope at a given extension of the worm-like chain (WLC)(53) curves (Figure 4). For these nonlinear curves, the values ranged from 0.1 to 40 pN/nm. A second estimate, the average extension stiffness, was obtained by simply taking the Fd to Dd ratio at detachment points of single molecule events: at 1.0–1.2 pN/nm. The low average stiffness suggests that the large 300 pN force required to stretch nebulin to 1 μm results from the very long extension of a compliant molecule. The large instantaneous stiffness, however, indicates that that many surges in resistance will be encountered and overcome along the stretch.
Discussion
For single-molecule force spectroscopy by AFM, the most challenging question remains: Is it really a single molecule measurement? Numerous technical innovations have been made in addressing this question. A common approach is to reduce the surface density of the tethered polymers on the substrate in order to enhance the hit rate for single molecule events. The use of a protein–resistant surface is effective in minimizing unwanted molecule–substrate interactions. To circumvent such technical obstacles, many investigators also adapted a force signature as an empirical indicator for nonartifactual, single molecule events.(31, 62) The signatures are typically determined by adsorbing dilute proteins onto a gold film, and the resulting force curves are selected that give peaks with contour lengths (as fitted by the WLC model) consistent with the size of the unfolded domains in the protein. Since this method is necessarily subjective and model-dependent, some groups have worked toward objective criteria for selecting force spectra.(31) This force signature method works well only for small tandem arrays of globular domains and not for giant complex proteins such as full-length titin and nebulin.
We therefore aimed to devise a single-molecule force spectroscopy platform that are suitable for giant elastic proteins. The current platform has four features: a SAM surface to eliminate or reduce protein adsorption; a low surface coverage to enhance single-molecule pickup; the use of antibodies for protein tethering at well-defined locations; and the bracketing of protein lengths to stretch various segments from the same target protein. We have used an oligo-ethyleneglycol SAM, and given the lack of binding of these protein components to the SAM surface (as determined by AFM imaging), it is likely that adsorption/desorption artifacts are negligible.(32, 70) Furthermore, force spectra taken in the absence of nebulin are featureless within the noise level of the instrument (Supporting Information, Figure S2), indicating no antibody–SAM or antibody–antibody adhesion. Site-specific attachment of the protein to substrate can generally be done through the covalent links via engineered thiols(70, 71) or noncovalently via antibodies.(30, 61) While engineering thiols to both termini works well for a small protein such as calmodulin,(71) it is impractical for giant proteins that contain numerous intrinsic thiols. Antibody bracketing technique, however, allows different fragments to be stretched from the same full length protein simply by preparing tenacious, high affinity antipeptide antibodies, thus alleviating the engineering and expression of each and every fragment with appropriate solubility and force tags.(72) Moreover, the unbinding force of the antibody/epitope pair acts as a mechanical delimiter above which the molecules become untethered without breakage of covalent bonds. As a result, single versus multiple molecule events can be identified by examining the detachment force, detachment distance, and the relative persistence length of major force peaks, as described above.
Detailed analysis of the complex craggy mountain range type of force spectra (Figures 3B and 4) by WLC models as well as the delineation of inherent periodicities by autocorrelation and an adaptive data analysis algorithm (HHT) led us to conclude that the stochastic unfolding of single nebulin modules are the fundamental structural events that generate the numerous minor peaks in the force curves of nebulin.
The mean persistence length of a single nebulin molecule at 0.36 nm is comparable to that of elastic titin PEVK at 0.3 nm(73) but shorter than those of unfolded globular proteins (e.g., titin immunoglobulin) at low (0.8 nm) and high (0.4 nm) forces.(37) This trend indicates that the unfolded nebulin and unwrapped titin PEVK are stiffer springs than the unfolded titin globular domains, as a longer persistence length takes less force to stretch. Additionally, the WLC formula is an interpolation that overestimates the fitted persistence lengths.(74, 75) However, the absolute values of persistence length are devoid of any molecular information since the WLC model describes statistical polymer configurations of a structureless, homogeneous material under the assumption that the chain is purely entropic (inextensible, no self-interaction, and no excluded volume effects, i.e., chain can cross itself).(76, 77) A recent experimental and theoretical study of the hydrophobic collapse of proteins has called the purely entropic model of protein elasticity into question.(78)
The average stiffness of single nebulin at 1 pN/nm indicates that single nebulin is compliant, with only 3–5% of the stiffness of the F-actin filament at 20–34 pN/nm.(79) Moreover, its low stiffness indicates that the large force 300 pN required to stretch nebulin to 1 μm length results from its unusually long extension. This stiffness value for the single molecule is higher than that reported for nebulin (or nebulin-containing) filaments in the sarcomere.(80) Yasuda et al.(80) measured the extension stiffness of engineered skeletal myofibrils in which the middle segments of the actin filaments in the I-band region had been selectively removed by gelsolin, thus freeing and exposing nebulin between the A- and Z-bands. On the assumption that titin does not contribute elasticity under the experimental conditions, the resulting extension stiffness of 0.01 to 0.1 pN/nm (as static elastic modulus) was attributed to nebulin alone.(79) Since it was unclear in that study whether nebulin was still associated with titin or other binding proteins that conceivably could affect the mechanical property of nebulin, further comparison with pure nebulin stiffness is premature. It is interesting that Yasuda et al.(80) reported that the myofibrils shortened when the associated actin was depolymerized, reflecting significant restoring force from nebulin.
A comparison of force signatures of proteins with distinct conformations provides useful clues on the conformational basis of nebulin elasticity. AFM-mediated stretching of titin's immunoglobulin and FN3 β-barrel domains,(1) spectrin's triple helical domains,(15) ankyrin's antiparallel α-helical spiral,(14) the myosin coiled-coil domain,(11, 12) bacteriorhodopsin α-helical loops,(13) and titin PEVK polyproline II helices(73, 81) revealed a progressive loss of the regular sawtooth patterns of titin and spectrin triple helices toward more irregular peaks (number, height, and spacing) as the structures change from β-barrels to various α-helical structures, to disordered coiled-coils, and to the smooth curves of intrinsically disordered titin PEVK. The force signature of nebulin resembles those of the disordered myosin α-helical coiled-coils and the ankyrin α-helical repeats with their irregularly spaced spikes. The structural basis of the force generating events in nebulin is being investigated in this lab.
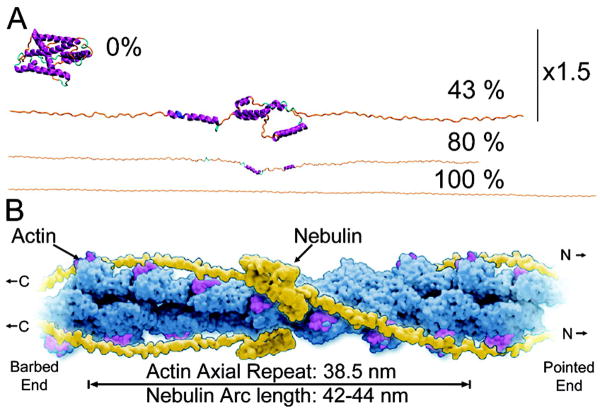
On the basis of sequence periodicity, immunolocalization and protein interaction evidence, we currently envision that in the sarcomere, two nebulin strands per thin filament wind around the outer edge of the actin helix at three potential actin sites (at subdomains 1, 2/3, and 1/3).(82) The elasticity of nebulin dictates that such intimate association occurs only after the nebulin molecules are mechanically loaded by at least 300 pN to elongate and to scaffold the 1 μm actin filaments. This loaded nebulin concept is illustrated in Figure 6 where a stretched seven-module nebulin super-repeat simulated by molecular dynamics (unpublished data) has been morphed onto an actin helix, with each super-repeat spanning one axial actin repeat of 38 nm with an arc length of 42–44 nm around actin. The exact conformation is currently unknown, and some portions of a super-repeat may be more extended than others. This model is therefore distinct from that of Phuhl et al.,(83) where a pair of unloaded, continuous α-helices fit snugly in the two central grooves of the actin filament.
Figure 6.
A stretch-to-match ruler hypothesis. (A) Structural simulation of stretching one nebulin superrepeat (modeled by ROSETTA) to 0, 43, 80, 100%. Note that the 0 and 43% extension models are enlarged by 50% for clarity. The 80% extension may be representative of the remaining structure at the detachment points in the current experiments (our unpublished data). (B) Full length nebulin associates along and around actin filaments in the sarcomere only when stretched to >1 μm, with the concomitant optimization of matching periodicity to helical parameters of actin. Each super-repeat spans one arc length around actin, with a single stretched full-length nebulin spanning and associating 26 actin axial repeats along the 1 μm long thin filament. Nebulin may exert significant compression on actin both longitudinally and radially. The exact structure of nebulin on the thin filament is unknown at present.
Since each myosin head generates 2–5 pN of force, it would take the collective effort of up to 100–200 myosin heads (out of the 300 myosin per thick filament) to stretch and prepare nebulin for incorporation into the 1 μm long thin filaments. This force requirement for thin filament assembly has interesting implications. First, the incorporation of nebulin into myofibrils during muscle development occurs only after the separate A-bands and the IZI bands are linked together and aligned.(84) We believe that significant force in the right orientation to stretch nebulin from the correct anchorage at the Z-band is possible only at this late stage of myofibril assembly. Second, since nebulin tethers myosin heads to actin and reversibly inhibits actoS1 ATPase in solution,(44) the possible contribution of an elastic and reversible nebulin tether to the active contraction between actin and myosin motors is implicated. Third, mechanically, the nebulin-based compression force would preload and stiffen the composite thin filament against lateral impact, resist stretching, and promote healing of severed actin filaments. Nebulin thus could endow thin filaments with mechanical attributes to manage stress and strain. This structural engineering feature may have evolved to meet the special needs of skeletal muscles that perform long-range shortening at high velocity. The absence of nebulin and its stabilizing influence in cardiac muscles distinguishes cardiac thin filaments both structurally and mechanically, and perhaps functionally.
In summary, we have developed a new protein nanomechanical platform that alleviates many of the problems associated with depending upon physisorption of the protein under study to the surface and/or the tip. This platform is generally applicable to protein force spectroscopy, especially to giant elastic proteins and biopolymers. An additional component of this platform is the use of adaptive data analysis methods that are ideally suited for force spectroscopy. On the basis of these advanced single molecule force techniques, we propose that nebulin may act as an elastic strain gauge that interacts optimally with actin and myosin only under appropriate stress and strain. Nebulin may also enhance the mechanical resistance of actin filaments and provide a restoring force and structural continuity.
Supplementary Material
Acknowledgments
We thank Gustavo Gutierrez-Cruz for technical assistance in preparing nebulin and Dr. Norden Huang for his insights in adaptive data analysis. We thank the reviewers of this manuscript for their insightful comments. This work was supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, and Department of Health and Human Services.
References
- 1.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 2.Kageshima M, Lantz MA, Jarvis SP, Tokumoto H, Takeda S, Ptak A, Nakamura C, Miyake J. Chem Phys Lett. 2001;343:77–82. doi: 10.1248/cpb.49.1512. [DOI] [PubMed] [Google Scholar]
- 3.Forbes JG, Lorimer GH. Science. 2000;288:63–64. doi: 10.1126/science.288.5463.63. [DOI] [PubMed] [Google Scholar]
- 4.Weisel JW, Shuman H, Litvinov RI. Curr Opin Struct Biol. 2003;19 doi: 10.1016/s0959-440x(03)00039-3. [DOI] [PubMed] [Google Scholar]
- 5.Hinterdorfer P, Baumgartner W, Gruber HJ, Schilcher K, Schindler H. Proc Natl Acad Sci USA. 1996;93:3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binnig G, Quate CF, Gerber C. Phys Rev Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 7.Stroh C, Wang H, Bash R, Ashcroft B, Nelson J, Gruber H, Lohr D, Lindsay SM, Hinterdorfer P. Proc Natl Acad Sci USA. 2004;101:12503–12507. doi: 10.1073/pnas.0403538101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansma H, Kim K, Laney D, Garcia R, Argaman M, Allen M, Parsons S. J Struct Biol. 1997;119:99–108. doi: 10.1006/jsbi.1997.3855. [DOI] [PubMed] [Google Scholar]
- 9.Frederix P, Akiyama T, Staufer U, Gerber C, Fotiadis D, Muller D, Engel A. Curr Opin Chem Biol. 2003;7:641–647. doi: 10.1016/j.cbpa.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Hinterdorfer P, Dufrene YF. Nature Methods. 2006;3:347–355. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 11.Root DD, Yadavalli VK, Forbes JG, Wang K. Biophys J. 2006;90:2852–2866. doi: 10.1529/biophysj.105.071597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwaiger I, Sattler C, Hostetter DR, Rief M. Nat Mater. 2002;1:232–235. doi: 10.1038/nmat776. [DOI] [PubMed] [Google Scholar]
- 13.Janovjak H, Struckmeier J, Hubain M, Kedrov A, Kessler M, Muller DJ. Structure. 2004;12:871–879. doi: 10.1016/j.str.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 14.Lee G, Abdi K, Jiang Y, Michaely P, Bennett V, Marszalek PE. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 15.Rief M, Pascual J, Saraste M, Gaub HE. J Mol Biol. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [DOI] [PubMed] [Google Scholar]
- 16.Wang K, Forbes JG, Jin AJ. Prog Biophys Mol Biol. 2001;77:1–44. doi: 10.1016/s0079-6107(01)00009-8. [DOI] [PubMed] [Google Scholar]
- 17.Carrion-Vazquez M, Li HB, Lu H, Marszalek PE, Oberhauser AF, Fernandez JM. Nat Struct Biol. 2003;10:738–743. doi: 10.1038/nsb965. [DOI] [PubMed] [Google Scholar]
- 18.Grandbois M, Beyer M, Rief M, Clausen-Schaumann H, Gaub HE. Science. 1999;283:1727–1730. doi: 10.1126/science.283.5408.1727. [DOI] [PubMed] [Google Scholar]
- 19.Rounsevell R, Forman JR, Clark J. Methods. 2004;34:100–111. doi: 10.1016/j.ymeth.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Forbes JG, Wang K. J Vac Sci Technol, A. 2004;22:1–5. [Google Scholar]
- 21.Borgia A, Williams PM, Clarke J. Annu Rev Biochem. 2008;77:101–125. doi: 10.1146/annurev.biochem.77.060706.093102. [DOI] [PubMed] [Google Scholar]
- 22.Muller DJ, Dufrene YF. Nat Nanotechnol. 2008;3:261–269. doi: 10.1038/nnano.2008.100. [DOI] [PubMed] [Google Scholar]
- 23.Sharma D, Perisic O, Peng Q, Cao Y, Lam C, Lu H, Li HB. Proc Natl Acad Sci USA. 2007;104:927–9283. doi: 10.1073/pnas.0700351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng Q, Zhuang SL, Wang MJ, Cao Y, Khor YA, Li HB. J Mol Biol. 2009;386:1327–1342. doi: 10.1016/j.jmb.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 25.Allen S, Davies J, Davies MC, Dawkes AC, Roberts CJ, Tendler SJB, Williams PM. Biochem J. 1999;341:173–178. [PMC free article] [PubMed] [Google Scholar]
- 26.Hugel T, Seitz M. Macromol Rapid Commun. 2001;22:989–1016. [Google Scholar]
- 27.Florin EL, Moy VT, Gaub HE. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 28.Leake MC, Wilson D, Gautel M, Simmons RM. Biophys J. 2004;87:1112–1135. doi: 10.1529/biophysj.103.033571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tskhovrebova L, Trinick J, Sleep JA, Simmons RM. Nature. 1997;387:308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 30.Kellermayer MSZ, Smith SB, Granzier HL, Bustamante C. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 31.Best RB, Brockwell DJ, Toca-Herrera JL, Blake AW, Smith DA, Radford SE, Clarke J. Anal Chim Acta. 2003;479:87–105. [Google Scholar]
- 32.Yadavalli VK, Forbes JG, Wang K. Langmuir. 2006;22:6969–6976. doi: 10.1021/la060320h. [DOI] [PubMed] [Google Scholar]
- 33.Butt HJ, Cappella B, Kappl M. Surf Sci Rep. 2005;59:1–152. [Google Scholar]
- 34.Rief M, Oesterhelt F, Heymann B, Gaub HE. Science. 1997;275:1295–1297. doi: 10.1126/science.275.5304.1295. [DOI] [PubMed] [Google Scholar]
- 35.Smith SB, Finzi L, Bustamante C. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 36.Carrion-Vazquez M, Oberhauser AF, Fowler SB, Marszalek PE, Broedel SE, Clarke J, Fernandez JM. Proc Natl Acad Sci USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rief M, Gautel M, Schemmel A, Gaub HE. Biophys J. 1998;75:3008–3014. doi: 10.1016/S0006-3495(98)77741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higgins MJ, Sader JE, Jarvis SP. Biophys J. 2006;90:640–647. doi: 10.1529/biophysj.105.066571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McElhinny AS, Kazmierski ST, Labeit S, Gregorio CC. Trends Cardiovasc Med. 2003;13:195–201. doi: 10.1016/s1050-1738(03)00076-8. [DOI] [PubMed] [Google Scholar]
- 40.Wallgren-Pettersson C, Pelin K, Nowak KJ, Muntoni F, Romero NB, Goebel HH, North KN, Beggs AH, Laing NG. Neuromusc Disord. 2004;14:461–470. doi: 10.1016/j.nmd.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 41.Kruger M, Wright J, Wang K. J Cell Biol. 1991;115:97–107. doi: 10.1083/jcb.115.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Labeit S, Gibson T, Lakey A, Leonard K, Zeviani M, Knight P, Wardale J, Trinick J. FEBS Lett. 1991;282:313–316. doi: 10.1016/0014-5793(91)80503-u. [DOI] [PubMed] [Google Scholar]
- 43.Root DD, Wang K. Biochemistry. 1994;33:12581–12591. doi: 10.1021/bi00208a008. [DOI] [PubMed] [Google Scholar]
- 44.Root DD, Wang K. Biochemistry. 2001;40:1171–1186. doi: 10.1021/bi0015010. [DOI] [PubMed] [Google Scholar]
- 45.Labeit S, Kolmerer B. J Mol Biol. 1995;248:308–315. doi: 10.1016/s0022-2836(95)80052-2. [DOI] [PubMed] [Google Scholar]
- 46.Wang K, Knipfer M, Huang QQ, van Heerden A, Hsu LC, Gutierrez G, Quian XL, Stedman H. J Biol Chem. 1996;271:4304–4314. doi: 10.1074/jbc.271.8.4304. [DOI] [PubMed] [Google Scholar]
- 47.Fowler VM, McKeown CR, Fischer RS. Curr Biol. 2006;16:R18–R20. doi: 10.1016/j.cub.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 48.Littlefield R, Fowler VM. Annu Rev Cell Dev Biol. 1998;14:487–525. doi: 10.1146/annurev.cellbio.14.1.487. [DOI] [PubMed] [Google Scholar]
- 49.Huang NE, Shen SSP. Hilbert Huang Transform and Its Applications. Vol. 5 World Scientific Publishing; River Edge, NJ: 2005. [Google Scholar]
- 50.Huang NE, Shen Z, Long SR, Wu MLC, Shih HH, Zheng QN, Yen NC, Tung CC, Liu HH. Proc R Soc London, Ser A. 1998;454:903–995. [Google Scholar]
- 51.Wu Z, Huang NE. Adv Adaptive Data Anal. 2009;1:1–41. [Google Scholar]
- 52.Hutter JL, Bechhoefer J. Rev Sci Instrum. 1993;64:1868–1873. [Google Scholar]
- 53.Bustamante C, Marko JF, Siggia ED, Smith S. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 54.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 55.Marchand P, Marmet L. Rev Sci Instrum. 1983;54:1034–1041. [Google Scholar]
- 56.Scott DW. Biometrika. 1979;66:605–610. [Google Scholar]
- 57.Flandrin P. http://perso.ens-lyon.fr/patrick.flandrin/emd.html.
- 58.Huang NE. http://rcada.ncu.edu.tw/research1.htm.
- 59.Wright J, Huang QQ, Wang K. J Muscle Res Cell Motil. 1993;14:476–483. doi: 10.1007/BF00297210. [DOI] [PubMed] [Google Scholar]
- 60.deGennes PG. Adv Colloid Interface Sci. 1987;27:189–209. [Google Scholar]
- 61.Tskhovrebova L, Trinick J, Sleep JA, Simmons RM. Nature. 1997;387:308–312. doi: 10.1038/387308a0. [DOI] [PubMed] [Google Scholar]
- 62.Carrion-Vazquez M, Oberhauser AF, Fisher TE, Marszalek PE, Li H, Fernandez JM. Prog Biophys Mol Biol. 2000;74:63–91. doi: 10.1016/s0079-6107(00)00017-1. [DOI] [PubMed] [Google Scholar]
- 63.Dammer U, Hegner M, Anselmetti D, Wagner P, Dreier M, Huber W, Guntherodt HJ. Biophys J. 1996;70:2437–2441. doi: 10.1016/S0006-3495(96)79814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao T, Wang AF, Liang XM, Tang HY, Auner GW, Salley SO, Ng KYS. Biotechnol Bioeng. 2007;98:1109–1122. doi: 10.1002/bit.21503. [DOI] [PubMed] [Google Scholar]
- 65.Berquand A, Xia N, Castner DG, Clare BH, Abbott NL, Dupres V, Adriaensen Y, Dufrene YF. Langmuir. 2005;21:5517–5523. doi: 10.1021/la050162e. [DOI] [PubMed] [Google Scholar]
- 66.Pfuhl M, Winder SJ, Morelli MAC, Labeit S, Pastore A. J Mol Biol. 1996;257:367–384. doi: 10.1006/jmbi.1996.0169. [DOI] [PubMed] [Google Scholar]
- 67.Florin EL, Moy VT, Gaub HE. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 68.Snyder PW, Lee G, Marszalek PE, Clark RL, Toone EJ. Proc Natl Acad Sci USA. 2007;104:2579–2584. doi: 10.1073/pnas.0606604104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Creighton TE. Proteins: Structures and Molecular Properties. 2nd. W. H. Freeman and Company; New York: 1992. [Google Scholar]
- 70.Ortiz C, Hadziioannou G. Macromolecules. 1999;32:780–787. [Google Scholar]
- 71.Hertadi R, Ikai A. Protein Sci. 2002;11:1532–1538. doi: 10.1110/ps.3600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rounsevell RW, Steward A, Clarke J. Biophys J. 2005;88:2022–2029. doi: 10.1529/biophysj.104.053744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Forbes JG, Jin AJ, Ma K, Gutierrez-Cruz G, Tsai WL, Wang K. J Muscle Res Cell Motil. 2005;26:291–301. doi: 10.1007/s10974-005-9035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bouchiat C, Wang MD, Allemand JF, Strick T, Block SM, Croquette V. Biophys J. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ogden RW, Saccomandi G, Sgura I. Comput Math Appl. 2007;53:276–286. [Google Scholar]
- 76.Kratky O, Porod G. Recl Trav Chim Pay B. 1949;68:1106–1122. [Google Scholar]
- 77.Yamakawa H. Helical Wormlike Chains in Polymer Solutions. Springer-Verlag; Berlin: 1997. [Google Scholar]
- 78.Walther KA, Grater F, Dougan L, Badilla CL, Berne BJ, Fernandez JM. Proc Natl Acad Sci USA. 2007;104:7916–7921. doi: 10.1073/pnas.0702179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu XM, Pollack GH. Biophys J. 2002;83:2705–2715. doi: 10.1016/S0006-3495(02)75280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yasuda K, Anazawa T, Ishiwata S. Biophys J. 1995;68:598–608. doi: 10.1016/S0006-3495(95)80221-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Watanabe K, Nair P, Labeit D, Kellermayer M, Greaser M, Labeit S, Granzier H. Biophys J. 2003;84:244A–244A. [Google Scholar]
- 82.Lukoyanova N, VanLoock MS, Orlova A, Galkin VE, Wang K, Egelman EH. Curr Biol. 2002;12:383–388. doi: 10.1016/s0960-9822(02)00678-4. [DOI] [PubMed] [Google Scholar]
- 83.Pfuhl M, Winder SJ, Pastore A. EMBO J. 1994;13:1782–1789. doi: 10.1002/j.1460-2075.1994.tb06446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moncman CL, Wang K. Cell Motil Cytoskel. 1996;34:167–184. doi: 10.1002/(SICI)1097-0169(1996)34:3<167::AID-CM1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.