Abstract
Tetrahymena ribozymes hold promise for repairing genetic disorders but are largely limited by their modest splicing efficiency and low production of final therapeutic proteins. Ribozyme evolution in intact living mammalian cells would greatly facilitate the discovery of new ribozyme variants with high in vivo activity, but no such strategies have been reported. Here we present a study using a new reporter enzyme, β-lactamase, to report splicing activity in single living cells and perform high-throughput screening with flow cytometry. The reporter ribozyme constructs consist of the self-splicing Tetrahymena thermophila group I intron ribozyme that is inserted into the ORF of the mRNA of β-lactamase. The splicing activity in single living cells can be readily detected quantitatively and visualized. Individual cells have shown considerable heterogeneity in ribozyme activity. Screening of Tetrahymena ribozymes with insertions in the middle of the L1 loop led to identification of better variants with at least 4-fold more final in vivo activity than the native sequence. Our work has provided a new reporter system that allows high-throughput screening with flow cytometry of single living mammalian cells for a direct and facile in vivo selection of desired ribozyme variants.
Variants of the Tetrahymena ribozyme have been shown to perform self-splicing and trans-splicing in mammalian cells, but the modest splicing efficiency of these ribozymes and low production of final proteins limit their ability to repair genetic disorders (1–5). Directed in vitro evolution of the Tetrahymena ribozyme has identified mutants with increased thermal stability (6) and efficacy in Escherichia coli (7), but ribozyme evolution in intact living mammalian cells has not yet been demonstrated. Such evolution would greatly facilitate the discovery of new active ribozyme variants that can work efficiently in vivo, especially in light of recent work showing that products of group I intron self-splicing are not translated efficiently in mammalian cells (1). Splicing-dependent reporter gene assays have been exploited to report ribozyme activity, including firefly luciferase and β-galactosidase (1, 8, 9), but they either lack single-cell resolution or require cell permeabilization, which greatly reduces survival rates. We now show that a newer reporter enzyme, β-lactamase (Bla), allows visualization of splicing activity in single living cells and high-throughput screening with flow cytometry. This improved readout reveals that insertions in the L1 loop of the ribozyme can cause at least 4-fold increases in the level of desired protein.
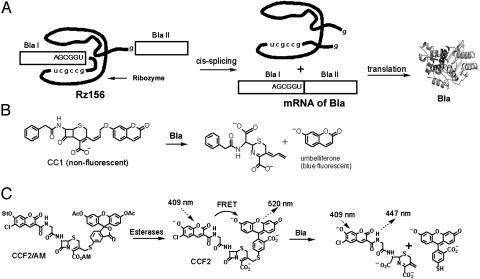
The 29-kDa TEM-1 isoform of Bla has become a popular “reporter” or “sensor” of biological processes and interactions, such as the activity of promoter/regulatory elements in living mammalian tissue culture cells (10, 11), constitutive and inducible protein associations (12–14), and HIV-1 virion fusion in primary T lymphocytes (15). Bla is relatively small, monomeric, and naturally absent from eukaryotes, but it is easily expressible in eukaryotic cells without noticeable toxicity. We have developed fluorogenic substrates CC1 and CCF2/AM, which enable nondestructive high-throughput screening of Bla levels in single living mammalian cells by increases in fluorescence intensity (16) and the ratio of blue-to-green emissions (10), respectively (Fig. 1).
Fig. 1.
Splicing-dependent Bla reporter gene in vitro and in vivo assays. (A) Schematic presentation of splicing-dependent Bla reporter gene strategy. The ribozyme reporter Rz156 consists of the Tetrahymena intron and a broken Bla ORF (BlaI and BlaII). Ribozyme self-splicing produces uninterrupted mRNA of Bla, which is translated into the reporter enzyme Bla. (B) Hydrolysis of nonfluorescent CC1 by Bla generates a blue fluorescent product that emits at 465 nm when excited at 360 nm. (C) The in vivo assay uses CCF2/AM (adapted from ref. 10). Membrane-permeable CCF2/AM is converted to CCF2 by intracellular esterases. When no Bla is present, CCF2 fluoresces green (at 520 nm) because of fluorescence resonance energy transfer (FRET) from the coumarin donor to the fluorescein acceptor. Bla hydrolysis splits off the fluorescein, disrupts FRET, and shifts the emission to blue (at 447 nm).
We now report constructs in which the self-splicing Tetrahymena thermophila group I intron ribozyme is inserted into the ORF of the mRNA of TEM-1 Bla (Fig. 1). The splicing activity can be readily detected in cell lysates quantitatively and sensitively by using CC1 and visualized in single living cells by using CCF2/AM. This reporter system was applied to screen variants of the Tetrahymena ribozyme for improved efficiency and led to the identification of variants with at least 4-fold more final activity than the native sequence. The compatibility of this system with high-throughput screening with flow cytometry should permit a direct and facile selection of desired ribozyme variants in mammalian cells.
Materials and Methods
Plasmid Construction of Ribozyme Constructs. All mammalian expression vectors were constructed by standard methods (17) and by PCR mutagenesis methods (18). PCR amplification was done by high-fidelity Pfu Turbo DNA polymerase (Stratagene) to avoid undesired mutations. The gene encoding Bla without a secretory signal was amplified from pUC19 (New England Biolabs). This amplified fragment was inserted in pDsRed2-N1 (Clontech) after removal of DsRed cDNA to make a cytomegalovirus (CMV) promoter-driven Bla expression vector, pCMV-Bla. Ribozyme sequence was derived from pTT1A3-T7 (a kind gift from Dr. Thomas Cech, University of Colorado, Boulder). PCR products of ribozyme were ligated at different positions within the Bla gene in pCMV-Bla. Addition and deletion of oligonucleotides were done by methods described in ref. 18. Bla and ribozyme sequences in all constructs were confirmed by DNA sequencing.
RT-PCR. A monkey kidney cell line, COS-1, was seeded in a six-well dish at a density of 4.0 × 105 per well 18–24 h before transfection with 4 μg of expression vector per well by Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, total RNAs were extracted by TRIzol (Invitrogen) according to the manufacturer's instructions. Before RNA extraction, EDTA was added to TRIzol at a final concentration of 45 mM to inhibit ribozyme activity. One microgram of total RNAs was treated by DNaseI (Invitrogen). After DNase treatment, 100 ng of total RNA was applied to reverse transcription (RT) by random primer (SuperScript First-Strand Synthesis System for RT-PCR, Invitrogen) in the presence of 50 mM l-argininamide to quench self-splicing during the RT reaction. The resulting cDNAs were amplified for 35 cycles by RT-1 forward primer (CAGAAACGCTGGTGAAAG) and RT-2 backward primer (CGTCAATACGGGATAATACC). PCR products were analyzed by electrophoresis in 2% agarose gel. The identities of the spliced products were confirmed by sequence analysis.
In Vitro Bla Assay by CC1. COS-1 cells were seeded in 12-well dishes at a density of 1.6 × 105 per well for 18–24 h before transfection with 1.6 μg of expression vector per well by Lipofectamine 2000. Forty-eight hours after transfection, cells were collected by centrifugation at room temperature at 2,600 × g for 1 min. After the cells were washed in 800 μl of PBS (GIBCO/BRL) twice, cell pellets were resuspended in 150 μl of 0.1 M phosphate buffer (pH 7.0). Cell lysates were prepared by three freezing and thawing cycles (freezing in dry ice/isopropanol for 10 min and thawing in 37°C water bath for 10 min). The supernatant was recovered by centrifugation at 16,000 × g for 2 min at 4°C. Assays were conducted in 96-well microtiter plates (Corning). To measure Bla activity, 45 μl of cell lysate and 5 μl of 1 mM CC1 were mixed in each well. Fluorescence was measured with a 360-nm excitation filter (42-nm bandwidth; Chroma Technology, Brattleboro, VT) and a 465-nm emission filter (35-nm bandwidth; Chroma Technology) at each time point in a GENios microplate reader (TECAN, Research Triangle Park, NC). Fluorescence data were normalized against total lysate protein contents determined by a Bradford assay (Bio-Rad).
Fluorescence Microscopy Imaging. COS-1 cells were split in a 12-well dish at a density of 1.6 × 105 per well 18–24 h before transfection. A total of 0.8 μg of expression vector per well was cotransfected with 0.8 μg of pDsRed2-N1 by Lipofectamine 2000. Twenty-four hours after transfection, cells were reseeded onto a 35-mm glass-bottom culture dish (MatTek, Ashland, MA). Forty-eight hours after reseeding, cells were washed twice with Hanks' balanced salt solution (HBSS, Sigma) and loaded for 1 h at room temperature with serum-free DMEM (GIBCO/BRL) containing 25 mM Hepes (Sigma), 2 μM CCF2/AM (Aurora Biosciences, San Diego), and 2.5 mM probenecid (Sigma). After a 1-h loading of CCF2/AM, cells were washed twice with HBSS and observed immediately under an Axiovert 200M fluorescence microscope (Zeiss) or incubated with standard medium containing 2.5 mM probenecid for 6 h at room temperature before fluorescence observation. The following filter set (Chroma Technology) was used for microscopic analysis of CCF2/AM: HQ405/20 for excitation, 425DCXR as the dichroic mirror, HQ460/40 for blue emission, and HQ530/30 for green emission. For the acquisition of DsRed images, the following filter set was used (Chroma Technology): excitation, HQ546/12; dichroic mirror, Q560LP; emission, HQ605/75. Images were analyzed with metamorph and metaf luor software (Universal Imaging, Downingtown, PA).
Flow Cytometry. Flow cytometry was conducted by using a two-laser FACS-DiVa system (Becton Dickinson). DsRed was measured with 150-mW excitation at 568 nm, with 610-nm short pass and 619- to 641-nm emission bandpass filters. CCF2 cleavage by Bla was assayed by exciting with 50 mW at 413 nm and monitoring emissions with 440- to 460-nm and 500- to 520-nm bandpass filters, with a 505-nm dichroic mirror to separate the two emissions. The healthy cells (≈95% of the population) were defined by forward and side scatter, then further restricted to those positive for the DsRed cotransfection marker. This subpopulation, typically ≈65% of the healthy cells, was analyzed by dividing the blue emission of cleaved CCF2 at 450 nm by the green emission of uncleaved CCF2 at 510 nm for each of ≈30,000 healthy DsRed-positive cells and plotting the frequency of occurrence vs. ratio. The flow cytometry was done on coded samples whose identities were not revealed until analysis was completed.
Results
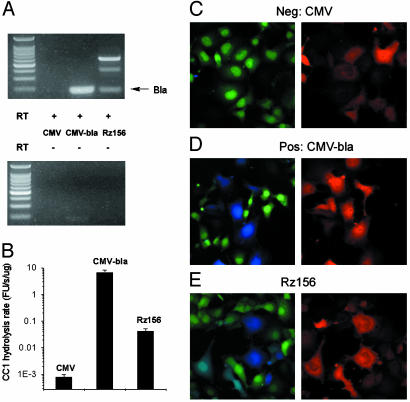
Our initial ribozyme reporters consisted of the self-splicing T. thermophila group I intron ribozyme with its original 5′ exon deleted, and the Bla ORF broken by the inserted intron into BlaI and BlaII (Fig. 1 A). The coding sequence of the first 23-aa secretory signal peptide of Bla was removed, so the expressed Bla resides in the cytosol. Several positions of the Bla ORF were chosen for the ribozyme insertion. For the self-splicing to proceed, the last nucleotide preceding the insertion site (which is also the splice site of the ribozyme) has to be uridine, and the last six nucleotides of the 5′ exon (which are the last six nucleotides of BlaI preceding the insertion site) must pair with the intronic internal guide sequences (IGS) of ribozyme (19). Consequently, the original IGS in the native ribozyme (GGAGGG) was altered to be complementary to these six nucleotides from BlaI. These intron-containing Bla genes were cloned into a plasmid vector with a CMV promoter for expression in mammalian cells. After transient transfection into COS-1 cells and incubation for 48 h, total RNAs were isolated for RT-PCR analysis. Cell lysates were collected for in vitro assay of Bla activity with a fluorogenic substrate CC1 (Fig. 1B). CC1 is much more sensitive than nitrocefin, a widely used colorimetric substrate, and the Bla activity in lysates was readily detectable in a <1-min incubation. Among three insertion sites examined (positions 92, 156, and 240), position 156 afforded a construct that produced the highest Bla enzymatic activity. This plasmid was named Rz156. Sequence analysis of the RT-PCR product confirmed that accurate ligation occurred at the splice site and a correct Bla mRNA was produced (Fig. 2A). Compared with the result from transfection of CMV-bla containing an uninterrupted Bla gene, the final Bla activity for Rz156 was ≈0.6% (Fig. 2B). A negative control (RzDead) with an insertion of 42 nucleotides after 312G within the ribozyme of Rz156 (where the guanosine-binding site is located) did not produce any RT-PCR product or Bla activity and left the ribozyme construct unspliced (data not shown).
Fig. 2.
In vitro and in vivo assays of Rz156. (A) RT-PCR analyses of RNA extracts from COS-1 cells transiently transfected with CMV, CMV-bla, and Rz156. The expected splicing product Bla was as indicated. The top bright band was unspliced Rz156, and the faint middle band might be mRNA spliced at the cryptic splice site (19). Samples in Upper contained reverse transcriptase, and, as a control, samples in Lower did not. Shown in the first lane of each panel are molecular mass markers. (B) In vitro Bla assay using CC1. Shown is the hydrolysis rate, determined from lysates of COS-1 cells transiently transfected with CMV, CMV-bla, or Rz156 in three independent transfections and normalized against total lysates protein content in a fluorescence units (FU) per s per μg of proteins. Note that hydrolysis rates are plotted in a log scale. (C–E) Fluorescence microscopy images of the COS-1 cells transfected with an empty vector (CMV) (C) or transiently transfected with CMV-bla (D) or with Rz156 (E). (C–E Left) An overlay of frames captured at 530 nm (green emission) and 460 nm (blue emission). (C–E Right) DsRed emission at 605 nm with excitation at 560 nm. Nontransfected cells show no Bla activity (green in Left and black in Right).
Bla activity from Rz156 could be readily visualized in single living cells with CCF2/AM. Consistent with the in vitro Bla activity assay, nearly 100% of the empty vector (CMV)-transfected cells emitted green light from uncleaved substrate (Fig. 2C), and COS-1 cells cotransfected with CMV-bla vector and a mammalian expression vector of a red fluorescent protein (DsRed) displayed the expected correlation between the DsRed fluorescence and Bla activity: only DsRed-positive (transfected) cells emitted blue signals from cleaved substrate (Fig. 2D). Cotransfection of Rz156 and DsRed revealed a similar correlation between the ribozyme activity and DsRed fluorescence: most DsRed-positive (transfected) cells showed blue emission after CCF2/AM incubation, whereas DsRed-negative (untransfected) cells emitted green due to lack of Bla activity (Fig. 2E). A low blue/green ratio (<0.4) was observed for untransfected cells, and a moderate (>1.5) to high (>3) ratio was observed for transfected cells. Although generally only DsRed-positive cells emitted blue, some cells with high blue/green ratio had low DsRed signal, and vice versa. Lack of correlation between the blue/green ratio and the intensity of the DsRed signal among individual cells suggests that the observed heterogeneity was not just due to variations in the amount of transfection but rather reflected cell–cell variations in efficiency of splicing and expression.
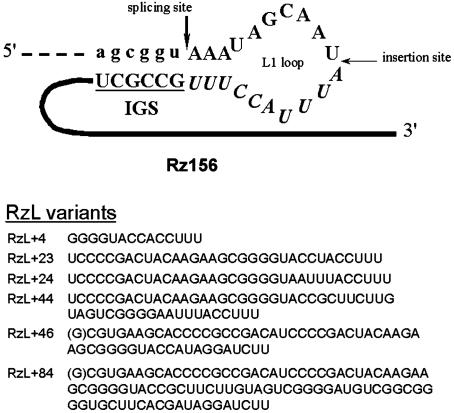
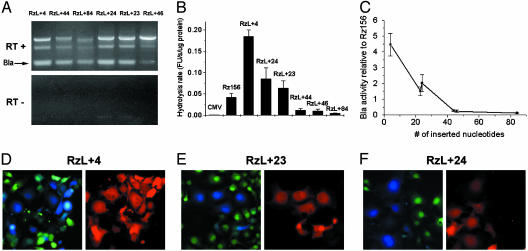
We then screened ribozyme variants with 4–84 oligonucleotides inserted into the middle of the L1 loop, located between the splice site and IGS, for increased Bla expression (Fig. 3). RT-PCR showed that all ribozyme variants produced the correctly spliced Bla mRNA (Fig. 4A). The intensities of the spliced product for RzL + 4, RzL + 23, and RzL + 24, which contained 18, 37, and 38 nucleotides in the loop, respectively, were the highest. The ratio of intensity between the unspliced ribozyme and spliced product varied for the different constructs, suggesting different efficiencies of self-splicing. In vitro Bla activity from these constructs also revealed a strong dependence of the Bla activity on the size of L1 loop (Fig. 4 B and C). All variants showed positive Bla activity, but RzL + 4, RzL + 24, and RzL + 23 exhibited higher Bla activity than Rz156, whose L1 loop was identical to that of the wild-type Tetrahymena intron. For example, RzL + 4 displayed up to a 4-fold increase in the Bla activity compared with Rz156. However, the Bla activity for constructs with a larger insertion (RzL + 44, RzL + 46, and RzL + 84) sharply decreased from 27% to <12% of that of Rz156. Although in both RzL + 46 and RzL + 84 the six nucleotides preceding the IGS (from position –7 to position –2) were altered to be complementary to nucleotides 158–163 of the Bla mRNA (which is called P10 interaction; ref. 1 and Fig. 3), RzL + 46 displayed almost the same Bla activity (24% of Rz156) as RzL + 44 (27% of Rz156).
Fig. 3.
Ribozyme variants with insertion of nucleotides at the indicated site of the L1 loop of the Tetrahymena ribozyme. Some have spontaneous deletions or mutations around the insertion site. The nucleotides displayed for each variant replace those in Rz156 starting from the indicated insertion site to the beginning of IGS (shown in italics). For RzL + 46 and RzL + 84, the third nucleotide after the splice site is G instead of A, shown in parentheses.
Fig. 4.
In vitro and in vivo assays of ribozyme variants with insertion at the L1 loop. (A) RT-PCR analyses of RNA extracts from COS-1 cells transiently transfected with RzL + 4, RzL + 23, RzL + 24, RzL + 44, RzL + 46, or RzL + 84. Reverse transcriptase was present in samples in Upper but not in Lower. The expected splicing product Bla was as indicated. (B) In vitro Bla assay using CC1 of lysates of COS-1 cells transiently transfected with CMV, Rz156, RzL + 4, RzL + 23, RzL + 24, RzL + 44, RzL + 46, or RzL + 84. Shown is the hydrolysis rate, determined from three independent transfections and normalized against total lysate protein contents in fluorescence units (FU) per s per μg of proteins. (C) The apparent dependence of the Bla activity of mutants on the number of inserted nucleotides. The relative Bla activity of each mutant is the ratio of its hydrolysis rate to that of Rz156. (D–F) Fluorescence microscopy of Cos-1 cells transiently transfected with ribozyme variant RzL + 4 (D), RzL + 23 (E), or RzL + 24 (F). (D–F Left) An overlay of frames captured at 530 nm and 460 nm with excitation at 405 nm after a 1-h loading of CCF2/AM. (D–F Right) Red emission at 605 nm of DsRed.
We cotransfected these ribozyme variant constructs with DsRed to identify single cells expressing ribozymes and displaying Bla activity under a fluorescence microscope. The percentage of the cells that emitted blue fluorescence varied among the constructs. When imaged immediately after CCF2/AM loading, more blue cells were observed from the dishes transfected with constructs of RzL + 4, RzL + 23, and RzL + 24 than those such as RzL + 44, RzL + 46, and RzL + 84 (Fig. 4 D–F); this finding is consistent with the results from both RT-PCR and in vitro Bla activity assay. For constructs with lower activity (RzL + 44, RzL + 46, and RzL + 84), a longer incubation time (6 h) resulted in readily detectable blue cells with a blue/green ratio from moderate (>1.5) to high (>3.0) (data not shown).
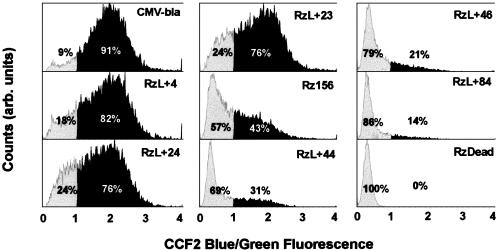
We next tested whether cells with splicing activity can be quantified by flow cytometry. COS-7 cells transfected with each of the ribozyme constructs and DsRed as a cotransfection marker were loaded with CCF2/AM and incubated for 1 h. Approximately 30,000 healthy DsRed-positive cells were analyzed by a fluorescence-activated cell sorting for each construct. In the negative control (RzDead) 100% of the cells fluoresced green, and in the positive control (CMV-bla) 91% of transfected (DsRed-positive) cells displayed a blue/green ratio of >1 (Bla-positive) (Fig. 5). For Rz156, 43% of transfected cells were Bla-positive (Fig. 5). It is noted that the splicing efficiency varied in individual cells even for the same construct, indicated by the range of the blue/green ratio. Again, the blue/green ratio did not show any clear correlation with the amount of DsRed within those cells that were DsRed-positive. RzL + 4, RzL + 23, and RzL + 24 gave 82%, 76%, and 76% Bla-positive transfected cells, respectively, all of which are larger than that of Rz156. For RzL + 44, RzL + 46, and RzL + 84, the percentages of Bla-positive transfected cells were 31%, 21%, and 14%, respectively, which are smaller than that of Rz156. This trend is consistent with the result from the CC1 Bla assay: the dependence of the splicing activity on the length of the inserted nucleotides at the L1 loop (Fig. 4C).
Fig. 5.
Fluorescence-activated cell sorter analysis of COS-7 cells transiently transfected with construct CMV-bla (positive control), RzL + 4, RzL + 24, RzL + 23, Rz156, RzL + 44, RzL + 46, RzL + 84, or a ribozyme dead mutant (RzDead, a negative control). DsRed cDNA was cotransfected as a transfection marker. Cells were incubated with CCF2/AM for 1 h before fluorescence-activated cell sorter analysis. The populations depicted consist of cells that were healthy (as judged by forward and side scatter) and DsRed-positive. For easy comparison of different constructs, the percentages of Bla-positive (dark area) and Bla-negative (light gray area) cells (defined as blue/green ratios >1 and <1, respectively) are indicated for each construct.
Discussion
We describe here an example of imaging of the splicing activity of ribozymes in single live mammalian cells. Splicing activity shows considerable heterogeneity among individual cells. Similar cell-to-cell heterogeneity also was reported in a recent study of ribozyme-mediated repairing of a mutant chloride channel by single-cell patch clamping (20). Flow cytometry is ideally suited to analyze this heterogeneity and to select the cells with the most efficient splicing. This approach should be most advantageous when screening large combinatorial libraries of ribozymes for maximum expression of protein resulting from correct splicing. A unique advantage of flow cytometry over other selection techniques (e.g., antibiotic resistance) is that flow cytometry can perform both negative and positive selections from the same reporter. This ability will be particularly important in future attempts to optimize trans-splicing RNAs to detect and transduce endogenous target mRNAs, which will require selecting not only for maximal activity in the presence of the target but also for minimal background in the absence of the target.
Our discovery of variants with insertion at the L1 loop of the Tetrahymena ribozyme suggested the important role of the L1 loop in the ribozyme cis-splicing activity. A slightly expanded loop (RzL + 4) displayed 4-fold better Bla activity and substantially more Bla-positive cells than Rz156 with the original loop, a finding that might support the hypothesis that some structural strain exists in the original sequence. It is not known whether this enhanced splicing activity is totally loop length-dependent. Although our study focused on the insertion at the L1 loop, insertions or mutations at other sites (7, 21–24) can be similarly examined with our assay. A more systematic approach would be a library of ribozyme variants with random insertions or mutations. Identification of variants with improved splicing activity may help us to better understand structure–function relationships of catalytic RNAs.
We have demonstrated our strategy with cis-splicing Tetrahymena ribozyme, but it should be applicable to trans-splicing ribozymes as well. Furthermore, insertions and/or mutations that result in enhanced in vivo splicing activity of cis-splicing ribozyme variants should be transferable to trans-splicing ribozymes. A trans-splicing ribozyme with high splicing efficiency should find a variety of applications, such as imaging tumor-specific oncogenes and repairing defective genes in inherited diseases.
Acknowledgments
We are grateful to Dr. Thomas Cech for the generous gift of T. thermophila group I intron plasmid. This work was supported in part by the Damon Runyon Cancer Research Foundation and the Burroughs Wellcome Fund (to J.R.), the University of California School of Medicine (to J.R.), and Department of Energy Grant DE-FG03-01ER63276 (to R.Y.T.). S.H. was supported in part by a medical fellowship from the Sumitomo Life Insurance Welfare Services Foundation.
Abbreviations: Bla, β-lactamase; IGS, internal guide sequences; RT, reverse transcription; CMV, cytomegalovirus.
References
- 1.Hagen, M. & Cech, T. R. (1999) EMBO J. 18, 6491–6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long, M. B. & Sullenger, B. A. (1999) Mol. Cell. Biol. 19, 6479–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lan, N., Howrey, R. P., Lee, S. W., Smith, C. A. & Sullenger, B. A. (1998) Science 280, 1593–1596. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe, T. & Sullenger, B. A. (2000) Proc. Natl. Acad. Sci. USA 97, 8490–8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullenger, B. A. & Gilboa, E. (2002) Nature 418, 252–258. [DOI] [PubMed] [Google Scholar]
- 6.Guo, F. & Cech, T. R. (2002) Nat. Struct. Biol. 9, 855–861. [DOI] [PubMed] [Google Scholar]
- 7.Guo, F. & Cech, T. R. (2002) RNA 8, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price, J. V. & Cech, T. R. (1985) Science 228, 719–722. [DOI] [PubMed] [Google Scholar]
- 9.Jones, J. T., Lee, S.-W. & Sullenger, B. A. (1996) Nat. Med. 2, 643–648. [DOI] [PubMed] [Google Scholar]
- 10.Zlokarnik, G., Negulescu, P. A., Knapp, T. E., Mere, L., Burres, N., Feng, L., Whitney, M., Roemer, K. & Tsien, R. Y. (1998) Science 279, 84–88. [DOI] [PubMed] [Google Scholar]
- 11.Whitney, M., Rockenstein, E., Cantin, G., Knapp, T., Zlokarnik, G., Sanders, P., Durick, K., Craig, F. F. & Negulescu, P. A. (1998) Nat. Biotechnol. 16, 1329–1333. [DOI] [PubMed] [Google Scholar]
- 12.Galarneau, A., Primeau, M., Trudeau, L.-E. & Michnick, S. W. (2002) Nat. Biotechnol. 20, 619–622. [DOI] [PubMed] [Google Scholar]
- 13.Wehrman, T., Kleaveland, B., Her, J.-H., Balint, R. F. & Blau, H. M. (2002) Proc. Natl. Acad. Sci. USA 99, 3469–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spotts, J. M., Dolmetsch, R. E. & Greenberg, M. E. (2002) Proc. Natl. Acad. Sci. USA 99, 15142–15147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavrois, M., de Noronha, C. & Greene, W. C. (2002) Nat. Biotechnol. 20, 1151–1154. [DOI] [PubMed] [Google Scholar]
- 16.Gao, W., Xing, B., Tsien, R. Y. & Rao, J. (2003) J. Am. Chem. Soc. 125, 11146–11147. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J. & Russell, D. W. (2001) in Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, New York), 3rd Ed.
- 18.Imai, Y., Matsushima, Y., Sugimura, T. & Terada, M. (1991) Nucleic Acids Res. 19, 2785–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Been, M. D. & Cech, T. R. (1986) Cell 47, 207–216. [DOI] [PubMed] [Google Scholar]
- 20.Rogers, C. S., Vanoye, C. G., Sullenger, B. A. & George, A. L. (2002) J. Clin. Invest. 110, 1783–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young, B., Herschlag, D. & Cech, T. R. (1991) Cell 67, 1007–1019. [DOI] [PubMed] [Google Scholar]
- 22.Treiber, D. K., Rook, M. S., Zarrinkar, P. P. & Williamson, J. R. (1998) Science 279, 1943–1946. [DOI] [PubMed] [Google Scholar]
- 23.Michel, F., Hanna, M., Green, R., Bartel, D. P. & Szostak, J. (1989) Nature 342, 391–395. [DOI] [PubMed] [Google Scholar]
- 24.Cate, J. H., Gooding, A. R., Podell, E., Zhou, K., Golden, B. L., Kundrot, C. E., Cech, T. R. & Doudna, J. A. (1996) Science 273, 1678–1685. [DOI] [PubMed] [Google Scholar]