Abstract
Mitotic spindle length control requires coordination between microtubule (MT) dynamics and motor-generated forces. To investigate how MT plus-end polymerization contributes to spindle length in Drosophila embryos, we studied the dynamics of the MT plus-end depolymerase, kinesin-8, and the effects of kinesin-8 inhibition using mutants and antibody microinjection. As expected, kinesin-8 was found to contribute to anaphase A. Furthermore, kinesin-8 depletion caused: (i) excessive polymerization of interpolar (ip) MT plus ends, which “overgrow” to penetrate distal half spindles; (ii) an increase in the poleward ipMT sliding rate that is coupled to MT plus-end polymerization; (iii) premature spindle elongation during metaphase/anaphase A; and (iv) an increase in the anaphase B spindle elongation rate which correlates linearly with the MT sliding rate. This is best explained by a revised “ipMT sliding/minus-end depolymerization” model for spindle length control which incorporates a coupling between ipMT plus end dynamics and the outward ipMT sliding that drives poleward flux and spindle elongation.
Keywords: Mitosis, spindle length control, microtubule dynamics, poleward flux, KLP67A
Introduction
The major purpose of mitosis is to coordinate the accurate distribution of genetic instructions, packaged into chromosomes, to the daughter products of each cell division. Chromosome segregation depends upon the action of the mitotic spindle, a subcellular protein machine that uses ensembles of kinesin and dynein motors plus microtubule (MT) dynamics to assemble itself and then to move separated chromatids polewards (anaphase A) and to separate the spindle poles, further pulling apart sister chromatids (anaphase B) (Cande and Hogan, 1989; Roostalu et al., 2010; Walczak et al., 2010). These events are accompanied by changes in spindle length (i.e. pole-to-pole spacing) that are controlled by a variety of factors that appear to vary in different systems (Goshima and Scholey, 2010). Following nuclear envelope breakdown in Drosophila syncytial embryos, for example, mitotic spindles are maintained at a constant length during early prometaphase (8μm in cycle 11) and again during metaphase and anaphase A (herein referred to as pre-anaphase B; length = 12 μm in cycle 11) and they undergo steady elongation (rate ≈ 0.1 μm/s) during the prometaphase-to-metaphase transition and subsequently during anaphase B (Brust-Mascher et al., 2004; Brust-Mascher and Scholey, 2002; Brust-Mascher et al., 2009; Cheerambathur et al., 2007; Civelekoglu-Scholey et al., 2010; Sharp et al., 2000a).
The control of mitotic spindle length is thought to require the coordination between MT polymer dynamics and motor-generated forces, and this coordination formed the basis of several models for the maintenance and adjustment of spindle length that occur during various stages of mitosis in several systems (Dumont and Mitchison, 2009; Goshima and Scholey, 2010; Tolic-Norrelykke, 2010). For example, models based on “interpolar (ip) MT sliding/minus-end depolymerization” mechanisms have been proposed for length control of mitotic spindles in Drosophila embryos and S2 cells (Brust-Mascher et al., 2004; Brust-Mascher and Scholey, 2002; Cheerambathur et al., 2007; Goshima et al., 2005; Wollman et al., 2008). It is proposed that the steady state length of the metaphase spindle is maintained by a balance between ipMT sliding and ipMT minus end depolymerization at the poles. In embryos, this balance is thought to be tipped by the cessation of ipMT depolymerization at spindle poles in response to cyclin B degradation, which triggers anaphase B spindle elongation, based on observations that the presence of non-degradable cyclin B extends the steady state length of the preanaphase B spindle indefinitely (Cheerambathur et al., 2007).
An alternative “slide-and-cluster” model has been proposed for anastral spindles based mainly on work done in Xenopus embryo extracts (Burbank et al., 2007; Dumont and Mitchison, 2009). In this model, ipMTs are nucleated around chromosomes, transported poleward, away from the chromosomes, and then they are clustered by minus-end directed motors to form a focus near the poles, while being lost by stochastic, catastrophic disassembly of their plus ends. It should be noted that, while the slide and cluster model was developed for anastral spindles, it may also apply to astral, centrosome-controlled spindles, some of which undergo loss of spindle pole focusing following loss of function of minus-end-directed clustering motors (Gaglio et al., 1996). The slide-and-cluster model is appealing because the mean MT sliding velocity and average lifetime of ipMTs lead naturally to the production of a dynamic, steady state spindle of constant length.
It should be noted that both the ipMT sliding/minus-end depolymerization and the slide-and-cluster models are based on the idea that pole-pole spacing is determined by forces generated within the spindle itself, but in addition to these “intrinsic forces”, it is also likely that in some systems, cortical force generators act on astral MTs to exert pushing and pulling forces on the spindle poles to influence mitotic spindle length (Saunders et al., 2007; Sharp et al., 2000a; Tolic-Norrelykke et al., 2004).
These models, supported by experimental data in different systems, make clear and testable predictions regarding the potential roles of several mitotic proteins in spindle length control, including MT-MT sliding motors, MT minus-end depolymerases, MT clustering proteins and force generators localized at the cell cortex. In addition, several experimental studies reveal that the perturbation of MT plus-end dynamics can also affect spindle length (Buster et al., 2007; Goshima and Vale, 2005; Goshima et al., 2005; Maiato et al., 2005; Rogers et al., 2002), and although microtubule plus end dynamics is a critical component of the slide-and cluster model, the role of MT plus end dynamics has not been very well addressed in “ipMT sliding/minus end depolymerization” models for Drosophila spindle length control.
One extensively studied mitotic motor that is thought to play an important role in controlling MT plus end dynamics is kinesin-8 (Stumpff and Wordeman, 2007; Walczak, 2006), which has both plus-end directed MT translocating and plus-end directed MT depolymerizing activities (Varga et al., 2006; Varga et al., 2009) and appears to control both spindle length and chromosome positioning in the spindle (Goshima et al., 2005; Stumpff et al., 2008). Previous studies reported that several kinesin-8 family proteins regulate MT plus end dynamics of cortical MTs where they mainly serve to modulate the MT catastrophe frequency (e.g. kip3 (Gupta et al., 2006), klp5/6 (Tischer et al., 2009), and Kif18A (Du et al., 2010)). Several complementary in vitro studies have suggested that kinesin-8 motors are length-dependent MT plus end depolymerases and processive MT plus end directed motors that bind to and translocate along the MT polymer lattice to reach the plus ends from which they remove tubulin subunits (Gupta et al., 2006; Mayr et al., 2007; Varga et al., 2006; Varga et al., 2009).
The Drosophila kinesin-8, KLP67A is thought to play essential roles in spindle length control and in the regulation of MT plus end dynamics. For example, klp67A hypomorphic mutants display abnormally long, curved spindles together with defects in chromosome congression and segregation in meiotic spindles (Gandhi et al., 2004; Savoian et al., 2004). Similarly, depletion of KLP67A using RNAi in S2 cells also resulted in longer spindles and defects in chromosome congression (Goshima et al., 2005; Savoian and Glover, 2010), which Savoian and Glover (2010) proposed may reflect the activity of KLP67A localized specifically at the kinetochore. Moreover, based on FRAP analysis, Buster et al. (2007) showed that KLP67A inhibition in S2 cells specifically affects MT plus-end dynamics within spindles in vivo, and also causes an increase in MT poleward flux rate, consistent with an increase in MT minus-end depolymerization. However, the precise mechanism of action of KLP67A, especially during embryonic mitoses, remains incompletely understood and requires further investigation.
Therefore, to test kinesin-8’s influence on mitotic spindle MT dynamics and to investigate the role of MT plus-end dynamics in spindle length control in Drosophila embryos, we decided to explore the mechanism of premature spindle elongation that occurs following the depletion of the kinesin-8, KLP67A (Gandhi et al., 2004). We observed that KLP67A depletion caused an increase in the MT sliding rate, which is correlated with the increase in pre-anaphase B and anaphase B spindle elongation rate, leading to a revision of our “ipMT sliding and minus-end depolymerization” model for spindle length control to include a coupling between MT plus end dynamics and MT sliding.
Materials and methods
Generation of transgenic flies, klp67A mutant and rescue experiments
A full-length Klp67A cDNA tagged with GFP was subcloned from a pMT vector, provided by G. Goshima (Nagoya University, Nagoya, Aichi, Japan) and R. Vale (University of California, San Francisco, San Francisco, CA), into the pWR-pUbq transformation vector (provided by N. Brown, University of Cambridge, Cambridge, England, UK) downstream of the polyubiquitin promoter. Transgenic flies with a single insertion at X chromosomes were generated with the above construct at Bestgene Inc. A klp67A hypomorphic, male sterile, partially female sterile mutant with genotype Klp67A332b24/Df(3L)29A6 was generated as previously described (Gandhi et al., 2004), using the germ lines Df(3L)29A6/TM6B and Klp67A332b24/TM6B provided by Andrea Pereira (University of Massachusetts, Amherst,MA). Rescue experiments were performed using standard genetic techniques. Briefly, partially rescued klp67A mutants (Klp67A-GFP/w;; Klp67A332b24/Df(3L)29A6) were obtained by crossing Klp67A-GFP/Klp67A-GFP;;Klp67A332b24/TM6B with Df(3L)29A6/TM6B. Fully rescued klp67A mutants (Klp67A-GFP/Klp67A-GFP;; Klp67A332b24/Df(3L)29A6) were obtained by crossing Klp67A-GFP/Y;; Klp67A332b24/TM6B with Klp67A-GFP/Klp67A-GFP;; Df(3L)29A6/TM6B. The rescued male and female flies, containing two copies of the Klp67A-GFP gene, appeared completely viable and fertile and displayed embryo mitotic spindle pole dynamics virtually identical to wild type flies (see results).
Drosophila stocks and embryo preparation
All flies were maintained according to standard procedures at 25°C and embryos were collected as described previously (Brust-Mascher and Scholey., 2009; Sharp et al., 1999). Transgenic flies co-expressing GFP-tubulin and RFP-histone were generated by crossing flies expressing GFP-tubulin (provided by T. Kaufman, Indiana University, Bloomington, IN) with flies expressing histone-RFP (from Bloomington Drosophila Stock Center) as described (Civelekoglu-Scholey et al. 2010). The GFP-cid transgenic flies were provided by Steven Henikoff (Fred Hutchinson Cancer Research). The EB1-GFP transgenic flies (expressed under the UASp promoter ) were provided by S. Rogers (University of North Carolina at Chapel Hill, Chapel Hill, NC), B. Eaton, and G. Davis (University of California, San Francisco, San Francisco, CA), and germ line expression of GFP-EB1 was driven by using the tubulin Gal4 drivers tub([matα4]GAL4-VP16-V2H) or tub([matα4]GAL4-VP16-V37). Embryos were microinjected with rhodamine tubulin (Cytoskeleton Inc, Denver, Colorado) to visualize tubulin.
Antibody generation and microinjection
Anti-KLP67A antibody was affinity purified with GST tagged KLP67A C-terminal fragment (602-781AA) from rabbit anti-KLP67A serum (provided by G. Goshima (Nagoya University, Nagoya, Aichi, Japan) and R. Vale (University of California, San Francisco, San Francisco, CA)) as described (Goshima and Vale, 2005). For embryo injection, we used two in-the-needle concentrations of purified anti-KLP67A: ~15mg/ml for mid anti-KLP67A injection and ~30mg/ml for high anti-KLP67A injection. Most embryos were injected in interphase of cycle 10 or 11 (a few in prometaphase of cycle 10 or 11).
Time-lapse and fluorescence speckle microscopy
Time-lapse images were taken on an inverted microscope (IX-70; Olympus) equipped with an UltraView spinning disk confocal head (PerkinElmer-Cetus) and a UPlan-Apochromat 100 × 1.35 NA or a Plan-Apochromat 60 × 1.4 NA oil immersion objective. Either a single confocal plane or a stack of seven planes spaced by 0.8 μm was acquired at time intervals of 0.5 to 15 s at room temperature (20–22°C). Images were acquired with a charge-coupled device camera (Orca II; Hamamatsu Photonics) using Volocity software (PerkinElmer-Cetus) and analyzed with Metamorph Imaging software (Universal Imaging, West Chester, PA). Pole to pole distance as a function of time was measured from the position of the poles in each image. Calculations and statistical analyses were performed on Excel (Microsoft). All error bars shown in Figures are SDs.
Online supplementary material
Fig. S1 shows that the microinjection of anti-KLP67A antibody caused dose-dependent mitotic defects in spindle length and chromosome segregation. In Fig. S2, a partially rescued klp67a mutant shows pre-anaphase B spindle elongation in the presence of kinetochore localized KLP67A-GFP. Video 1 shows the localization of KLP67A-GFP in fully rescued Drosophila embryo mitotic spindles. Video 2 shows the dynamics of GFP-cid in a control embryo. Video 3 shows the dynamics of GFP-cid in an embryo injected with “high anti-KLP67A”. Video 4 shows the dynamics of GFP-tubulin and RFP-histone in a control embryo. Video 5 shows the dynamics of GFP-tubulin and RFP-histone in an embryo injected with “mid anti-KLP67A”. Video 6 shows the dynamics of GFP-tubulin and RFP-histone in an embryo injected with “high anti-KLP67A”.
Results
Dynamics of KLP67A and depletion of its function in Drosophila syncytial embryos
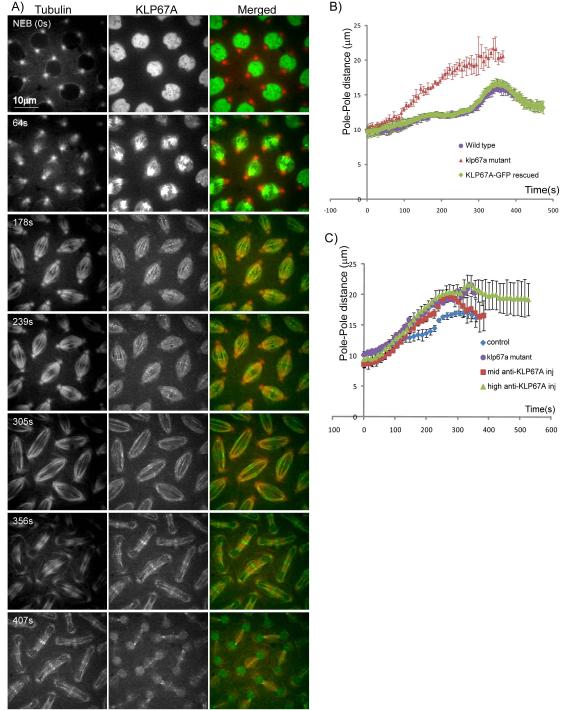
To understand the role of the kinesin-8, KLP67A, in Drosophila syncytial embryonic mitosis, we made transgenic flies expressing KLP67A-GFP in a corresponding mutant background using standard fly genetic techniques. Expression of the KLP67A-GFP transgene restored fertility, and viability to the klp67A mutants. Significantly, the major mitotic defect caused by depletion of KLP67A function in the mutant embryos, i.e. the formation of long and curved spindles (Gandhi et al., 2004), was rescued by KLP67A-GFP expression (Fig. 1). The KLP67A-GFP transgene restored wild type spindle morphology and geometry (Fig. 1a) and the dynamics of mitotic spindle poles in the rescued flies was virtually identical to that seen in wild type embryos (Fig. 1b). Thus the expression of KLP67A-GFP functionally compensates for the loss of KLP67A in mutants.
Figure 1. Localization and dynamics of KLP67A-GFP in Drosophila embryo mitotic spindles.
(A) Micrographs from a time lapse video of a representative embryo showing rhodamine-tubulin (left) KLP67A-GFP (center) and merged (right) (tubulin:red; KLP67A-GFP:green). Time in each frame is given in seconds from nuclear envelope breakdown (NEB). In interphase and prophase, KLP67A-GFP was sequestered in the nucleus. Starting after NEB through telophase, KLP67A-GFP localized all along spindle MTs and faintly decorated astral MTs. During metaphase and anaphase, we also observed distinct spots of KLP67A-GFP that lay near the metaphase spindle equator and separated with the chromosomes during anaphase A. This is likely to be kinetochore-associated KLP67A as reported previously by others (Savoian and Glover, 2010). During anaphase B and telophase, KLP67A-GFP was concentrated at the spindle midzone. Once the nuclear envelope reformed, KLP67A-GFP was re-recruited into the nucleus. (B) Spindle pole dynamics in (i) wild type embryos; (ii) klp67A mutant embryos; and (iii) KLP67A-GFP-rescued klp67A mutant embryos showing that the expression of KLP67A-GFP fully compensated for the depletion of KLP67A in the mutants. (C) Spindle pole dynamics in wild type embryos; klp67A mutant embryos; the “mid anti-KLP67A”- injected embryos; and the “high anti-KLP67A” injected embryos showing that KLP67A depletion by anti-KLP67A microinjection produced premature spindle pole separation, virtually identical to the klp67A hypomorphic mutants. See Fig. S1 for still images.
We monitored the localization and dynamics of KLP67A-GFP throughout mitosis in living, rescued embryos (Fig. 1, video 1). KLP67A accumulated in the nucleus during interphase and late telophase (Fig. 1a, top and bottom rows) and localized along spindle MTs (including weakly along astral MTs), throughout mitosis (Fig. 1a). We observed distinct puncta of KLP67A-GFP at the kinetochores during metaphase and anaphase A (Fig. 1a, rows 3 and 4) and then, during anaphase B and telophase, KLP67A-GFP was observed to concentrate at the spindle midzone (Fig. 1a, bottom three rows), as reported previously (Goshima and Vale, 2005; Savoian and Glover, 2010).
The effect of loss of KLP67A function in Drosophila syncytial embryo mitotic spindles was evaluated using previously described loss-of-function klp67A mutants (Gandhi et al., 2004) and also by the microinjection of an affinity purified anti-KLP67A antibody (Goshima and Vale, 2005). The anti-KLP67A injection successfully depleted KLP67A-GFP from the spindle and caused premature and persistent spindle elongation (Fig. 1c and Fig. S1), very similar to that observed in the mutants. This suggests that the microinjection of this anti-KLP67A antibody efficiently depleted KLP67A from mitotic spindles in Drosophila syncytial embryos leading to a corresponding loss of function, and did not produce any detectable “dominant” effects on spindle dynamics beyond those seen in mutants (e.g. as a consequence of artifactually crosslinking spindle MTs into a rigor-like network).
Anti-KLP67A injection into embryos co-expressing GFP-tubulin and RFP-histone displayed a gradient of mitotic defects. Proximal to the antibody injection site, where the concentration of anti-KLP67A was highest, we observed defective chromosome segregation and the formation of abnormally long and curved spindles. Further away from the injection site, where the concentration of anti-KLP67A was lower, we observed abnormally long spindles but no defects in chromosome segregation were evident.
Based on this, we suspected that the mitotic defects produced by anti-KLP67A injection were dose-dependent, that is, a lower dose of anti-KLP67A injection (away from the injection site) affected spindle length, while a higher dose (close to the injection site) affected both spindle length and chromosome segregation. To test this, we adjusted the injected concentration of the antibody. The lower concentration, herein referred to as “mid anti-KLP67A” (in-the-needle concentration ~15mg/ml), on average displaced 46% of KLP67A-GFP from the spindle, as quantified by fluorescence intensity (Fig. S1b). In these embryos we routinely observed persistent spindle elongation but complete chromosome segregation accompanied by only minor defects (Fig. S1a middle and Video 5). The higher concentration, herein called “high anti-KLP67A” injection (in-the-needle concentration ~30 mg/ml), on average depleted 76% of KLP67A-GFP from mitotic spindles, as quantified by fluorescence intensity (Fig. S1b). In these embryos, we routinely observed incomplete chromosome segregation and persistent spindle elongation (Fig. S1a right and Video 6). Antibody injection at the two different concentrations allowed us to discriminate between the effects of KLP67A depletion on spindle length control and chromosome segregation (Fig. S1 and Videos 4-6).
KLP67A-dependent control of MT plus end dynamics in one half spindle prevents MT “overgrowth” into the opposite half spindle
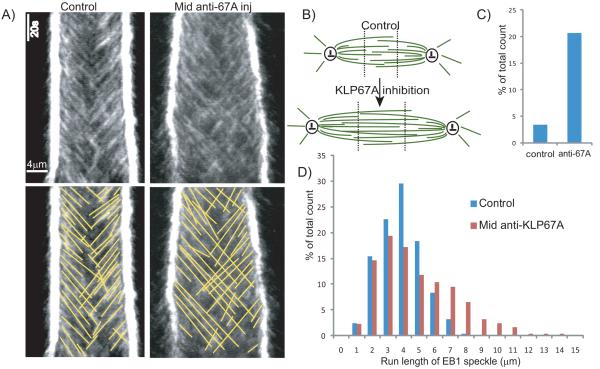
To test the effect of KLP67A depletion on MT plus-end dynamics within the spindle, we injected “mid anti-KLP67A” with rhodamine tubulin into Drosophila embryos expressing GFP-EB1, a marker of growing MT plus end tips (Rogers et al., 2002). In control spindles, MT plus ends moving away from their proximal pole seldom crossed the spindle mid-zone into the distal half spindle. Following KLP67A inhibition, however, a large number of MT plus ends were observed to translocate across the spindle equator into the opposite half spindle, with some of them even reaching the opposing pole (Figs. 2a, b). This suggests that KLP67A normally limits the extent of polymerization of spindle MTs, so that their plus ends are confined mainly to their proximal half spindles and do not grow too far into their opposite half spindles.
Figure 2. KLP67A depletion affects the dynamics of growing MT plus-ends.
(A) Comparison of representative EB1-GFP kymographs from control and “mid anti-KLP67A” injected embryos showing that KLP67A depletion allows more growing MTs to cross the spindle equator and enter the distal half spindle to reach the opposite pole. (B) Cartoon showing change in spindle MT organization after KLP67A inhibition. MTs are shown in green and centrosomes are shown in black. Dotted black line shows the arbitrary division of the spindle into three domains along the pole-pole axis which were used to semi-quantify the extent of penetration of one half spindle by MT plus-ends from the other half spindle. (C) Comparison of the percentage of growing MT plus ends that penetrate into the distal one-third spindle region, adjacent to the distal pole (division into the three regions as shown in B) - control and “mid” anti-KLP67A injected embryos are shown. (D) Histogram of EB1 speckle run lengths in control and “mid” anti-KLP67A injected embryos. Average length and total counts for C and D are shown in Table 1.
To quantify the effect of KLP67A inhibition on the distribution of growing MT plus ends within spindles, we arbitrarily divided the spindle into three equal-sized regions along the pole-pole axis (Fig. 2b), and counted the number of moving EB1 speckle trajectories that ended in each region. Strikingly, KLP67A inhibition caused a 6-fold increase (from 3.4% to 20.7%) in the percentage of EB1 speckles that reached the region corresponding to the distal one-third of the spindle near the opposing pole (Fig. 2c). This strongly supports the hypothesis that KLP67A confines growing MTs to their proximal half spindles by limiting the polymerization of MT plus ends in wild type embryos.
The slope and run length (along the x-axis) of EB1 speckles in the kymographs provide estimates of the rate of spindle MT polymerization and the distance over which the polymerizing plus end translocates (average length, (Lpoly)). On average, KLP67A inhibition increased the run length of an EB1 speckle 1.5-fold (from 3.38±1.45μm to 4.97±2.48 μm), reflecting an increase in the translocation distance, Lpoly, but had no apparent effect on the MT growth rate (i.e. dynamic instability parameter, Vg) (Table 1). Since the average length of MT polymerization, Lpoly , is proportional to the MT growth rate (unchanged parameter, Vg) and is inversely proportional to the MT catastrophe frequency (fcat=1/Tpoly ≈Vg/Lpoly, (Shelden and Wadsworth, 1993)), the increase in the average EB1 run length may reflect a decrease in the catastrophe frequency after KLP67A inhibition. The histogram of EB1 speckle run lengths (Fig. 2d) further documents that KLP67A inhibition allowed many MTs to polymerize to longer lengths than in controls, however, the distribution of speckles with shorter run lengths (<4μm) was less affected. This suggests that KLP67A selectively restricts the growth of longer MTs in a length-dependent manner, possibly through the regulation of the MT catastrophe rate.
Table 1.
Quantitation of spindle dynamic parameters following KLP67A inhibition.
| Control | Mid Anti-KLP67A injection |
High Anti-KLP67A injection |
|
|---|---|---|---|
|
MT polymerization rate
(μm/s) a |
0.26±0.10 (2/8/1016) |
0.25±0.07 (3/13/1476) |
NAb |
|
Run length of EB1 speckle
(μm) |
3.38±1.45 (2/8/1016) |
4.97±2.48 (3/13/1476) |
NAb |
| MT sliding rate (μm/s) | 0.048±0.020 (2/10/1270) |
0.072±0.030 (3/13/1551) |
0.099±0.035 (1/3/1222) |
| MT poleward flux (μm/s) | 0.044±0.020 (2/10/1270) |
0.054±0.030 (3/13/1551) |
0.075±0.035 (1/3/1222) |
| Anaphase A rate(μm/s) | 0.097±0.013 (2/3/20) |
0.110±0.023 (2/3/16) | 0.053±0.010 (1/3/18) or NAc |
|
Interkinetochore distance
(μm) |
0.73±0.12 (3/22/68) | 0.76±0.13 (2/11/25) | 0.88±0.14 (2/11/25) |
Notes: The number of 1) embryos, 2) spindles and 3) speckles, kinetochores or kinetochore pairs analyzed are indicated in parentheses.
MT polymerization rates were calculated by subtracting pole movement from raw EB1 speckle velocity.
The EB1 data for high concentration of anti-KLP67A was not available because we could not fully track most MT plus ends within the same confocal plane of spindle poles.
Anaphase A rates were not available for the spindles that showed halted chromosome segregation by high anti-KLP67A injection.
KLP67A inhibition increased the rates of both MT sliding and poleward flux
In wild type Drosophila syncytial embryos, the length of the pre-anaphase B spindle is proposed to be maintained constant by a balance between outward ipMT translocation generated by ipMT sliding motors and ipMT minus-end depolymerization at the poles (Brust-Mascher et al., 2004; Brust-Mascher and Scholey, 2002; Brust-Mascher and Scholey, 2009). Since KLP67A inhibition caused premature pre-anaphase B spindle elongation, we hypothesized that it tipped the balance between ipMT sliding and ipMT minus-end depolymerization in favor of sliding. To test this, we measured the velocity of translocation of tubulin speckles away from the spindle equator (MT sliding rate) and towards the poles (MT poleward flux) in Drosophila embryos injected with the “mid” and “high” concentrations of anti-KLP67A.
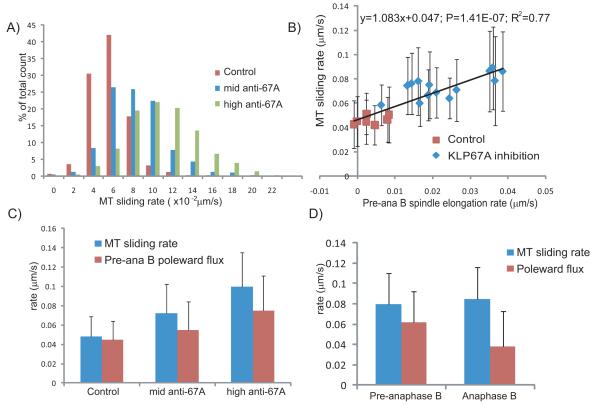
We observed a significant increase in the rate of tubulin speckle translocation away from the equator (MT sliding rate) after KLP67A inhibition (Figs. 3a, c & Table 1), supporting the hypothesis that KLP67A inhibition increased the outward force that drives ipMT sliding. In wild type Drosophila embryos, it is generally believed that the outward pushing force is solely generated by the antiparallel ipMT sliding motor, kinesin-5 (Brust-Mascher et al., 2004; Brust-Mascher et al., 2009; Sharp et al., 1999; Tao et al., 2006). However, this model is complicated by the change in ipMT plus-end distribution that results from KLP67A inhibition (discussed later).
Figure 3. KLP67A depletion increased the rates of (i) ipMT sliding away from the spindle equator; and (ii) poleward MT flux.
(A) Histogram of MT sliding rate (tubulin speckle velocity away from the equator) in control (red), the “mid” anti-KLP67A injected embryos (blue) and the “high” anti-KLP67A injected embryos (green). Total counts are shown in Table 1. (B) Linear correlation between the MT sliding rate and the premature pre-anaphase B spindle elongation rate in each spindle from control and anti-KLP67A injected embryos. P-value =1.41499E-07. R squared = 0.77. For each spindle, 50-200 speckles were measured. (C) Comparison of the MT sliding rate (blue) and pre-anaphase B MT poleward flux rate (red) in controls compared to “mid” and “high” anti-KLP67A injected embryos. P<2.2e-16 for all pairwise t-tests. Total counts are shown in Table 1. (D) Comparison of MT sliding rate (blue) and MT poleward flux rate (red) during pre-anaphase B (911 speckles in 8 spindles measured) and anaphase B (877 speckles in the same 8 spindles measured) in a representative embryo injected with “mid” anti-KLP67A antibody.
Interestingly, the increase in MT sliding rate was linearly correlated with the increase in (premature) pre-anaphase B spindle elongation rate in each spindle (Fig. 3b), suggesting that an increase in the rate of ipMT sliding could reflect an intrinsic mechanism that drives premature pre-anaphase B spindle elongation after KLP67A inhibition. However, the change in spindle elongation rate was far less than twice the change in MT sliding rate (Fig. 3b), indicating the involvement of another mechanism. We propose that the increase in pre-anaphase B spindle elongation rate after KLP67A inhibition may be explained by an increase in the net difference between MT sliding rate and MT minus end depolymerization, because, after KLP67A inhibition, we also observed an increase in MT minus-end depolymerization as reflected in an increase in poleward MT flux (Fig. 3c & Table 1). Thus, in control embryos, there is little difference between the MT sliding rate and MT poleward flux, consistent with the idea that outward ipMT sliding is balanced by MT depolymerization at the poles to keep the pre-anaphase B spindle length constant (Brust-Mascher et al., 2004; Brust-Mascher and Scholey, 2002; Brust-Mascher et al., 2009), but after anti-KLP67A inhibition, the difference between the ipMT sliding rate and ipMT poleward flux increases, leading to an increase in the pre-anaphase B spindle elongation rate.
The increased ipMT sliding rate is correlated with an increase in the anaphase B spindle elongation rate
Several studies support the hypothesis that anaphase B spindle elongation is driven by an ipMT “sliding filament” mechanism (Brust-Mascher and Scholey, 2002; Hogan et al., 1993; Mastronarde et al., 1993; Masuda et al., 1988; McIntosh and McDonald, 1989; Saxton and McIntosh, 1987; Sharp et al., 1999). In Drosophila embryo mitosis, we have proposed that outwardly sliding ipMTs are depolymerized at their minus-ends prior to anaphase B, thereby preventing spindle elongation. However, this depolymerization is downregulated at the onset of anaphase B in response to cyclin B degradation, which allows ipMT sliding to then drive spindle elongation (Brust-Mascher et al., 2004; Brust-Mascher and Scholey, 2002; Cheerambathur et al., 2007). In addition to ipMT sliding forces generated by the spindle midzone, cortical dynein mediated pulling forces acting on astral MTs could also be involved in spindle elongation, though in Drosophila embryos, such cortical forces contribute little to anaphase B (Brust-Mascher et al., 2009; Sharp et al., 2000a). We reasoned that if anaphase B spindle elongation is mainly driven by the sliding apart of ipMTs, the increase in ipMT sliding after KLP67A inhibition might lead to an increase in anaphase B spindle elongation rate.
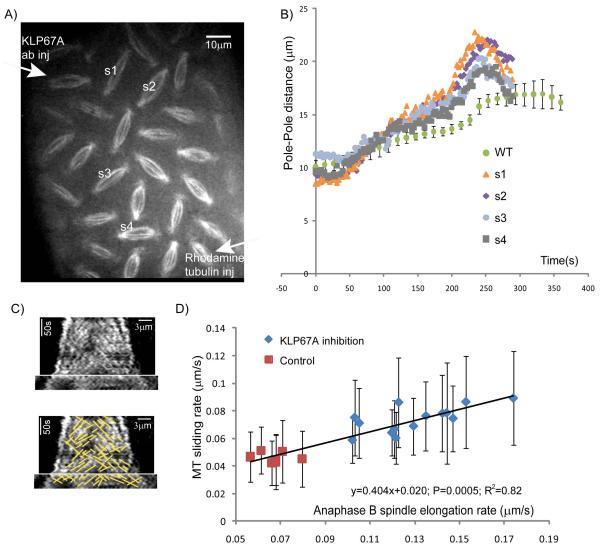
Following KLP67A inhibition either by injection of “mid” anti-KLP67A, or in “semi-rescued” klp67A mutants which contain only one copy of the Klp67A-GFP gene (the aforementioned fully rescued mutants need two copies of the Klp67A-GFP gene), we observed a population of mitotic spindles that displayed a higher anaphase B spindle elongation rate than control spindles. For example, Fig. 4b shows the dynamics of spindle poles in four spindles in an embryo injected with a “mid” anti-KLP67A concentration, and the positions of these spindles relative to the injection sites are shown in Fig. 4a. In this embryo, all measured spindles showed higher anaphase B spindle elongation rates than control spindles, but the rate of elongation correlated with the position of the spindle relative to the injection site – the rate of spindle elongation was highest closer to the anti-KLP67A injection site and decreased in more distal spindles (Figs. 4a and 4b).
Figure 4. Higher anaphase B spindle elongation rate after KLP67A depletion is correlated with an increase in the MT sliding rate.
(A) A still image from a time lapse video of a representative embryo injected with anti-KLP67A antibody (at the upper left corner) showing a gradient of mitotic defects. (B) Spindle pole dynamics of the four spindles shown in A compared to controls (WT) showing that KLP67A inhibition caused an increase in anaphase B spindle elongation rate. (C). Representative tubulin kymograph from an embryo partially depleted of KLP67A, showing that tubulin speckles move away from the spindle equator at a similar rate to the rate of spindle pole separation during anaphase B. (D) Linear correlation between the MT sliding rate and anaphase B spindle elongation rate in spindles. Data from two representative embryos displaying a higher anaphase B spindle elongation rate after partial KLP67A depletion compared to a control embryo. P-value=0.0005.R square=0.82. For each spindle, 50-200 speckles were measured.
The increase in anaphase B rate after KLP67A inhibition was linearly correlated with the increase in MT sliding rate, in support of a long proposed “ipMT sliding filament” model for anaphase B spindle elongation (Brust-Mascher et al., 2004; Brust-Mascher and Scholey, 2002; Masuda et al., 1988; Sharp et al., 1999). We measured ipMT sliding rates for each spindle in two KLP67A inhibited embryos that showed higher than control anaphase B rates (e.g. Fig. 4c). A linear correlation (Fig. 4d) was observed between the anaphase B spindle elongation rate and average MT sliding rate in each spindle, suggesting that the faster anaphase B spindle elongation observed in KLP67A inhibited spindles was driven by the same mechanism that caused the increase in MT sliding rates. This mechanism could be “intrinsic”, i.e. generated by molecules residing within the spindle or it could be generated outside the spindle (discussed later).
While the rate of MT sliding in control and KLP67A-inhibited embryos differed, we observed that there is no significant change in the MT sliding rate before and after anaphase B onset within any individual spindle. We compared the MT sliding rate during pre-anaphase B and anaphase B in each measured embryo and did not detect a significant difference (e.g. Fig. 3d). Although there is no significant change in average MT sliding rate at anaphase B onset, the average MT poleward flux was much lower during anaphase B than during pre-anaphase B (Fig. 3d), supporting the hypothesis that the switch leading to suppression of ipMT poleward flux at anaphase B onset that is seen in wild type embryos is also present after KLP67A inhibition (Brust-Mascher et al., 2004; Brust-Mascher and Scholey, 2002; Brust-Mascher et al., 2009).
KLP67A is required for anaphase A chromosome segregation
Roles for kinesin-8 motors in chromosome dynamics during mitosis have been documented in several systems, but not for KLP67A in Drosophila embryos (Mayr et al., 2007; Savoian et al., 2004; Savoian and Glover, 2010; Stumpff et al., 2008; Stumpff and Wordeman, 2007). Moreover, it is unclear if KLP67A contributes to anaphase A during mitosis, and indeed Buster et al. (2007) concluded that “KLP67A appears to impact chromatid-to-pole motion in meiosis but not mitosis”. Here we have confirmed previous observations on KLP67A’s role in chromosome positioning and also clarified its role in anaphase A chromosome-to-pole movement during mitosis (Fig. 5 & Fig. S1), by quantifying the effect of KLP67A depletion on chromosome segregation using transgenic flies expressing a fluorescent kinetochore marker, GFP-CID, the Drosophila CENP-A homolog.
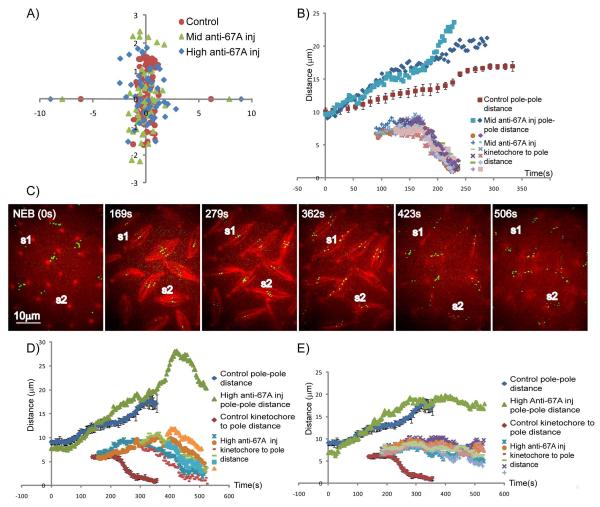
Figure 5. Chromosome congression and anaphase A are perturbed by anti-KLP67A antibody injection.
(A) Positions of metaphase kinetochores in: (i) control (red); (ii) “mid” anti-KLP67A (green); and (iii) “high” anti-KLP67A (blue) injected embryo spindles, showing that KLP67A inhibition causes kinetochores to scatter over a wider range relative to the spindle equator. Average pole positions are plotted along x-axis. (B) Pole-pole distance and kinetochore-to-pole distances as a function of time for two representative spindles injected with “mid” concentrations of anti-KLP67A in comparison with controls (red). After “mid” anti-KLP67A injection, anaphase A started at the normal time, kinetochores moved synchronously toward the pole, and chromosome segregation was completed. Based on their different behavior in anaphase B spindle elongation, spindles injected with mid concentration of anti-KLP67A could be categorized into two groups. In the first group (resembled by pole dynamics plotted in blue), the onset of anaphase B is blurred because there’s no significant difference between pre-anaphase B and anaphase B spindle elongation rate. In the second group (resembled by the pole dynamics plotted in light blue), the anaphase B spindle elongation rate is usually higher than control, the timing of anaphase A and anaphase B is perturbed. (C, D ,E) show that “high” anti-KLP67A injection caused slow or incomplete kinetochore to pole movement (C, MT: red; CID-GFP: green), which could be categorized into two groups. In the first group (exemplified by spindle S1 in C and the data plotted in D), kinetochores moved toward the poles in a non-synchronous manner at a slower rate than normal but were nonetheless able to reach the poles to complete their separation. In the second group (exemplified by spindle S2 in C and the data plotted in E), kinetochore-to-pole movement stopped prematurely after an initial phase of separation. Noticeably, in both groups, anaphase A started later than in control spindles.
In metaphase, KLP67A inhibition caused kinetochores to occupy a wider area around the spindle equator (Fig. 5a), indicating that KLP67A may be required for proper chromosome congression. Compared to controls, there is an increase in interkinetochore distance after KLP67A injection, especially after high anti-KLP67A injection (Table 1). The increase in interkinetochore distances indicates either improper kMT attachments or an increase in interkinetochore tension generated by higher MT poleward flux/MT sliding rate following KLP67A inhibition.
While the anaphase A kinetochore to pole movement rate after “mid” anti-KLP67A injection (0.110±0.023 μm/s) was close to control rates (0.097±0.013 μm/s) (Figure 5b, Table 1), “high” anti-KLP67A injection caused slow, unsynchronized kinetochore-to-pole movement and incomplete chromosome segregation (Table 1, Fig. S1 right, Figs 5c-e and Video 3). The latter defects in kinetochore-to-pole motility were of two types. The first type (exemplified by spindle S1 in Fig 5c and the data plotted in Fig 5d), included kinetochores which moved toward the poles in a non-synchronous manner at a slower rate (0.053±0.010 μm/s) than normal but were nonetheless able to reach the poles to complete their separation. In the second type (exemplified by spindle S2 in Fig 5c and the data plotted in Fig 5e), kinetochore-to-pole movement stopped prematurely after an initial phase of separation. Noticeably, in both groups, anaphase A started later than in control spindles. .
Discussion
In the current study, we have tested kinesin-8’s influence on mitotic spindle MT dynamics and investigated the role of MT plus-end dynamics in spindle length control in Drosophila embryos by exploring the mechanism of premature spindle elongation that occurs following the depletion of the kinesin-8, KLP67A. We observed that KLP67A depletion caused more persistent growth of MT plus ends, allowing them to translocate across distal half-spindles and an increase in the MT sliding rate which is correlated with the increase in pre-anaphase B and anaphase B spindle elongation rates. The premature pre-anaphase B spindle elongation that occurs after KLP67A inhibition appears to be caused by a differential increase in both the ipMT sliding rate and MT minus-end depolymerization rate, where the increase in sliding exceeds that of minus-end depolymerization. The increase in anaphase B rate after KLP67A inhibition was linearly correlated with the increase in ipMT sliding rate, in support of the “sliding filament” model for anaphase B spindle elongation. Additionally, KLP67A was found to be essential for anaphase A chromosome separation and synchrony during anaphase A independent of its effect on poleward flux in Drosophila embryonic mitosis.
Mutant rescue and correlation of mutant phenotype with antibody inhibition
The experimental strategy employed here involved: (i) analyzing the dynamics of GFP-tagged KLP67A in mitotic spindles of living embryos in rescued flies; and (ii) inhibiting the function of the kinesin-8, KLP67A in embryonic spindles using antibody inhibition and through the analysis of existing mutants. The fact that the KLP67A-GFP protein rescued mitosis in loss-of-function mutants supports the hypothesis that the tagged motor has retained its normal physiological functions. Antibody inhibition experiments must be critically evaluated and carefully controlled in order to rule out possible experimental artifacts (Brust-Mascher and Scholey, 2009; Scholey, 1998). In the current study, our observation that the inhibitory antibody, so far as we can tell, appears to exactly phenocopy the loss-of-function mutant provides compelling evidence that it yields reliable information and does not have any dominant (or other artifactual) effects.
Effects on chromosome dynamics
We found that KLP67A inhibition in Drosophila embryos by anti-KLP67A injection caused defects in metaphase congression and incomplete and unsynchronized anaphase A kinetochore segregation. Similarly, the inhibition of kinetochore-associated dynein, which is thought to be essential for stable kinetochore MT attachments (Varma et al., 2008), also caused defects in metaphase kinetochore alignment and slower anaphase A rates (Sharp et al., 2000b). The results suggest that KLP67A is essential for metaphase congression as well as anaphase chromosome segregation and synchrony, possibly by acting via one or both of two possible mechanisms. It is possible that it may be required for proper kinetochore MT interaction, so that its inhibition causes kinetochore misattachments that indirectly affect chromosome segregation. For example, it may be involved in the prevention and/or correction of merotelic kinetochore-microtubule attachments (Salmon et al., 2005) by switching MT plus end growth to shrinkage, as suggested for Drosophila meiosis (Savoian et al., 2004). Another possibility is that KLP67A contributes directly to the pacman mechanism of anaphase A chromosome segregation through its plus-end depolymerase activity, so that its inhibition slows the anaphase A chromosome segregation rates. In support of this, KLP67A localized to kinetochores from metaphase to anaphase A (Fig. 1a). Further studies are needed to understand its exact role in chromosome dynamics.
Prevention of MT overgrowth
Our results suggest that KLP67A plays an important role in controlling the dynamics of MT plus ends to limit MT polymerization thereby preventing the plus ends from extending far enough to reach the opposite pole. Consequently, the loss of KLP67A function allows significant numbers of MT plus ends to invade the opposite half spindle, sometimes impinging on the distal pole where they could act as polymer ratchets to temporarily exert an outward pushing force on the distal centrosome, thereby elongating the spindle. Indeed, this could contribute to the mechanism of premature or fast spindle elongation seen in spindles depleted of kinesin-8 function. In wild type embryos, it appears that KLP67A may restrict the overgrowth of spindle MTs by increasing the MT catastrophe frequency of longer MTs rather than by affecting the MT growth rate. This result is consistent with previous studies showing that other kinesin-8s control the plus end catastrophe frequency of cortical and interphase MTs (kip3 (Gupta et al., 2006) and klp5/6 (Tischer et al., 2009) ). In the current study, we did not test directly if KLP67A also regulates the MT shrinkage rate or rescue frequency of spindle MTs, a question which requires further investigation.
Is KLP67A-regulated kinetochore MT plus-end dynamics essential for spindle length control?
It has been shown that only kinetochore-associated KLP67A determines mitotic spindle length in Drosophila S2 cells (Savoian and Glover, 2010), but our data suggest that this is unlikely to be the case in Drosophila syncytial embryos. In embryos, as in S2 cells, KLP67A depletion very likely promotes the polymerization of kMT plus ends and it is plausible that this could contribute to premature pre-anaphase B spindle elongation. However, it is unlikely to contribute to the increase in anaphase B spindle elongation rate, because kinetochore MTs are largely disassembled by the time of anaphase B onset (Brust-Mascher and Scholey, 2002). Because the rate of outward sliding of MTs monitored by FSM did not change significantly between pre-anaphase B and anaphase B (Fig. 3d), and we observed an increase in anaphase B spindle elongation rate, it is likely that the change in ipMTs dynamics by itself is sufficient to cause faster spindle pole separation.
Challenges to current models of spindle length control
After KLP67A inhibition, the increase in both the pre-anaphase B and anaphase B spindle elongation rates were linearly correlated with the increase in MT sliding rate, suggesting that the increase in ipMT sliding rate reflects an intrinsic mechanism that drives both pre-anaphase B and anaphase B spindle elongation. However, our observations challenge several theoretical models for spindle length control (Fig. 6a).
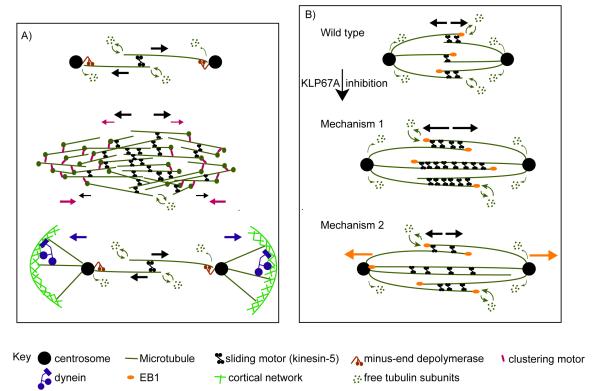
Figure 6. Models for spindle length control.
(A) Diagram of current models for spindle length control. Forces generated by different motors are shown by arrows with corresponding colors. According to the “ipMT sliding and minus-end depolymerization” model (top panel) (Brust-Mascher et al., 2004; Brust-Mascher and Scholey, 2002; Goshima et al., 2005), ipMT sliding driven by MT sliding motors (e.g. kinesin-5) is balanced by MT minus-end depolymerization at the poles to keep the metaphase spindle length constant. At anaphase B onset, downregulation of MT minus-end depolymerization allows ipMT sliding to drive spindle elongation. According to the “slide and cluster” model (center panel) (Burbank et al., 2007; Dumont and Mitchison, 2009), while MT sliding and clustering motors cooperate to drive outward MT movement near the equator (where MT overlaps are mostly anti-parallel), the clustering motors counteract the outwardly sliding MTs near the poles (where MT overlaps are mostly parallel), so the rate of MT sliding gradually slows down with increasing distance from the equator, and the spindle pole is formed when the MT sliding velocity slows to zero. This model focuses on the assembly and maintenance of anastral metaphase spindles rather than anaphase B spindle elongation. The astral MT pulling model (bottom panel) is proposed as a mechanism for spindle elongation (Saunders et al., 2007; Sharp et al., 2000a). According to this model, the cortical forces exerted on astral MTs by dynein generate pulling forces for pole-pole separation and spindle elongation. (B) Possible mechanisms by which excessive ipMT polymerization leads to faster ipMT sliding after KLP67A inhibition. Compared to control spindles (top panel), KLP67A inhibition caused prolonged and excessive ipMT plus end polymerization. In the first mechanism (center panel), we assume that the engaged force-generating sliding motors, regulated by the small and rapidly turning over antiparallel overlaps, are loaded. The abnormally large antiparallel MT overlap zone allows the recruitment of more MT sliding motors, which reduces the load per motor, thereby increasing their sliding velocity. In the second mechanism (bottom panel), we assume that the MT sliding motors work at their unloaded velocity. KLP67A depletion allows “overgrowing” ipMT polymer ratchets to push the distal centrosome outwards (orange arrows). Presumably the MT sliding motors may then serve as brakes that control the faster, MT polymer ratchet dependent ipMT sliding.
First, the observation that KLP67A depletion caused an increase in MT sliding rate challenges our “ipMT sliding and minus-end depolymerization” model (Fig. 6a, top panel) which predicts that the ipMT sliding motors, notably kinesin-5 motors, normally function at their unloaded free sliding velocity throughout pre-anaphase B and anaphase B (Brust-Mascher et al., 2004; Brust-Mascher et al., 2009). One explanation for our observation that KLP67A inhibition elevates the rate of sliding is that it effectively decreases the load on these sliding motors, by allowing more motors to bind to increased antiparallel ipMT overlap region, thereby allowing them to slide ipMTs apart at a faster rate, which would be inconsistent with this prediction of our model (Brust-Mascher et al., 2004).
Second, the increase in MT sliding/poleward flux rate and persistent spindle elongation observed after KLP67A inhibition appears to contradict the “slide-and-cluster” model in which MT polymerization and depolymerization is assumed to occur at the plus ends (Burbank et al., 2007; Dumont and Mitchison, 2009). According to the “slide and cluster model” (Fig. 6a center panel), sliding (kinesin-5) and clustering (e.g. dynein) motors cooperate near the spindle equator where most MTs overlap in an anti-parallel orientation, giving rise to rapid MT sliding rates in this region. These motors antagonize one another at increasing distances away from the spindle equator where MTs overlap in a parallel orientation, giving rise to a gradual slowing down of MT sliding to eventually stall at the poles. This is not consistent with our observations since we detect similar sliding rates throughout the spindle (while we are confident that the optics we are using are of high enough quality to detect such spatial variations, we cannot formally rule out the possibility that this could be detected using superior optics (Needleman et al. 2010)). In addition, in the context of the slide-and-cluster model, inhibition of KLP67A would presumably only increase the anti-parallel overlap region where sliding and clustering motors cooperate, leading to the formation of a longer spindle, in which the same (maximal) sliding rate at the equatorial region would be maintained, since there is no motor antagonism in this region. Moreover, the observed persistent spindle elongation in response to KLP67A inhibition can not be accounted for by this model, which predicts that the spindle should simply reach a longer steady-state length.
Third, the formation of curved spindles following KLP67A inhibition (Fig. S1) (Gandhi et al., 2004) suggests that it is unlikely that premature spindle elongation and outward MT sliding are driven mainly by cortical pulling forces acting on astral MTs (Fig. 6a bottom panel) – we can envisage intrinsic spindle forces pushing the half spindles against the cortical furrows to the extent that the spindles buckle, but it is unclear how astral pulling forces could do this. Moreover, cortical pulling mechanisms require that the rate of pole-pole separation should be twice the MT sliding rate but no less, whereas we observed that the rate of pre-anaphase B spindle pole separation in KLP67A-inhibited spindles was less than twice the ipMT sliding rate, suggesting the pre-anaphase B pole separation was driven by intrinsic forces within spindle rather than by cortical pulling forces, in agreement with previous experimental studies (Brust-Mascher et al., 2009).
Of these models, the “ipMT sliding and minus-end depolymerization” model can be modified to conform to the observations described in the current study through the incorporation of a coupling between ipMT sliding and MT plus end dynamics. We propose two possible mechanisms by which excessive ipMT polymerization might be coupled to faster ipMT sliding after KLP67A inhibition (Fig. 6b).
It is plausible that our (non-essential) model prediction that the kinesin-5 motors are normally working at their unloaded free sliding rate is wrong, and that they are, in fact, slowed down e.g. by opposing motors or MAPs. In this case, the number of actively engaged kinesin-5 motors determines the load per motor, and this in turn is determined by rapid antiparallel ipMT overlap turnover. After KLP67A inhibition, excessive MT plus end polymerization might lead to the formation of a larger antiparallel MT overlap zone, which will naturally incorporate larger numbers of kinesin-5 motors due to their high affinity for antiparallel MT polarity patterns (van den Wildenberg et al., 2008). This increase in the number of outward ipMT sliding motors may in turn lead to a decrease in the load per motor so that their sliding velocity would increase (Fig. 6b center panel).
Alternatively, another possible scenario is that the rate of ipMT sliding increased via its coupling with other mechanism. For example, it is possible that, after KLP67A inhibition, the overgrowth of MTs nucleated at one pole allows them to reach the other, opposite pole and exert transient outward forces on this distal pole via a polymer ratchet mechanism. Assuming MTs throughout the spindle are crosslinked into a mechanical continuum by dynamic MT crosslinkers (Brust-Mascher et al., 2009), the ratcheting force exerted by individual overgrowing MTs may be transmitted throughout spindle MTs, producing a global increase in the rate of ipMT sliding. Furthermore, it is possible that when the proposed polymerization ratchet force augments the sliding rate of ipMTs occurring at the unloaded velocity of kinesin-5 motors (Brust-Mascher et al., 2009), the sliding motors may enter a braking regime (Crevel et al., 2004; Kaseda et al., 2009; Saunders et al., 2007) (Fig. 6b bottom panel).
Thus, in our modified “ipMT sliding and minus-end depolymerization” model, the coupling between MT plus end dynamics and ipMT sliding provides a mechanism for MT plus end dynamics to be converted to spindle elongation. When ipMT sliding increases to the extent that it can no longer be balanced by minus-end depolymerization at the poles, spindle elongation occurs, providing a suitable explanation for premature pre-anaphase B spindle elongation and for the increase in the rate of anaphase B spindle elongation after KLP67A depletion. The KLP67A inhibition experiments also revealed a coupling between MT plus-end polymerization, MT sliding and MT minus-end depolymerization that is required to maintain the pre-anaphase B spindle at a constant length, but the detailed mechanism by which this coupling occurs needs further investigation. In addition, which of the aforementioned mechanisms underlies the fast sliding, premature elongation and anaphase A defects observed in response to KLP67A inhibition requires further investigation.
Supplementary Material
Acknowledgments
We thank members of the Scholey laboratory, especially Dr Gul Civelekoglu-Scholey for valuable suggestions and discussion, and the many colleagues listed in the materials and methods who generously provided reagents, especially Drs Andrea Pereira and Gohta Goshima. This work was supported by NIH grant GM 55507 to JMS.
Abbreviations list
- MTs
Microtubules
- ipMTs
interpolar Microtubules
- kMTs
kinetochore Microtubules
References
- Brust-Mascher I, Civelekoglu-Scholey G, Kwon M, Mogilner A, Scholey JM. Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc Natl Acad Sci U S A. 2004;101:15938–15943. doi: 10.1073/pnas.0407044101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher I, Scholey JM. Microtubule flux and sliding in mitotic spindles of Drosophila embryos. Mol Biol Cell. 2002;13:3967–3975. doi: 10.1091/mbc.02-05-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher I, Scholey JM. Microinjection Techniques for Studying Mitosis in the Drosophila melanogaster Syncytial Embryo. JoVE. 2009;31 doi: 10.3791/1382. http://www.jove.com/index/details.stp?id=1382, DOI: 10.3791/1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher I, Sommi P, Cheerambathur DK, Scholey JM. Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Mol Biol Cell. 2009;20:1749–1762. doi: 10.1091/mbc.E08-10-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbank KS, Mitchison TJ, Fisher DS. Slide-and-cluster models for spindle assembly. Curr Biol. 2007;17:1373–1383. doi: 10.1016/j.cub.2007.07.058. [DOI] [PubMed] [Google Scholar]
- Buster DW, Zhang D, Sharp DJ. Poleward tubulin flux in spindles: regulation and function in mitotic cells. Mol Biol Cell. 2007;18:3094–3104. doi: 10.1091/mbc.E06-11-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cande WZ, Hogan CJ. The mechanism of anaphase spindle elongation. Bioessays. 1989;11:5–9. doi: 10.1002/bies.950110103. [DOI] [PubMed] [Google Scholar]
- Cheerambathur DK, Civelekoglu-Scholey G, Brust-Mascher I, Sommi P, Mogilner A, Scholey JM. Quantitative analysis of an anaphase B switch: predicted role for a microtubule catastrophe gradient. J Cell Biol. 2007;177:995–1004. doi: 10.1083/jcb.200611113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, Tao L, Brust-Mascher I, Wollman R, Scholey JM. Prometaphase spindle maintenance by an antagonistic motor-dependent force balance made robust by a disassembling lamin-B envelope. J Cell Biol. 2010;188:49–68. doi: 10.1083/jcb.200908150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevel IM, Alonso MC, Cross RA. Monastrol stabilises an attached low-friction mode of Eg5. Curr Biol. 2004;14:R411–412. doi: 10.1016/j.cub.2004.05.030. [DOI] [PubMed] [Google Scholar]
- Du Y, English CA, Ohi R. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr Biol. 2010;20:374–380. doi: 10.1016/j.cub.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Dumont S, Mitchison TJ. Force and length in the mitotic spindle. Curr Biol. 2009;19:R749–761. doi: 10.1016/j.cub.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Bonaccorsi S, Wentworth D, Doxsey S, Gatti M, Pereira A. The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol Biol Cell. 2004;15:121–131. doi: 10.1091/mbc.E03-05-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Scholey JM. Control of Mitotic Spindle Length. Annu Rev Cell Dev Biol. 2010 July 6th; doi: 10.1146/annurev-cellbio-100109-104006. epub ahead of print; DOI: 10.1146/annurev-cellbio-100109-104006. [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol Biol Cell. 2005;16:3896–3907. doi: 10.1091/mbc.E05-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Length control of the metaphase spindle. Curr Biol. 2005;15:1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Gupta ML, Jr., Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Hogan CJ, Wein H, Wordeman L, Scholey JM, Sawin KE, Cande WZ. Inhibition of anaphase spindle elongation in vitro by a peptide antibody that recognizes kinesin motor domain. Proc Natl Acad Sci U S A. 1993;90:6611–6615. doi: 10.1073/pnas.90.14.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaseda K, McAinsh AD, Cross RA. Walking, hopping, diffusing and braking modes of kinesin-5. Biochem Soc Trans. 2009;37:1045–1049. doi: 10.1042/BST0371045. [DOI] [PubMed] [Google Scholar]
- Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN, McDonald KL, Ding R, McIntosh JR. Interpolar spindle microtubules in PTK cells. J Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H, McDonald KL, Cande WZ. The mechanism of anaphase spindle elongation: uncoupling of tubulin incorporation and microtubule sliding during in vitro spindle reactivation. J Cell Biol. 1988;107:623–633. doi: 10.1083/jcb.107.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- McIntosh JR, McDonald KL. The mitotic spindle. Sci Am. 1989;261:48–56. doi: 10.1038/scientificamerican1089-48. [DOI] [PubMed] [Google Scholar]
- Needleman DJ, Groen A, Ohi R, Maresca T, Mirny L, Mitchison T. Fast microtubule dynamics in meiotic spindles measured by single molecule imaging: evidence that the spindle environment does not stabilize microtubules. Mol Biol Cell. 2010;21:323–333. doi: 10.1091/mbc.E09-09-0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Schiebel E, Khmelinskii A. Cell cycle control of spindle elongation. Cell Cycle. 2010;9:1084–1090. doi: 10.4161/cc.9.6.11017. [DOI] [PubMed] [Google Scholar]
- Salmon ED, Cimini D, Cameron LA, DeLuca JG. Merotelic kinetochores in mammalian tissue cells. Philos Trans R Soc Lond B Biol Sci. 2005;360:553–568. doi: 10.1098/rstb.2004.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Powers J, Strome S, Saxton WM. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr Biol. 2007;17:R453–454. doi: 10.1016/j.cub.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoian MS, Gatt MK, Riparbelli MG, Callaini G, Glover DM. Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J Cell Sci. 2004;117:3669–3677. doi: 10.1242/jcs.01213. [DOI] [PubMed] [Google Scholar]
- Savoian MS, Glover DM. Drosophila Klp67A binds prophase kinetochores to subsequently regulate congression and spindle length. J Cell Sci. 2010;123:767–776. doi: 10.1242/jcs.055905. [DOI] [PubMed] [Google Scholar]
- Saxton WM, McIntosh JR. Interzone microtubule behavior in late anaphase and telophase spindles. J Cell Biol. 1987;105:875–886. doi: 10.1083/jcb.105.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM. Functions of motor proteins in echinoderm embryos: an argument in support of antibody inhibition experiments. Cell Motil Cytoskeleton. 1998;39:257–260. doi: 10.1002/(SICI)1097-0169(1998)39:4<257::AID-CM1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000a;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat Cell Biol. 2000b;2:922–930. doi: 10.1038/35046574. [DOI] [PubMed] [Google Scholar]
- Shelden E, Wadsworth P. Observation and quantification of individual microtubule behavior in vivo: microtubule dynamics are cell-type specific. J Cell Biol. 1993;120:935–945. doi: 10.1083/jcb.120.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Wordeman L. Chromosome congression: the kinesin-8-step path to alignment. Curr Biol. 2007;17:R326–328. doi: 10.1016/j.cub.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L, Mogilner A, Civelekoglu-Scholey G, Wollman R, Evans J, Stahlberg H, Scholey JM. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol. 2006;16:2293–2302. doi: 10.1016/j.cub.2006.09.064. [DOI] [PubMed] [Google Scholar]
- Tischer C, Brunner D, Dogterom M. Force- and kinesin-8-dependent effects in the spatial regulation of fission yeast microtubule dynamics. Mol Syst Biol. 2009;5:250. doi: 10.1038/msb.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolic-Norrelykke IM. Force and length regulation in the microtubule cytoskeleton: lessons from fission yeast. Curr Opin Cell Biol. 2010;22:21–28. doi: 10.1016/j.ceb.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Tolic-Norrelykke IM, Sacconi L, Thon G, Pavone FS. Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Curr Biol. 2004;14:1181–1186. doi: 10.1016/j.cub.2004.06.029. [DOI] [PubMed] [Google Scholar]
- van den Wildenberg SM, Tao L, Kapitein LC, Schmidt CF, Scholey JM, Peterman EJ. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr Biol. 2008;18:1860–1864. doi: 10.1016/j.cub.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Varga V, Leduc C, Bormuth V, Diez S, Howard J. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- Varma D, Monzo P, Stehman SA, Vallee RB. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment. J Cell Biol. 2008;182:1045–1054. doi: 10.1083/jcb.200710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE. Kinesin-8s: motoring and depolymerizing. Nat Cell Biol. 2006;8:903–905. doi: 10.1038/ncb0906-903. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Cai S, Khodjakov A. Mechanisms of chromosome behaviour during mitosis. Nat Rev Mol Cell Biol. 2010;11:91–102. doi: 10.1038/nrm2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman R, Civelekoglu-Scholey G, Scholey JM, Mogilner A. Reverse engineering of force integration during mitosis in the Drosophila embryo. Mol Syst Biol. 2008;4:195. doi: 10.1038/msb.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.