Abstract
DiGeorge syndrome, or velocardiofacial syndrome (DGS/VCFS), is a rare and usually sporadic congenital genetic disorder resulting from a constitutional microdeletion at chromosome 22q11.2. While rare cases of malignancy have been described, likely due to underlying immunodeficiency, central nervous system tumors have not yet been reported. We describe an adolescent boy with DGS/VCFS who developed a temporal lobe pleomorphic xanthoastrocytoma. High-resolution single nucleotide polymorphism array studies of the tumor confirmed a constitutional 22q11.21 deletion, and revealed acquired gains, losses and copy number neutral loss of heterozygosity of several chromosomal regions, including a homozygous deletion of the CDKN2A/B locus. The tumor also demonstrated a common V600E mutation in the BRAF oncogene. This is the first reported case of a patient with DiGeorge syndrome developing a CNS tumor of any histology and expands our knowledge about low-grade CNS tumor molecular genetics.
Keywords: 22q11.2 deletion syndrome, DiGeorge syndrome, Pleomorphic xanthoastrocytoma, BRAF, SNP array
Introduction
DiGeorge syndrome, or velocardiofacial syndrome (DGS/VCFS), is a rare and usually sporadic congenital genetic disorder resulting from a constitutional microdeletion at chromosome 22q11.2 [1–3]. The clinical aspects of the disorder were first described in 1968 by pediatric endocrinologist Angelo DiGeorge, with discovery of the associated molecular genetics ensuing in the following decades [1, 2]. It has an estimated prevalence of 1:4,000 live births and is typically diagnosed in early childhood [4]. The physical and laboratory features vary widely between patients. The more common findings include congenital structural heart disease, absent or hypoplastic thymus-related hypoparathyroidism (hypocalcemia) and T-cell immunodeficiency (recurrent infections), palate/speech problems, learning disabilities and psychiatric disease [1, 2]. The exact mechanism by which the syndrome's features arise is unclear, but may involve migrational defects of neural crest-derived tissues somehow related to the deletion at chromosome 22q11.2. Patients also have a malignancy predisposition, typically non-CNS lymphoma, often associated with Epstein-Barr virus infection. Other tumors, such as hepatoblastoma, Wilms’ tumor and acute lymphoblastic leukemia have also been reported. The exact mechanism by which neoplasia occurs is uncertain, but may involve T-lymphocyte abnormalities, chronic viral infection-associated malignant transformation and aberrations in the ability to detoxify certain environmental carcinogens [5]. Central nervous system tumors are not typically associated with the syndrome.
The diagnosis of DGS/VCFS is made by recognizing the typical clinical features along with detection of the 22q11.2 deletion by fluorescence in situ hybridization analysis, multiplex ligation dependent probe amplification (MLPA) or SNP array testing [2, 6, 7]. The majority of patients represent a spontaneous de novo deletion event, with only a small minority being inherited in an autosomal fashion from a parent. Historical descriptions of the syndrome from the past decades have not observed neoplasia as an association or risk factor. Recently, however, there have been case reports of patients with DGS/VCFS developing certain non-CNS tumors, such as renal cell carcinoma and hepatoblastoma [8].
To our knowledge, a CNS tumor has never been reported in a patient with DGS/VCFS. We describe here the clinical and molecular genetic features of the first-ever reported case of a temporal lobe pleomorphic xanthoastrocytoma (PXA) developing in a patient with DGS/VCFS.
Case report
A 16 year-old boy with known DGS/VCFS and a past history notable for repaired structural heart disease, developmental delay and intractable epilepsy (previously normal MRI brain) was found to have asymptomatic papilledema on routine examination. Gadolinium-infused MRI revealed a 5.8 × 8.5 × 6.3 cm, enhancing, heterogenous, right temporal lobe mass with subfalcine herniation (Fig. 1). Spine imaging was negative for evidence of metastatic tumor. A standard right temporal craniotomy with an uncomplicated gross total resection of the tumor was performed, using intraoperative MRI technology. Histopathology demonstrated a non-anaplastic, W.H.O. grade II, pleomorphic xanthoastrocytoma, with intermingled spindle and multinucleated giant cells, scattered lymphocytes and eosinophilic granular bodies. The cells had a low cycling index (low MIB-1), were GFAP positive, demonstrated prominent reticulin fibers and SMARCB1/BAF47 nuclear immunochemistry staining was retained (Fig. 2). Neuronal marker staining with synaptophysin was positive in some of the pleomorphic tumor cells, consistent with the pattern typically seen in PXA. The patient recovered well from neurosurgery, no adjuvant chemotherapy or radiation therapy was warranted and there has been no evidence of tumor recurrence more than 1 year after resection.
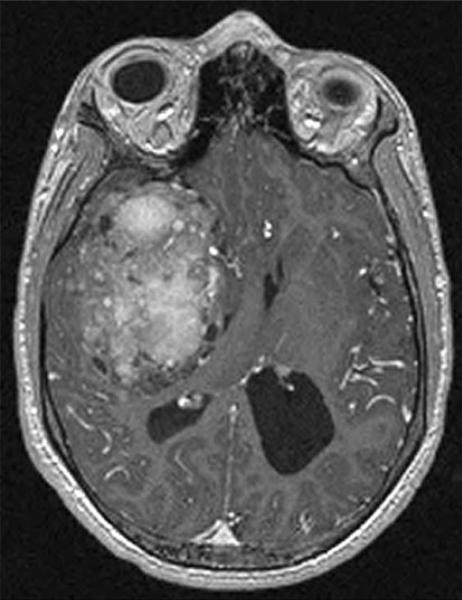
Fig. 1.
Pre-operative axial T1-weighted, post-Gadolinium, MRI showing a heterogeneous enhancing right temporal lobe-area tumor with associated midline shift
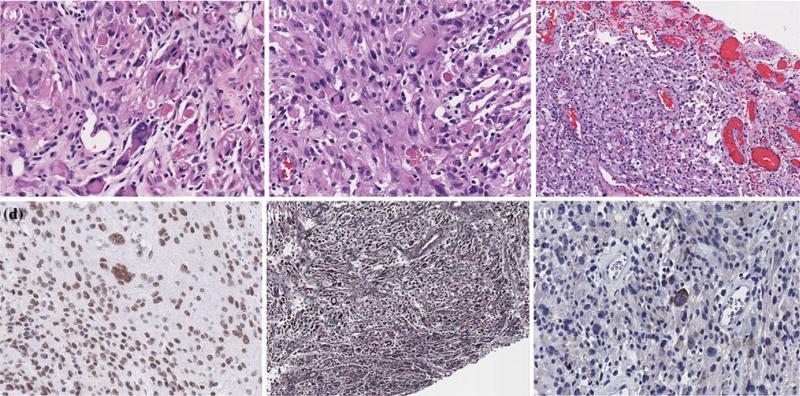
Fig. 2.
Photomicrographs of the right temporal lobe pleomorphic xanthoastrocytoma, showing compact regions with cellular pleomorphism and scattered eosinophilic granular bodies (a,b), a leptomeningeal compact region with perivascular and interstitial lymphocytes, xanthoma cells and pleomorphic cells (c), retained SMARCB1/BAF47 (INI-1) nuclear staining (d), reticulin staining of the superficial component with reticulin fibers around individual cells and clusters of cells (e) and synaptophysin staining showing positivity in some of the large pleomorphic cells (f). Original magnification, 200×
Tumor tissue was screened for a chromosome 22q11.2 deletion using MLPA and further characterized for copy number abnormalities (CNAs) and loss of heterozygosity (LOH) with an Illumina 610 K high-resolution SNP-based oligonucleotide array (Illumina, Inc., San Diego, CA) according to previously reported methods [9]. A constitutional 22q11.2 deletion was confirmed by MLPA and mapped by SNP array studies to encompass the region from 17,257,787 to 19,792,353 (NCBI36/hg18 March 2006 human reference sequence) in 22q11.21 (Table 1 and Fig. 3). SNP array studies of parental blood samples confirmed that the deletion was de novo. As shown in Table 1, additional CNAs and one region of copy number neutral LOH were identified in the tumor DNA sample. These included whole chromosome gains of 7 and 10 in the majority of cells, and gain of chromosome 21 in a minor population of tumor cells. As shown in Fig. 3, there was a copy number neutral LOH event involving all of chromosome 9, with a homozygous deletion within 9p21.3 that encompassed the CDKN2A/B (p16) tumor suppressor locus. There were three discontinuous deletions in chromosome 21, one of which included the ERG oncogene.
Table 1.
SNP array results from the tumor DNA
| Chromosome | Start | End | Abnormality |
|---|---|---|---|
| 7 | Whole chromosome | Whole chromosome | Gain |
| 9 | Whole chromosome | Whole chromosome | CN LOH |
| 9p21.3 | 19,930,143 | 20,420,468 | Duplication with LOH |
| 9p21.3 | 20,451,775 | 23,120,500 | Homozygous deletion |
| 10 | Whole chromosome | Whole chromosome | Gain |
| 21 | Whole chromosome | Whole chromosome | Gain |
| 21q21.1q21.2 | 22,668,778 | 24,107,000 | Heterozygous deletion |
| 21q22.13 | 37,028,079 | 37,104,999 | Heterozygous deletion |
| 21q22.13q22.2 | 37,727,512 | 38,987,173 | Heterozygous deletion |
| 22q11.21 | 17,257,787 | 19,792,353 | Heterozygous deletion |
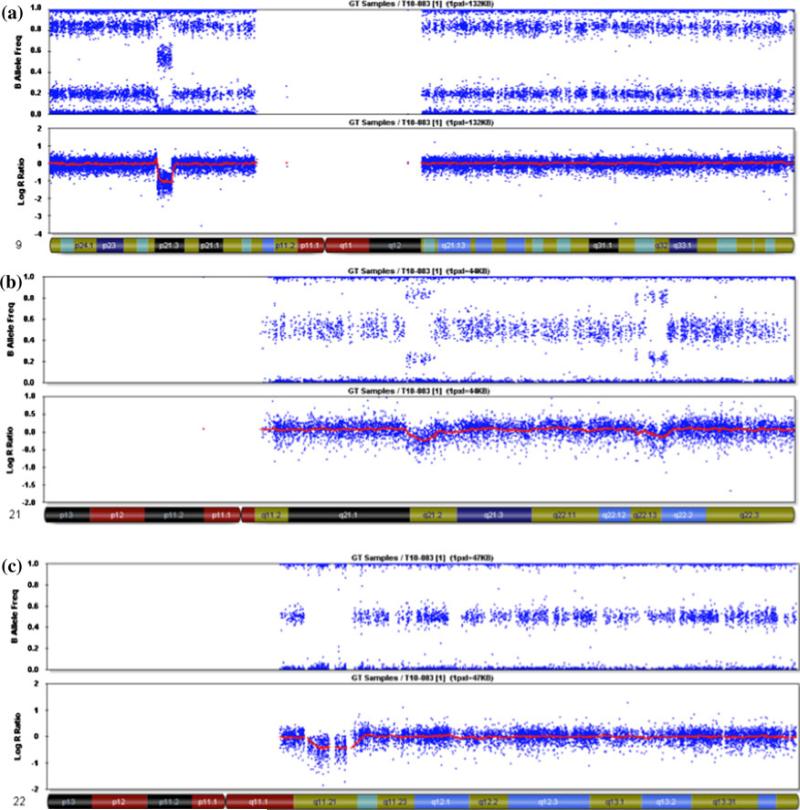
Fig. 3.
Illumina BeadStudio results for the tumor DNA. a Chromo-some 9 shows a split in the B allele frequency with no change in the log R ratio, consistent with a CN LOH event for the whole chromosome. There is also a homozygous deletion in 9p21.3 that contains the CDKN2A locus. b Chromosome 21 shows a split in the B allele frequency and an increase in the log R ratio consistent with a duplication of the whole chromosome in very few cells. There are also three regions of deletion. c Chromosome 22 has a 2.5 Mb region of heterozygous deletion in q11.21 that is present in all cells, and represents the underlying germline deletion
Mutation analysis of BRAF exons 11 and 15 was performed by polymerase chain reaction (PCR) and sequencing as described [10]. The tumor demonstrated a single base mutation in exon 15 that results in a V600E amino acid change (data not shown).
Discussion
DiGeorge syndrome is a well-recognized chromosome microdeletion deletion syndrome associated with a variety of phenotypic findings. Chromosome band 22q11.2 contains a series of low copy repeats that are prone to recombination, deletion and duplication. The typical 3 Mb deletion associated with DGS/VCFS is located between repeats A to D [3] and contains approximately 30–40 genes, many of which have not been characterized. There are only sporadic reported cases of DGS/VCFS patients developing non-CNS tumors [8], which could arise due to deletions and/or mutations of genes in the typical deleted region of 22q11.2, or by chance due to alterations in another genomic location. In contrast, the SMARCB1 locus is just distal to this region in 22q11.2, between repeats F and G, and patients with constitutional deletions that encompass this region are predisposed to rhabdoid tumors of the CNS, kidney and extra renal sites [7]. The present case is the first report of a patient with DGS and a proximal deletion in 22q11.2 who has developed a primary CNS neoplasm.
Pleomorphic xanthoastrocytoma is a low-grade (WHO grade II) neuroepithelial tumor that occurs in both children and adults, with a predilection for the supratentorial brain, particularly the temporal lobe [11]. It is a rare neoplasm, accounting for less than one percent of CNS tumors. The majority of patients have a seizure history as part of their presenting symptom complex. While resection is typically curative, PXA is more likely to undergo spontaneous malignant transformation compared to other low-grade tumors (e.g., pilocytic astrocytoma), suggesting that vigilant long-term follow-up is important, with the potential need for adjuvant radiation therapy or chemotherapy at tumor recurrence [12, 13].
Our patient had the typical DGS/VCFS constitutional (and tumor) deletion 22q11.2 as detected by SNP array testing, but no evidence of the SMARCB1 alteration that is associated with both non-CNS (e.g., kidney) and CNS (e.g., atypical teratoid/rhabdoid tumor) malignant rhabdoid tumors observed in very young children [7, 9]. Rather, the tumor had evidence of a BRAF oncogene mutation, whole gains of chromosomes 7, 10 and 21 and homozygous loss of 9p, the latter of which is often associated with aggressive CNS tumors [14]. Mutations of the BRAF oncogene have been described in a variety of malignant and benign tumors [15–17].
The majority of pilocytic astrocytomas (WHO grade I) and some fibrillary astrocytomas (WHO grade II) have activation of the BRAF oncogene due to a tandem duplication in 7q34 and novel KIAA1549-BRAF fusion [17–19]. These tumors are generally associated with an excellent survival rate when they are amenable to surgical resection. In contrast, our recent SNP array and BRAF mutation studies of various low-grade, non-pilocytic, gliomas have shown that activating mutations in the BRAF oncogene, as opposed to the duplication and BRAF fusion, occur in a variety of other histologic subtypes, e.g., ganglioglioma and PXA [10]. In addition to BRAF mutations, whole chromosome gains and losses, along with deletions of chromosome 9p have been reported on several occasions [14, 20, 21], which may signify a greater likelihood of recurrence.
This DGS patient's PXA likely represented a sporadic tumor event. With no evidence of a deletion or LOH, and retained expression in tumor cells for the SMARCB1 gene, we speculate that the BRAF mutation and/or the additional copy number changes and CDKN2A/B deletions were responsible for development of the tumor. This is the first time a CNS tumor has been diagnosed in a DGS patient and represents an additional tumor type that might be observed in this syndrome. The relationship to the underlying immunodeficiency of DGS is uncertain. The novel observations described here warrant ongoing reporting of these phenomena so as to permit better understanding of both CNS tumorigenesis and DiGeorge syndrome.
Acknowledgments
Supported in part by grants from the National Institutes of Health, CA 133173 and CA 46274 (JAB) and the N.B. Carter Neuro-Oncology Endowment Fund at Cook Children's Medical Center, Fort Worth, TX (JCM).
Abbreviations
- BRAF
B-Raf proto-oncogene serine/threonine-protein kinase
- CNS
Central nervous system
- MRI
Magnetic resonance image
- PXA
Pleomorphic xanthoastrocytoma
- SNP
Single nucleotide polymorphism
- WHO
World Health Organization
- DGS/VCFS
DiGeorge syndrome/velocardiofacial syndrome
Contributor Information
Jeffrey C. Murray, Neurosciences Program, Hematology and Oncology Center, Cook Children's Medical Center, 901 Seventh Avenue, Suite 220, Fort Worth, TX 76104, USA
David J. Donahue, Neurosciences Program, Hematology and Oncology Center, Cook Children's Medical Center, 901 Seventh Avenue, Suite 220, Fort Worth, TX 76104, USA
Saleem I. Malik, Neurosciences Program, Hematology and Oncology Center, Cook Children's Medical Center, 901 Seventh Avenue, Suite 220, Fort Worth, TX 76104, USA
Yvette B. Dzurik, Department of Pathology, Cook Children's Medical Center, 801 Seventh Avenue, Fort Worth, TX 76104, USA
Emily Z. Braly, Neurosciences Program, Hematology and Oncology Center, Cook Children's Medical Center, 901 Seventh Avenue, Suite 220, Fort Worth, TX 76104, USA
Margaret J. Dougherty, Department of Pediatrics, The Children's Hospital of Philadelphia, Abramson Research Building, Room 1002, 3615 Civic Center Boulevard, Philadelphia, PA 19104, USA
Katherine W. Eaton, Department of Pediatrics, The Children's Hospital of Philadelphia, Abramson Research Building, Room 1002, 3615 Civic Center Boulevard, Philadelphia, PA 19104, USA
Jaclyn A. Biegel, Departments of Pediatrics and Pathology, The Children's Hospital of Philadelphia, Abramson Research Building, Room 1002, 3615 Civic Center Boulevard, Philadelphia, PA 19104, USA
References
- 1.DiGeorge AM. Congenital absence of the thymus and its immunologic consequences: concurrence with congenital hypoparathyroidism. IV(1) March of Dimes-Birth Defects Foundation; White Plains, NY: 1968. pp. 116–121. [Google Scholar]
- 2.Emanuel BS, McDonald-McGinn D, Saitta SC, Zackai EH. The 22q11.2 deletion syndrome. Adv Pediatr. 2001;48:39–73. [PubMed] [Google Scholar]
- 3.Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, Emanuel BS. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 4.Oskarsdottir S, Vujic M, Fasth A. Incidence and prevalence of the 22q11 deletion syndrome: a population-based study in Western Sweden. Arch Dis Child. 2004;89(2):148–151. doi: 10.1136/adc.2003.026880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald-McGinn DM, Reilly A, Wallgren-Pettersson C, Hoyme HE, Yang SP, Adam MP, Zackie EH, Sullivan KE. Malignancy in chromosome 22q11.2 deletion syndrome (Di-george syndrome/velocardiofacial syndrome). Am J Med Genet. 2006;140A:906–909. doi: 10.1002/ajmg.a.31199. [DOI] [PubMed] [Google Scholar]
- 6.Jalali GR, Vorstman JA, Errami A, Vijzelaar R, Biegel J, Shaikh T, Emanuel BS. Detailed analysis of 22q11.2 with a high density MLPA probe set. Hum Mutat. 2008;29(3):433–440. doi: 10.1002/humu.20640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson EM, Shaikh TH, Gururangan S, Jones MC, Malkin D, Nikkel SM, Zuppan CW, Wainwright LM, Zhang F, Biegel JA. High-density single nucleotide polymorphism array analysis in patients with germline deletions of 22q11.2 and malignant rhabdoid tumor. Hum Genet. 2007;122:117–127. doi: 10.1007/s00439-007-0386-3. [DOI] [PubMed] [Google Scholar]
- 8.Scattone A, Caruso G, Marzullo A, Piscitelli D, et al. Neoplastic disease and deletion 22q11.2: a multicentric study and report of two cases. Pediatr Pathol Mol Med. 2003;22:323–341. doi: 10.1080/pdp.22.4.323.341. [DOI] [PubMed] [Google Scholar]
- 9.Jackson EM, Sievert AJ, Gai X, Hakonarson H, Judkins AR, Tooke L, Perin JC, Xie H, Shaikh TH, Biegel JA. Genomic analysis using high-density single nucleotide polymorphism-based oligonucleotide arrays and multiplex ligation-dependent probe amplification provides a comprehensive analysis of INI1/SMARCB1 in malignant rhabdoid tumors. Clin Cancer Res. 2009;15(6):1923–1930. doi: 10.1158/1078-0432.CCR-08-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dougherty MJ, Santi M, Brose MS, Ma C, Resnick AC, Sievert AJ, Storm PB, Biegel JA. Activating mutations in BRAF characterize a spectrum of pediatric low-grade gliomas. Neuro Oncol. 2010 doi: 10.1093/neuonc/noq007. doi: 10.1093/neuonc/noq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. W.H.O. Classification of tumors of the central nervous system. IARC; Lyon: 2007. [Google Scholar]
- 12.Kepes JJ, Rubinstein LJ, Ansbacher L, Schreiber DJ. Histopathological features of recurrent pleomorphic xanthoastrocytomas: Further corroboration of the glial nature of the neoplasm: A study of 3 cases. Acta Neuropathol. 1989;78:585–593. doi: 10.1007/BF00691285. [DOI] [PubMed] [Google Scholar]
- 13.Fouladi M, Jenkins J, Burger P, Langston J, Merchant T, Heideman R, Thompson S, Sanford A, Kun L, Gajjar A. Pleomorphic xanthoastrocytoma: favorable outcome after complete surgical resection. Neuro-Oncol. 2001;3:184–192. doi: 10.1093/neuonc/3.3.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber RG, Hoischen A, Ehrler M, Zipper P, Kaulich K, Blaschke B, Becker AJ, Weber-Mangal S, Jauch A, Radlwimmer B, Schramm J, Wiestler OD, Lichter P, Reifenberger G. Frequent loss of chromosome 9, homozygous CDKN2A/p14ARF/CDKN2B deletion and low TSC1 mRNA expression in pleomorphic xanthoastrocytomas. Oncogene. 2007;26:1088–1097. doi: 10.1038/sj.onc.1209851. [DOI] [PubMed] [Google Scholar]
- 15.Davies H, Bignelli GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 16.Michaloglou C, Vredeveld LCW, Mooi WJ, Peeper DS. BRAFE600 in benign and malignant human tumors. Oncogene. 2008;27:877–895. doi: 10.1038/sj.onc.1210704. [DOI] [PubMed] [Google Scholar]
- 17.Sievert AJ, Jackson EM, Gai X, et al. Duplication of 7q34 in pediatric low-grade astrocytomas detected by high-density single nucleotide polymorphism-based genotype arrays results in a novel BRAF fusion gene. Brain Pathol. 2009;19(3):449–458. doi: 10.1111/j.1750-3639.2008.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones DTW, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forshew T, Tatvossian RG, Lawson ARJ, et al. Activation of the ERK/MAPK pathway: a signature genetic defect in posterior fossa pilocytic astrocytomas. J Pathol. 2009;218(2):172–181. doi: 10.1002/path.2558. [DOI] [PubMed] [Google Scholar]
- 20.Sawyer JR, Roloson GJ, Chadduck WM, Boop FA. Cytogenetic findings in a pleomorphic xanthoastrocytoma. Cancer Genet Cytogenet. 1991;55:225–230. doi: 10.1016/0165-4608(91)90081-5. [DOI] [PubMed] [Google Scholar]
- 21.Yin X, Hui AB, Lion EC, Ding M, Chang AR, Ng H. Genetic imbalances in pleomorphic xanthoastrocytoma detected by comparative genomic hybridization and literature review. Cancer Genet Cytogenet. 2002;132:14–19. doi: 10.1016/s0165-4608(01)00512-x. [DOI] [PubMed] [Google Scholar]